Abstract
To better understand seed germination, a complex developmental process, we developed a proteome analysis of the model plant Arabidopsis for which complete genome sequence is now available. Among about 1,300 total seed proteins resolved in two-dimensional gels, changes in the abundance (up- and down-regulation) of 74 proteins were observed during germination sensu stricto (i.e. prior to radicle emergence) and the radicle protrusion step. This approach was also used to analyze protein changes occurring during industrial seed pretreatments such as priming that accelerate seed germination and improve seedling uniformity. Several proteins were identified by matrix-assisted laser-desorption ionization time of flight mass spectrometry. Some of them had previously been shown to play a role during germination and/or priming in several plant species, a finding that underlines the usefulness of using Arabidopsis as a model system for molecular analysis of seed quality. Furthermore, the present study, carried out at the protein level, validates previous results obtained at the level of gene expression (e.g. from quantitation of differentially expressed mRNAs or analyses of promoter/reporter constructs). Finally, this approach revealed new proteins associated with the different phases of seed germination and priming. Some of them are involved either in the imbibition process of the seeds (such as an actin isoform or a WD-40 repeat protein) or in the seed dehydration process (e.g. cytosolic glyceraldehyde-3-phosphate dehydrogenase). These facts highlight the power of proteomics to unravel specific features of complex developmental processes such as germination and to detect protein markers that can be used to characterize seed vigor of commercial seed lots and to develop and monitor priming treatments.
The new plant formed by sexual reproduction starts as an embryo within the developing seed, which arises from the ovule. Desiccation is the final phase of maturation for most seeds growing in temperate climates, enabling them to survive for many years. Therefore, the seed occupies a central position in the higher plant life cycle. Dry mature seeds are resting organs, having low moisture content (5%–15%) with metabolic activity almost at a standstill. For germination to occur, they need to be hydrated under conditions that encourage metabolism, e.g. a suitable temperature and the presence of oxygen. This water uptake is triphasic, including an initial rapid period (phase I), followed by a plateau phase with little change in water content (phase II), and a subsequent increase in water content coincident with radicle emergence and resumption of growth (phase III). Germination sensu stricto refers to phases I and II of this process, during which imbibed seeds maintain their desiccation tolerance (Bradford, 1990; Bewley and Black, 1994; Bewley, 1997).
From a biochemical and molecular point of view, studying germination is difficult because a population of seeds does not complete the process synchronously (Still et al., 1997). Priming treatments (i.e. pregermination treatments) are used to synchronize the germination of individual seeds (Heydecker et al., 1973). They initiate germination-related processes, but prevent emergence of the radicle and are followed by a drying for storage and marketing of the treated seeds. Seed priming generally causes faster germination and faster field emergence, which have practical agronomic implications, notably under adverse germination conditions (McDonald, 2000). Optimization of such treatments actually rests on carrying out subsequent germination assays, which only provide retrospective indications of the effectiveness of the priming conditions. Therefore, there is strong interest in identifying molecular markers of germination and/or priming for use by the seed industry (Job et al., 2000). The few processes already described to play a role during seed priming include cell cycle-related events (De Castro et al., 2000), endosperm weakening by hydrolase activities (Groot et al., 1988; Bradford et al., 2000), and mobilization of storage proteins (Job et al., 1997).
Germination sensu stricto consists of many processes; some can be completed, whereas others may have just been started, notably during priming. Proteomics and cDNA microarray technology may prove valuable by providing simultaneous information over a multitude of processes (Cahill et al., 2001). Arabidopsis contains the smallest plant genome (120 Mb) of which sequencing is now completed (The Arabidopsis Genome Initiative, 2000) and is, therefore, the model for studying plant genetics. In the present study, we developed a proteome analysis of seed germination and priming using this model plant. Improvements in two-dimensional (2-D) gel electrophoresis techniques now offer highly reproducible resolution for protein separation. Moreover, the recent advances in mass spectrometry permit analysis of low amounts of proteins separated in 2-D gels. The long-term objective of this work is to provide reference maps of seed proteins to focus on the effects of environmental changes and developmental stages during seed maturation, desiccation, and germination. Such an approach already proved successful to investigate differential protein expression in Arabidopsis upon environmental changes or mutations (Meurs et al., 1992; Leymarie et al., 1996; Santoni et al., 1997, 1998, 1999; for review, see Jacobs et al., 2000).
RESULTS
Preparation of Seed Samples
Under optimal conditions (25°C), dry mature Arabidopsis seeds started to germinate at 1.6 d of imbibition and it took almost 2.2 d for 50% of the seeds to germinate (Table I). Dry mature seeds were treated using either a hydro- or an osmopriming treatment. Both methods showed significant advancement in start of germination and reduced substantially the time to reach 50% of germination (Table I).
Table I.
Effects of 1-d hydropriming and 7-d osmopriming on germination performance of Arabidopsis seeds
| Seeds | T1 | T50 | Gmax |
|---|---|---|---|
| d | % | ||
| Dry mature | 1.60 ± 0.04 | 2.17 ± 0.01 | 98.0 ± 1.2 |
| Hydroprimed | 0.80 ± 0.04 | 1.58 ± 0.02 | 98.0 ± 1.2 |
| Osmoprimed | 1.24 ± 0.17 | 1.58 ± 0.10 | 96.7 ± 1.8 |
T1 represents the start of germination (time to reach 1% of germination ± sd); T50 represents the time to reach 50% of germination (±sd); and Gmax represents the final percentage of germination (±sd).
Proteome Analyses
Proteins were extracted from the various seed samples (dry mature seeds, 1-d- and 2-d-imbibed seeds, and hydroprimed and osmoprimed seeds) and analyzed by 2-D gel electrophoresis as described in “Materials and Methods.” A systematic comparison of 2-D gels for the various total protein extracts (examples are shown in Fig. 1) allowed classifying seed proteins from their specific accumulation patterns according to the following types (see Tables II, III, and IV; Fig. 2). The type-0 proteins corresponded to proteins present in dry mature seeds and whose abundance remained constant throughout the germination process (i.e. up to 2 d of imbibition). Most proteins from the dry mature state belong to this group: there were 1,251 type-0 proteins (Table II) out of 1,272 proteins in 2-D gels from the dry mature seeds. Type-1 and -2 (Table II) corresponded to proteins whose abundance varied (up- and down-regulation, respectively) during germination sensu stricto (i.e. prior to radicle emergence). Type-3 and -4 proteins (Table II) showed an accumulation pattern characteristic of the radicle emergence step. Type-5 proteins (Table III) were mostly detected in the imbibed seeds; their abundance was very low in dry mature seeds, increased in the 1-d imbibed seeds, and decreased upon drying these seeds back to the original moisture content of the mature dry seeds. Therefore, the type-5 proteins are characteristic of the imbibed state of the seed tissues. The type-6 proteins (Table III) conversely corresponded to proteins present in the dry mature seeds, whose abundance first strongly decreased following 1-d imbibition and then re-increased up to about the same level as in mature dry seeds upon subsequent drying. Therefore, these type-6 proteins are associated specifically with the desiccated state of the seeds. For the primed samples (see Table IV), type-9 and -10 proteins can be considered as being osmopriming specific, whereas type-11 and -12 proteins are hydropriming specific. Finally, type-7 and -8 proteins corresponded to intrinsic priming markers because these proteins exhibited the same accumulation pattern during both hydro- and osmopriming treatments.
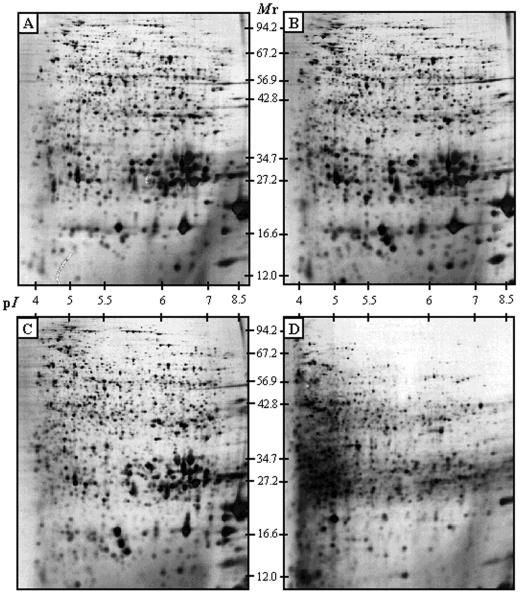
Figure 1.
2-D analysis of the Arabidopsis proteome during seed germination. 2-D gel profiles of total proteins from: A, dry mature seeds (1,272 proteins detected in 2-D gels); B, 1-d imbibed seeds (1,338 proteins detected); C, 2-d imbibed seeds (1,461 proteins detected); and D, 3-d imbibed seeds (1,133 proteins detected). An equal amount (200 μg) of total protein extracts was loaded in each gel.
Table II.
Synopsis of protein types observed during germination
| Protein Type | Characteristics | No. of Proteins Detected | No. of Proteins Identified |
|---|---|---|---|
| 0 | Present at all stages of germination | 1,251 | 38 |
| 1 | Increased level during germination sensu stricto | 32 | 13 |
| 2 | Decreased level during germination sensu stricto | 7 | 1 |
| 3 | Increased level during radicle protrusion | 21 | 7 |
| 4 | Decreased level during radicle protrusion | 14 | 8 |
The no. of proteins in each type detected on 2-D gels and identified by matrix-assisted laser-desorption ionization time of flight (MALDI-TOF) analysis is indicated.
Table III.
Synopsis of protein types observed during imbibition drying
| Protein Type | Characteristics | No. of Proteins Detected | No. of Proteins Identified |
|---|---|---|---|
| 5 | Imbibition specific | 19 | 7 |
| 6 | Desiccation specific | 3 | 1 |
Imbibition-specific proteins correspond to proteins whose level increased during 1-d imbibition of the dry mature seeds and then decreased upon subsequent drying. Desiccation-specific proteins correspond to proteins present in the dry mature seeds and whose level decreased during 1-d imbibition, then re-increased upon subsequent drying. The no. of proteins in each type detected on 2-D gels and identified by MALDI-TOF analysis is indicated.
Table IV.
Synopsis of protein types observed during priming
| Protein Type | Characteristics | No. of Proteins Detected | No. of Proteins Identified |
|---|---|---|---|
| 7 | Increased level during OP and HP | 3 | 3 |
| 8 | Decreased level during OP and HP | 2 | – |
| 9 | Increased level during OP | 3 | 2 |
| 10 | Decreased level during OP | 3 | 1 |
| 11 | Increased level during HP | 10 | 3 |
| 12 | Decreased level during HP | 2 | – |
The no. of proteins in each type detected on 2-D gels and identified by MALDI-TOF analysis is indicated. OP, Osmopriming; HP, hydropriming.
Figure 2.
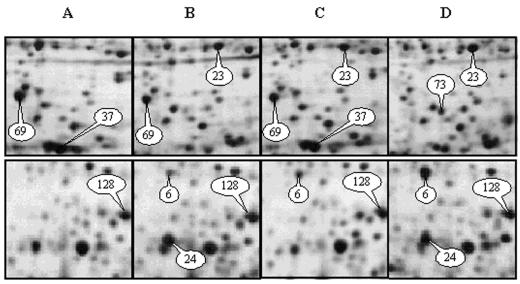
Characterization of some Arabidopsis seed proteins whose abundance vary during germination and priming. A, Dry mature seeds; B, 1-d imbibed seeds (germination sensu stricto); C, redried 1-d imbibed seeds (hydroprimed seeds); D, 2-d imbibed seeds (radicle emergence step). Proteins shown in each top window are within the pI and size ranges: 6.2 < pI < 7.0 and 40.0 kD < Mr < 58.0 kD; labeled proteins (experimental molecular mass in kD, experimental pI) are type-4 protein number 69 (43.67 and 6.29), type-6 protein number 37 (40.29 and 6.49), type-3 protein number 73 (42.28 and 6.47); and type-11 protein number 23 (56.48 and 6.64). Proteins shown in each bottom window are within the pI and size ranges: 4.8 < pI < 5.5 and 41.0 kD < Mr < 57.0 kD; labeled proteins (experimental molecular mass in kD, experimental pI) are type-5 protein number 24 (42.94 and 5.06), type-11 protein number 6 (54.71 and 5.06), and type-0 protein number 128 (47.55 and 5.44). The protein types are depicted in Tables II through IV. Identified proteins (by MALDI-TOF and/or post source decay [PSD]) are listed in Tables V through IX. Protein spot quantitation was carried out as described in “Materials and Methods,” from at least three gels for each condition. For example, in C the following spot volumes were measured: spot 23 (435, 429, and 466; 444 ± 16); spot 69 (575, 546, and 540; 554 ± 15); spot 37 (1,141, 1,054, and 1,097; 1097 ± 36); spot 6 (78, 64, and 74; 73 ± 6); and spot 128 (244, 254, and 259; 252 ± 6).
The protein maps produced in triplicate from two independent protein extractions showed a high level of reproducibility (for examples of protein spot quantitation, see Fig. 2). Some of the protein spots resolved by 2-D gel electrophoresis were excised from gels, digested with trypsin, and analyzed by matrix-assisted laser-desorption ionization time of flight (MALDI-TOF) mass spectrometry (Tables V–IX). These spots were chosen because they were well resolved when visualized after Coomassie or silver staining and included abundant proteins within the different protein types listed in Tables II through IV. Water-soluble proteins identified by MALDI-TOF analysis are also listed in Table V. Randomly chosen proteins (type-3 protein no. 67 in Table VII, type-5 proteins nos. 24 and 41 in Table VIII, and type-0 protein no. 102 in Table V) were submitted to sequencing by the PSD approach. There was excellent agreement between the MALDI-TOF and PSD data.
Table V.
Arabidopsis polypeptides whose abundance remained constant during germination
| No. | Exp. Mr | Exp. pI | Arabidopsis Protein Name | Cov. % | Theo. Mr | Theo. pI | Accession No. |
|---|---|---|---|---|---|---|---|
| 80 | 32.64 | 5.85 | α-Subunit of 12S cruciferinab | 33 | 34.68 | 6.42 | T04623 |
| 81 | 28.95 | 5.89 | α-Subunit of 12S-1 seed storage proteinc | 40 | 29.11 | 6.49 | P15455 |
| 82 | 33.89 | 6.24 | α-Subunit of 12S cruciferinc | 42 | 34.68 | 6.42 | T04623 |
| 83 | 34.35 | 6.42 | α-Subunit of 12S cruciferin | 33 | 34.68 | 6.42 | T04623 |
| 84 | 30.46 | 6.61 | α-Subunit of 12S-1 seed storage protein | 42 | 29.11 | 6.49 | P15455 |
| 85 | 27.20 | 6.50 | α-Subunit of 12S-2 seed storage protein | 32 | 27.24 | 6.34 | P15456 |
| 97 | 28.89 | 6.17 | α-Subunit of 12S-1 seed storage proteinc | 40 | 29.11 | 6.49 | P15455 |
| 117 | 25.42 | 5.78 | α-Subunit of 12S-2 seed storage proteind | 33 | 27.24 | 6.34 | P15456 |
| 118 | 29.21 | 6.04 | α-Subunit of 12S cruciferine | 26 | 34.68 | 6.42 | T04623 |
| 119 | 30.27 | 6.10 | α-Subunit of 12S-1 seed storage proteinc | 36 | 29.11 | 6.49 | P15455 |
| 120 | 30.40 | 6.16 | α-Subunit of 12S-1 seed storage proteinc | 28 | 29.11 | 6.49 | P15455 |
| 133 | 29.28 | 8.22 | α-Subunit of 12S-1 seed storage proteinc | 30 | 29.11 | 6.49 | P15455 |
| 134 | 26.35 | 6.34 | α-Subunit of 12S-2 seed storage proteinbf | 21 | 27.24 | 6.34 | P15456 |
| 141 | 29.35 | 7.42 | α-Subunit of 12S-1 seed storage proteinc | 21 | 29.11 | 6.49 | P15455 |
| 142 | 27.90 | 7.40 | α-Subunit of 12S-1 seed storage proteing | 40 | 29.11 | 6.49 | P15455 |
| 86 | 18.00 | 5.68 | β-Subunit of a putative seed storage proteinb | 26 | 19.78 | 5.61 | 4204298 |
| 87 | 18.43 | 6.36 | β-Subunit of 12S-2 seed storage proteinb | 33 | 20.80 | 7.03 | P15456 |
| 98 | 18.08 | 7.90 | β-Subunit of 12S-1 seed storage protein | 45 | 20.90 | 8.68 | P15455 |
| 115 | 32.51 | 5.76 | Similar to Brassica oleracea aspartic protease subunit | 19 | 35.22 | 5.90 | 2160151 |
| 125 | 28.69 | 5.06 | Subunit of aspartic proteinase | 28 | 33.34 | 5.06 | 1354272 |
| 90 | 64.11 | 4.81 | Similar to disulfide isomerase protein from alfalfa (Medicago sativa) | 39 | 55.60 | 4.81 | 5263328 |
| 144 | 15.87 | 6.82 | Peptidylprolyl isomerase ROC1 | 48 | 18.60 | 7.83 | T06073 |
| 94 | 34.92 | 5.21 | Late embryogenesis abundant (LEA)-like protein | 29 | 29.55 | 5.21 | 4140256 |
| 102 | 60.51 | 5.22 | LEA protein AtECP63h | 17 | 48.49 | 5.43 | 4415909 |
| 136 | 77.87 | 4.97 | Heat shock protein 70 cognate | 19 | 71.37 | 5.03 | P22953 |
| 105 | 48.63 | 5.61 | Hypothetical protein similar to elongation factor 1-gamma | 38 | 46.40 | 5.55 | 9994099 |
| 127 | 93.92 | 5.89 | Translation elongation factor EF-2 | 31 | 94.25 | 5.89 | 6056373 |
| 129 | 47.20 | 5.50 | Eucaryotic initiation factor 4α-1 | 28 | 46.70 | 5.47 | 1170503 |
| 103 | 57.95 | 5.58 | Enolase | 13 | 47.72 | 5.54 | P25696 |
| 106 | 41.64 | 5.77 | Alcohol dehydrogenase | 22 | 41.84 | 6.11 | P06525 |
| 108 | 49.11 | 6.16 | Putative indole-3-glycerol phosphate synthase | 13 | 44.58 | 6.98 | 4587610 |
| 109 | 64.79 | 6.35 | Malate oxidoreductase | 15 | 64.28 | 6.32 | 3687228 |
| 114 | 39.56 | 5.87 | Similar to cytosolic malate dehydrogenase | 21 | 35.57 | 6.11 | 2341034 |
| 121 | 40.92 | 5.45 | Member of the phosphoglycerate kinase family | 16 | 42.19 | 5.43 | 4835754 |
| 124 | 31.91 | 5.04 | Similar to lactoylglutathione lyase (glyoxalase) | 45 | 31.93 | 5.19 | 3157947 |
| 126 | 27.16 | 5.51 | Cytosolic triosephosphate isomerase | 26 | 27.15 | 5.24 | 414550 |
| 131 | 38.48 | 6.22 | Aldose reductase-like protein | 14 | 36.38 | 6.23 | 7327826 |
| 145 | 46.76 | 5.87 | Citrate synthase | 45 | 52.78 | 6.41 | 7436692 |
The Arabidopsis polypeptides whose abundance remained constant during germination are defined as type-0 proteins in Table II. Cov., Coverage; Exp., experimental; Theo., theoretical. In some cases, although MALDI-TOF analysis unambiguously identified polypeptides as to corresponding to 12S cruciferin subunits, there was disagreement between experimental and theoretical Mr and pI values. The possibility that the identified polypeptides corresponded to proteolytic fragments of the cruciferin subunits was assessed by removing amino acids sequentially at http://www.expasy.ch/tools/peptide-mass.html, either from the N- or C-terminal portion of the subunit sequence. The process was reiterated until the theoretical Mr and pI values matched the experimental values.
A good fit was obtained by removing eight amino acids from the C-terminal sequences and 11 amino acids from the N-terminal sequences.
Some of these fragments were found present in the water-soluble protein extract.
Cruciferin subunits for which theoretical Mr and pI values did not match the experimental values, even after amino acid removal.
A good fit was obtained by removing 27 amino acids from the C-terminal sequences.
A good fit was obtained by removing 28 amino acids from the C-terminal sequences and 13 amino acids from the N-terminal sequences.
A good fit was obtained by removing three amino acids from the C-terminal sequences and four amino acids from the N-terminal sequences.
A good fit was obtained by removing 18 amino acids from the N-terminal sequences.
Protein that was identified by both MALDI-TOF and PSD analyses.
Table IX.
Arabidopsis polypeptides whose abundance specifically varied during priming
| Protein Type | No. | Exp. Mr | Exp. pI | Arabidopsis Protein Name | Cov. | Theo. Mr | Theo. pI | Accession No. | Rel Abund HPa | Rel Abund OPa |
|---|---|---|---|---|---|---|---|---|---|---|
| % | ||||||||||
| 7 | 4 | 57.35 | 4.89 | β-2 Tubulin | 36 | 50.73 | 4.70 | 166898 | 4.2 ± 0.3 | 4.1 ± 0.4 |
| 7 | 89 | 15.80 | 5.72 | β-Subunit of 12S-2 seed storage proteinb | 36 | 20.80 | 7.03 | P15456 | 4.0 ± 0.1 | 2.6 ± 0.2 |
| 7 | 12 | 16.14 | 5.71 | β-Subunit of 12S-2 seed storage proteinc | 30 | 20.80 | 7.03 | P15456 | 3.4 ± 0.1 | 2.0 ± 0.1 |
| 9 | 8 | 17.93 | 4.99 | Class I heat shock protein 17.4 | 26 | 17.45 | 5.20 | P19036 | 1.1 ± 0.1 | 4.5 ± 0.5 |
| 9 | 9 | 17.94 | 5.50 | Heat shock protein 17.7 | 42 | 17.77 | 5.36 | S71248 | 1.3 ± 0.1 | 4.5 ± 0.2 |
| 10 | 1 | 79.36 | 5.08 | Luminal-binding protein | 20 | 71.17 | 5.08 | 1695719 | 0.9 ± 0.1 | 0.3 ± 0.03 |
| 11 | 17 | 73.40 | 6.58 | Phosphoenolpyruvate carboxykinase | 17 | 73.40 | 6.61 | 7449802 | 4.0 ± 0.2 | 1.0 ± 0.1 |
| 11 | 23 | 56.48 | 6.64 | Catalase 2 | 34 | 56.91 | 6.63 | P25819 | 3.4 ± 0.1 | 1.3 ± 0.1 |
| 11 | 6 | 54.71 | 5.06 | Tubulin α-Chain | 58 | 49.54 | 4.93 | 320183 | 2.7 ± 0.2 | 0.9 ± 0.1 |
Type-7 proteins, proteins whose level specifically increased during both hydropriming (HP) and osmopriming (OP); type-9 and type-10 proteins, proteins whose level specifically increased or decreased during osmopriming, respectively; type-11 proteins, proteins whose level specifically increased during hydropriming. Cov., Coverage; Exp., experimental; Rel Abund, relative abundance; Theo., theoretical.
Data obtained from densitometric analysis of individual spots (see Fig. 2): normalized spot volume in the primed seeds divided by the normalized spot volume in the dry mature seeds ± sd.
Theoretical Mr and pI values matched the experimental values by removing 15 amino acids from the C-terminal sequences.
Theoretical Mr and pI values matched the experimental values by removing 14 amino acids from the C-terminal sequences.
Table VII.
Arabidopsis polypeptides whose abundance specifically varied during radicle protrusion
| Protein Type | No. | Exp. Mr | Exp. pI | Arabidopsis Protein Name | Cov. | Theo. Mr | Theo. pI | Accession No. | Relative Abundancea |
|---|---|---|---|---|---|---|---|---|---|
| % | |||||||||
| 3 | 58 | 36.28 | 5.49 | Jasmonate-inducible myrosinase-binding protein | 41 | 32.16 | 5.46 | 9279641 | >200 |
| 3 | 67 | 63.25 | 6.47 | Thioglucosidase (myrosinase)b | 20 | 60.26 | 6.91 | S57621 | >200 |
| 3 | 63 | 57.09 | 6.11 | Isocitrate lyase | 30 | 50.42 | 6.29 | 113024 | >200 |
| 3 | 49 | 42.67 | 5.51 | S-adenosylmethionine synthetase | 19 | 42.79 | 5.51 | 9229983 | 18 ± 1 |
| 3 | 50 | 42.94 | 5.43 | Chloroplastic translation elongation factor EF-Tu (mature form) | 23 | 44.72 | 5.32 | S09152 | 5.0 ± 1.0 |
| 3 | 73 | 42.28 | 6.47 | β-Glucosidase | 18 | 57.83 | 6.02 | 9294684 | 4.0 ± 0.4 |
| 3 | 66 | 62.82 | 6.33 | Similar to jasmonate-induced protein homologue | 16 | 62.00 | 6.00 | 8777418 | 3.1 ± 0.2 |
| 4 | 110 | 57.00 | 6.39 | 12S Cruciferin precursor | 20 | 55.86 | 6.36 | T04623 | 0.5 ± 0.02 |
| 4 | 61 | 66.68 | 5.78 | Putative seed maturation protein | 43 | 67.19 | 5.78 | 4559335 | 0.4 ± 0.02 |
| 4 | 69 | 43.67 | 6.29 | 12S-2 Seed storage protein precursor | 18 | 48.03 | 6.56 | P15456 | 0.16 ± 0.07 |
| 4 | 56 | 34.02 | 5.36 | LEA protein in group 5 | 61 | 26.82 | 5.46 | 1526422 | 0.12 ± 0.01 |
| 4 | 71 | 50.44 | 7.67 | 12S-1 Seed storage protein precursor | 22 | 49.99 | 7.26 | P15455 | 0.09 ± 0.01 |
| 4 | 25 | 23.32 | 6.89 | β-Subunit of 12S cruciferin | 43 | 21.20 | 6.19 | T04623 | 0.05 ± 0.006 |
| 4 | 45 | 76.38 | 5.24 | Identical to heat shock protein 70 | 24 | 70.91 | 5.30 | 6587810 | 0.04 ± 0.005 |
| 4 | 11 | 16.61 | 5.75 | EM-like protein GEA1 | 30 | 16.61 | 5.75 | Q07187 | <0.005 |
Type-3 and -4 proteins, proteins whose accumulation level specifically increased or decreased during the radicle emergence step (2-d imbibed seeds), respectively. Cov., Coverage; Exp., experimental; Theo., theoretical.
Data obtained from densitometric analysis of individual spots (see Fig. 2): normalized spot volume in the 2-d imbibed seeds (Fig. 1C) divided by the normalized spot volume in the 1-d imbibed seeds (Fig. 1B) ± sd; >200 means that the accumulation level of the corresponding protein in the 1-d imbibed seeds was close to background; <0.005 means that the accumulation level of the corresponding protein in the 2-d imbibed seeds was close to background.
Protein identified by both MALDI-TOF and PSD analyses.
Table VIII.
Arabidopsis polypeptides whose abundance specifically varied during imbibition drying
| Protein Type | No. | Exp. Mr | Exp. pI | Arabidopsis Protein Name | Cov. | Theo. Mr | Theo. pI | Accession No. | Relative Abundancea |
|---|---|---|---|---|---|---|---|---|---|
| % | |||||||||
| 5 | 44 | 16.14 | 7.42 | β-Subunit of a 12S seed storage protein | PSD | 20.72 | 6.36 | 4204299 | >200 |
| 5 | 24 | 42.94 | 5.06 | Actin 7b | 28 | 41.96 | 5.26 | P53492 | >200 |
| 5 | 77 | 25.42 | 5.78 | α-Subunit of 12S cruciferinc | 30 | 34.68 | 6.42 | T04623 | >200 |
| 5 | 32 | 15.16 | 5.75 | β-Subunit of 12S-2 seed storage proteind | 18 | 20.80 | 7.03 | P15456 | 198 ± 9 |
| 5 | 26 | 96.38 | 5.75 | Cytoplasmic aconitate hydratase (aconitase) | 19 | 98.15 | 5.79 | 4586021 | 12 ± 1 |
| 5 | 76 | 23.94 | 5.58 | α-Subunit of 12S cruciferine | 27 | 34.68 | 6.42 | T04623 | 8.5 ± 1.7 |
| 5 | 41 | 37.72 | 6.74 | WD-40 repeat proteinb | 42 | 35.78 | 7.61 | 2289095 | 6.8 ± 0.5 |
| 6 | 37 | 40.29 | 6.49 | Cytosolic GAPDH | 26 | 36.99 | 6.34 | P25858 | 0.27 ± 0.03 |
Type-5 (imbibition-specific proteins), proteins whose accumulation level specifically increased during 1-d imbibition and decreased upon subsequent drying; type-6 (desiccation-specific proteins), proteins that were present in the dry mature seeds and whose level decreased during 1-d imbibition, then re-increased upon subsequent drying. Cov., Coverage; Exp., experimental; Theo., theoretical.
Data obtained from densitometric analysis of individual spots (see Fig. 2): normalized spot volume in the 1-d imbibed seeds (Fig. 1B) divided by the normalized spot volume in the dry mature seeds (Fig. 1A) ± sd; >200 means that the accumulation level of the corresponding protein in the dry mature seeds was close to background.
Also PSD, proteins that were identified by both MALDI-TOF and PSD analyses.
Theoretical Mr and pI values matched the experimental values by removing 66 amino acids from the C-terminal sequences and 13 amino acids from the N-terminal sequences.
Theoretical Mr and pI values matched the experimental values by removing 16 amino acids from the C-terminal sequences.
Theoretical Mr and pI values matched the experimental values by removing 88 acids from the C-terminal sequences and 13 amino acids from the N-terminal sequences.
The study was extended to the analysis of Arabidopsis seed proteome after 3 d of germination corresponding to phase III of the developmental process, by which nearly 100% of the seeds had completed germination (Table I). A dramatic modification of the protein pattern was observed (Fig. 1). There were too many spot changes to reference them all.
DISCUSSION
Seed germination is a key developmental process in the plant life cycle. Upon imbibition, embryonic cells switch from quiescence to highly active metabolism. In the present study, we initiated a broad proteomic analysis of the germination process using the model plant Arabidopsis, which is well defined genetically. Our general aim is to identify characteristic proteins of the various developmental phases, which will help understanding the biochemical and molecular processes underlying germination. In addition, these specific proteins might help characterizing germination vigor and optimizing industrial germination enhancement treatments.
For the dry mature seeds, there were about 1,300 proteins isolated in reproducible 2-D gels (Fig. 1). Most of them corresponded to type-0 proteins, whose abundance did not change significantly during germination. This suggests an important function for these proteins, at least up to radicle emergence (e.g. storage proteins and enzymes involved in storage lipid mobilization or in protein metabolism; see Table V).
The results will be discussed in the following text.
Mobilization of Stored Seed Reserves (Includes Type-0, -1, -3, -4, -5, -7, and -11)
11-12S Globulins are abundant seed storage proteins, being widely distributed among higher plants. They are synthesized during seed maturation on the mother plant in a precursor form consisting of a single protein chain of about 60 kD. At later stages, the precursor form is cleaved, yielding the mature globulins generally found in dry mature seeds. These are composed of six subunit pairs that interact noncovalently, each of which consists of an acidic α-subunit of Mr ≈ 40,000 and a basic β-subunit of Mr ≈ 20,000 covalently joined by a single disulfide group. These storage proteins are subsequently broken down during germination and used by the germinating seedling as an initial food source (Bewley and Black, 1994; Shewry et al., 1995). Contrasting with these well-accepted findings, dry mature Arabidopsis seeds were found to contain three forms of 12S globulins (cruciferins): (a) residual precursor forms (type-4 proteins nos. 69, 71, and 110 [Table VII and Fig. 2]), (b) α- and β-subunits (type-0 proteins nos. 83, 84, 85, 86, 87, and 98 in Table V; type-4 protein no. 25 in Table VII; and type-5 protein no. 44 in Table VIII), and (c) proteolyzed forms of α-subunits (type-0 proteins nos. 80, 117, 118, and 142 in Table V). The presence of some residual precursor forms in the dry mature Arabidopsis seeds was unexpected (Bewley and Black, 1994). A possible explanation could be that the maturation process giving rise to the formation of the α- and β-chains was not fully completed when developing seeds entered into quiescence. Another unexpected finding was to recover β-subunits (type-0 proteins nos. 86 and 87) and two fragments deriving from α-chains (type-0 proteins nos. 80 and 134) in the water-soluble protein fraction from the dry mature Arabidopsis seeds (Table V). Because by definition globulins are not soluble in water (Bewley and Black, 1994; Shewry and Casey, 1999), this behavior presumably reflects an early mobilization of the cruciferins during the maturation phase. This suggests that the anabolic processes that occur before germination and the catabolic processes that normally occur during germination are not fully separated developmentally in Arabidopsis. An examination of the data in Table V suggests that this initial mobilization of the cruciferins preferentially begins with proteolysis of the α-chains (type-0 proteins). This hypothesis is consistent with the hypothetical model of the 11S globulin subunits structure derived from the consensus structure model for 7S globulin subunits (Lawrence et al., 1994). It is also consistent with the determination of cleavage sites under limited trypsinolysis of 11S globulins, all of these sites being located within the α-chains (Müntz et al., 1999). Likewise, during germination sensu stricto fragments of α- and β-chains (type-1 proteins nos. 12, 32, 76, and 89 in Table VI) were released. Furthermore, during the radicle emergence step a 12S β-subunit was completely degraded (type-4 protein no. 25 in Table VII). Thus, once initiated at the level of α-subunits, mobilization of cruciferins continues during germination at the level of the β-subunits, indicating that the initial proteolysis of α-chains increased the sensitivity of β-chains toward further proteolytic attacks.
Table VI.
Arabidopsis polypeptides whose abundance specifically varied during germination sensu stricto
| Protein Type | No. | Exp. | Exp. | Arabidopsis Protein Name | Cov. | Theo. Mr | Theo. pI | Accession No. | Relative Abundancea |
|---|---|---|---|---|---|---|---|---|---|
| Mr | pI | % | |||||||
| 1 | 44 | 16.14 | 7.42 | β-Subunit of a 12S seed storage protein | PSD | 20.72 | 6.36 | 4204299 | >200 |
| 1 | 24 | 42.94 | 5.06 | Actin 7b | 28 | 41.96 | 5.26 | P53492 | >200 |
| 1 | 77 | 25.42 | 5.78 | α-Subunit of 12S cruciferinc | 30 | 34.68 | 6.42 | T04623 | >200 |
| 1 | 32 | 15.16 | 5.75 | β-Subunit of 12S-2 seed storage proteind | 18 | 20.80 | 7.03 | P15456 | 198 ± 9 |
| 1 | 26 | 96.38 | 5.75 | Cytoplasmic aconitate hydratase (aconitase) | 19 | 98.15 | 5.79 | 4586021 | 12 ± 1 |
| 1 | 76 | 23.94 | 5.58 | α-Subunit of 12S cruciferine | 27 | 34.68 | 6.42 | T04623 | 8.5 ± 1.7 |
| 1 | 41 | 37.72 | 6.74 | WD-40 repeat proteinb | 42 | 35.78 | 7.61 | 2289095 | 6.8 ± 0.5 |
| 1 | 4 | 57.35 | 4.89 | β-2 Tubulin | 36 | 50.73 | 4.70 | 166898 | 4.2 ± 0.3 |
| 1 | 89 | 15.80 | 5.72 | β-Subunit of 12S-2 seed storage proteinf | 36 | 20.80 | 7.03 | P15456 | 4.0 ± 0.1 |
| 1 | 17 | 73.40 | 6.58 | Phosphoenolpyruvate carboxykinase | 17 | 73.40 | 6.61 | 7449802 | 4.0 ± 0.2 |
| 1 | 12 | 16.14 | 5.71 | β-Subunit of 12S-2 seed storage proteing | 30 | 20.80 | 7.03 | P15456 | 3.4 ± 0.1 |
| 1 | 23 | 56.48 | 6.64 | Catalase 2 | 34 | 56.91 | 6.63 | P25819 | 3.4 ± 0.1 |
| 1 | 6 | 54.71 | 5.06 | Tubulin alpha chain | 58 | 49.54 | 4.93 | 320183 | 2.7 ± 0.2 |
| 2 | 37 | 40.29 | 6.49 | Cytosolic GAPDH | 26 | 36.99 | 6.34 | P25858 | 0.27 ± 0.03 |
Type-1 and -2 proteins, proteins whose accumulation level specifically increased or decreased during 1-d imbibition (germination sensu stricto), respectively. Cov., Coverage; Exp., experimental; Theo., theoretical.
Data obtained from densitometric analysis of individual spots (see Fig. 2): normalized spot volume in the 1-d imbibed seeds (Fig. 1B) divided by the normalized spot volume in the dry mature seeds (Fig. 1A) ± sd; >200 means that the accumulation level of the corresponding protein in the dry mature seeds was close to background.
Also PSD, proteins that were identified by PSD approach.
Theoretical Mr and pI values matched the experimental values by removing 66 amino acids from the C-terminal sequences and 13 amino acids from the N-terminal sequences.
Theoretical Mr and pI values matched the experimental values by removing 16 amino acids from the C-terminal sequences.
Theoretical Mr and pI values matched the experimental values by removing 88 amino acids from the C-terminal sequences and 13 amino acids from the N-terminal sequences.
Theoretical Mr and pI values matched the experimental values by removing 15 amino acids from the C-terminal sequences.
Theoretical Mr and pI values matched the experimental values by removing 14 amino acids from the C-terminal sequences.
Triacylglycerols are the major storage lipids in seeds (Bewley and Black, 1994; Miquel and Browse, 1995). In the present work, several enzymes playing a role in their catabolism were identified, including phosphoenolpyruvate carboxykinase (type-1 protein no. 17, Table VI), catalase (type-1 protein no. 23, Table VI), aconitase (type-5 protein no. 26, Table VIII), isocitrate lyase (type-3 protein no. 63, Table VII), malate dehydrogenase (type-0 protein no. 114, Table V), phosphoglycerate kinase (type-0 protein no. 121, Table V), and citrate synthase (type-0 protein no. 145, Table V). As has been shown previously (Comai et al., 1989; Turley and Trelease, 1990; Bewley and Black, 1994), some of them were already detected from the dry mature stage, whereas others accumulated during later stages of seed germination, before radicle emergence, or following this event. Such a progressive buildup of the pathway is presumably required for proper timing of storage lipid mobilization. More studies are needed to dissect the regulatory elements involved in this differential accumulation.
Germination Sensu Stricto (Includes Type-1, -2, -5, -6, -7, and -11)
By the 1st d of germination, corresponding to germination sensu stricto (i.e. none of the seeds germinated during this period; Table I), the abundance of 39 proteins varied (up- and down-regulation), belonging to the type-1 and-2 proteins described in Table II. Because about 1,300 proteins could be isolated in reproducible 2-D gels from the dry mature seeds, it appears that early phases of the germination process are associated only with modifications in the abundance of a limited number of proteins. This suggests either that resumption of metabolic activity during germination relies mainly upon proteins that are synthesized de novo but remain undetectable by the present technique or upon proteins that are stored during seed maturation on the mother plant. These type-1 and -2 proteins (Table II) had different behaviors upon submitting the 1-d imbibed seeds to a desiccation step, back to the original moisture content of the dry mature seeds. In one case, the accumulation patterns were maintained, indicating that these proteins could be considered as priming markers; if this occurred, these proteins were listed in Table IX. In the other case, the accumulation profiles were not maintained, but resembled those observed from the dry mature seeds: Here, the type-1 and -2 proteins were considered to represent specific proteins for the imbibed or desiccated state of the seeds. Therefore, they were referred as imbibition-desiccation proteins and are listed in Table VIII. The nature of these proteins is discussed in more detail below. These proteins, whose level vary globally during germination sensu stricto, correlate with initial events in the mobilization of protein and lipid reserves and the resumption of cell cycle activity (Table VI).
Radicle Emergence (Type-3 and -4)
Among the proteins associated with later stages of germination, a myrosinase (type-3 protein no. 67 in Table VII) was identified. This enzyme, whose activity was found highest in seeds and seedlings (Bones, 1990), catalyzes the hydrolysis of glucosinolates, a group of sulfur-containing glycosides. This occurs when plant tissue is damaged; for example, by herbivory. Although intact glucosinolates are relatively nontoxic, their breakdown products have important biological effects, as for the goitrogenic species that perturb thyroid function or the very reactive isothiocyanates that present antibacterial and antifungal properties (Rask et al., 2000). It has been reported that the level of total glucosinolates strongly decreases during germination and the early stages of plant development (Clossais-Besnard and Larher, 1991). Altogether, the data reinforce the finding that myrosinase accumulates in high quantity by the end of the germination process to hydrolyze glucosinolates, as a defense mechanism to protect the future seedling against herbivores and pathogens. Myrosinases have been shown to associate with other proteins, including myrosinase-binding proteins, presumably to regulate hydrolysis of glucosinolates (Rask et al., 2000). Consistent with this view, the abundance of two jasmonate-inducible myrosinase-binding proteins (type-3 proteins nos. 58 and 66, Table VII) strongly increased at the radicle emergence step.
Another type-3 protein associated with radicle protrusion, number 49 in Table VII, corresponded to S-adenosyl-Met synthetase (Ado-Met synthetase) that catalyzes the formation of S-adenosyl-Met (Ado-Met) from Met and ATP. Besides its well-known role as a methyl donor in a myriad of transmethylation reactions, Ado-Met is involved in several reactions that are essential for plant growth and development, such as the biosynthesis of ethylene, spermidine, spermine, and biotin (Ravanel et al., 1998). Owing to these housekeeping functions, this enzyme is presumably required for germination. Furthermore, its absence in the dry mature seeds helps to understand how embryonic cells can repress their metabolic activity to maintain quiescence. It is interesting that in germinating wheat embryos, Ado-Met synthetase is synthesized from the pool of stored mRNAs (Mathur et al., 1991), the quality of which is an important factor of seed vigor (Osborne, 1980).
Several water stress-related proteins were also identified, including LEA proteins and heat shock proteins (HSPs). LEA proteins accumulate late during embryogenesis, coincident with acquisition of desiccation tolerance of the developing seeds, and disappear during germination. They are presumed to be involved in binding or replacement of water, in sequestering ions that will accumulate under dehydration conditions, or in maintaining protein and membrane structure (Dure, 1993; Cuming, 1999). HSPs participate in diverse cellular processes by acting as molecular chaperones (Hong and Vierling, 2000). They are also described as being developmentally regulated, being abundant in dry mature seeds, and disappearing during germination (Wehmeyer et al., 1996). In agreement with these findings, the level of two LEAs (type-4 proteins nos. 11 and 56, Table VII) and of the HSP70 (type-4 protein no. 45, Table VII) decreased by the end of the germination process. Also, the type-10 protein number 1 (Table IX), whose level specifically decreased during osmopriming, was identified as a luminal-binding protein (BiP), a widely distributed and highly conserved group of proteins belonging to the HSP70 family (Munro and Pelham, 1987; Kalinski et al., 1995; Cascardo et al., 2000). Yet, the abundance of two LEAs (type-0 proteins nos. 94 and 102, Table V) and of the HSP70 cognate (type-0 protein no. 136, Table V) remained constant throughout the germination process. This suggests that some of the LEAs and HSPs may afford a protective function, not only during seed maturation, but also throughout germination. Such behavior was already observed during pea (Pisum sativum) seed germination (DeRocher and Vierling, 1995).
A putative seed maturation protein (SMP; type-4 protein no. 61 in Table VII) was present in dry mature seeds and disappeared by the 2nd d of germination. It showed high sequence homology with an SMP from soybean (Glycine max) (Hsing et al., 1998) and a pea protein called SBP65 (for seed biotinylated protein of 65 kD; Duval et al., 1994b). The two latter proteins exhibit a very peculiar biochemical feature in that they are biotinylated at a specific Lys residue within the atypical tetrapeptide sequence VGKF (Duval et al., 1994a; for review, see Alban et al., 2000). Sequence analysis disclosed that the Arabidopsis protein number 61 does contain this conserved tetrapeptide sequence, strongly suggesting that it might be biotinylated in vivo. Biotin is a vitamin required by all forms of life, being the cofactor of several housekeeping carboxylases (Patton et al., 1998; Alban et al., 2000). Because soybean SMP (Hsing et al., 1998) and pea SBP65 (Duval et al., 1994b) exhibit strictly seed-specific accumulation patterns and do not exhibit any known biotin-dependent enzyme activity, it has been proposed that they may play a role in sequestering biotin late in embryogenesis for subsequent use during germination (Alban et al., 2000). The present data suggest that such a role could be extended to the Arabidopsis homologue of the soybean and pea proteins.
Finally, the chloroplast translation elongation factor EF-Tu (type-3 protein no. 50 in Table VII) was found to specifically accumulate at the radicle emergence stage. EF-Tu is encoded by a nuclear gene in a precursor form containing an N-terminal transit peptide for plastid localization (Baldauf and Palmer, 1990). The protein identified in our study corresponds to the mature, functional, form of EF-Tu because experimental Mr and pI values indicated that it was devoid of the transit peptide sequence. It is interesting that three cytosolic translation factors, numbers 105, 127, and 129, were already present from the dry mature stage (type-0 proteins in Table V). Our results are in general agreement with those of Harrak et al. (1995) showing that the plastid translational apparatus is established early during plant development, presumably to allow the buildup of the photosynthetic system of which several components are encoded by plastid genes.
Imbibition (Type-5) and Desiccation (Type-6)
As pointed out by Kermode (1990, 1995), desiccation tolerance of seeds is a complex multifactorial trait involving a multitude of genes whose expression ultimately leads to mechanisms of both cellular protection, to sustain limited changes during drying itself, and cellular repair, to reverse any desiccation-induced changes when the appropriate hydrated conditions are reestablished. To better understand the mechanisms involved in adaptation/tolerance of seeds to water stress, we looked at changes in 2-D protein patterns that occurred in dry mature seeds upon 1-d imbibition and following subsequent drying of the imbibed seeds back to the original water content of the dry mature seeds. This comparative analysis led to the identification of two types of proteins. One corresponded to imbibition-associated polypeptides, whose levels increased during the 1-d imbibition and then decreased during subsequent drying. These proteins are referred as type-5 proteins in Table III. The other corresponded to desiccation-associated polypeptides, whose level decreased during the 1-d imbibition and then re-increased during subsequent drying. These proteins are referred as type-6 proteins in Table III.
Among the 19 imbibition-associated proteins of type-5, seven proteins, corresponding to the most abundant proteins within this type, were identified by MALDI-TOF analysis (see Table VIII). The imbibition-associated type-5 protein 24 (Fig. 2) was identified as actin 7 (ACT 7). Actin is a fundamental component of the cytoskeleton that participates in a number of cellular processes, such as cytoplasmic streaming, cell division, cell elongation, tip growth, nuclear positioning, and establishment of cell polarity (Kost et al., 1999). The Arabidopsis actin gene family comprises 10 members that are differentially expressed during development (McDowell et al., 1996). In particular, ACT7 is the only active actin gene in the hypocotyl and seed coat, suggesting that ACT7 is the sole source of actin for all actin-based processes in these tissues. In addition, the ACT7 promoter linked to a reporter gene showed strong expression in germinating seeds, which led McDowell et al. (1996) to propose that ACT7 expression is required for germination and/or hypocotyl growth. Our data, established at the protein level, are in perfect agreement with this proposal. Also, Santoni et al. (1994) reported that an actin isoform might have a role in the elongation process because the expression of this protein was consistently correlated with the hypocotyl elongation process.
The imbibition-associated protein 41 (type-5 protein in Table VIII) was found to belong to the large family of WD-40 repeat proteins that contain a structurally repetitive segment of 40 amino acid residues usually ending with the sequence Trp-Asp (Neer et al., 1994). It is interesting that this protein exhibits very high identity (>80%) with members of the receptor of activated C kinase subfamily, notably the protein encoded by the alfalfa Msgbl gene (McKhann et al., 1997). These proteins have a function in signal transduction and hormone-controlled plant cell division (McKhann et al., 1997). Because one of the specific features of early germination is the resumption of cell cycle activity (Georgieva et al., 1994; De Castro et al., 2000), it is tempting to propose that the WD-40 protein number 41 is somehow involved in this process. Furthermore, because its accumulation level was strongly and reversibly affected by the hydration status of the seeds, it also provides a good candidate to define the cascade emanating from imbibition and leading to resumption of cell cycle activity during germination. To our knowledge, this is the first evidence for accumulation of such a protein in the germination process.
It is remarkable that all protein changes associated with the desiccated state of seeds corresponded to proteins already present in the dry mature seeds. Their abundance readily diminished during imbibition and then re-increased during drying, up to about the same level as in the dry mature seeds (type-6 proteins, Table III). Thus, dehydration stress induces specific and reversible protein changes in seeds. In the present work, one of the three detected desiccation-specific type-6 proteins (no. 37, Table VIII and Fig. 2) was identified as cytosolic glyceraldehyde-3-phosphate dehydrogenase (GAPDHc). A previous study reported that dehydration strongly increases the GAPDHc protein level in leaves and callus tissue of the resurrection plant Craterostigma plantagineum that can withstand very severe desiccation (Velasco et al., 1994). The induction of GAPDHc during desiccation apparently is a conserved feature among different tissues and organs in plants. Other environmental stress conditions have been reported to induce increased levels of GAPDHc, not only in plants but also in animals. For example, this occurs during heat shock in Arabidopsis plants (Yang et al., 1993) and Xenopus laevis embryos (Nickells and Browder, 1988) and during anaerobic stress in maize (Zea mays; Chang et al., 2000), rice (Oryza sativa; Ricard et al., 1989), soybean (Russell et al., 1990), and Arabidopsis (Yang et al., 1993) plants. Leprince and Hoekstra (1998) reported that dehydration strongly increases the cytoplasmic viscosity of cowpea cotyledons, thereby impeding oxygen diffusion through the tissues and imposing anoxic stress conditions. Thus, a common feature of heat shock and anaerobic stresses might be oxygen deprivation, which could be the signal responsible for the observed increased level of GAPDHc during both types of stresses. In addition to catalyzing a reaction in glycolysis and gluconeogenesis, GAPDHc has also been shown to exhibit protein kinase activity, to bind RNA, and to enhance ribozyme and phosphotransferase activities (Chang et al., 2000, and references therein). We do not know yet which of the multiple activities of GAPDHc plays a role during dehydration stress. An elucidation of the multifaceted properties of such proteins synthesized during dehydration will help in understanding the mechanisms of desiccation tolerance in living cells.
Priming (Includes Type-7, -9, -10, and -11)
Priming of seeds has been shown to have beneficial effects on the germination and emergence of many species (Bradford, 1986). Therefore, the characterization of specific protein markers for improvement of seed quality is both of academic and economical interest. In the present study, we wished to exploit Arabidopsis as a model system for molecular characterization of seed priming. Based on the techniques currently used for a number of commercial crops (McDonald, 2000), we developed two priming treatments for Arabidopsis seeds: a hydro- and an osmopriming treatment. Hydropriming consists in soaking seeds in water and redrying them before they complete germination. Osmopriming is the process of soaking seeds in osmotica of low water potential to control the amount of water they imbibe. Both treatments improved Arabidopsis seed performance (Table I).
Three priming-associated polypeptides, whose abundance increased during both hydro- and osmopriming treatments, were detected and referred as type-7 proteins in Table IV. Two of these intrinsic priming proteins, numbers 12 and 89 (type-7 in Table IX), were identified as degradation products of 12S-cruciferin β-subunits. The same behavior has been observed during priming of sugar beet seeds (Job et al., 1997). This highlights the similarity, concerning storage protein mobilization during priming, between seeds from different plant families.
Tubulin subunits were found to accumulate during priming. This was the case for both α- (type-11 protein no. 6 in Table IX; Fig. 2) and β- (type-7 protein no. 4 in Table IX) chains. Such an accumulation of β-tubulin during priming has repeatedly been observed in many species, in relation with reactivation of cell cycle activity (see De Castro et al., 2000). In contrast, α-tubulin accumulation during priming has never been reported before.
A hydropriming-specific protein was identified as a catalase isoform (type-11 protein no. 23 in Table IX; Fig. 2). Its abundance increased during hydropriming and continued to increase at the radicle emergence stage. This observation is in agreement with the results of Gidrol et al. (1994) showing an accumulation of peroxidase activities, including catalase, during soybean seed germination. It is presumed that hydropriming initiates an oxidative stress, which generates reactive oxygen species, and therefore catalase must be present to minimize cell damage.
The abundance of low-Mr HSPs (LMW HSPs), of 17.4 kD (type-9 protein no. 8) and of 17.7 kD (type-9 protein no. 9; see Table IX) was found to specifically increase in osmoprimed seeds. Because LMW HSPs have molecular chaperone activity (Lee et al., 1995), these data suggest that LMW HSPs act by maintaining the proper folding of other proteins during the incomplete hydration resulting from soaking of the seeds in the polyethylene glycol (PEG) solution. In contrast, in the absence of osmotic stress LMW HSPs decline quickly during germination (Wehmeyer and Vierling, 2000, and references therein). In agreement with our data, the abundance of LMW HSPs was also observed to increase during osmopriming of soybean and maize seeds (Czarnecka et al., 1984; Heikkila et al., 1984). The detection of such proteins also confirms that water stress generated by high osmotic potential induces specific changes in protein synthesis (Davison and Bray, 1991; Jin et al., 2000).
CONCLUSIONS
In conclusion, our results reinforce recent findings (Chang et al., 2000; Jacobs et al., 2000; Peltier et al., 2000; Thiellement et al., 2001) that proteomic approaches can be successfully applied globally to investigate protein expression patterns in plants, even when working with rather limited amounts of material as in the case of Arabidopsis seeds. Most proteins resolved in 2-D gels gave high quality MALDI-TOF spectra, and most of these spectra allowed identification. Some of the presently identified proteins had previously been shown to play a role during germination and/or priming in several plant species, a finding that underlines the usefulness of using Arabidopsis as a model system. Furthermore, the present approach revealed new proteins associated with the imbibed state of the seeds (such as an actin isoform or a WD-40 repeat protein) or with the dehydrated state of the seeds (e.g. GAPDHc). Because Arabidopsis is amenable to reverse genetics, the role of these proteins during the germination process can be further assessed.
MATERIALS AND METHODS
Germination Experiments
Nondormant seeds of Arabidopsis, ecotype Landsberg erecta, were used in all experiments. Germination assays were carried out on three replicates of 50 seeds. Seeds were incubated at 25°C, with 8-h light daily, on two sheets of absorbent paper wetted with 4.5 mL of distilled water in covered plastic boxes. A seed was regarded as germinated when the radicle protruded through the seed coat.
Priming Treatments
Hydroprimed seeds were prepared by incubating dry mature seeds on wetted paper, for 1 d at 25°C, as described above for the germination experiments. Then, treated seeds were redried at room temperature. Osmoprimed seeds were prepared by incubating dry mature seeds in the dark, for 7 d at 20°C, in a −0.75 MPa PEG 6000 solution (Michel and Kaufmann, 1973), then briefly washing to remove adhering PEG, and drying in circulating air at 20°C during 3 d at 32% (v/v) of relative humidity.
Preparation of Total and Water-Soluble Protein Extracts
Total and water-soluble protein extracts were prepared from dry mature seeds, seeds at different stages of germination, and from primed seeds. Total proteins were extracted in the thiourea/urea lysis buffer as described (Harder et al., 1999) containing the protease inhibitor cocktail “complete Mini” from Roche Molecular Biochemicals (Meylan, France): 5,300 units Dnase I, 1,600 units Rnase A, and 0.2% (v/v) Triton X-100. After 10 min at 4°C, 14 mm dithiothreitol was added and the total protein extracts were stirred for 20 min at 4°C. Soluble proteins were extracted in chilled-distilled water.
2-D Electrophoresis
Proteins were first separated by electrophoresis according to charge. Isoelectrofocusing was carried out with 200 μg of proteins of the various extracts using gel strips forming an immobilized nonlinear pH gradient from 3 to 10. Strips were rehydrated for 14 h at 22°C with the thiourea/urea lysis buffer as described (Harder et al., 1999) containing 2% (v/v) Triton X-100, 20 mm dithiothreitol, and the protein extracts. Isoelectrofocusing was performed at 22°C in the Multiphor II system (Amersham Pharmacia Biotech, Orsay, France) for 1 h at 300 V and 7 h at 3,500 V. Proteins were then separated according to size. Equilibrated gel strips (Görg et al., 1987) were placed on top of vertical 10% (w/v) polyacrylamide gels. A denaturing solution (1% [w/v] low-melting agarose, 0.4% [w/v] SDS, 0.15 m bis-Tris, and 0.1 m HCl) was loaded on gel strips. After agarose solidification, electrophoresis was performed at 10°C in a buffer (pH 8.0) containing 25 mm Trizma base, 200 mm taurine, and 0.1% (w/v) SDS for 1 h at 35 V and 110 V overnight. Ten gels were run in parallel (Isodalt system from Amersham Pharmacia Biotech). For each condition analyzed, 2-D gels were made in triplicate and from two independent protein extractions.
Protein Staining and Analysis of 2-D Gels
Gels were stained with either silver nitrate as described (Blum et al., 1987) or the GelCode blue stain from Pierce (Rockford, IL), using the Hoefer Automated Gel Stainer apparatus from Amersham Pharmacia Biotech. Silver-stained gels were scanned with the Sharp JX-330 scanner equipped with the Labscan version 3.00 from Amersham Pharmacia Biotech. Image analysis was carried out with the ImageMaster 2-D Elite version 3.01 software (Amersham Pharmacia Biotech), according to the instruction booklet from Amersham Pharmacia Biotech. After spot detection and background subtraction (mode: average on boundary), 2-D gels were aligned, matched, and the quantitative determination of the spot volumes was performed (mode: total spot volume normalization). For each analysis, statistical data showed a high level of reproducibility between normalized spot volumes of gels produced in triplicate from the two independent protein extractions. Specific spots were described as showing variations during germination and priming when their volumes were significantly different (at least 2-fold in relative abundance) in the three analyzed silver-stained gels from each extraction. The most abundant proteins were identified by MALDI-TOF analysis.
Protein Identification by Mass Spectrometry
Spots of interest were excised from GelCode-stained 2-D gels. Proteins were characterized after in-gel trypsin cleavage by mass spectrometry and peptide mass fingerprinting, using a Bruker Reflex II MALDI-TOF mass spectrometer (Pappin et al., 1993). Proteins were identified by searching protein sequence databases (SWISS-PROT, the Protein Identification Resource, and GenPept) using tryptic-peptide molecular masses. To denote a protein as unambiguously identified, the following criteria were used: Coverage of the protein by the matching peptides must reach a minimum of 12% and at least four independent peptides should match within a stringent 0.001% maximum deviation of mass accuracy. As much as possible, protein identities were further confirmed from PSD spectra, generated from selected peptides. The searches with the PSD data were carried out using an in-house-developed algorithm, correctly assigning proteins from raw PSD spectra in less than 1 s. Theoretical masses and pI of identified proteins were predicted by sequence entry at http://www.expasy.ch/tools/peptide-mass.html. Search for sequence homologies was carried out at http://www.Arabidopsis.org/cgi-bin/blast/TAIRblast.pl.
ACKNOWLEDGMENT
We are grateful to Viviane Brozek for her help in setting up the 2-D technique.
Footnotes
This work was supported by the European Community (Fisheries, Agriculture and Agro-Industrial Research project grant no. CT97–3711, “Genetic and Molecular Markers for Seed Quality”), by the Region Rhône-Alpes (Programme “Biotechnologies: La Semence”), and by the Fund for Scientific Research of Flanders.
LITERATURE CITED
- Alban C, Job D, Douce R. Biotin metabolism in plants. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:17–47. doi: 10.1146/annurev.arplant.51.1.17. [DOI] [PubMed] [Google Scholar]
- Baldauf SL, Palmer JD. Evolutionary transfer of the chloroplast tufA gene to the nucleus. Nature. 1990;344:262–265. doi: 10.1038/344262a0. [DOI] [PubMed] [Google Scholar]
- Bewley JD. Seed germination and dormancy. Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD, Black M. Seeds: Physiology of Development and Germination. New York: Plenum Press; 1994. [Google Scholar]
- Blum H, Beier H, Gross HJ. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- Bones AM. Distribution of β-thioglucosidase activity in intact plants, cell and tissue cultures and regenerant plants of Brassica napus L. J Exp Bot. 1990;41:737–744. [Google Scholar]
- Bradford KJ. Manipulation of seed water relations via osmotic priming to improve germination under stress conditions. HortScience. 1986;21:1105–1112. [Google Scholar]
- Bradford KJ. A water relation analysis of seed germination rates. Plant Physiol. 1990;94:840–849. doi: 10.1104/pp.94.2.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford KJ, Chen F, Cooley MB, Dahal P, Downie B, Fukunaga KK, Gee OH, Gurusinghe S, Mella RA, Nonogaki H. Gene expression prior to radicle emergence in imbibed tomato seeds. In: Black M, Bradford KJ, Vázquez-Ramos J, editors. Seed Biology: Advances and Applications. Wallingford, UK: CABI International; 2000. pp. 231–251. [Google Scholar]
- Cahill DJ, Nordhoff E, O'Brien J, Klose J, Eickhoff H, Lehrach H. Bridging genomics and proteomics. In: Pennington SR, Dunn MJ, editors. Proteomics: from Protein Sequence to Function. Oxford: BIOS Scientific Publishers; 2001. pp. 1–22. [Google Scholar]
- Cascardo JCM, Almeida RS, Buzeli RAA, Carolino SMB, Otoni WC, Fontes EPB. The phosphorylation state and expression of soybean BiP isoforms are differentially regulated following abiotic stresses. J Biol Chem. 2000;275:14494–14500. doi: 10.1074/jbc.275.19.14494. [DOI] [PubMed] [Google Scholar]
- Chang WWP, Huang L, Shen M, Webster C, Burlingame AL, Roberts JKM. Patterns of protein synthesis and tolerance of anoxia in root tips of maize seedlings acclimated to a low-oxygen environment, and identification of proteins by mass spectrometry. Plant Physiol. 2000;122:295–317. doi: 10.1104/pp.122.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clossais-Besnard N, Larher F. Physiological role of glucosinolates in Brassica napus: concentration and distribution pattern of glucosinolates among plant organs during a complete life cycle. J Sci Food Agric. 1991;56:25–38. [Google Scholar]
- Comai L, Dietrich RA, Maslyar DJ, Baden CS, Harada JJ. Coordinate expression of transcriptionally regulated isocitrate lyase and malate synthase genes in Brassica napus L. Plant Cell. 1989;1:293–300. doi: 10.1105/tpc.1.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuming AC. LEA proteins. In: Shewry PR, Casey R, editors. Seed Proteins. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 753–780. [Google Scholar]
- Czarnecka E, Edelman L, Schöffl F, Key JL. Comparative analysis of physical stress responses in soybean seedlings using cloned heat shock cDNAs. Plant Mol Biol. 1984;3:45–58. doi: 10.1007/BF00023415. [DOI] [PubMed] [Google Scholar]
- Davison PA, Bray CM. Protein synthesis during osmopriming of leek (Allium porrum) seeds. Seed Sci Res. 1991;1:29–35. [Google Scholar]
- De Castro RD, van Lammeren AAM, Groot SPC, Bino RJ, Hilhorst HWM. Cell division and subsequent radicle protrusion in tomato seeds are inhibited by osmotic stress but DNA synthesis and formation of microtubular cytoskeleton are not. Plant Physiol. 2000;122:327–335. doi: 10.1104/pp.122.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRocher A, Vierling E. Cytoplasmic HSP70 homologues of pea: differential expression in vegetative and embryonic organs. Plant Mol Biol. 1995;27:441–456. doi: 10.1007/BF00019312. [DOI] [PubMed] [Google Scholar]
- Dure L. The LEA proteins of higher plants. In: Verma DPS, editor. Control of Plant Gene Expression. New York: CRC Press; 1993. pp. 325–335. [Google Scholar]
- Duval M, DeRose RT, Job C, Faucher D, Douce R, Job D. The major biotinyl protein from Pisum sativum seeds covalently binds biotin at a novel site. Plant Mol Biol. 1994a;26:265–273. doi: 10.1007/BF00039537. [DOI] [PubMed] [Google Scholar]
- Duval M, Job C, Alban C, Douce R, Job D. Developmental patterns of free and protein-bound biotin during maturation and germination of seeds of Pisum sativum: characterization of a novel seed-specific biotinylated protein. Biochem J. 1994b;299:141–150. doi: 10.1042/bj2990141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva EI, Lòpez-Rodas G, Hittmair A, Feichtinger H, Brosch G, Loidl P. Maize embryo germination. Planta. 1994;192:118–124. [Google Scholar]
- Gidrol X, Lin WS, Dégousée N, Yip SF, Kush A. Accumulation of reactive oxygen species and oxidation of cytokinin in germinating soybean seeds. Eur J Biochem. 1994;224:21–28. doi: 10.1111/j.1432-1033.1994.tb19990.x. [DOI] [PubMed] [Google Scholar]
- Görg A, Postel W, Weser J, Günther S, Strahler JR, Hanash SM, Somerlot L. Elimination of point streaking on silver stained two-dimensional gels by addition of iodoacetamide to the equilibration buffer. Electrophoresis. 1987;8:122–124. [Google Scholar]
- Groot SPC, Kieliszewska-Rokicha B, Vermeer E, Karssen CM. Gibberellin-induced hydrolysis of endosperm cell walls in gibberellin-deficient tomato seeds prior to radicle protrusion. Planta. 1988;174:500–504. doi: 10.1007/BF00634479. [DOI] [PubMed] [Google Scholar]
- Harder A, Wildgruber R, Nawrocki A, Fey SJ, Larsen PM, Görg A. Comparison of yeast cell protein solubilization procedures for two-dimensional electrophoresis. Electrophoresis. 1999;20:826–829. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<826::AID-ELPS826>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Harrak H, Lagrange T, Bisanz-Seyer C, Lerbs-Mache S, Mache R. The expression of nuclear genes encoding plastid ribosomal proteins precedes the expression of chloroplast genes during early phases of chloroplast development. Plant Physiol. 1995;108:685–692. doi: 10.1104/pp.108.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkila JJ, Papp JET, Schultz GA, Bewley JD. Induction of heat shock protein messenger RNA in maize mesocotyls by water stress, abscisic acid, and wounding. Plant Physiol. 1984;76:270–274. doi: 10.1104/pp.76.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydecker W, Higgins J, Gulliver RL. Accelerated germination by osmotic seed treatment. Nature. 1973;246:42–44. [Google Scholar]
- Hong S-W, Vierling E. Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci USA. 2000;97:4392–4397. doi: 10.1073/pnas.97.8.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsing YC, Tsou CH, Hsu TF, Chen ZY, Hsieh KL, Hsieh JS, Chow TY. Tissue- and stage-specific expression of a soybean (Glycine max L.) seed-maturation, biotinylated protein. Plant Mol Biol. 1998;38:481–490. doi: 10.1023/a:1006079926339. [DOI] [PubMed] [Google Scholar]
- Jacobs DI, van der Heijden R, Verpoorte R. Proteomics in plant biotechnology and secondary metabolism research. Phytochem Anal. 2000;11:277–287. [Google Scholar]
- Jin S, Chen CCS, Plant AL. Regulation by ABA of osmotic-stress-induced changes in protein synthesis in tomato roots. Plant Cell Environ. 2000;23:51–60. [Google Scholar]
- Job C, Kersulec A, Ravasio L, Chareyre S, Pépin R, Job D. The solubilization of the basic subunit of sugarbeet seed 11-S globulin during priming and early germination. Seed Sci Res. 1997;7:225–243. [Google Scholar]
- Job D, Capron I, Job C, Dacher F, Corbineau F, Côme D. Identification of germination-specific protein markers and their use in seed priming technology. In: Black M, Bradford KJ, Vázquez-Ramos J, editors. Seed Biology: Advances and Applications. Wallingford, UK: CAB International; 2000. pp. 449–459. [Google Scholar]
- Kalinski A, Rowley DL, Loer DS, Foley C, Buta G, Hermann EM. Binding-protein expression is subject to temporal, developmental and stress-induced regulation in terminally differentiated soybean organs. Planta. 1995;195:611–621. doi: 10.1007/BF00195722. [DOI] [PubMed] [Google Scholar]
- Kermode AR. Regulatory mechanisms involved in the transition from seed development to germination. Plant Sci. 1990;9:155–195. [Google Scholar]
- Kermode AR. Regulatory mechanisms in the transition from seed development to germination: interactions between the embryo and the seed environment. In: Kigel J, Galili G, editors. Seed Development and Germination. New York: Marcel Dekker; 1995. pp. 273–332. [Google Scholar]
- Kost B, Mathur J, Chua N-H. Cytoskeleton in plant development. Curr Opin Plant Biol. 1999;2:462–470. doi: 10.1016/s1369-5266(99)00024-2. [DOI] [PubMed] [Google Scholar]
- Lawrence MC, Izard T, Beuchat M, Blagrove RJ, Coleman PM. Structure of phaseolin at 2.2 Å resolution: implications for a common vicilin/legumin structure and the genetic engineering of seed storage proteins. J Mol Biol. 1994;238:748–776. doi: 10.1006/jmbi.1994.1333. [DOI] [PubMed] [Google Scholar]
- Lee GJ, Pokala N, Vierling E. Structure and in vitro molecular chaperone activity of cytosolic small heat shock proteins from pea. J Biol Chem. 1995;270:10432–10438. doi: 10.1074/jbc.270.18.10432. [DOI] [PubMed] [Google Scholar]
- Leprince O, Hoekstra FA. The responses of cytochrome redox state and energy metabolism to dehydration support a role for cytoplasmic viscosity in desiccation tolerance. Plant Physiol. 1998;118:1253–1264. doi: 10.1104/pp.118.4.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leymarie J, Damerval C, Marcotte L, Combes V, Vartanian N. Two-dimensional protein patterns of Arabidopsis wild-type and auxin insensitive mutants, axr1, axr2, reveal interactions between drought and hormonal responses. Plant Cell Physiol. 1996;37:966–975. doi: 10.1093/oxfordjournals.pcp.a029046. [DOI] [PubMed] [Google Scholar]
- Mathur M, Saluja D, Sachar RC. Post-transcriptional regulation of S-adenosylmethionine synthetase from its stored mRNA in germinated wheat embryos. Biochim Biophys Acta. 1991;1078:161–170. doi: 10.1016/0167-4838(91)99005-d. [DOI] [PubMed] [Google Scholar]
- McDonald MB. Seed priming. In: Black M, Bewley JD, editors. Seed Technology and Its Biological Basis. Sheffield, UK: Sheffield Academic Press Ltd.; 2000. pp. 287–325. [Google Scholar]
- McDowell JM, An Y-Q, Huang S, McKinney EC, Meagher RB. The Arabidopsis ACT7 actin gene is expressed in rapidly developing tissues and responds to several external stimuli. Plant Physiol. 1996;111:699–711. doi: 10.1104/pp.111.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann HI, Frugier F, Petrovics G, de la Peña TC, Jurkevitch E, Brown S, Kondorosi E, Kondorosi A, Crespi M. Cloning of a WD-repeat-containing gene from alfalfa (Medicago sativa): a role in hormone-mediated cell division? Plant Mol Biol. 1997;34:771–780. doi: 10.1023/a:1005899410389. [DOI] [PubMed] [Google Scholar]
- Meurs C, Basra AS, Karssen CM, van Loon LC. Role of abscisic acid in the induction of desiccation tolerance in developing seeds of Arabidopsis thaliana. Plant Physiol. 1992;98:1484–1493. doi: 10.1104/pp.98.4.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel BE, Kaufmann MR. The osmotic potential of polyethylene glycol 6000. Plant Physiol. 1973;51:914–916. doi: 10.1104/pp.51.5.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel M, Browse J. Lipid biosynthesis in developing seeds. In: Kigel J, Galili G, editors. Seed Development and Germination. New York: Marcel Dekker; 1995. pp. 169–193. [Google Scholar]
- Munro S, Pelham HRB. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- Müntz K, Horstmann C, Schlesier B. Vicia globulins. In: Shewry PR, Casey R, editors. Seed Proteins. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 259–284. [Google Scholar]
- Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- Nickells RW, Browder LW. A role for glyceraldehyde-3-phosphate dehydrogenase in the development of thermotolerance in Xenopus laevis embryos. J Cell Biol. 1988;107:1901–1909. doi: 10.1083/jcb.107.5.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne DJ. Senescence in seeds. In: Thimann KV, editor. Senescence in Plants. Boca Raton, FL: CRC Press; 1980. pp. 13–37. [Google Scholar]
- Pappin DJC, Hojrup P, Bleasby AJ. Rapid identification of proteins by peptide-mass fingerprinting. Curr Biol. 1993;3:327–332. doi: 10.1016/0960-9822(93)90195-t. [DOI] [PubMed] [Google Scholar]
- Patton DA, Schetter AL, Franzmann LH, Nelson K, Ward ER, Meinke DW. An embryo-defective mutant of Arabidopsis disrupted in the final step of biotin synthesis. Plant Physiol. 1998;116:935–946. doi: 10.1104/pp.116.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier JB, Friso G, Kalume DE, Roepstorff P, Nilsson F, Adamska I, van Wijk KJ. Proteomics of the chloroplast: systematic identification and targeting analysis of luminal and peripheral thylakoid proteins. Plant Cell. 2000;12:319–341. doi: 10.1105/tpc.12.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask L, Andréasson E, Ekbom B, Eriksson S, Pontoppidan B, Meijer J. Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol Biol. 2000;42:93–113. [PubMed] [Google Scholar]
- Ravanel S, Gakière B, Job D, Douce R. The specific features of methionine biosynthesis and metabolism in plants. Proc Natl Acad Sci USA. 1998;95:7805–7812. doi: 10.1073/pnas.95.13.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard B, Rivoal J, Pradet A. Rice cytosolic glyceraldehyde 3-phosphate dehydrogenase contains two subunits differentially regulated by anaerobiosis. Plant Mol Biol. 1989;12:131–139. doi: 10.1007/BF00020498. [DOI] [PubMed] [Google Scholar]
- Russell DA, Wong DML, Sachs MM. The anaerobic response of soybean. Plant Physiol. 1990;92:401–407. doi: 10.1104/pp.92.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni V, Bellini C, Caboche M. Use of two-dimensional protein-pattern analysis for the characterization of Arabidopsis thaliana mutants. Planta. 1994;192:557–566. [Google Scholar]
- Santoni V, Delarue M, Caboche M, Bellini C. A comparison of two-dimensional electrophoresis data with phenotypical traits in Arabidopsis leads to the identification of a mutant (cri1) that accumulates cytokinins. Planta. 1997;202:62–69. doi: 10.1007/s004250050103. [DOI] [PubMed] [Google Scholar]
- Santoni V, Rabilloud T, Doumas P, Rouquié D, Mansion M, Kieffer S, Garin J, Rossignol M. Towards the recovery of hydrophobic proteins on two-dimensional electrophoresis gels. Electrophoresis. 1999;20:705–711. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<705::AID-ELPS705>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Santoni V, Rouquié D, Doumas P, Mansion M, Boutry M, Degand H, Dupree P, Pack L, Sherrier J, Prime T. Use of a proteome strategy for tagging proteins present at the plasma membrane. Plant J. 1998;16:633–641. doi: 10.1046/j.1365-313x.1998.00335.x. [DOI] [PubMed] [Google Scholar]
- Shewry PR, Casey R. Seed proteins. In: Shewry PR, Casey R, editors. Seed Proteins. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 1–10. [Google Scholar]
- Shewry PR, Napier JA, Tatham AS. Seed storage proteins: structures and biosynthesis. Plant Cell. 1995;7:945–956. doi: 10.1105/tpc.7.7.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Still DW, Dahal P, Bradford KJ. A single-seed assay for endo-β-mannanase activity from tomato endosperm and radicle tissues. Plant Physiol. 1997;113:13–20. doi: 10.1104/pp.113.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Thiellement H, Plomion C, Zivy M. Proteomics as a tool for plant genetics and breeding. In: Pennington SR, Dunn MJ, editors. Proteomics: from Protein Sequence to Function. Oxford: BIOS Scientific Publishers; 2001. pp. 289–309. [Google Scholar]
- Turley RB, Trelease RN. Development and regulation of three glyoxysomal enzymes during cotton seed maturation and growth. Plant Mol Biol. 1990;14:137–146. doi: 10.1007/BF00018555. [DOI] [PubMed] [Google Scholar]
- Velasco R, Salamini F, Bartels D. Dehydration and ABA increase mRNA levels and enzyme activity of cytosolic GAPDH in the resurrection plant Craterostigma plantagineum. Plant Mol Biol. 1994;26:541–546. doi: 10.1007/BF00039567. [DOI] [PubMed] [Google Scholar]
- Wehmeyer N, Hernandez LD, Finkelstein RR, Vierling E. Synthesis of small heat-shock proteins is part of the development program of late seed maturation. Plant Physiol. 1996;112:747–757. doi: 10.1104/pp.112.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehmeyer N, Vierling E. The expression of small heat shock proteins in seeds responds to discrete developmental signals and suggests a general protective role in desiccation tolerance. Plant Physiol. 2000;122:1099–1108. doi: 10.1104/pp.122.4.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Kwon HB, Peng HP, Shih MC. Stress responses and metabolic regulation of glyceraldehyde-3-phosphate dehydrogenase genes in Arabidopsis. Plant Physiol. 1993;101:209–216. doi: 10.1104/pp.101.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]