Abstract
The generalist insect herbivore Trichoplusia ni (cabbage looper) readily consumes Arabidopsis and can complete its entire life cycle on this plant. Natural isolates (ecotypes) of Arabidopsis are not equally susceptible to T. ni feeding. While some are hardly touched by T. ni, others are eaten completely to the ground. Comparison of two commonly studied Arabidopsis ecotypes in choice experiments showed that Columbia is considerably more resistant than Landsberg erecta. In no-choice experiments, where larvae were confined on one or the other ecotype, weight gain was more rapid on Landsberg erecta than on Columbia. Genetic mapping of this difference in insect susceptibility using recombinant inbred lines resulted in the discovery of the TASTY locus near 85 cM on chromosome 1 of Arabidopsis. The resistant allele of this locus is in the Columbia ecotype, and an F1 hybrid has a sensitive phenotype that is similar to that of Landsberg erecta. The TASTY locus is distinct from known genetic differences between Columbia and Landsberg erecta that affect glucosinolate content, trichome density, disease resistance, and flowering time.
The interactions between plants and insect herbivores comprise a complex, co-evolved natural system. Plants put up an array of chemical and physical barriers to keep from being eaten and insects do their best do circumvent these defenses. Broadly, plant-feeding insects can be classified as either generalist or specialist herbivores (Bernays and Chapman, 1994). Generalist herbivores tend to consume the least defended parts of a variety of plants. Specialist herbivores, on the other hand, have developed a tolerance for a particular species or group of plants and often co-opt plant chemical defenses as attractive signals. Plants are not passive players in their interaction with insects, but mount induced defense responses when they are under attack. Such responses can include the production of feeding deterrents such as protease inhibitors (Broadway and Colvin, 1992) and the release of volatiles that attract predators of the insect herbivores (Mattiacci et al., 1995). Both methyl jasmonate (McConn et al., 1997) and ethylene (Kahl et al., 2000; Stotz et al., 2000) have been implicated as molecules that mediate induced insect defenses in plants.
While chemical ecology of plant-insect interactions is a well-developed field, molecular genetic analysis has become possible only relatively recently. The small crucifer Arabidopsis has been used extensively to study plant-microbe interactions, and it is also an excellent model system for studying the genetic basis of insect defense in plants. A variety of insects, both generalist herbivores and crucifer-feeding specialists, have been shown to feed on Arabidopsis in nature or in the lab. These include Lepidoptera: Trichoplusia ni, Pieris rapae, Plutella xylostella, Spodoptora exigua, Spodoptera littoralis, Helicoverpa zea, Pseudoplusia includens, Heliothis virescens; Coleoptera: Phyllotreta zimmermani, Psylliodes convexior; Homoptera: Brevicoryne brassicae, Myzus persicae; Diptera: Bradysia impatiens; and Thysanoptera: Frankliniella occidentalis (Grant-Peterson, 1993; Singh et al., 1994; Grant-Petersson and Renwick, 1996; Rashotte and Feldmann, 1996; McConn et al., 1997; Santos et al., 1997; Mauricio, 1998; Reymond et al., 2000; Stotz et al., 2000). We chose to use T. ni for our experiments because this insect is commercially available, can be raised easily in the lab on a defined medium, and can complete its entire life cycle feeding on Arabidopsis.
T. ni is a generalist herbivore, whose larvae feed on a wide variety of plant species, including Brassica crops, sugar beets, beans, cotton, potatoes, tomatoes, lettuce, celery, alfalfa, melons, cucumbers, squash, and citrus (Shorey et al., 1962). However, not all of these plants are equally suitable as hosts for T. ni and there is also intraspecies variation in host plant attractiveness. For instance, both leaf chemical content (Khan et al., 1986a; Khan et al., 1987) and trichome density (Khan et al., 1986b) affect the susceptibility of different soybean cultivars to T. ni feeding. While Brassicaceae are generally quite attractive host plants for T. ni (hence the common name cabbage looper), they also produce deterrent signals. For example, cabbage leaf extracts can inhibit oviposition by adult moths on an otherwise attractive substrate (Renwick and Radke, 1982). The most extensively studied insect defense mechanism of the Brassicaceae is the glucosinolate-myrosinase system. Upon plant damage, myrosinase cleaves the thioglucoside linkage of glucosinolates, enabling further breakdown and the release of a variety of thiocyanates, isothiocyanates, and nitriles that are distasteful to many herbivores (Bones and Rossiter, 1996). In particular, T. ni larval feeding is inhibited and weight gain is reduced with increased glucosinolates in the diet (Grant-Peterson, 1993; Shields and Mitchell, 1995; Stowe, 1998).
Natural accessions of Arabidopsis, which have been collected from a variety of habitats around the world, show considerable variation at the DNA sequence level and represent an important resource for discovering genetic variation affecting a large variety of phenotypes. A comparison of genomic sequences of the two most commonly studied ecotypes, Columbia (Col) and Landsberg erecta (Ler), reveals a polymorphism roughly every 2,000 bp (Arabidopsis Genome Initiative, 2000). Many examples of phenotypic variation among ecotypes have been documented, including disease resistance (Kunkel, 1996), leaf trichome density (Larkin et al., 1996), glucosinolate content (Magrath et al., 1994; Mithen et al., 1995), and epicuticular wax composition (Rashotte et al., 1997). Frequently such phenotypic variation is quantitative (continuous) and is the result of genetic changes in multiple genes. Statistical methods that enable the mapping of multiple genes or quantitative trait loci (QTLs; Tanksley, 1993; Jansen, 1996; Kearsey and Farquar, 1998) have been used to map a variety of QTLs in Arabidopsis (Alonso-Blanco and Koornneef, 2000).
In this work we characterize T. ni feeding on Arabidopsis and begin a genetic dissection of the variation in insect susceptibility among Arabidopsis ecotypes. In particular, we present the novel finding of a highly significant QTL on chromosome 1 of Arabidopsis, which causes the Col ecotype to be much more resistant to insect feeding than the Ler ecotype. This locus is distinct from known Arabidopsis QTLs and other genes that might be expected to affect insect feeding by altering glucosinolate content, wax composition, trichome density, flowering time, or disease resistance.
RESULTS
In initial experiments, we observed the feeding habits of T. ni on the rosette leaves of 4-week-old Arabidopsis plants. As is typical of generalist herbivores, T. ni larvae usually began feeding on the oldest (least well defended) parts of the Arabidopsis rosette and worked their way toward the center (Bernays and Chapman, 1994). This behavior is similar to what has been observed with T. ni larvae feeding on cabbage (Broadway and Colvin, 1992). In many cases the larvae reached a point toward the center of the Arabidopsis rosette where they stopped feeding, even if they were confined on the plant and were not given any other food. We used choice experiments to test whether this preference for older leaves is due to their inherent structure or chemistry, or is simply the result of their position at the outside of the rosette. Leaf plugs made from older Col leaves that were beginning to senesce were compared with ones made from young leaves at the center of the rosette of the same plants. In 72 experiments with individual larvae, 51 larvae consumed more of the older leaves, 17 consumed more of the younger leaves, and four did not consume any leaf material. Overall, there was a significant preference for older leaves (P < 0.01, chi-square test).
As inducible responses are known to play an important role in plant defense against insects (McConn et al., 1997; Kahl et al., 2000), we looked at the effects of leaf damage on the feeding preferences of T. ni. Leaf plugs from damaged (squeezed with pliers 2 d earlier) and undamaged leaves of the Col ecotype were compared in choice experiments. In 36 runs of the experiment, 26 larvae showed a preference for undamaged leaves, eight showed a preference for damaged leaves, and two larvae did not consume any leaf material. Among the larvae that consumed leaf plugs, there was a significant preference (P < 0.01, chi-square test) for leaf plugs made from undamaged leaves. These results indicate that there are damage-inducible responses in Arabidopsis that occur over 2 d that cause plants to be less attractive for cabbage looper feeding.
The size of the remaining leaf rosette, i.e. the point at which T. ni larvae stopped feeding, differs among Arabidopsis ecotypes. While some ecotypes are eaten all the way to the ground, others are hardly touched at all. We assessed T. ni feeding on 34 ecotypes in a randomized array by measuring the diameter of the rosette that was left when T. ni larvae stopped feeding. There was little or no variation among the 16 plants of each ecotype that were grown together in one pot in these experiments. The ecotypes were given numerical scores based on the diameter of the remaining rosettes: 1, <1 cm; 2, 1 to 2 cm; and 3, >2 cm. Averaged results of two (22 ecotypes) or three (12 ecotypes) independent runs of the experiment are presented in Table I. About one-half of the lines tested were almost completely consumed by the T. ni larvae. Among the remainder there was a continuum of resistance levels culminating with four ecotypes that were hardly touched by T. ni under our growing conditions. Non-quantitative observations of the larval feeding indicated that Shahdara was the most consistently resistant ecotype among those that were tested.
Table I.
T. ni feeding on Arabidopsis ecotypes
| Stock Center No. | Ecotype Name | Feeding Scorea |
|---|---|---|
| CS934 | Aa-0 | 1.0 |
| CS3109 | Ber | 1.0 |
| CS917 | Da(1)-12 | 1.0 |
| CS910 | Di-G | 1.0 |
| CS919 | Di-M | 1.0 |
| CS920 | En-D | 1.0 |
| CS1148 | Est-0 | 1.0 |
| CS20 | Ler-0 | 1.0 |
| CS925 | Litva | 1.0 |
| CS1338 | LL | 1.0 |
| CS1390 | Nd-0 | 1.0 |
| CS3081 | No-0 | 1.0 |
| CS1643 | Oy-1 | 1.0 |
| CS3110 | Wei | 1.0 |
| CS915 | Ws | 1.0 |
| CS921 | En-T | 1.3 |
| CS3179 | Gr3 | 1.3 |
| CS904 | Mh-0 | 1.3 |
| CS913 | RLD1 | 1.3 |
| CS1640 | Tsu-1 | 1.3 |
| CS1096 | Cvi-0 | 1.5 |
| CS1320 | Li-5 | 1.7 |
| CS972 | Bla-2 | 2.0 |
| CS1637 | Ema-0 | 2.0 |
| CS923 | H55 | 2.0 |
| CS6195 | Wu-0 | 2.3 |
| CS3180 | Co | 2.5 |
| CS933 | Col-0 | 2.5 |
| CS1635 | Cnt-1 | 2.7 |
| CS903 | Kas-1 | 2.7 |
| CS916 | Condara | 3.0 |
| CS922 | Hodja | 3.0 |
| CS905 | Ms-0 | 3.0 |
| CS6180 | Shahdara | 3.0 |
1, Most sensitive; 3, most resistant.
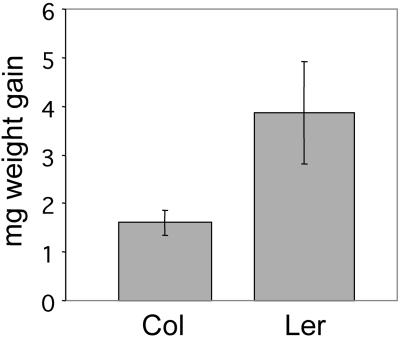
We concentrated on the Col and Ler ecotypes in further experiments. Even though Col and Ler do not represent the extremes of sensitivity to T. ni feeding (scores of 2.5 and 1, respectively, in Table I), the wealth of genetic and biochemical information that is available for this pair of ecotypes greatly simplifies subsequent genetic work. In whole plant feeding experiments, T. ni larvae show a clear preference for Col over Ler plants when given a choice. This feeding preference was also reflected in the weight gain of T. ni larvae in a no-choice feeding experiment. Newly hatched larvae were allowed to feed on Col or Ler plants for 4 d and their dry weight was recorded. Mean and sd of the weight of 113 larvae feeding on Col and 119 larvae feeding on Ler are shown in Figure 1. The mean weight gain of T. ni on the Col plants was significantly less than on the Ler plants (1.61 mg versus 3.87 mg, P < 0.01, Student's t test).
Figure 1.
Mean and sd of weight gain (dry weight) of newly hatched cabbage looper larvae feeding for 1 week on Col (n = 113) and Ler (n = 119) ecotypes in a no-choice experiment (P < 0.01, Student's t test).
We attempted to quantify the T. ni sensitivity difference between Col and Ler using a leaf plug choice experiment similar to the ones that were used to compare old versus young and damaged versus undamaged leaves. In 22 runs of this experiment, 10 larvae showed a preference for Col leaf plugs and 12 showed a preference for Ler leaf plugs, an insignificant difference (P = 0.42, chi-square test). This result is in contrast to both the whole-plant feeding and weight gain experiments, where there was a significant preference for Ler. Perhaps not surprisingly, there are physiological differences between detached leaf plugs consumed by T. ni over a period of hours and whole plants consumed over a period of days.
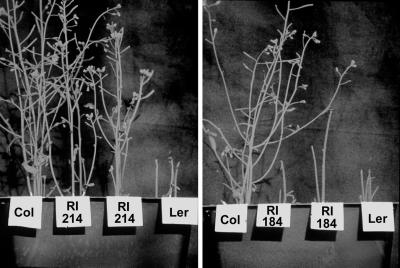
Although Col plants are more resistant to T. ni feeding at all growth stages, this difference is most apparent in the early flowering stage of the plants (about 5 weeks old). Ler flowers, siliques, and the flower stalk are readily consumed, but the inflorescences are hardly touched on Col plants (Fig. 2). At the end of this experiment (3 d after T. ni addition), the larvae wandered off rather than consume any more Col material. Therefore, we chose this growth stage for subsequent experiments designed to determine the genetic basis of the difference in insect susceptibility of Col and Ler. The large number of markers and well-defined genetic map available for the Col and Ler ecotypes enable the genetic mapping of complex traits like insect feeding. We measured T. ni feeding preferences on flowering plants of 100 recombinant inbred (RI) lines derived from Col and Ler (Lister and Dean, 1993). Each individual RI plant was given a score of 0 (Ler-like), 1 (Col-like), or 0.5 (indeterminate), resulting in a total score of 0 to 8 for the eight plants of each RI line tested. Figure 2 shows examples of plants at the end of two such feeding experiments. RI line 214 has a completely Col-like phenotype and received a score of 8, whereas RI line 184 has a completely Ler-like phenotype and received a score of 0 in this assay.
Figure 2.
Five-week-old plants of Col, Ler, and RI lines derived from these ecotypes were exposed to cabbage looper larvae for 3 d. Larvae were allowed to feed at will until they stopped feeding. The inflorescences and leaves of Col and RI line 214 are resistant to cabbage looper feeding, whereas Ler and RI line 184 are sensitive and are almost completely consumed.
Data from the RI line feeding experiments were analyzed by interval mapping with the QGene program (Nelson, 1997; http://www.QGene.org) using the Col/Ler marker data available from the Nottingham Arabidopsis Stock Center (NASC) Web site (http://nasc.nott.ac.uk). A QTL with a significant effect (log of the odds [LOD] = 5.05) on insect feeding is located between markers Tag1 and p4 at approximately 85 cM on chromosome 1 of Arabidopsis (Fig. 3). The 99% significance threshold was determined to be a LOD score of 3.2 and the peak on chromosome 1 is the only one above this threshold. A similar result was obtained in a second run of this experiment using a subset of 30 RI lines chosen for their high level of recombination (NASC Web site; data not shown). We have named this locus TASTY because one or more genes in this interval affect the palatability of Arabidopsis for insect larvae.
Figure 3.
LOD scores for cabbage looper feeding on RI lines derived from Col and Ler ecotypes relative to markers on the Arabidopsis genetic map. A locus between the markers Tag1 and p4 on chromosome I has a significant effect on cabbage looper feeding, with the resistant allele coming from the Col ecotype. The dashed line represents the 99% confidence threshold for this data set.
Analysis of marker and phenotype data from individual RI lines shows the expected result that the resistant (less tasty) allele of TASTY is derived from the Col ecotype. F1 hybrids of Col and Ler are larger and more vigorous than either parental ecotype. However, T. ni larvae feeding on flowering F1 hybrid plants consumed the entire inflorescence, leaving only a bit of the flower stalk. At the end of the experiment, the F1 hybrids looked like the Ler plants in Figure 2, an indication that the sensitive TASTY allele from Ler is at least phenotypically dominant.
The glucosinolate-myrosinase system is thought to play an important role in the insect defense of the Brassicaceae and high glucosinolate concentrations are known to have a negative effect on T. ni feeding (Grant-Peterson, 1993; Shields and Mitchell, 1995; Mitchell et al., 1996; Stowe, 1998). Thus, we were interested in determining whether the TASTY locus was involved in glucosinolate biosynthesis. We measured the glucosinolate and myrosinase concentrations in Col and Ler plants grown under the same conditions as the RI lines that were used for QTL mapping (Fig. 4). There was no significant difference in the mean total glucosinolate content of Col and Ler (342 versus 402 μm g−1 plant material, P = 0.086, Student's t test). If anything, the glucosinolate levels were slightly higher in the Ler plants. At the 95% significance threshold, there was less myrosinase activity in Col than in Ler (1.76 versus 2.45 μm Glc released/min/g plant material, P = 0.011, Student's t test). Given the known aversion of T. ni for glucosinolates, these results are the opposite of what one would expect if the TASTY locus were involved in determining total glucosinolate content.
Figure 4.
A, Total glucosinolate content of Col and Ler plants expressed as micromolar glucosinolates per gram of plant material. Bars show mean and sd of eight samples, P = 0.08 (Student's t test). B, Myrosinase activity in Col and Ler plants expressed as micromolar Glc released from sinigrin per minute per gram of plant material. Bars show mean and sd of eight samples, P = 0.01 (Student's t test).
DISCUSSION
In this work we used T. ni feeding on Arabidopsis as a model for studying insect-plant interactions. While T. ni has not been reported to feed on Arabidopsis in the wild, it is not unreasonable to think that it would do so if given the opportunity. As a polyphagous insect herbivore that has been introduced in many parts of the world, cabbage looper feeds on many species that were not its original hosts. The feeding behavior of cabbage looper on Arabidopsis is similar to that on the natural host, Brassica sp. (older leaves are consumed first). In addition, adult moths will readily lay their eggs on Arabidopsis, and larvae can develop to pupation with no other food source (G. Jander, unpublished results).
While T. ni showed a strong preference for the Ler ecotype in both whole plant feeding experiments and weight gain experiments, there was no significant difference between Col and Ler in leaf plug experiments. In the whole plant assays, the T. ni larvae were allowed to feed on growing plants over a period of days. In contrast, leaf plugs were consumed over a period of about 3 h. Perhaps the difference between T. ni feeding on whole Col and Ler plants is a consequence of induced defense responses that develop relatively slowly and are expressed more strongly in Col than in Ler. It is also possible that Ler could become more sensitive as a result of T. ni feeding. Such a response has been reported for T. ni feeding on wild radish (Raphanus sativus), where plants that been previously damaged by T. ni allowed somewhat better larval growth than uninduced controls (Agrawal, 2000).
F1 hybrids derived from Col and Ler have a T. ni-sensitive phenotype that is similar to Ler. This could be the result of either genetic dominance of insect sensitivity or haplo-insufficiency in the heterozygous plants. Dominant sensitivity is most easily imagined if the TASTY locus has a regulatory function. For instance, a negative regulator of insect defenses that is present in Ler but not Col could result in dominant sensitivity in the F1 plants. Haplo-insufficiency in the heterozygous plants could occur if one instead of two copies of the Col allele of TASTY result in lower levels of an insect deterrent found in the Col ecotype. F1 plants would be insect-sensitive if T. ni larvae are able to tolerate this reduced deterrent concentration. It will be difficult to completely rule out or confirm either model until the TASTY gene has been cloned and can be used to perform experiments in transgenic plants.
Glucosinolates are thought to be an important part of the insect defense of Arabidopsis and other crucifers. However, our results showing no difference or perhaps slightly higher myrosinase and glucosinolate levels in Ler than in Col (Fig. 4) support the conclusion that TASTY locus is not involved in determining total glucosinolate content. We also considered the possibility that T. ni larvae are responding to qualitative differences in the profile of the more than 20 kinds of glucosinolates that have been identified in Arabidopsis (Haughn et al., 1991). The same population of Col/Ler RI lines that we used in our study has been used to genetically map qualitative differences in glucosinolate content (Magrath et al., 1994; Mithen et al., 1995; Campos de Quiros et al., 2000). The loci that were identified are on chromosomes 4 and 5 of Arabidopsis and are thus distinct from the TASTY locus on chromosome 1. However, we cannot entirely rule out the possibility that T. ni is responding to some Col/Ler difference in the glucosinolate profile that was not apparent in HPLC assays.
In addition to glucosinolate content, we also considered four other phenotypic traits that vary among Arabidopsis ecotypes and are known to influence insect feeding in Arabidopsis or other plants: trichome density, flowering time, disease resistance, and surface wax content (Alonso-Blanco and Koornneef, 2000). These traits either do not vary significantly between Col and Ler or known QTLs affecting the traits map at genetic loci that are distinct from TASTY.
The presence of trichomes in the leaf surface is associated with resistance to cabbage looper in soybeans (Khan et al., 1986b), and it is conceivable that the lower frequency of trichomes on the surface of Ler compared to Col could cause the T. ni feeding preference that we have observed. However, the major locus that affects trichome density is found near the erecta gene on chromosome 2 (Larkin et al., 1996) and thus is distinct from TASTY.
Choice experiments with leaf plugs made from young or senescing leaves of Arabidopsis showed that T. ni exhibited a significant preference for the older leaves. Flowering time and leaf senescence vary among ecotypes, and genes affecting these traits could be responsible for the feeding preference. Although a number of loci affecting flowering time differ between Col and Ler (Jansen et al., 1995; Mitchell-Olds, 1996), none of these coincide with TASTY.
There is increasing evidence for cross-talk between the pathways involved in disease and insect resistance in plants. Some of the many known Arabidopsis disease-related genes differ between Col and Ler (Kunkel, 1996; Buell and Somerville, 1997), but these again are at loci that are distinct from TASTY.
Finally, the presence of surface waxes on plants affects the feeding of many insect herbivores (Eigenbrode and Espelie, 1995). However, while there is variation in surface wax content among Arabidopsis ecotypes, Col and Ler do not have significant differences in either the total wax load or the surface wax profile (Rashotte et al., 1997).
The Arabidopsis ecotypes in our study varied greatly in their resistance to T. ni. Many ecotypes were eaten completely to the ground, but others were hardly touched. It is perhaps significant that the four central Asian ecotypes (Kas-1, Condara, Hodja, and Shahdara) were all very resistant to cabbage looper feeding and included three of the four most resistant accessions. Arabidopsis is a recently introduced species in many parts of the world and genetic relatedness of ecotypes generally does not follow a geographic pattern (Innan et al., 1997). However, Arabidopsis accessions from central Asia as a group do appear to be genetically distinct from those in Europe, the origin of most of the other ecotypes in our study (Sharbel et al., 2000). Using one of the central Asian ecotypes as a parent in a mapping cross might yield other loci affecting insect feeding in addition to the TASTY locus that we have discovered.
In this work we present the novel finding of TASTY, a genetic locus on chromosome 1 of Arabidopsis that strongly affects the feeding behavior of T. ni. While ecotype-specific variation of many different plant traits has been identified genetically in Arabidopsis, we believe that this is the first example of the mapping of a natural locus affecting the feeding of a chewing insect herbivore. Further work with the TASTY locus may lead to the discovery of new mechanisms of insect resistance in plants.
MATERIALS AND METHODS
Growth Conditions
Trichoplusia ni (cabbage looper) eggs from a highly inbred population were purchased from Entopath, Inc. (Easton, PA). Larvae for weight gain experiments were moved to plants immediately after hatching. Larvae for choice and no-choice feeding experiments were reared at room temperature in Petri dishes on a pinto bean diet (Guy et al., 1985). Approximately 50 eggs were placed in each Petri dish and larvae were removed when they reached 80 to 100 mg wet weight. Seeds of Arabidopsis ecotypes and recombinant inbred lines were obtained from the Arabidopsis Biological Resource Center (Columbus, OH). All seeds were cold-stratified at 4°C in 0.1% (w/v) agar for 4 d before planting. Plants were grown in Metromix 200 potting soil (Scotts, Marysville, OH) in standard nursery flats. Natural sunlight was supplemented with metal halide lights to produce a 16:8 d-night cycle.
Ecotype Comparisons
Sixteen plants of each of 34 ecotypes were grown in 10-cm pots, eight pots to a flat in a randomized array. After 4 weeks of growth, eight T. ni larvae weighing 80 to 100 mg each were placed in each pot and were allowed to feed and roam at will. After 3 d the diameter of the remaining rosette of leaves of each plant was measured with a ruler. Each ecotype was given a score of 1 to 3: 1, most sensitive, <1 cm diameter of remaining rosette leaves; 2, intermediate, 1 to 2 cm diameter of remaining rosette leaves; 3, resistant, >2 cm diameter of remaining rosette leaves.
QTL Mapping with RI Lines
Sixteen plants were raised in a 10-cm pot, a row of four Col and Ler plants on either side and two rows with a total of eight plants from a particular Col/Ler RI line (Lister and Dean, 1993) in between. Pots with different RI lines were placed in a randomized array, eight to a flat. When the plants were 5 weeks old (early flowering stage), eight T. ni larvae were placed on each container of 16 plants and were allowed to roam and feed at will. After 3 d the RI line plants were assessed in comparison to the Col and Ler plants in the same pot. Each individual plant was given a score of 0 (sensitive, Ler-like), 1 (resistant, Col-like), or 0.5 (indeterminate) for a cumulative score of 0 to 8 for each RI line. Plants where no flowers, siliques, or cauline leaves remained were given a score of 0; plants where most of these parts of the inflorescence remained uneaten received a score of 1; and plants that were not clearly in either category received a score of 0.5. Marker data for the Col/Ler RI lines were obtained from the NASC Web page and QTL data analysis for cabbage looper feeding was done using QGene (Nelson, 1997). The threshold value for 99% significance was determined empirically (Churchill and Doerge, 1994) by randomizing the phenotypic data and assigning them to RI lines. This was done 200 times and the LOD score of the highest peak determined by interval analysis with QGene was recorded in each case. These 200 LOD scores were ordered and the 198th-highest score was taken for the significance threshold.
Choice Experiments
Three comparisons of pairs of Arabidopsis leaf material were made using choice feeding experiments: (a) Old leaves that were beginning to senesce versus young leaves about 1 cm long; (b) undamaged leaves versus leaves that had been damaged 2 d earlier by squeezing once across the midrib with pliers; and (c) Mature leaves of Col versus mature leaves of Ler. A cork borer was used to make leaf plugs 7 mm in diameter from the two types of plant material that were to be compared (old versus young, damaged versus undamaged, Col versus Ler). In the case of the damaged leaves, care was taken to obtain a leaf plug from a part of the leaf that had not actually been damaged. Eight leaf plugs, four of each kind, were arranged in a 5-cm diameter circle on moistened Whatman filter paper, alternating the two types of leaf plugs. To reduce the effect of outside visual cues, each experimental setup was surrounded by a 10-cm diameter, 15-cm-high white paper cylinder, and the entire setup was placed in a 23°C incubator with fluorescent lighting. T. ni larvae were removed from pinto bean diet before the experiment and were starved for 8 h. One larva was placed in the center of the circle of leaf plugs and was observed until approximately 50% of the total leaf material was eaten (usually about 3 h). If a larva had not consumed any leaf material after 6 h, the experiment was also ended. A digital image was taken of the leaf plugs at the end of the experiment and the leaf area remaining was calculated using the NIH Image computer program (developed at the United States National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image). To account for leaf shrinkage during the experiment, the area consumed was calculated by comparison to leaf plugs from a control without larvae.
Weight Gain Experiments
Newly hatched T. ni larvae were placed onto 4- to 5-week-old Arabidopsis plants, one per plant. Larvae were confined to one plant by placing the entire plant inside a nylon mesh bag before moving it back to the greenhouse. After 5 d, the larvae were removed from the plant, desiccated by drying overnight in an oven at 65°C, and weighed individually.
Glucosinolate Assays
Total glucosinolate content of Arabidopsis extracts was measured by the release of Glc after treatment with myrosinase, adapted from Heaney and Fenwick (1981). Leaves and inflorescences of plants were harvested, frozen in liquid nitrogen, and ground to a fine powder. Three volumes of boiling water were added to the samples and they were boiled for an additional 2 min to inactivate endogenous myrosinase. Plant particles were allowed to settle out and 0.5 mL of the supernatant was mixed with 0.5 mL of lead acetate-barium acetate solution (11.3 g of lead acetate, 7.65 g of barium acetate, 0.29 mL of glacial acetic acid in 100 mL of water). Samples were spun in a microcentrifuge and 0.4 mL of the supernatant was loaded on a column made by packing 1 mL DEAE Sephadex (Sigma, St. Louis) into a Pasteur pipette plugged with glass wool. The column was washed with 2 mL of 4 m acetic acid and 6 mL of water. Myrosinase (Sigma) solution (500 μL, 3 mg mL−1) was added to the column and it was left overnight at room temperature. Control samples did not receive any myrosinase. The column was eluted with 3 mL of water, and Glc concentration was measured with a hexokinase-based Glc detection kit (Sigma) used according to the manufacturer's instructions.
Myrosinase Assays
Myrosinase activity in Arabidopsis extracts was measured by the release of Glc from the commercially available glucosinolate sinigrin, an adaptation of a previously published protocol (Mitchell-Olds and Pedersen, 1998). Leaves and inflorescences of plants were harvested, frozen in liquid nitrogen, and ground to a fine powder. Three volumes of 100 mm Tris, pH 7.4, 1 μg mL−1 leupeptin, and 0.1 mm phenylmethylsulfonyl fluoride were added and the samples were vortexed briefly. Plant particles were spun out in a microcentrifuge and the supernatant was passed through a desalting column (Bio-Rad Micro Biospin 6). Fifty microliters of the desalted extract was added to 500 μL of 0.3 mm ascorbate, 0.1 m NaAcetate, pH 5.5, and 5 mg mL−1 sinigrin (Sigma). Control samples were as follows: (a) Plant extracts and assay buffer without sinigrin, and (b) assay buffer and sinigrin without plant extracts. The reaction was allowed to proceed at 28°C for 30 min and then the myrosinase was inactivated by boiling for 5 min. Released Glc was measured with a hexokinase-based Glc detection kit (Sigma) used according to the manufacturer's instructions.
Footnotes
This work was supported by a National Research Service Award (no. GM18735 to G.J.) and by the National Institutes of Health (grant no. GM48707 to F.M.A.).
LITERATURE CITED
- Agrawal AA. Specificity of induced resistance in wild radish: causes and consequences for two specialist and two generalist herbivores. Oikos. 2000;89:493–500. [Google Scholar]
- Alonso-Blanco C, Koornneef M. Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends Plant Sci. 2000;5:22–29. doi: 10.1016/s1360-1385(99)01510-1. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;108:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Bernays EA, Chapman RF. Host-Plant Selection by Phytophagous Insects. New York: Chapman and Hill; 1994. [Google Scholar]
- Bones AM, Rossiter JM. The myrosinase-glucosinolate system, its organisation and biochemistry. Physiol Plant. 1996;97:194–208. [Google Scholar]
- Broadway RM, Colvin AA. Influence of cabbage protease inhibitors in situ on the growth of larval Trichoplusia ni and Pieris rapae. J Chem Ecol. 1992;18:1009–1023. doi: 10.1007/BF00980059. [DOI] [PubMed] [Google Scholar]
- Buell CR, Somerville SC. Use of Arabidopsis recombinant inbred lines reveals a monogenic and a novel digenic resistance mechanism to Xanthomonas campestris pv. campestris. Plant J. 1997;12:21–29. doi: 10.1046/j.1365-313x.1997.12010021.x. [DOI] [PubMed] [Google Scholar]
- Campos de Quiros R, Magrath R, McCallum D, Kroymann J, Scnabelrauch D, Mitchell-Olds T, Mithen R. Alpha-keto acid elongarion and glucosinolate biosynthesis in Arabidopsis thaliana. Theor Appl Genet. 2000;101:429–437. [Google Scholar]
- Churchill GA, Doerge RW. Empirical Threshold Values for Quantitative Trait Mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigenbrode SD, Espelie KE. Effects of plant epicuticular lipids on insect herbivores. Ann Rev Entomol. 1995;40:171–194. [Google Scholar]
- Grant-Peterson J. The effect of allelochemical differences in Arabidopsis thaliana on the responses of the herbivores Trichoplusia ni and Pieris rapae. PhD thesis. Ithaca, NY: Department of Entomology, Cornell University; 1993. [Google Scholar]
- Grant-Petersson J, Renwick JAA. Effects of ultraviolet B exposure of Arabidopsis thaliana on herbivory by two crucifer-feeding insects. Environ Entomol. 1996;25:135–142. [Google Scholar]
- Guy RH, Leppla NC, Rye JR, Green CW, Barrette SL, Hollien KA. Trichoplusia ni. In: Singh P, editor. Handbook of Insect Rearing. New York: Elsevier; 1985. pp. 487–493. [Google Scholar]
- Haughn GW, Davin L, Giblin M, Underhill EW. Biochemical genetics of plant secondary metabolites in Arabidopsis thaliana. Plant Physiol. 1991;97:217–226. doi: 10.1104/pp.97.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaney RK, Fenwick GR. A micro-column method for the rapid determination of total glucosinolate content of cruciferous material. Z Pflanzenzuech. 1981;87:89–95. [Google Scholar]
- Innan H, Terauchi R, Miyashita NT. Microsatellite polymorphism in natural populations of the wild plant Arabidopsis thaliana. Genetics. 1997;146:1441–1452. doi: 10.1093/genetics/146.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RC. Complex plant traits: time for polygenic analysis. Trends Plant Sci. 1996;1:89–94. [Google Scholar]
- Jansen RC, Van Ooijen JW, Stam P, Lister C, Dean C. Genotype-by-environment interaction in genetic mapping of multiple quantitative trait loci. Theor Appl Genet. 1995;91:33–37. doi: 10.1007/BF00220855. [DOI] [PubMed] [Google Scholar]
- Kahl J, Siemens D, Aerts RJ, Gaebler R, Kuehnemann F, Preston CA, Baldwin IT. Herbivore-induced ethylene suppresses a direct defense but not a putative indirect defense against an adapted herbivore. Planta. 2000;210:336–342. doi: 10.1007/PL00008142. [DOI] [PubMed] [Google Scholar]
- Kearsey MJ, Farquar GL. QTL analysis in plants: where are we now? Heredity. 1998;80:137–142. doi: 10.1046/j.1365-2540.1998.00500.x. [DOI] [PubMed] [Google Scholar]
- Khan ZR, Ciepiela A, Norris DM. Behavioral and physiological responses of cabbage looper, Trichoplusia ni (Huebner), to steam distillates from resistant versus susceptible soybean varieties. J Chem Ecol. 1987;13:1903–1915. doi: 10.1007/BF01013239. [DOI] [PubMed] [Google Scholar]
- Khan ZR, Norris DM, Chiang HS, Weiss NE, Oosterwyk AS. Light-induced susceptibility in soybean to cabbage looper Trichoplusia ni (Lepidoptera: Noctuidae) Environ Entomol. 1986a;15:803–808. [Google Scholar]
- Khan ZR, Ward JT, Norris DM. Role of trichomes in soybean resistance to cabbage looper Trichoplusia ni. Entomol Exp Appl. 1986b;42:109–117. [Google Scholar]
- Kunkel BN. A useful weed put to work: Genetic analysis of disease resistance in Arabidopsis thaliana. Trends Genet. 1996;12:62–69. doi: 10.1016/0168-9525(96)81402-8. [DOI] [PubMed] [Google Scholar]
- Larkin JC, Young N, Prigge M, Marks MD. The control of trichome spacing and number in Arabidopsis. Development. 1996;122:997–1105. doi: 10.1242/dev.122.3.997. [DOI] [PubMed] [Google Scholar]
- Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- Magrath R, Bano F, Morgner M, Parkin I, Sharpe A, Lister C, Dean C, Turner J, Lydiate D, Mithen R. Genetics of aliphatic glucosinolates: I. Side chain elongation in Brassica napus and Arabidopsis thaliana. Heredity. 1994;72:290–299. [Google Scholar]
- Mattiacci L, Dicke M, Posthumus MA. β-Glucosidase: an elicitor of herbivore-induced plant odor that attracts host-searching parasitic wasps. Proc Natl Acad Sci USA. 1995;92:2036–2040. doi: 10.1073/pnas.92.6.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauricio R. Costs of resistance to natural enemies in field populations of the annual plant Arabidopsis thaliana. Am Nat. 1998;151:20–27. doi: 10.1086/286099. [DOI] [PubMed] [Google Scholar]
- McConn M, Creelman RA, Bell E, Mullet JE, Browse J. Jasmonate is essential for insect defense in Arabidopsis. Proc Natl Acad Sci USA. 1997;94:5473–5477. doi: 10.1073/pnas.94.10.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell BK, Justus KA, Asaoka K. Deterrency and the variable caterpillar: Trichoplusia ni and sinigrin. Entomol Exp Appl. 1996;80:27–31. [Google Scholar]
- Mitchell-Olds T. Genetic constraints on life history and evolution: quantitative trait loci influencing growth and flowering time in Arabidopsis. Evolution. 1996;50:140–145. doi: 10.1111/j.1558-5646.1996.tb04480.x. [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T, Pedersen D. The molecular basis of quantitative genetic variation in central and secondary metabolism of Arabidopsis. Genetics. 1998;149:739–747. doi: 10.1093/genetics/149.2.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithen R, Clarke J, Lister C, Dean C. Genetics of aliphatic glucosinolates: III. Side chain structure of aliphatic glucosinolates in Arabidopsis thaliana. Heredity. 1995;74:210–215. [Google Scholar]
- Nelson JC. QGene: software for marker-based genomic analysis and breeding. Mol Breed. 1997;3:239–245. [Google Scholar]
- Rashotte A, Feldmann K. Epicuticular waxes and aphid resistance in Arabidopsis cer mutants and ecotypes. Plant Physiol Suppl. 1996;111:87. [Google Scholar]
- Rashotte AM, Jenks MA, Nguyen TD, Feldmann KA. Epicuticular wax variation in ecotypes of Arabidopsis thaliana. Phytochemistry. 1997;45:251–255. doi: 10.1016/s0031-9422(96)00792-3. [DOI] [PubMed] [Google Scholar]
- Renwick JAA, Radke CD. Activity of cabbage extracts in deterring oviposition by the cabbage looper, Trichoplusia ni. In: Visser JH, Minks AK, editors. Proceedings of the 5th International Symposium on Insect-Plant Relationships. Wageningen, The Netherlands: Centre for Agricultural Publication and Documentation; 1982. pp. 139–143. [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell. 2000;12:707–719. doi: 10.1105/tpc.12.5.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos MO, Adang MJ, All JN, Boerma HR, Parrott WA. Testing transgenes for insect resistance using Arabidopsis. Mol Breed. 1997;3:183–194. [Google Scholar]
- Sharbel TM, Haubold B, Mitchell-Olds T. Genetic isolation by distance in Arabidopsis thaliana: biogeography and postglacial colonization of Europe. Mol Ecol. 2000;9:2109–2118. doi: 10.1046/j.1365-294x.2000.01122.x. [DOI] [PubMed] [Google Scholar]
- Shields VDC, Mitchell BK. Sinigrin as a feeding deterrent in two crucifer-feeding, polyphagous lepidopterous species and the effects of feeding stimulant mixtures on deterrrency. Philos Trans R Soc Lond B. 1995;347:439–446. [Google Scholar]
- Shorey HH, Andres LA, Hale RL. The biology of Trichoplusia ni (Lepidoptera: Noctuidae): I. Life history and behavior. Ann Entomol Soc Am. 1962;55:591–597. [Google Scholar]
- Singh R, Ellis PR, Pink DAC, Phelps K. An investigation of the resistance to cabbage aphid in Brassica species. Ann Appl Biol. 1994;125:457–465. [Google Scholar]
- Stotz HU, Pittendrigh BR, Kroyman J, Weniger K, Fritsche J, Bauke A, Mitchell-Olds T. Induced plant defense responses against chewing insects: ethylene signaling reduces resistance of Arabidopsis against Egyptian cotton worm, but not diamondback moth. Plant Physiol. 2000;124:1007–1017. doi: 10.1104/pp.124.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe KA. Realized defense of artificially selected lines of Brassica rapa: effects of quantitative genetic variation in foliar glucosinolate concentration. Environ Entomol. 1998;27:1166–1174. [Google Scholar]
- Tanksley SD. Mapping polygenes. Annu Rev Genet. 1993;27:205–233. doi: 10.1146/annurev.ge.27.120193.001225. [DOI] [PubMed] [Google Scholar]