Abstract
The inositol triphosphate (IP3)-signaling pathway has been associated with several developmental and physiological processes in plants, but we currently know little about the regulation of this pathway. Inositol 5′ phosphatases (5PTases) are enzymes that remove a 5′ phosphate from several potential second messengers, including IP3. In catalyzing the removal of a 5′ phosphate from second messenger substrates, 5PTases can act to terminate signal transduction events. We describe the molecular analysis of At5PTase1, a 5PTase gene from Arabidopsis. When expressed transiently in Arabidopsis leaf tissue or ectopically in transgenic plants, At5PTase1 allowed for the increased hydrolysis of I(1,4,5)P3 and I(1,3,4,5)P4 substrates. At5PTase1 did not hydrolyze I(1)P, I(1,4)P2, or PI(4,5)P2 substrates. This substrate specificity was similar to that of the human Type I 5PTase. We identified 14 other potential At5PTase genes and constructed an unrooted phylogenetic tree containing putative Arabidopsis, human, and yeast 5PTase proteins. This analysis indicated that the Arabidopsis 5PTases were grouped in two separate branches of the tree. The multiplicity of At5PTases indicates that these enzymes may have different substrate specificities and play different roles in signal termination in Arabidopsis.
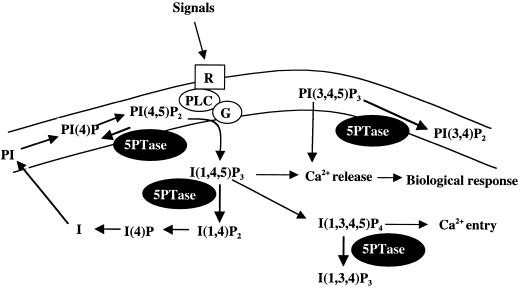
All organisms require the ability to respond to their environment to adapt and survive. In response to extracellular signals, many organisms utilize the inositol triphosphate (IP3)-signaling pathway that utilizes membrane-bound receptors coupled to the production of the second messenger IP3 (Berridge, 1993). The pathway is initiated by signal-induced activation of phospholipase C, which cleaves a phosphatidylinositide (4, 5) bisphosphate (PIP2) substrate producing the second messenger IP3 (see Fig. 1). IP3, which accumulates rapidly and transiently, binds to intracellular receptors, resulting in release of calcium (Ca2+) from intracellular stores. Ample evidence shows that this signaling pathway is used by plants (for review, see Munnik et al., 1998; Stevenson et al., 2000), and Ca2+ release in response to signals has been particularly well studied in plants (Trewavas and Mahlo, 1998). For example, gravity stimulates a rapid increase in IP3 in maize pulvini (Perera et al., 1999). Red light elicits a rapid Ca2+ intracellular release in etiolated wheat protoplasts, which can be mimicked by microinjection of IP3 (Shacklock et al., 1992). Other signals likely to generate IP3 include plant hormones. In the case of abscisic acid stimulation, it has been shown that microinjection of IP3 into stomata initiates closure (Gilroy et al., 1990), and that endogenous IP3 levels increase within 2 min of abscisic acid addition to stomata (Lee et al., 1996).
Figure 1.
Signaling via IP3 and signal termination via 5PTases. Extracellular signals are perceived by putative receptors (R) that stimulate phospholipase C (PLC)/G protein complexes (G) to convert substrate PIP2 into the second messenger IP3. IP3 and related second messengers alter intracellular Ca2+ levels, thereby triggering downstream biological events. Termination of signaling can occur by hydrolysis of four different second messengers by 5PTase enzymes (black ovals).
Legendre et al. (1993) demonstrated a role for IP3 in the defense response by documenting a rapid increase in IP3 in soybean suspension cells in response to elicitor. Tobacco stimulated with elicitor also produces a rapid intracellular release of Ca2+ (Chandra and Low, 1997). The induction of IP3 and Ca2+ in response to elicitor suggests that these second messengers provide a means to amplify the pathogenic signal.
The signal induced by IP3 is terminated through the sequential dephosphorylation of IP3 to free inositol. This hydrolysis is catalyzed by a group of specific inositol phosphatases. Removal of the first 5′ phosphate from second messenger IP3 has recently come into focus because enzymes that catalyze this hydrolysis can terminate IP3 signal transduction (Fig. 1). Animal and yeast enzymes with the ability to hydrolyze IP3 have been purified, and genes encoding some of these enzymes have been cloned (for review, see Erneux et al., 1998). These enzymes are referred to as 5′ phosphatases (5Ptases), as they have the ability to remove a 5′ phosphate from a group of substrates that includes IP3. Evidence that 5PTases can terminate signaling events comes from experiments with the human Type I 5PTase (HsTypeI) gene (De Smedt et al., 1994). When HsTypeI 5PTase protein levels are depleted in animal tissue culture cells, IP3 and Ca2+ levels are elevated and cells display a transformed phenotype (Speed et al., 1996, 1999). In contrast, when HsTypeI 5PTase is overexpressed, Ca2+ oscillations do not occur (De Smedt et al., 1997).
We have previously cloned and characterized genes from tomato encoding inositol monophosphatases (IMPs), which act to remove the final 1′ or 4′ phosphate from an IP substrate (Gillaspy et al., 1995). We report here the molecular characterization of At5PTase1, an Arabidopsis 5PTase that catalyzes the hydrolysis of IP3 and IP4. We have utilized transient expression and stable transformation of Arabidopsis to show that At5PTase1 encodes an inositol phosphatase capable of hydrolyzing IP3 and IP4 second messengers. At5PTase1 is part of a large conserved protein family that contains many enzymes with the potential to metabolize second messengers. These data indicate that At5PTase1 is an excellent candidate for an enzyme that functions to terminate IP3-mediated signaling in plants.
RESULTS
Identification of Arabidopsis 5PTase Proteins
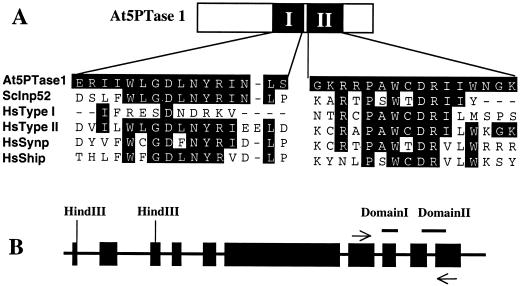
To identify potential IP3 signal-terminating enzymes in plants we identified an Arabidopsis expressed sequence tag (EST; clone 254GT7) encoding a protein homologous to the conserved catalytic domain of animal 5PTases. We utilized this clone to obtain the full-length cDNA clone, At5PTase1 (accession no. AF117062). Alignment of the predicted protein encoded by At5PTase1 with various animal and yeast 5PTases indicated that At5PTase1 shares amino acid identity in the catalytic region (domains I and II as defined by Majerus et al., 1999) required for 5PTase activity (Communi and Erneux, 1996; Communi et al., 1996; Jefferson and Majerus, 1996; see Fig. 2A and supplemental data available at www.plantphysiol.org). At5PTase1 shares 19.2% and 42.4% identity in the catalytic domain with human Type I (De Smedt et al., 1994) and II (Ross et al., 1991) 5Ptases, respectively. In contrast, identity over the entire protein is 10% (HsTypeI) and 17.8% (HsTypeII).
Figure 2.
Structure of At5PTase1 predicted protein sequence and alignment with known 5PTases. A, Schematic representation of At5PTase1 protein. The catalytic region consists of two conserved domains (I and II) found in all identified 5PTase proteins (Communi and Erneux, 1996; Communi et al., 1996; Jefferson and Majerus, 1996). B, Genomic structure of the At5PTase1-coding region. The genomic sequence and cDNA corresponding to At5PTase1 were compared using BLAST and Lasergene software. The black boxes correspond to the 10 identified exons. Exon sequences corresponding to domains I and II of the catalytic region are indicated by the bars on top; the location of PCR primers is noted by the arrows.
We identified the genomic sequence of At5PTase1 present in the database and determined that At5PTase1 is encoded by 10 exons (see Fig. 2B). The catalytic region is encoded by exons 8 through 10, with domain I being contained entirely on exon 8. Domain II, however, requires the proper splicing of exons 9 and 10. This genomic arrangement suggests the possibility of exon shuffling, which could result in the acquisition of new catalytic properties.
Analysis of Inositol Phosphate Substrate Specificity of At5PTase1 in Vitro
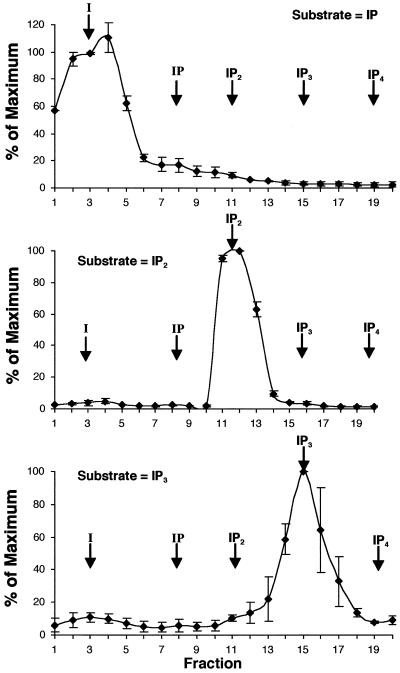
To determine if the At5PTase1 gene encodes an active 5PTase enzyme, we first established an assay to measure hydrolysis of radiolabeled I(1,3,4,5)P4, I(1,4,5)P3, I(1,4)P2, and I(1)P. We incubated radiolabeled inositol phosphates alone or in conjunction with a known inositol phosphatase and separated the products by chromatography. The inositol phosphates incubated without added enzyme eluted reproducibly with a triethylammonium buffer gradient as described in Maslanski and Busa (1990; see Figs. 3–5). To test whether this procedure would reproducibly separate inositol phosphate hydrolysis products, we incubated each radiolabeled inositol phosphate substrate with the previously cloned tomato inositol monophosphatase gene products (LeIMP1, LeIMP2, and LeIMP3; Gillaspy et al., 1995). These IMPs had been shown to hydrolyze 14C-labeled I(1)P, but had not been characterized with respect to hydrolysis of higher inositol phosphate substrates (Gillaspy et al., 1995). We incubated bacterial extracts expressing LeIMP1, 2, or 3, or control bacterial extracts with 3H-I(1,3,4,5)P4, 3H-I(1,4,5) P3, 3H-I(1,4)P2, and 14C-I(1)P substrates and separated the products as before. Incubation of control bacterial extracts with each substrate did not result in significant hydrolysis of any of the substrates (data not shown). As expected, all three LeIMPs removed a phosphate from the I(1)P substrate, resulting in a shift of label from the IP peak to one corresponding to free inositol (see Fig. 3A, data shown for LeIMP2 only). These data corresponded well with results obtained in previous assays using Dowex column chromatography (Gillaspy et al., 1995). However, I(1,4)P2 and I(1,4,5)P3 substrates, even though they contain a phosphate group at the 1 position, were not hydrolyzed by any of the LeIMPs (Fig. 3, data shown for LeIMP2 only). These data indicate that the LeIMP genes encode specific monophosphatases.
Figure 3.
LeIMP-2 is a specific monophosphatase. Bacterial extracts expressing LeIMP2 were incubated with radiolabeled inositol phosphate substrates as described. Inositol and inositol phosphates were separated with SepPak columns using a triethylammonium buffer gradient. Twenty-one-milliliter fractions were collected and a portion was analyzed by scintillation counting. The mean value from three experiments was determined and normalized by comparison with the maximal value obtained. Arrows indicate the position of inositol phosphate peaks obtained when non-recombinant bacterial extracts were incubated with radiolabeled substrates.
Figure 5.
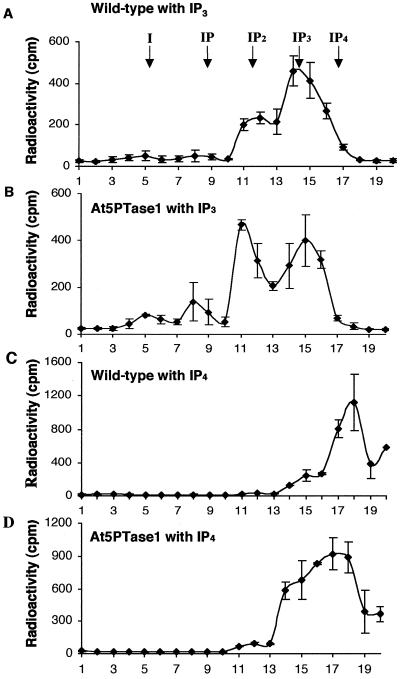
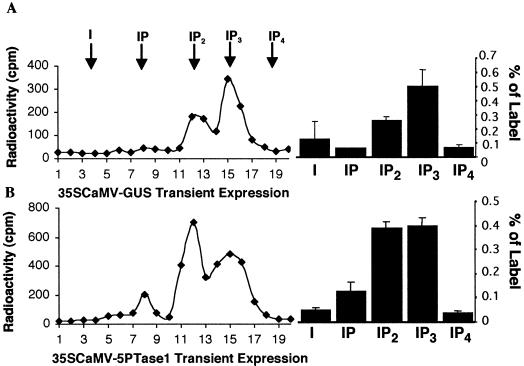
Ectopic expression of At5PTase1 yields an active 5PTase. Protein extracts were isolated from Arabidopsis wild type (A and C) and transgenic plants expressing At5PTase1 under control of the 35SCaMV promoter (B and D) and were incubated with H3-I(1,4,5)P3 (A and B) or H3-I(1,3,4,5)P4 (C and D) substrate. Products were separated over SepPak columns with a triethylammonium buffer gradient as described. The total radioactivity present in each fraction was calculated for two separate experiments and is presented as mean and sd. The arrows indicate the peak position of inositol and inositol phosphates.
To compare the catalytic activity of At5PTase1 with the LeIMP gene products, we constructed A. tumefaciens strains carrying a 35S promoter of the cauliflower mosaic virus (35SCaMV)-At5PTase1 expression cassette to use in plant transient expression assays. Arabidopsis plants were vacuum infiltrated with the Agrobacterium strains carrying the At5PTase1 construct or a 35SCaMV-β-glucuronidase (GUS) construct (35SCaMV-GUS). Twenty-four hours after vacuum infiltration, plant tissue extracts were prepared for enzyme activity assays. As GUS was highly expressed 24 h after vacuum infiltration with the 35SCaMV-GUS construct, we chose this time point to analyze 35SCaMV-At5PTase1 constructs. To determine if At5PTase1 could hydrolyze the 5′ phosphate of I(1,4,5)P3, we incubated transient assay plant extracts with individual radiolabeled substrates as before. Chromatograms from 35S-GUS-infiltrated plants contained two peaks corresponding to IP3 and IP2 and indicated that significant levels of endogenous IP3 hydrolytic activity existed in these extracts (Fig. 4A). Chromatograms from plant tissue infiltrated with 35S-At5PTase1 contained three peaks corresponding to IP3, IP2, and IP (Fig. 4B). In contrast to results using 35SCaMV-GUS-expressing tissue, At5PTase1-expressing tissue converted a greater proportion of the I(1,4,5)P3 substrate to IP2, as seen by an increase in radioactivity eluting as IP2. This indicated that transient expression of At5PTase1 increased the breakdown of I(1,4,5)P3 (Fig. 4B). At5PTase1 transient expression also increased the proportion of radiolabel eluting as IP (Fig. 4B). This is likely due to an endogenous IP2-hydrolyzing activity, as similar results were seen when 35SCaMV-GUS transient expression extracts were incubated with I(1,4)P2 (data not shown).
Figure 4.
Transient expression of At5PTase1 yields an active 5PTase. Arabidopsis leaf tissue was vacuum infiltrated with Agrobacterium tumefaciens containing a 35SCaMV-GUS construct (A) or a 35SCaMV-At5PTase1 construct (B) and tissue was extracted 24 h later. Extracts were incubated with H3-I(1,4,5)P3 substrate and the products separated over SepPak columns with a triethylammonium buffer gradient as described. Chromatograms are presented on the left. The arrows indicate the peak position of inositol and inositol phosphates as determined in Figure 3. The total radioactivity present in each peak was calculated for two separate experiments and is presented as mean and sd on the right.
We also tested the ability of these extracts to hydrolyze radiolabeled (1,3,4,5)P4. We found that transient expression of At5PTase1, but not GUS, led to hydrolysis of I(1,3,4,5)P4 (data not shown). The hydrolysis of I(1,4,5)P3 and I(1,3,4,5)P4 by At5PTase1 was specific, as seen by the inability to hydrolyze I(1,4)P2 or I(1)P (data not shown). We conclude from these experiments that At5PTase1 is a specific 5PTase that does not act on phosphoryl groups esterified at other positions. This indicates that At5PTase1 is capable of terminating IP3-generated signaling events in plants.
Analysis of Inositol Phosphate Substrate Specificity of At5PTase1 in Vivo
To determine whether At5PTase1 could be ectopically expressed in transgenic plants and function to degrade IP3, we constructed ectopically expressing transgenic At5PTase1 plants under control of the 35S promoter. Kanamycin-resistant seedlings were isolated from T0 plants and were characterized by PCR amplification to verify that the ectopic expression construct was present. Four independent lines of ectopic-expressing At5PTase1 plants were grown to the T2 generation and were characterized with respect to their development and 5PTase activity. All of these At5PTase1 transgenic lines developed normally. Plants from one ectopically expressing line (5P-e4) showed an increased ability to hydrolyze I(1,4,5)P3 and I(1,3,4,5)P4 as compared with wild-type Arabidopsis plants (see Fig. 5). Chromatograms from wild-type plants incubated with radiolabeled IP3 indicated, once again, that endogenous levels of 5PTase activity existed (Fig. 5A). Chromatograms from At5PTase1transgenic plants indicated that ectopic expression of this gene resulted in an increased ability to breakdown IP3 (Fig. 5B). In a similar manner, these transgenic plants hydrolyzed I(1,3,4,5)P4, as indicated by the greater proportion of the radioactivity eluting at the IP3 position in At5PTase1-expressing plants as compared with wild-type plants (compare Fig. 5, C with D). In contrast, At5PTase1 plants did not hydrolyze either I(1,4)P2 or I(1)P (data not shown). We conclude that ectopic expression of At5PTase1 results in an increased ability to hydrolyze IP3 and IP4 molecules.
In addition to their ability to catalyze the hydrolysis of IP3 and IP4, some animal and yeast 5PTases have the ability to act on other 5′ phosphoinositol-containing substrates such as PIP2 and PIP3. We tested whether At5PTase1 transgenics could also hydrolyze a 3H-PIP2 substrate. As a positive control we used purified Inp52 protein from yeast in activity assays. Separation of reaction products by thin-layer chromatography (TLC) indicated that in contrast to Inp52, At5PTase1 did not catalyze the hydrolysis of this substrate (data not shown).
Analysis of Differential Expression of At5PTase1
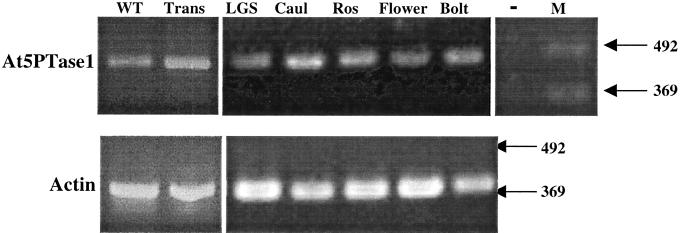
To examine expression of At5PTase1 we utilized semi-quantitative reverse transcriptase-PCR (RT-PCR) amplification. Oligonucleotide primers from exons 7 and 10 of At5PTase1 and actin (ACT2) primers were used to amplify cDNA products synthesized from RNAs of different tissues. Contaminating At5PTase1 genomic DNA present would yield a 660-bp PCR product, whereas amplification of At5PTase1 cDNA is predicted to give rise to a 450-bp product. The expected 450-bp product from cDNA was detected in several Arabidopsis tissues, including cauline leaf, rosette leaf, flower, and light-grown seedlings (see Fig. 6). We also analyzed transgenic At5PTase1 plants by RT-PCR and confirmed that these plants contain a 2.5-fold higher level of At5PTase1 mRNA (Fig. 6).
Figure 6.
RT-PCR analysis of At5PTase1 expression. cDNA was synthesized from 5 μg of wild-type rosette leaves (WT), At5PTase1 transgenic leaves (Trans), light-grown seedling (LGS), cauline leaf (Caul), rosette leaf (Ros), flower, and bolt RNAs with Murine Moloney Leukemia Virus-RT and oligo dT primer. After dilution, cDNAs were amplified with Taq polymerase using At5PTase1 or actin primers for 30 cycles (94oC, 1 min, 56°C 1.5 min, and 72°C, 1 min) and were analyzed by gel electrophoresis. The At5PTase1 negative control (−) contained no RNA template. In addition, no products were amplified in reactions using each At5PTase 1 or actin primer alone. A 100-bp DNA ladder was used as a marker (M).
The At5PTase Protein Family
To determine if At5PTase1 is part of a multigene family we used the At5PTase1 gene to probe a genomic DNA blot. At high stringency the At5PTase1 cDNA probe hybridized to a single genomic fragment from EcoRI and HindIII digests. At low stringency we found a second putative At5PTase gene (see online supplemental data). Database searches performed on October 5, 2000 indicated that 14 additional Arabidopsis genomic sequences had the ability to encode 5PTase enzymes. Alignment of the predicted coding regions from these genes, At5PTase1, and representatives from human and yeast 5PTases indicated conservation of the 5PTase catalytic region (see Table I). Amino acid identity between catalytic domains of the At5PTases, animal, and yeast 5PTases ranges from 13% to 45%. Within the At5PTases, identity in this region ranges from 32% to 94%. Thus, in general, the At5PTases are more similar to one another than to any particular animal or yeast 5PTase.
Table I.
Substrate preferences of 5PTases
| Species | No. of Amino Acids | Substrate(s) | Special Domains | Reference | Identity to At5Ptase1 | |
|---|---|---|---|---|---|---|
| % | ||||||
| Human | Type I 5PTase | 412 | IP3, IP4 | CAAX | Connolly et al. (1987) | 10.0 |
| Human | Type II 5PTase | 942 | IP3, PIP2 | CAAX, GAP | Ross et al. (1991) | 17.8 |
| Human | OCRL | 813 | PIP2 | None | Zhang et al. (1995) | 15.8 |
| Human | Synaptojanin 1 | 1,575 | IP4, PIP3 | Sac1, Pro-rich | McPherson et al. (1996) | 18.0 |
| Human | Ship 1 | 1,188 | PIP2 | SH2, Pro-rich | Damen et al. (1996) | 15.1 |
| Yeast | Inp51 | 946 | PIP2 | Sac1, Pro-rich | Stolz et al. (1998a) | 19.8 |
| Yeast | Inp52 | 1,183 | PIP2 | Sac1, Pro-rich | Stolz et al. (1998b) | 18.0 |
| Yeast | Inp53 | 1,107 | PIP2 | Sac1, Pro-rich | Stolz et al. (1998b) | 14.9 |
| Yeast | Inp54 | 384 | Unknown | Unknown | Database only | 11.7 |
| Arabidopsis | At5PTase2 | 646 | Unknown | None predicted | Database only | 34.4 |
| Arabidopsis | At5PTase1 | 590 | IP3, IP4 | None predicted | Database only | – |
| Arabidopsis | AAF43224 | 670 | Unknown | None predicted | Database only | 29.9 |
| Arabidopsis | CAB86425 | 574 | Unknown | None predicted | Database only | 31.4 |
| Arabidopsis | BAB11645 | 569 | Unknown | None predicted | Database only | 29.9 |
| Arabidopsis | AAF79735 | 585 | Unknown | None predicted | Database only | 32.6 |
| Arabidopsis | AAD15403 | 501 | Unknown | None predicted | Database only | 34.9 |
| Arabidopsis | AAC98062 | 401 | Unknown | None predicted | Database only | 34.3 |
| Arabidopsis | AAD21781 | 417 | Unknown | None predicted | Database only | 36.0 |
| Arabidopsis | BAB11520 | 366 | Unknown | None predicted | Database only | 34.2 |
| Arabidopsis | AAD46036 | 331 | Unknown | None predicted | Database only | 21.1 |
| Arabidopsis | CAB41466 | 1,101 | Unknown | None predicted | Database only | 16.8 |
| Arabidopsis | AAC23399 | 1,305 | Unknown | None predicted | Database only | 16.3 |
| Arabidopsis | AAD30615 | 1,136 | Unknown | None predicted | Database only | 16.1 |
| Arabidopsis | AAD32289 | 1,144 | Unknown | None predicted | Database only | 15.9 |
Accession nos. are given for all except Type I 5PTase (CAA54676), Type II 5PTase (P32019), OCRL (AAB03839), Synaptojanin 1 (AAC51921), Ship 1 (AAB53573), Inp51(NP_012264), Inp52 (NP_014293), Inp53 (NP_014752), and Inp54 (NP_014576). At5PTase2 and CAB41466 were previously identified in the database as putative 5PTase genes.
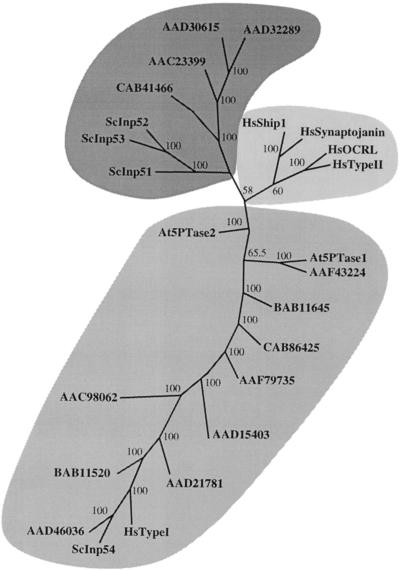
We were interested in predicting evolutionary relationships of plant, animal, and yeast 5PTases to determine if proteins with similar substrate specificity were phylogenetically related. We constructed an unrooted phylogenetic tree using a ClustalW alignment and the ProtPars program in the Phylip software package (see Fig. 7). This tree was similar to one constructed using the neighbor-joining method. The tree contained three main branches of 5PTases (Fig. 7). A group of 11 putative At5PTases, including At5PTase1, is found on the lower branch of this tree. These proteins range in size from 36 to 75 kD (see Table I). Included in this branch are the human Type I 5PTase (HsTypeI) and yeast Inp54 (ScInp54) proteins. Although the substrate specificity of ScInp54 is unknown, the human Type I 5PTase protein catalyzes the hydrolysis of IP3 and IP4 only (Laxminarayan et al., 1993).
Figure 7.
Phylogenetic analysis of 5PTase proteins. 5PTase protein sequences were aligned with ClustalW software. Alignments were analyzed with the ProtPars function of Phylip available by web interface on the Institute Pasteur Server (http://bioweb.pasteur.fr/seqanal/phylogeny/intro-uk.html) to generate an unrooted tree. Percentage bootstrap values of 500 replicates are given at each branch point. Branch lengths are to scale. Accession numbers are given for all except Type I 5PTase (CAA54676), Type II 5PTase (P32019), OCRL (AAB03839), Synaptojanin 1 (AAC51921), Ship 1 (AAB53573), Inp51(NP 012264), Inp52 (NP 014293), Inp53 (NP 014752), and Inp54 (NP 014576). At5PTase2 and CAB41466 were previously identified in the database as putative 5PTase genes. Accompanying information on size and substrate specificity can be found online in the supplemental data.
The other animal 5PTases, including the human Type II 5PTase (HsTypeII), a 107-kD protein, were contained on an upper branch of the tree (Fig. 7). These proteins have various substrate specificities, but have in common the ability to remove a 5′ phosphate from a lipid-containing substrate (i.e. PIP2 or PIP3). A third branch contained the yeast Inp51, Inp52, and Inp53 proteins (ScInp51, ScInp52, and ScInp53, respectively), and four At5PTases (Fig. 7). These proteins range in size from 104 to 143 kD. The yeast 5PTases have been shown to catalyze the hydrolysis of lipid-containing substrates, and they contain a separate N terminal domain with phosphoinositide polyphosphatase activity (Guo et al., 1999).
An unrooted phylogenetic tree of the putative catalytic region of 5PTase proteins yielded similar results in that the four larger At5PTases comprised one branch, whereas the 11 smaller At5PTases comprised a separate branch (data not shown). We conclude that these analyses identify two types of At5PTases, and that the larger At5PTases may be more functionally related to the yeast 5PTases.
DISCUSSION
The 5PTases comprise a large group of proteins generally thought of as signal terminators (Majerus et al., 1999). We report here the first plant 5PTase gene and show that the encoded protein can act to hydrolyze IP3 and IP4 substrates. This substrate specificity is similar to the HsTypeI 5PTase, a Group 1 5PTase (as defined by Majerus et al., 1999), which has been shown to function as an IP3 signal terminator (Speed et al., 1996, 1999).
We defined the substrate specificity of At5PTase1 by analyzing transient and ectopic expression of At5PTase1 in planta. It is interesting that expression in both of these systems yielded almost identical results in terms of level of 5PTase activity (Figs. 4 and 5). In our assays we were able to measure endogenous activities of putative 5PTases, as well as IP2- and IP4-hydrolyzing enzymes. Plant IP3 and IP2 hydrolytic activities have been documented in carrot cells where radiolabeled I(1,4,5)P3 substrate was metabolized to IP2 and IP by soluble and microsomal fractions of cultured carrot cells (Memon et al., 1989). Other investigators have also characterized IP3 hydrolytic activities in plant cells where the initial metabolic product was shown to be I(1,4)P2 or I(4,5)P2 (Joseph et al., 1989; Drobak et al., 1991; Martinoia et al., 1993). The removal of a 5′ or 1′ phosphate from I(1,4,5)P3 is probably catalyzed by two different enzymes, as the 5PTase was identified as a cytosolic activity and the 1′ phosphatase was found associated with the vacuole (Martinoia et al., 1993). Our data provides evidence that At5PTase1 is a 5′-specific inositol phosphatase, in that I(1,4,5)P3 is specifically broken down to IP2.
Endogenous levels of I(1,4)P2 hydrolytic activity are much lower then IP3 hydrolytic activity. A large amount of IP2 must be made available before this endogenous activity becomes apparent (Figs. 4B and 5B). The AtSAL1 and AtSAL2 genes from Arabidopsis encode proteins capable of hydrolyzing I(1,4)P2 and are referred to as inositol polyphosphatases (Quintero et al., 1996; Gil-Mascarell et al., 1999). The higher endogenous levels of 5PTases as compared with inositol polyphosphatases may be due to the multiplicity of At5PTases present in the cell or to the necessity of this protein in terminating signaling events.
An endogenous I(1,3,4,5)P4 hydrolytic activity was also present in our wild-type plant extracts (Fig. 5C). Enzymes with this catalytic activity have not been characterized in plant cells to date. I(1,3,4,5)P4 has been identified as a second messenger in animal hippocampal neurons where it is proposed to promote Ca2+ influx (Tsubokawa et al., 1996). Although I(1,3,4,5)P4 has not yet been linked to plant signaling events, our data demonstrate that At5PTase1 has the capacity to terminate I(1,3,4,5)P4 signaling in plant cells. A recent role for I(1,4,5,6)P4 has been delineated in yeast where this IP4 isomer is required for proper transcription factor assembly stability and/or function (Odom et al., 2000).
With the completion of the Arabidopsis genome sequencing project near we have the opportunity to analyze the genome for large protein families such as the 5PTases. Our analysis, beginning with the predicted amino acid sequence of At5PTase1, yielded the identification of 14 more putative At5PTases. Our phylogenetic analysis indicated that these proteins can be placed into two groups that correlate with size. The group of smaller proteins (including At5PTase1) may be more related to the HsTypeI enzyme, which catalyzes the hydrolysis of IP3 and IP4 second messengers. The group of larger At5PTases may be more similar to the yeast 5PTases that catalyze the hydrolysis of lipid-containing substrates such as PIP2 and PIP3. It is of note that recent database searches (October, 2000) reveal the presence of multiple, putative 5PTases from maize, rice, tomato, and Medicago truncatula that also fall into two groups according to size and homology to the At5PTases. Therefore, it is likely that our tentative identification of two groups of At5PTases will be extended to crop species.
To determine what signaling events these At5PTases function in will require physiological and genetic approaches. A clue to the function of these genes could be derived from looking at regulation of the transcribed messages and proteins. At5PTase1 is expressed at low levels in several tissues. A recent report detailing the identification of up-regulated mRNAs in nematode-infected Arabidopsis roots provides a physiological process in which At5PTase1 may be involved (Hermsmeier et al., 2000). As the At5PTase1 gene becomes expressed 68 times greater upon nematode infection, it is possible that signal termination via At5PTase1-catalyzed IP3 breakdown is important for this plant-animal interaction. It will be of interest to investigate whether At5PTase1 or the other At5PTase genes are differentially regulated in response to other stimuli in Arabidopsis. This approach may delineate specific physiological pathways that require At5PTase involvement.
MATERIALS AND METHODS
Plant Growth and Transformation
Arabidopsis ecotype Columbia plants were maintained in Sunshine Mix in growth chambers at 24°C under a 16-h regime of proximal fluorescent illumination. Agrobacterium tumefaciens carrying a pSLJAt5PTase1 construct was used in vacuum infiltration transformation of Arabidopsis as described in Clough and Bent (1998). The resulting seeds were selected on 0.5× Murashige-Skoog media containing 50 μg mL−1 kanamycin for 10 d. Transformants were identified, grown to maturity, and selfed. Plants were transferred at the eight-leaf stage to soil and were grown as indicated above.
Gene Cloning and Sequence Analysis
Clone 245GT7 (EST) was obtained from the Arabidopsis Resource Center at Ohio State University, Columbus. A fragment of this clone was used to screen a size-selected Arabidopsis (Columbia) cDNA library, and it yielded a full-length cDNA named At5PTase1. An identical sequence was deposited into GenBank by M. Parzer, S. deVos, and M. Lookeren Campagne (accession no. AF117062).
A 1.8-kb HindIII fragment of At5PTase1 was subcloned into the HindIII site of pBluescript in frame behind the β-galactosidase leader peptide, resulting in clone pβgal:At5PTase1. The HindIII fragment was also subcloned into the shuttle vector pBluescript 316 between the 35SCaMV promoter and Nos 3′ sequences, resulting in the clone pBS316:At5PTase1. A 35SCaMV-At5PTase1-Nos fragment was subcloned into the binary vector pSLJ7292, resulting in pSLJAt5PTase1, which was transferred into A. tumefaciens by a triparental mating.
Alignments were constructed with ClustalW software. Unrooted phylogenetic trees were created with the PHYLIP package (Felsenstein, 1989) using ProtPars with maximum parsimony. Five hundred bootstrap replicates were performed and a consensus tree was produced in the DrawTree program, which was accessed from the Institute Pasteur server (http://bioweb.pasteur.fr/seqanal/phylogeny/intro-uk.html).
DNA Gel-Blot Analyses
Ten micrograms of Arabidopsis genomic DNA was digested with restriction enzymes, separated by agarose gel electrophoresis, and transferred to nylon membranes as described by Narita and Gruissem (1989), except that transfer of DNA occurred in the presence of 0.4 m NaOH. A randomly primed At5PTase1 fragment was used as a probe in an aqueous hybridization performed at 65°C. Low stringency washes were performed at 42°C in 0.1× SSC.
RT-PCR
Total RNA was isolated from Arabidopsis tissues with TRI Reagent (Sigma, St. Louis) following the manufacturer's instructions. Five micrograms of total RNA from rosette leaves of mature wild-type or transgenic plants, and rosette leaves, cauline leaves, bolts, flowers, and 5-d-old light-grown seedlings was used in each RT-PCR assay. Oligo dT15 (0.03 μg) was incubated with RNA at 70°C for 5 min and then placed on ice. Murine Moloney Leukemia Virus reverse transcriptase enzyme (Promega, Madison, WI) and buffer along with dNTPs and RNasin (Promega) were added and incubated for 1 h at 42°C. The reaction was heated to 90°C for 5 min and stored ò −20°C until further use. Twelve and one-half picomoles each of At5PTase1-specific primers, 5PT1for2 (ACTGGGCGCGTATTGTTCT) and 5PT1rev2 (ACT CGGTTTAAGGCATCACG) or actin primers, ACT327S (ATGAAGATTAAGGTCGTGGCAC) and ACT8–3N1 (GTTTTTATCCGAGTTTGAAGAGGC) was added to 5 μL of RT-derived template in a total reaction volume of 25 μL. The location of the 5PTase1 primers used for PCR with respect to the conserved domains are shown in Figure 2. Taq DNA polymerase and supplied buffer (Promega), 0.5 mm dNTPs, and 2.5 mm final MgCl2 were added and heated to 94°C for 3 min. PCR amplification consisted of 30 cycles of a 1-min denaturation at 94°C, followed by a 1.5-min annealing period at 56°C, and a 1-min extension period at 72°C. Eight microliters of each reaction was analyzed by electrophoresis on a 1.5% (w/v) agarose gel in the presence of ethidium bromide and was visualized with a UV light box, a digital camera (Eastman-Kodak, Rochester, NY), and the Photoenhancer software package.
Expression of Phosphatase Proteins and Activity Analysis
For transient expression of At5PTase1, A. tumefaciens carrying the pSLJAt5PTase1 construct or a 35SCaMV:GUS construct (RG2, kindly provided by Dr. Paul Bottino) was vacuum infiltrated into 3-week-old unbolted Arabidopsis plants. To visualize GUS transient expression, leaf tissue was incubated with 0.025 μm of ferrocyanide, 0.025 μm of ferrous cyanide, 0.1 mm sodium phosphate buffer, pH 7.0, 0.01% (v/v) Triton X-100, and 0.5 μg mL−1 5-bromo-4-chloro-3-indoyl-β-d-GlcUA for 4 h at 37°C.
Twenty-four hours after vacuum infiltration, crude extracts were prepared from infected leaf tissue by grinding the tissue in liquid nitrogen. The tissue was added to extraction buffer (50 mm Tris, pH 8.0, 1 mm EDTA, 1 μm phenylmethylsulfonyl fluoride, and 10% [w/v] Suc), and Plant Protease Inhibitor Cocktail (Sigma) was added to a final concentration of 1 mL/30 g fresh tissue. The extract was homogenized in a Dounce homogenizer, Triton X-100 was added to a final concentration of 0.1% (v/v), and the homogenate was centrifuged at 14,000 rpm. Protein was quantitated utilizing a kit (Bio-Rad, Hercules, CA) and equal amounts of protein were added to enzyme assays.
The construction and induction of β-gal:LeIMP expression constructs has been described (Gillaspy et al., 1995). Expression was induced by addition of 10 mm isopropyl- thio-β-galactoside. Crude protein extracts were added to phosphatase assays as previously described with the following modifications: at the end of the reaction, 1 mL of water was added and the reaction was applied to a SepPak column (Waters, Milford, MA). Inositol was eluted with 5 mL of water and 4 mL of 0.02 m triethylammonium buffer (TEAB). IP, IP2, and IP3 were eluted with 4 mL of 0.1, 0.3, and 0.4 m TEAB, respectively. IP4 was eluted with 6 mL of 0.6 m TEAB. One-milliliter fractions were collected and analyzed by liquid scintillation counting. Substrates used were 14C-IP (25 μCi mL−1; ARC, St. Louis), 3H-I(1,4)P2 (10 μCi mL−1; NEN, Boston), 3H-I(1,4,5)P3 (10 μCi mL−1; NEN), and 3H-I(1,3,4,5)P4 (10 μCi mL−1; NEN).
For analysis of lipid-containing substrate hydrolysis, 3H- PI(4,5)P2 (10 μCi mL−1; ARC) was added to assays and reaction products were separated by TLC as described by Guo et al. (1999). Phosphatidylinositol, phosphatidylinositol (4) phosphate, and phosphatidylinositol (4,5) bisphosphate were identified by comigration with unlabeled standards visualized by iodine staining of the TLC plate. Lanes of the plate were sectioned into 1-cm blocks that were scraped and quantitated by liquid scintillation counting.
ACKNOWLEDGMENTS
The authors acknowledge Michael Goley and Jean Styer for general assistance, and John McDowell, Elizabeth Grabau, and Jim Westwood for critical review of the manuscript. Yeast Inp52 protein was generously provided by John York. We also thank the Arabidopsis Resource Center at Ohio State University, Columbus, for supplying EST clones and cDNA libraries.
Footnotes
This work was supported by a Jeffress Trust (award to G.E.G.) and by the Hatch Project (no. VA–135583).
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
LITERATURE CITED
- Berridge MJ. Inositol trisphosphate and calcium signaling. Nature. 1993;361:315–325. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- Chandra S, Low PS. Measurement of Ca2+fluxes during elicitation of the oxidative burst in aequorin-transformed tobacco cells. J Biol Chem. 1997;272:28274–28280. doi: 10.1074/jbc.272.45.28274. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Communi D, Erneux C. Identification of an active site cysteine residue in human type I Ins(1,4,5) P35-phosphatase by chemical modification and site-directed mutagenesis. Biochem J. 1996;320:181–186. doi: 10.1042/bj3200181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Communi D, Lecocq R, Erneux C. Arginine 343 and 350 are two active residues involved in substrate binding by human Type I D-myo-inositol 1,4,5,-trisphosphate 5-phosphatase. J Biol Chem. 1996;271:11676–11683. doi: 10.1074/jbc.271.20.11676. [DOI] [PubMed] [Google Scholar]
- Connolly T, Bansal V, Bross T, Irvine RF, Majerus PW. The metabolism of tris- and tetraphosphates of inositol by 5-phosphomonoesterase and 3-kinase enzymes. J Biol Chem. 1987;262:2146–2149. [PubMed] [Google Scholar]
- De Smedt F, Missiaen L, Parys JB, Vanweyenberg V, De Smedt H, Erneux C. Isoprenylated human brain type I inositol 1,4,5-trisphosphate 5-phosphatase controls Ca2+oscillations induced by ATP in Chinese hamster ovary cells. J Biol Chem. 1997;272:17367–17375. doi: 10.1074/jbc.272.28.17367. [DOI] [PubMed] [Google Scholar]
- De Smedt F, Verjans B, Mailleux P, Erneux C. Cloning and expression of human brain type I inositol 1,4,5-trisphosphate 5-phosphatase: high levels of mRNA in cerebellar Purkinje cells. FEBS Lett. 1994;347:69–72. doi: 10.1016/0014-5793(94)00509-5. [DOI] [PubMed] [Google Scholar]
- Drobak BK, Watkins PAC, Chattaway JA, Roberts K, Dawson AP. Metabolism of inositol(1,4,5) trisphophate by a soluble enzyme fraction from pea (Pisum sativum) roots. Plant Physiol. 1991;95:412–419. doi: 10.1104/pp.95.2.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erneux C, Govaerts C, Communi D, Pesesse X. The diversity and possible functions of the inositol 5-polyphosphatases. Biochim Biophys Acta. 1998;1436:185–199. doi: 10.1016/s0005-2760(98)00132-5. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP: phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Gillaspy GE, Keddie JS, Oda K, Gruissem W. Plant inositol monophosphatase is a lithium-sensitive enzyme encoded by a multigene family. Plant Cell. 1995;7:2175–2185. doi: 10.1105/tpc.7.12.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Mascarell R, Lopez-Coronado JM, Belles JM, Serrano R, Rodriguez PL. The ArabidopsisHAL2-like gene family includes a novel sodium-sensitive phosphatase. Plant J. 1999;17:373–383. doi: 10.1046/j.1365-313x.1999.00385.x. [DOI] [PubMed] [Google Scholar]
- Gilroy S, Read ND, Trewavas AJ. Elevation of cytoplasmic calcium by caged calcium or caged inositol triphosphate initiates stomatal closure. Nature. 1990;346:769–771. doi: 10.1038/346769a0. [DOI] [PubMed] [Google Scholar]
- Guo S, Stolz LE, Lemrow SM, York JD. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- Hermsmeier D, Hart JK, Byzova M, Rodermel SR, Baum TJ. Changes in mRNA abundance within Heterodera schachtii-infected roots of Arabidopsis thaliana. Mol Plant-Microbe Interact. 2000;13:309–315. doi: 10.1094/MPMI.2000.13.3.309. [DOI] [PubMed] [Google Scholar]
- Jefferson AB, Majerus PW. Mutation of the conserved domains of two inositol polyphosphate 5-phosphatases. Biochemistry. 1996;35:7890–7894. doi: 10.1021/bi9602627. [DOI] [PubMed] [Google Scholar]
- Joseph SK, Esch T, Bonner WD. Hydrolysis of inositol phosphates by plant extracts. Biochem J. 1989;264:851–856. doi: 10.1042/bj2640851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxminarayan KM, Matzaris M, Speed CJ, Mitchell CA. Purification and characterization of a 43-kDa membrane-associated inositol polyphosphate 5-phosphatase from human placenta. J Biol Chem. 1993;268:4968–4974. [PubMed] [Google Scholar]
- Lee Y, Choi Y, Suh S, Lee J, Assman S, Joe C, Kelleher J, Crain R. Abscisic acid-induced phosphoinositide turnover in guard cell protoplasts of Vicia faba. Plant Physiol. 1996;110:987–996. doi: 10.1104/pp.110.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre L, Yueh YG, Crain R, Haddock N, Heinstein PF, Low PS. Phospholipase C activation during elicitation of the oxidative burst in cultured plant cells. J Biol Chem. 1993;268:24559–24563. [PubMed] [Google Scholar]
- Majerus P, Kisseleva M, Norris FA. The role of phosphatases in inositol signaling reactions. J Biol Chem. 1999;274:10669–10672. doi: 10.1074/jbc.274.16.10669. [DOI] [PubMed] [Google Scholar]
- Martinoia E, Locher R, Vogt E. Inositol trisphosphate metabolism in subcellular fractions of barley (Hordeum vulgareL.) mesophyll cells. Plant Physiol. 1993;102:101–105. doi: 10.1104/pp.102.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslanski J, Busa WB. A sensitive and specific mass assay for myo inositol and inositol phosphates. In: Irvine RF, editor. Methods In Inositide Research. New York: Raven Press; 1990. pp. 113–126. [Google Scholar]
- McPherson PS, Garcia EP, Slepnev VI, David C, Zhang X, Grabs D, Sossin WS, Bauerfeind R, Nemoto Y, De Camilli PA. Presynaptic inositol-5-phosphatase. Nature. 1996;379:353–357. doi: 10.1038/379353a0. [DOI] [PubMed] [Google Scholar]
- Memon AR, Rincon M, Boss WF. Inositol trisphosphate metabolism in carrot (Daucus carotaL.) cells. Plant Physiol. 1989;91:477–480. doi: 10.1104/pp.91.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T, Irvine RF, Musgrave A. Phospholipid signalling in plants. Biochim Biophys Acta. 1998;1389:222–272. doi: 10.1016/s0005-2760(97)00158-6. [DOI] [PubMed] [Google Scholar]
- Narita JO, Gruissem W. Tomato hydroxymethylglutaryl-CoA reductase is required early in fruit development but not during ripening. Plant Cell. 1989;1:181–190. doi: 10.1105/tpc.1.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- Perera IY, Heilmann I, Boss WF. Transient and sustained increases in inositol 1,4,5-trisphosphate precede the differential growth response in gravistimulated maize pulvini. Proc Natl Acad Sci USA. 1999;96:5838–5843. doi: 10.1073/pnas.96.10.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Garciadeblas B, Rodriguez-Navarro A. The SAL1 gene of Arabidopsis, encoding an enzyme with 3′(2′),5′-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase activities, increases salt tolerance in yeast. Plant Cell. 1996;8:529–537. doi: 10.1105/tpc.8.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross TS, Jefferson AB, Mitchell C, Majerus PW. Cloning and expression of human 75-kDa inositol polyphosphate-5-phosphatase. J Biol Chem. 1991;266:20283–20289. [PubMed] [Google Scholar]
- Shacklock PS, Read ND, Trewavas AJ. Cytosolic free calcium mediates red light-induced photomorphogenesis. Nature. 1992;358:753–755. [Google Scholar]
- Speed CJ, Little PJ, Hayman JA, Mitchell CA. Underexpression of the 43-kDa inositol polyphosphate 5-phosphatase is associated with cellular transformation. EMBO J. 1996;15:4852–4861. [PMC free article] [PubMed] [Google Scholar]
- Speed CJ, Neylon CB, Little PJ, Mitchell CA. Underexpression of the 43-kDa inositol polyphosphate 5-phosphatase is associated with spontaneous calcium oscillations and enhanced calcium responses following endothelin-1 stimulation. J Cell Sci. 1999;112:669–679. doi: 10.1242/jcs.112.5.669. [DOI] [PubMed] [Google Scholar]
- Stevenson JM, Perera IY, Heilmann I, Persson S, Boss WF. Inositol signaling and plant growth. Trends Plant Sci. 2000;5:252–258. doi: 10.1016/s1360-1385(00)01652-6. [DOI] [PubMed] [Google Scholar]
- Stolz L, Kuo W, Longchamps J, Sekhon M, York J. INP51, a yeast inositol polyphosphate 5-phosphatase required for phosphatidylinositol 4,5-bisphosphate homeostasis and whose absence confers a cold-resistant phenotype. J Biol Chem. 1998a;273:11852–11861. doi: 10.1074/jbc.273.19.11852. [DOI] [PubMed] [Google Scholar]
- Stolz LE, Huynh CV, Thorner J, York JD. Identification and characterization of an essential family of inositol polyphosphate 5-phosphatases (INP51, INP52 and INP53 gene products) in the yeast Saccharomyces cerevisiae. Genetics. 1998b;148:1715–1729. doi: 10.1093/genetics/148.4.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewavas AJ, Mahlo R. Ca2+ signalling in plant cells: the big network! Curr Opin Plant Biol. 1998;5:428–433. doi: 10.1016/s1369-5266(98)80268-9. [DOI] [PubMed] [Google Scholar]
- Tsubokawa H, Oguro K, Robinson HP, Masuzawa T, Kawai N. Intracellular inositol 1,3,4,5-tetrakisphosphate enhances the calcium current in hippocampal CA1 neurones of the gerbil after ischemia. J Physiol. 1996;497:67–78. doi: 10.1113/jphysiol.1996.sp021750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Hartz PA, Philip E, Racusen LC, Majerus PW. Cell lines from kidney proximal tubules of a patient with Lowe syndrome lack OCRL inositol polyphosphate 5-phosphatase and accumulate phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 1998;273:1574–1582. doi: 10.1074/jbc.273.3.1574. [DOI] [PubMed] [Google Scholar]