Abstract
An α-xylosidase active against xyloglucan oligosaccharides was purified from cabbage (Brassica oleracea var. capitata) leaves. Two peptide sequences were obtained from this protein, the N-terminal and an internal one, and these were used to identify an Arabidopsis gene coding for an α-xylosidase that we propose to call AtXYL1. It has been mapped to a region of chromosome I between markers at 100.44 and 107.48 cM. AtXYL1 comprised three exons and encoded a peptide that was 915 amino acids long, with a potential signal peptide of 22 amino acids and eight possible N-glycosylation sites. The protein encoded by AtXYL1 showed the signature regions of family 31 glycosyl hydrolases, which comprises not only α-xylosidases, but also α-glucosidases. The α-xylosidase activity is present in apoplastic extractions from Arabidopsis seedlings, as suggested by the deduced signal peptide. The first eight leaves from Arabidopsis plants were harvested to analyze α-xylosidase activity and AtXYL1 expression levels. Both increased from older to younger leaves, where xyloglucan turnover is expected to be higher. When this gene was introduced in a suitable expression vector and used to transform Saccharomyces cerevisiae, significantly higher α-xylosidase activity was detected in the yeast cells. α-Glucosidase activity was also increased in the transformed cells, although to a lesser extent. These results show that AtXYL1 encodes for an apoplastic α-xylosidase active against xyloglucan oligosaccharides that probably also has activity against p-nitrophenyl-α-d-glucoside.
Xyloglucan is the main hemicellulosic polysaccharide present in the primary cell walls of dicotyledonous plants (Fry, 1989; Hayashi, 1989). It consists of a linear (1→4)-β-linked d-glucan backbone that carries α-d-xylosyl, β-d-galactosyl-(1→2)-α-d-xylosyl and α-l-fucosyl-(1→2)-β-d-galactosyl-(1→2)-α-d-xylosyl sidechains attached to the OH-6 of β-glucosyl residues. Xyloglucan is thought to be the load-bearing component in the primary cell walls because of its proposed cross-linking of the cellulose microfibrils, this cross-linking being the major factor controlling the rate of cell expansion (Fry, 1989). In addition to its structural role, xyloglucan may act as a source of signaling molecules. Oligosaccharides derived from xyloglucan have been found to be formed in vivo (Fry, 1986) and they have been shown to regulate auxin- (McDougall and Fry, 1988, 1990) and acid pH-induced (Lorences et al., 1990) growth.
Enzymes that modify xyloglucan oligosaccharides have been detected in plant cell walls (Fry, 1995). An α-fucosidase that removes the α-l-fucosyl residue from XXFG has been purified from pea epicotyls (Farkas et al., 1991; Augur et al., 1993) and has later been cloned (Augur et al., 1995). A nasturtium β-d-galactosidase able to remove the terminal β-d-galactosyl residue from xyloglucan side-chains has also been characterized (Edwards et al., 1988). Finally, an α-d-xylosidase has been purified from auxin-treated pea epicotyls (O'Neill et al., 1989), and a second one from cotyledons of nasturtium seedlings (Fanutti et al., 1991). Both enzymes showed relatively high substrate specificity. They acted on xyloglucan oligosaccharides and specifically released the unsubstituted side chain xylosyl residue attached to the backbone glucosyl residue situated farthest from the reducing end of the molecule.
It has been shown by Lorences and Fry (1993) that oligosaccharides lacking the Xyl at the non-reducing terminus are unable to act as acceptors for xyloglucan endotransglycosylase in bean and pea leaves. α-Xylosidase, by removing that particular xylosyl residue, would therefore reduce the concentration of oligosaccharides available for transglycosylation reactions. Xyloglucan endotransglycosylase, when the acceptors are oligosaccharides, has the same effect on xyloglucan as endo-β-glucanase, i.e. a reduction of the cross-linking chains. Thus, α-xylosidase could be playing an important role in cell expansion (Fry, 1995). Although the action of α-xylosidase on polymeric xyloglucan has not yet been proved, there is some indirect evidence that supports this possibility (Guillén et al., 1995). It should also be noted that exo-β-glucosidase cannot act on the Glc at the non-reducing end of the oligosaccharide backbone unless α-xylosidase has previously removed the Xyl attached to it (Koyama et al., 1983).
In the present paper we report the purification of an α-xylosidase from cabbage (Brassica oleracea var. capitata) leaves. Two peptide sequences were obtained from this protein. They were used to identify an α-xylosidase coding gene from Arabidopsis that we propose to call AtXYL1. It was sequenced and its expression was shown to be higher in younger leaves. α-Xylosidase activity displayed a similar pattern. AtXYL1 was expressed in Saccharomyces cerevisiae, and a protein with α-xylosidase activity was produced.
RESULTS AND DISCUSSION
Purification of an α-Xylosidase from Cabbage Leaves
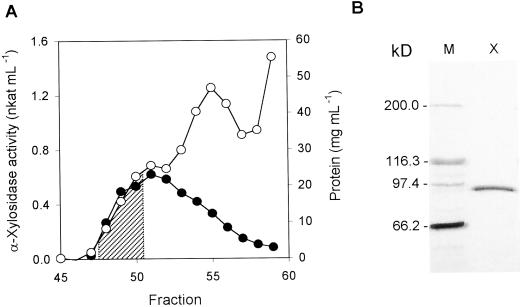
The extracts obtained from cabbage leaves were able to release Xyl from xyloglucan oligosaccharides, as measured by pentose release. This α-xylosidase activity was purified after four consecutive purification steps. Along this procedure the activity always appeared as a single peak. The purified extract lacked β-galactosidase and β-glucosidase activities, the only other enzymes known to act on tamarind xyloglucan oligosaccharides, thus doing away with possible interferences in α-xylosidase measurements. The amount of Xyl released by the purified enzyme was never higher than the amount of Xyl linked to the terminal glucosyl residue, as expected from the previous characterizations of plant α-xylosidases (O'Neill et al., 1989; Fanutti et al., 1991). The specific activity of the purified α-xylosidase was 27.7 nkat mg−1 (Table I). The optimum pH for this enzyme was 4.5 and its pI was 8.3, as determined by isoelectric focusing (IEF; data not shown). Three selected fractions from a Sephacryl S-100 HR column were pooled and subjected to SDS-PAGE, revealing a single band of 89 kD (Fig. 1).
Table I.
Purification of an α-xylosidase from young cabbage leaves
| Step | Activity | Protein | Specific Activity | Recovery | Purification Index |
|---|---|---|---|---|---|
| nkat | mg | nkat mg−1 | % | ||
| (NH4)2SO4 precipitation | 144 | 300 | 0.48 | 100 | 1 |
| SP-Sepharose | 37.5 | 10.3 | 3.1 | 26 | 7.6 |
| (Concanavaline-A [Con-A]) Sepharose | 25.8 | 5.5 | 4.7 | 18 | 9.8 |
| Sephacryl S-100 (high resolution [HR]) | 1.94 | 0.07 | 27.7 | 1.4 | 57.7 |
Figure 1.
Purification of α-xylosidase from young cabbage leaves. A, Selected region of gel permeation chromatography on Sephacryl S-100 HR column. Protein content (○) and α-xylosidase activity (●) of each fraction were measured. B, SDS-PAGE of fractions 48 to 50 from Sephacryl S-100 HR chromatography. The selected fractions (stripped area on A) were pooled, concentrated, and separated by SDS-PAGE. Left lane, Molecular markers; right lane, α-xylosidase-containing fractions.
Two different types of α-xylosidases had been previously purified to homogeneity. Those from Aspergillus niger (Matsushita et al., 1985) and Bacillus sp. (Zong and Yasui, 1989; Zong et al., 1989) were able to hydrolyze simple xylosides such as p-nitrophenyl-α-d-xylopyranoside or isoprimeverose and they were also able to remove the terminal Xyl from xyloglucan oligosaccharides. The other type, the plant α-xylosidases, is comprised of the enzymes purified from pea epicotyls (O'Neill et al., 1989) and from nasturtium cotyledons (Fanutti et al., 1991). These showed higher substrate specificity. They did not hydrolyze p-nitrophenyl-α-d-xylopyranoside or isoprimeverose, but they did specifically cleave the unsubstituted side chain xylosyl residue attached to the backbone glucosyl residue situated farthest from the reducing end of the xyloglucan oligosaccharides. Both enzymes had an apparent molecular mass of 85 kD on SDS-PAGE, and the optimum pH were in the range 4.9 to 5.1. The pI was 7.35 to 7.7 for the pea enzyme and 5.0 to 7.1 for the nasturtium.
An Arabidopsis Gene Coding for α-Xylosidase
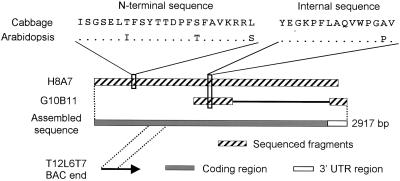
The 89-kD protein band from cabbage leaves (Fig. 1) was excised from PAGE gels and the N-terminal and an internal peptide sequences were obtained from it (Fig. 2). In the public databases, two Arabidopsis expressed sequence tags (ESTs; H8A7T7 and G10B11T7; Newman et al., 1994) were found that could be translated to protein fragments, including regions highly similar (34 out of 38 identical amino acids) to the sequences obtained from cabbage α-xylosidase (Fig. 2). These ESTs had been obtained from two partial cDNA clones that were derived from the same gene, as we established by completely sequencing the longer clone (H8A7; GenBank accession no. AF087483) and partially sequencing the shorter one (G10B11). The assembled sequence was 2,917 bp long. We propose AtXYL1 as the name for this Arabidopsis gene.
Figure 2.
Diagram showing the identification of two Arabidopsis ESTs (H8A7T7 and G10B11T7) and a bacterial artificial chromosome (BAC) clone (T12L6) containing sequences highly similar to those obtained from cabbage α-xylosidase. For Arabidopsis homologous sequences, dots mean conserved amino acids.
In addition, an Arabidopsis BAC end sequence (T12L6T7, accession no. AL094155) was found that showed total identity with H8A7 in a region 193 bp long. This sequence indicated that clone T12L6 from the Texas A&M University BAC library included only a fragment of AtXYL1. Fingerprinting data from the Genomic Sequencing Center showed that this clone most likely overlapped with two Institut für Genbiologische Furschung (Berlin) BAC clones (Mozo et al., 1998b), F2A8 and F28A15, with a Fingerprinted Contigs score value respectively of 2 × 10−17 and 1 × 10−13 (Marra et al., 1997). Both clones had been mapped by the Max Planck Institut für Moleculare Pflanzenphysiologie to a region of chromosome I between markers pCITf117 (100.44 cM) and mi462 (107.48 cM; Mozo et al., 1998a). We screened by PCR a set of overlapping IGF-BAC clones from this region and found that F22C21 was the only one that included AtXYL1 (Fig. 3).
Figure 3.
A, Tiling path of the region in chromosome I where F28A15 and F2A8 had been mapped. This diagram is based on Max Planck Institut für Moleculare Pflanzenphysiologie hybridization data. The initial F has been omitted from clone names. Five clones, including the two already mentioned, were selected for screening and are marked in black. Another clone, F24J5, has been recently sequenced by the SPP consortium (Stanford DNA sequencing and Technology Center, Plant Gene Expression Center at University of California-Berkeley, and the University of Pennsylvania). It includes the entire AtXYL1 gene (GenBank accession no. AC008075). B, Screening of the five IGF-BAC clones by PCR in search for AtXYL1.
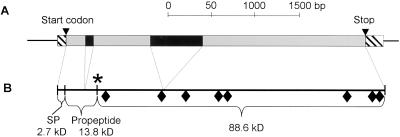
A 4,288-bp region from clone F22C21 was sequenced (Fig. 4) and submitted to GenBank (accession no. AF144078). It encompassed the entire coding sequence. According to the cDNA sequence, AtXYL1 had two introns (91 and 622 bp long); the splice sites for both were found to be standard (data not shown). The AtXYL1 transcription start site was located by 5′-RACE using RNA from young Arabidopsis leaves. The fragment thus amplified included a 5′-cap and overlapped with the sequence previously obtained from clone H8A7, nearly completing the known AtXYL1 mRNA, which was now 3,037 bp long and went from the transcription start site to 18 bp downstream of a potential polyA signal in the 3′-untranslated region. A complete IGF-BAC clone (F24J5, accession no. AC008075), which included AtXYL1, was later sequenced by the Plant Gene Expression Center.
Figure 4.
Structure of AtXYL1 gene and its protein. A, Sequenced fragment from clone F22C21. Transcripted region is drawn as a set of boxes, stripped (5′- and 3′-untranslated regions), black (introns), or gray (coding regions). B, Protein diagram. ♦, Potential glycosylation sites. The asterisk marks the position of active cabbage α-xylosidase N terminus. SP, Signal peptide.
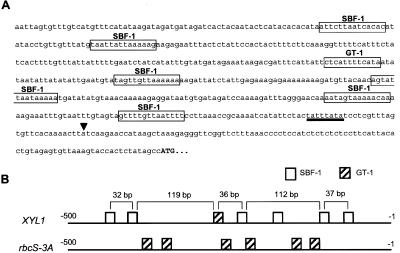
Analysis of AtXYL1 promoter was based on F24J5 sequence and showed a TATA-like motif centered at position −30. What was more interesting was that several potential binding sites for transcription factors of the GT family were observed to be arranged in a pattern reminiscent of the one found in promoters where these transcription factors are known to be active (Fig. 5). Six SBF-1 binding sites and one GT-1 motif have been found in the AtXYL1 promoter. Both kinds of sites are functionally related and their sequences are quite similar (Villain et al., 1994). In fact, three of the SBF-1 sites were also recognized by the software as GT-1 sites. What is more interesting is that five of the SBF-1 motifs and the GT-1 site are arranged in three regularly spaced pairs, as are the GT-1 motifs in the promoters of the rbcS multigene family encoding the small subunit of Rubisco (Gilmartin and Chua, 1990). These results suggest a possible regulation by light, but further studies will be necessary.
Figure 5.
Schematic representation of AtXYL1 promoter. A, Sequence from −500 to the start codon. ▾, Transcription start site. A TATA-like sequence is underlined. Potential binding sites for SBF-1 and GT-1, transcription factors of the GT-family, are boxed. B, Concise view of binding sites arrangement in AtXYL1 and rbcS-3 promoters.
Family 31 of Glycoside Hydrolases
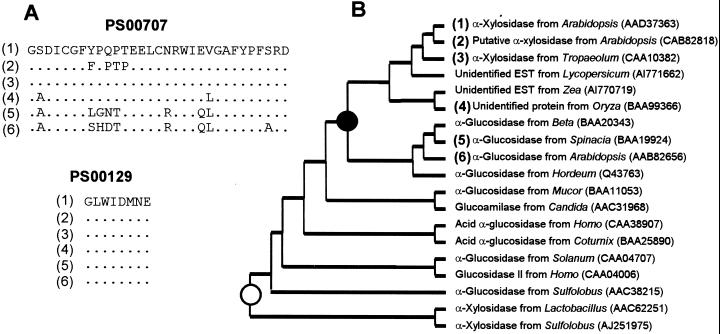
The amino acid sequence identity between the protein coded by AtXYL1 (AtXYL1) and the two signature regions of family 31 glycosyl hydrolases (Henrissat, 1991; Frandsen and Svensson, 1998) showed that the α-xylosidase from Arabidopsis, whose gene we had sequenced, was a member of that family (Fig. 6). More recently, an α-xylosidase cDNA sequence from nasturtium cotyledons has been submitted to the public databases (accession no. AJ131520). When the translation product was compared with AtXYL1, it showed 75% identity over an 899-amino acids consensus length. Searching the Arabidopsis genome, a hypothetical protein (accession no. CAB82818) was found that presented an even higher identity with AtXYL1, 75% identity over 915 amino acids. We propose AtXYL2 as its provisional name. Cabbage peptides showed lower similarity with AtXYL2 than with AtXYL1.
Figure 6.
Partial alignment and phylogenetic relationship among family 31 α-glycosyl hydrolases. A, Alignment of the family 31 PROSITE signature sequences (labeled with the PROSITE accession no.) from known and putative plant α-xylosidases and two plant α-glucosidases. Dots mean conserved amino acids. B, Concise cladogram for family 31 glycosyl hydrolases. ●, The probable point where plant cell wall α-xylosidases started to diverge from α-glucosidases. ○, The independent origin of recently discovered prokaryotic α-xylosidases. Figures between brackets show sequence correspondence between A and B.
AtXYL2 gene includes two stop codons in what should be the coding sequence. This can only be solved assuming, as The Arabidopsis Genome Project has done, two introns that would create two serious gaps in the protein, 45 and 15 amino acids long, when aligned with the other plant family 31 protein sequences. The sequences of these hypothetical introns are as highly conserved with respect to AtXYL1 as the putative coding regions. Thus, it seems likely that AtXYL2 is really a pseudogene that has recently lost its function.
Furthermore, several ESTs from different plant species and one unidentified rice gene that presented a high similarity with AtXYL1 were also found (Fig. 6). Most of the characterized enzymes in family 31 excise α-Glc residues, although two α-xylosidases from prokaryotes (accession nos. AAC62251 and AJ251975) have recently been discovered (Chaillou et al., 1998; Moracci et al., 2000). These α-xylosidases are not closely related to their plant counterparts and seem to have evolved independently.
Arabidopsis α-Xylosidase Is an Apoplastic Enzyme
AtXYL1 protein presented a potential signal peptide 22 amino acids long (Fig. 4), suggesting an apoplastic localization. Since the traditional centrifugation method to obtain the apoplastic fluid was not useful for Arabidopsis seedlings, a different experimental approach has been assayed. The seedlings were grown on water with shaking at 120 rpm to retard the development of the cuticle as described by Monroe et al. (1999) for mustard seedlings.
The apoplastic fluid was partially extracted with 1 m NaCl. When using extraction buffer at pH 4.5, α-xylosidase activity detected in the apoplastic fraction accounted for 37% of the activity in the whole plant. At pH 6, just 16% of the total activity was extracted, possibly due to a higher charge in cell wall pectins. In contrast, Glc-6-P dehydrogenase (G6PDH) was not detected in any fraction when extracted at pH 4.5 due to its instability, but when buffer at pH 6 was used for extraction, the apoplastic fraction accounted for less than 0.2% of the total G6PDH activity. This absence of G6PDH activity extracted with 1 m NaCl proved the lack of cytoplasmic contamination obtained with this mild extraction method. It follows then that at least a fraction of α-xylosidase activity is located in the apoplast. The remaining activity might be inside the protoplast or, more likely, it might not have been able to diffuse into the medium due to the extremely gentle extraction procedure.
α-Xylosidase Is Developmentally Regulated
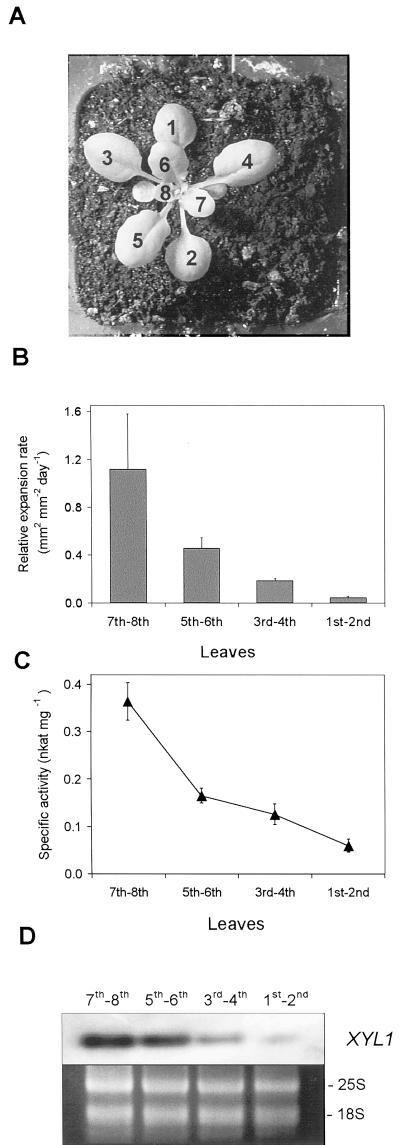
Arabidopsis plants grown for 19 d had eight or more leaves (Fig. 7). At this time there was a clear growth gradient, with younger leaves growing faster than older ones. Although the two oldest leaves had almost achieved their maximum size, the youngest pair was growing 24 times faster. The specific activity of α-xylosidase showed a good positive relationship with growth rate, because it was six times higher in the fastest growing leaves than in the slowest growing ones. AtXYL1 mRNA levels were also lower the older the leaves were. Thus, it seems apparent a positive relationship among growth, α-xylosidase activity, and AtXYL1 gene expression. This is not surprising at all since xyloglucan depolymerization increases during plant growth (Nishitani and Masuda, 1983; Hoson, 1993). This would raise the concentration of xyloglucan oligosaccharides, and α-xylosidase activity should, therefore, be higher in fast-growing walls.
Figure 7.
Developmental regulation of α-xylosidase. A, Photography of a typical 19-d-old Arabidopsis plant. The first eight true leaves were numbered according to their appearance order. B, Leaf relative expansion rate as function of developmental stage. The first eight leaves were sorted into four categories. Data are mean values with sd for 10 plants. C, α-Xylosidase-specific activity extracted from different leaf groups. D, Analysis of AtXYL1 expression using reverse transcriptase (RT)- PCR followed by Southern hybridization. rRNA stained with ethidium bromide was used as control.
Regarding the possible expression of AtXYL2, it should be noted that in the public databases there are at least 56 ESTs corresponding to AtXYL1 and none to AtXYL2. This would suggest that AtXYL2 is not expressed at all or only at low levels.
Heterologous Expression
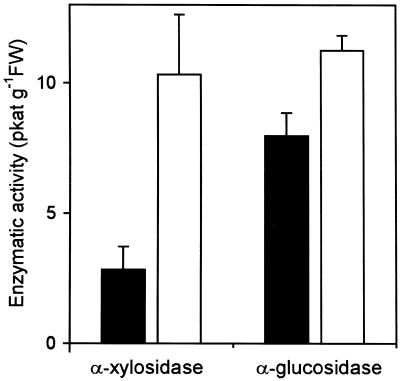
AtXYLI was used to transform S. cerevisiae cells and these cells were grown for 48 h in YP4 high stability expression medium. α-Xylosidase activity was not detected in the culture medium, even though the expression plasmid included the α-factor signal peptide. On the other hand, when the cells were disrupted with glass beads, low levels of α-xylosidase activity were present in the extract (Fig. 8). Cells transformed with AtXYL1 show 3.63 times the activity of cells transformed with a control plasmid that had no insert, this increase being statistically significant (P < 0.01). Similar results were found when cells were grown in YP Expression medium (data not shown). These results support the assignation of α-xylosidase activity to AtXYL1. It should be noted that an α-glucosidase from barley that belongs to family 31 has also been heterologously expressed in yeast and its specific activity was 312 times lower than that of the native enzyme (Frandsen et al., 2000).
Figure 8.
Heterologous expression of AtXYL1. α-Xylosidase and α-glucosidase activities of extracts from S. cerevisiae cells transformed with AtXYL1 (white bars) or with a control plasmid that had no insert (black bars) grown in YP4 High Stability Expression medium. Both activities are referred to the fresh weight of harvested cells.
α-Glucosidase activity was also detected in transformed and control cells, the activity being 1.41 times higher in the former. This difference was also statistically significant (P < 0.01). An N-terminal sequence identical to the one we obtained from cabbage α-xylosidase was recently assigned to a broccoli enzyme with α-glucosidase activity (Monroe et al., 1999). Our results are in agreement with the possibility of AtXYL1 having, in addition to its α-xylosidase activity, an α-glucosidase activity against p-nitrophenyl-α-d-glucopyranoside. No activity against p-nitrophenyl-α-d-xyloside was detected in the transformed or the control cells.
Since the recombinant protein should have incorporated a FLAG marker peptide, an ANTI-FLAG affinity gel was used to purify AtXYL1 protein. A single band of 120 kD, corresponding to the expected Mr, was observed on SDS-PAGE gels (data not shown), but no activity could be detected, probably due to the pH 7.4 used during purification, which had been observed to inactivate cabbage α-xylosidase.
Concluding Remarks
Our study identified an Arabidopsis α-xylosidase gene (AtXYL1) that encodes an apoplastic enzyme able to release Xyl from xyloglucan oligosaccharides. AtXYL1 amino acid sequence showed 75% identity with the sequence of nasturtium α-xylosidase, the only other sequenced plant α-xylosidase. Aglu-2 had been previously proposed by Monroe's group as the name for the Arabidopsis gene we have called AtXYL1 on the basis of its homology to the broccoli enzyme partially purified by them (Monroe et al., 1999). It is important to note that although this enzyme had α-glucosidase activity against p-nitrophenyl-α-d-glucopyranoside, it was not tested for possible α-xylosidase activity against xyloglucan oligosaccharides. The results from our heterologous expression experiments show that it is probable that Arabidopsis and cabbage α-xylosidases have in vitro activity against certain α-glucosidase substrates. α-Xyl and α-Glc are structurally very similar with their hydroxyl groups in the equatorial positions. Sulfolobus sulfataricus α-xylosidase, a family 31 enzyme, has low activity against maltose and 4-p-nitrophenyl-α-glucoside (Moracci et al., 2000). However, we think AtXYL1 is a more appropriate name, since α-Glc containing polysaccharides have not been found on cell walls and xyloglucan is one of the main components of primary walls (Carpita and Gibeaut, 1993). The high similarity with nasturtium α-xylosidase also supports the idea that the physiologically relevant activity of AtXYL1 protein is α-xylosidase, although other possibilities cannot be ruled out at present.
AtXYL1 polypeptide was found to be 915 amino acids long, including a potential signal peptide of 22 amino acids (Fig. 4). Without the signal peptide AtXYL1 would be almost 100 kD in size. The maturation process for α-xylosidase in Arabidopsis is presently unknown, but if it can be assumed that the N-terminal sequence of the mature Arabidopsis protein is placed in a similar position as on the purified cabbage α-xylosidase, then an 11-kD pro-peptide must be cleaved during processing. This would reduce the molecular mass to 88.6 kD, which would fit our data from PAGE gels (Fig. 1). The position in the sequence where cabbage α-xylosidase N terminus is located is very similar to that of 70-kD human lysosomal α-glucosidase, the best known enzyme of family 31 (Wisselaar et al., 1993). It is interesting that this enzyme also loses a C-terminal fragment of more than 7.6 kD. Moreover, the presence of eight potential N-glycosylation sites in AtXYL1 sequence and the fact that cabbage α-xylosidase bound to Con-A Sepharose suggests that the mature enzyme is probably glycosylated, as is also the case for lysosomal α-glucosidase (Wisselaar et al., 1993). In this instance, the mass of the enzyme would be clearly increased over the 88.6 kD mentioned above, but this could be compensated if α-xylosidase lost a C-terminal fragment during maturation.
The family 31 cladogram suggests that a gene duplication event took place in this family at an early phase of plant evolution (Fig. 6). Both known α-xylosidases appear in the same branch, which includes several unidentified proteins. Since xyloglucan occurs in all higher plants (Hayashi, 1989), it seems reasonable to presume that an α-xylosidase gene has always been necessary. Thus, it would not be surprising to find that all the genes in the same branch as AtXYL1 are also α-xylosidase genes.
The presence of possible light-regulated elements in AtXYL1 promoter is also consistent with this idea of a growth-related enzyme. α-Xylosidase role might be more interesting than just degrading oligosaccharides as soon as they are produced. A particular oligosaccharide concentration depends on its formation and degradation rates. As we noted before, there are several of them that show a powerful effect on growth rate (McDougall and Fry, 1988, 1990; Lorences et al., 1990), and in some cases this effect requires a Xyl residue on the non-reducing end of the oligosaccharide molecule (Lorences and Fry, 1993). For these oligosaccharides the level of α-xylosidase activity would be critical. It is, therefore, likely that a more detailed regulation exists for this enzyme. Having identified an α-xylosidase gene in Arabidopsis means that many of the genetic tools developed for this species can now be employed to better understand the regulation of a key step in xyloglucan metabolism.
MATERIAL AND METHODS
Plant Material
Cabbages (Brassica oleracea var. capitata) were purchased at a local market and were used in the same day.
For RNA and protein extraction, Arabidopsis ecotype Columbia was grown for 16 or 19 d at 22°C under a 16-/8-h light/dark photoperiod. Seeds were cold treated at 4°C for 2 d and then directly sown on soil.
To separate apoplastic and cytoplasmic fractions, Arabidopsis seeds were superficially sterilized with 1% (w/v) NaClO for 10 min, thoroughly rinsed with sterile water, and grown for 3 d in 100-mL flasks containing 20 mL of sterile distilled water at 25°C, with orbital shaking at 120 rpm under a 16-/8-h, light/dark photoperiod.
Enzyme and Protein Assays
Tamarind (Tamarindus indica) xyloglucan was digested with Trichoderma viride endoglucanase (Sigma, St. Louis) to obtain an oligosaccharide mixture that was employed as substrate for α-xylosidase determination. This activity was determined as pentose release (Xyl being the only pentose in xyloglucan), according to the method of Roe and Rice (1948) as modified by Edwards et al. (1985). Aliquots of α-xylosidase containing extracts were incubated with 400 μg of oligosaccharides in 0.1 m Na acetate, pH 4.5, at 40°C for a variable period according to the extract activity.
The activity of extracts against p-nitrophenyl-α-d-glucopyranoside and p-nitrophenyl-α-d-xylopyranoside was also measured in 0.1 m Na acetate, pH 4.5, at 40°C, but with 2 mm substrate concentration. In the case of p-nitrophenyl-β-d-galactopyranoside and p-nitrophenyl-β-d-glucopyranoside, the substrate concentration was 5 mm. After incubation, the reactions were stopped by adding 200 mm Na2CO3 and the A400 was measured as described elsewhere.
Protein concentration was quantified using Coomassie protein assay reagent (Pierce, Rockford, IL).
Enzyme Purification
Leaves from cabbage centers (2 kg) were cut into small pieces, frozen with liquid nitrogen, and homogenized to a fine powder with an Omnimixer (Sorvall, Newton, CT). The powder was suspended in 5 L of 50 mm sodium acetate buffer, pH 4.5, and was maintained in suspension with magnetic stirring at 4°C for 2 h. The unextracted material was collected by vacuum filtration through muslin, resuspended with the same volume of 1 m sodium acetate buffer, pH 4.5, and left overnight at 4°C with magnetic stirring. The suspension was then centrifuged at 7,000g for 40 min, and the supernatant was considered as the crude extract.
The crude extract was fractionated by ammonium sulfate precipitation. The protein precipitated between 40% and 80% of saturation was dissolved in 20 mm imidazole-HCl buffer, pH 6.5, containing 50 mm NaCl and dialyzed against the same buffer. Afterward, the extract was chromatographed on an SP-Sepharose (Amersham Pharmacia Biotech AB, Uppsala) column (30 × 2.5 cm). It was eluted in 20 mm imidazole-HCl buffer, pH 6.5, containing a linear gradient of NaCl (50 mm to 1 m), and 4.5-mL fractions were collected. The active fractions were pooled and brought to 90% saturation with (NH4)2SO4.
In preparation for affinity chromatography, the precipitate was dissolved in binding buffer (20 mm sodium acetate buffer, pH 5.0, containing 200 mm NaCl, 1 mm CaCl2, and 1 mm Mn2Cl) and then was dialyzed against the same buffer. The partially purified extract was then applied to a Con-A-Sepharose (Amersham Pharmacia Biotech) column (3 × 1.5 cm) and was washed with binding buffer. Retained proteins were then eluted with 80 mm methyl-O-d-glucopyranoside, dissolved in the same buffer, and immediately brought to 90% saturation with (NH4)2SO4.
The precipitate was dissolved in 20 mm sodium acetate buffer, pH 4.5, containing 200 mm NaCl and was then dialyzed against the same buffer. The protein extract was applied on a Sephacryl S-100-HR (Sigma) column (100 × 1 cm) and 1.5-mL fractions were collected. All the purification procedures were performed at 4°C.
SDS-PAGE
Gel permeation selected fractions containing α-xylosidase activity and 70 μg of total protein was pooled and divided in two similar aliquots. These were then concentrated with a Centricon-10 (Millipore Corporation, Bedford, MA), vacuum dried, and analyzed by SDS-PAGE on two 7.5% (w/v) gels using Mini-Protean II Electrophoresis Cell (Bio-Rad, Hercules, CA), following the manufacturer directions. After a brief staining with Coomassie Blue, the only visible band was sliced from both gels. Eurosequence (Groningen, Netherlands) performed N-terminal sequence with one band and tryptic peptide sequencing with the other.
Analytical electrophoresis was done as described above, except for staining where Silver Stain Kit, Protein (Amersham Pharmacia Biotech) was used. Silver Stain SDS-PAGE standards, high range (Bio-Rad), were employed to determine apparent molecular weights.
IEF
IEF was conducted using a 110-mL focusing column (AMPHOLINE 8100, Amersham Pharmacia Biotech) according to the manufacturer specifications. AMPHOLINE ampholites (Amersham Pharmacia Biotech) were employed to create the pH gradient (3.5–10). After 24 h at 500 V, 5-mL fractions were collected.
Genetic Material
Genomic DNA clones from IGF-BAC library (Mozo et al., 1998b) and cDNA clones from Kieber and Ecker Columbia library (Kieber et al., 1993) were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus).
PCR Screening
BACs suspected to include AtXYL1 were purified from Escherichia coli cultures (500 mL) using Plasmid Maxi Kit (Qiagen GmbH, Hilden, Germany). Primers SCREEN1 (5′-TTCTCACCCACCCAATCCTA-3′) and SCREEN2 (5′-GATAAGTTCGGATCCGGAGA-3′) were employed for PCR in the following conditions: 94°C for 2 min followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, followed by 72°C for 5 min.
Nucleotide Sequencing
DNA was sequenced using T7 Sequenase Quick-denature Plasmid Sequencing Kit (Amersham Pharmacia Biotech). Gels were run in Sequi-Gen GT Sequencing Cell (Bio-Rad).
RACE
Enhanced Avian Reverse Transcriptase (Sigma) was employed according to the manufacturer instructions to produce single-stranded cDNA from 1.7 μg of RNA extracted from the leaves of 16-d-old Arabidopsis plants. RACE1 primer (5′-CTACATTGATGAAATCCTAAAG-3′), annealing to the region encompassing exons 2 and 3 in AtXYL1 RNA, was employed. A 5′-end polyA tail was then incorporated to the cDNA using Terminal Deoxynucleotidyl Transferase (Amersham Pharmacia Biotech). Double-stranded cDNA was subsequently produced with RACE2 primer (5′-GACTCGAGTCGACATCGATTTTTTTTTTTTTTTTT-3′) and Accu Taq LA DNA Polymerase (Sigma) in the following conditions: 94°C for 3 min, 30°C for 30 s, and 45°C for 5 min. RACE3 (5′-GACTCGAGTCGACATCGA-3′) and a second gene-specific primer, SCREEN2, were then added to the reaction mixture and 40 cycles of PCR amplification were performed (94°C for 1 min, 50°C for 1 min, and 68°C for 1 min). A clear band (approximately 550 bp) was observed in agarose electrophoresis, purified with Wizard PCR preps (Promega, Madison, WI), reamplified, and then finally sequenced.
Computer Analysis
Family 31 cladogram was based on multiple alignments done with CLUSTAL W (1.5). Trees were then drawn with Windows Easy Tree v 1.32 (Joaquin Dopazo, Glaxo Wellcome, Spain) using the Neighbor Joining Method. EST placement was based in a separate tree that only included data from the region where they all overlap. Complete sequences were used to place the other branches. Similarity searches were done with the National Center for Biotechnology Information BLAST 2.0 set of programs (Altschul et al., 1997). Promoter analysis was done with MatInspector V 2.2 (Genomatix Software, Munich) and PatSearch 1.1 using the TRANSFAC 4.0 plant matrices and the TRANSFAC 3.5 sites (Wingender et al., 2000).
Apoplastic Fraction Extraction
The apoplastic fluid from Arabidopsis seedlings was obtained as described by Monroe et al. (1999). Seedlings, grown on water for 3 d, were transferred to 10 mL of 0.1 m sodium acetate buffer, pH 4.5, containing 1 m NaCl, and were shaken for 6 h at 120 rpm. The saline extract was considered as the apoplastic fraction.
Seedlings were then homogenized in 5 mL of 0.1 m sodium acetate buffer, pH 4.5, containing 1 m NaCl. The homogenate was shaken for 2 h and was centrifuged at 15,000g for 20 min. The supernatant was considered as the cytoplasmic fraction. Both extracts were concentrated with a Centricon-10 (Millipore) to approximately 200 μL and were then diluted to 2 mL with 100 mm sodium acetate, pH 4.5. This procedure was repeated twice. The extract was concentrated and α-xylosidase activity was then measured.
The absence of cytoplasmic contamination was assessed according to Sánchez et al. (1997) by measuring G6PDH. To measure this activity, extracts were obtained as stated above except for the extraction buffer that was 0.1 m MES [2-(N-morpholino)-ethanesulfonic acid]-HCl, pH 6, containing 1 m NaCl. G6PDH activity was determined measuring the absorption increase at 340 nm, in a reaction mixture (1 mL) containing 100 μL protein extract, 1 mm Glc-6-P, 0.2 mm NADP+, 1 mm MgCl2, and 100 mm Tricine {N-[2-hydroxy-1,1-Bis(hydroxymethyl)ethyl]glycine}-NaOH, pH 8.0 (Takahama, 1993). α-Xylosidase activity was also determined in these extracts by hydrolysis of xyloglucan oligosaccharides, as described earlier.
RNA and Protein Extraction in Arabidopsis
The first eight true leaves of 19-d-old plants were harvested, sorted into four groups according to their appearance order, and immediately frozen in liquid nitrogen. For each pair of leaves, protein was extracted from 200 mg of tissue (100 mg for 7th–8th leaves) in 800 μL of 1 m sodium acetate, pH 4.5, with the addition of 4 μL of 100 mm phenylmethylsulfonyl fluoride stock solution. After 2.5 h of shaking at 4°C, the homogenate was centrifuged twice at 10,000g for 10 min. In each case the supernatant was collected. The extract was then concentrated with a Centricon-10 (Millipore) to approximately 100 μL and was diluted to 2 mL with 100 mm sodium acetate, pH 4.5. This procedure was carried out three times. After a last concentration step the volume was adjusted to 400 μL with the same buffer.
RNA was extracted from 200 mg of each category of leaves (60 mg for 7th–8th leaves) with Rneasy Plant Mini Kit (Qiagen, Valencia, CA) and was quantified through A260.
RT-PCR
Enhanced Avian RT-PCR Kit (Sigma) was employed as recommended by the manufacturer to perform one-step RT-PCR. RACE1 primer was chosen to avoid amplifying genomic DNA, and RT1 (5′-TCCATCGAAGAATCTCCCGA-3′) was the second primer. The expected product was 782 bp long. For each leaf category, a reaction was set up that included 500 ng of RNA, 50 pmol of each primer, and 2.75 mm MgCl2 in a total volume of 50 μL. After 45 min at 45°C, PCR tubes were heated at 94°C for 3 min, followed by 12 cycles of 94°C for 1 min, 50°C for 1 min, and 68°C for 1 min, followed by 68°C for 4 min.
RACE1 and RT1 primers show 20% and 14% mismatch when aligned with AtXYL2. To unequivocally identify the amplified mRNA, a longer amplification (40 cycles) was performed in similar conditions with RNA from the youngest leaves. The resulting band was purified and identified as an AtXYL1 amplification product by restriction mapping.
Southern Hybridization
A digoxigenin-labeled probe was produced with digoxigenin-High Prime DNA Labeling and Detection Starter Kit II (Roche Diagnostics GmbH, Mannheim, Germany) using as template a 593-bp DNA fragment amplified by PCR from clone H8A7, using primers RACE1 and HYB1 (5′-GAACAACCACCACAAGTCGG-3′).
RT-PCR products from 12 cycles of reactions (15 μL) were electrophoresed in a 0.9% (w/v) agarose gel containing ethidium bromide. After the gel was photographed, it was blotted to a positively charged nylon membrane (Roche) using VacuGene XL Vacuum blotting System (Amersham Pharmacia Biotech) as recommended by the manufacturer.
Probe detection was done with DIG-High Prime DNA Labeling and Detection Starter Kit II. In brief, after blocking the membrane, hybridization was carried out overnight and then membranes were washed, blocked, incubated with anti-DIG-AP antibodies, washed again, and finally incubated with disodium 3-(4-methoxyspiro[1,2-dioxetane-3,2′-{5′-chloro}tricyclo{3.3.1.13,7}decan]4-yl)-Star chemiluminescent reagent.
Heterologous Expression
FLAG Yeast N-Terminal Expression System (Sigma) was employed as suggested by the manufacturer. AtXYL1 coding region, excluding the potential signal peptide, was amplified by PCR using H8A7 clone as a template and EXP-1 (5′-GAATTCTACAAAACCATCGGCAAAGGCTA-3′) and EXP-2 (5′-GCGGCCGCTTAATTGATACCCATTTTCCAGGA-3′) as primers. These primers include restriction sites for EcoRI and NotI, respectively. PCR conditions were as follows: 94°C for 2 min followed by 25 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 2 min, followed by 72°C for 5 min. Ex-Taq polymerase (Takara, Shiga, Japan) was used, according to the manufacturer's recommendations. The reaction included 25 pmol of each primer and 50 ng of template.
A unique PCR product of correct size was observed on agarose electrophoresis, purified, and ligated to YEpFLAG expression Vector, using T4DNA ligase (Promega). XL1-Blue E. coli was transformed with the recombinant plasmid (Sambrook et al., 1989). BJ3505 Saccharomyces cerevisiae was then transformed with plasmid purified from E. coli as indicated by the Expression System manufacturer.
Fresh saturated cultures of two independently transformed colonies were used to inoculate 500-mL flasks containing 100 mL of YP4 high stability expression medium (1% [w/v] Glc, 3% [w/v] glycerol, 1% [w/v] yeast extract, 8% [w/v] peptone, and 20 mm CaCl2) or YP expression medium (1% [w/v] Glc, 3% [w/v] glycerol, 1% [w/v] yeast extract, and 2% [w/v] peptone). Cultures were grown for 48 or 96 h, respectively, at 30°C and 200 rpm. Cells were harvested by centrifugation (4,500g for 10 min), disrupted with glass beads, and extracted in 5 mL of 1 m sodium acetate buffer, pH 4.5, for 2 h. The extracts were brought to 90% saturation with (NH4)2SO4 and the precipitate were resuspended in 0.1 m sodium acetate buffer, pH 4.5. Control transformations were performed as indicated by the manufacturer and cells were grown and treated as above.
Purification of the expressed protein was performed as indicated by the manufacturer using a 1-mL column packed with ANTI-FLAG M1 Affinity Gel. The extract containing α-xylosidase activity was filtered, concentrated, and diluted with water repeatedly and then finally diluted with buffer to achieve a final concentration of 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 10 mm CaCl2. This sample was loaded in the affinity column and after several washes with the same buffer, the retained protein was eluted in 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 2 mm EDTA.
ACKNOWLEDGMENTS
We are grateful to Dr. Ester P. Lorences for her helpful comments and to Mr. D.A. Lindsay for correcting the English version.
Footnotes
This work was supported by the Dirección General de Enseñanza Superior e Investigación Científica (grant no. PB98–0640) and by Xunta de Galicia (grant no. PGIDT00PXI20002PN).
LITERATURE CITED
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augur C, Benhamou N, Darvill A, Albersheim P. Purification, characterization, and cell wall localization of an α-fucosidase that inactivates a xyloglucan oligosaccharin. Plant J. 1993;3:415–426. doi: 10.1046/j.1365-313x.1993.t01-24-00999.x. [DOI] [PubMed] [Google Scholar]
- Augur C, Stiefel V, Darvill A, Albersheim P, Puigdomenech P. Molecular cloning and pattern of expression of an α-l-fucosidase gene from pea seedlings. J Biol Chem. 1995;270:24839–24843. doi: 10.1074/jbc.270.42.24839. [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Chaillou S, Lokman BC, Leer RJ, Posthuma C, Postma PW, Pouwels PH. Cloning, sequence analysis, and characterization of the genes involved in isoprimeverose metabolism in Lactobacillus pentosus. J Bacteriol. 1998;180:2312–2320. doi: 10.1128/jb.180.9.2312-2320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M, Bowmann YJL, Dea ICM, Reid JSG. A β-d-galactosidase from nasturtium (Tropaeolum majusL.) cotyledons: purification, properties, and demonstration that xyloglucan is the natural substrate. J Biol Chem. 1988;263:4333–4337. [PubMed] [Google Scholar]
- Edwards M, Dea ICM, Bulpin PV, Reid JSG. Xyloglucan mobilization in the cotyledons of Tropaeolum majusL. seeds following germination. Planta. 1985;163:133–140. doi: 10.1007/BF00395907. [DOI] [PubMed] [Google Scholar]
- Fanutti C, Gidley MJ, Reid JSG. A xyloglucan-oligosaccharide-specific α-d-xylosidase or exo-oligoxyloglucan-α-xylohydrolase from germinated nasturtium (Tropaeolum majus L.) seeds: purification, properties and its interaction with a xyloglucan-specific endo-(1-4)-β-d-glucanase and other hydrolases during storage-xyloglucan mobilization. Planta. 1991;184:137–147. doi: 10.1007/BF00208247. [DOI] [PubMed] [Google Scholar]
- Farkas V, Hanna R, Maclachlan G. Xyloglucan oligosaccharide α-l-fucosidase activity from growing pea stems and germinating nasturtium seeds. Phytochemistry. 1991;30:3203–3207. doi: 10.1016/0031-9422(91)83176-l. [DOI] [PubMed] [Google Scholar]
- Frandsen TP, Lock F, Mirgorodskaya E, Roepstorff P, Svensson B. Purification, enzymatic characterization, and nucleotide sequence of a high isoelectric-point α-glucosidase from barley malt. Plant Physiol. 2000;123:275–286. doi: 10.1104/pp.123.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen TP, Svensson B. Plant α-glucosidases of the glycoside hydrolase family 31: molecular properties, substrate specificity, reaction mechanism, and comparison with family members of different origin. Plant Mol Biol. 1998;37:1–13. doi: 10.1023/a:1005925819741. [DOI] [PubMed] [Google Scholar]
- Fry SC. In vivo formation of xyloglucan nonasaccharide: a possible biologically active cell wall fragment. Planta. 1986;169:443–453. doi: 10.1007/BF00392143. [DOI] [PubMed] [Google Scholar]
- Fry SC. The structure and function of xyloglucan. J Exp Bot. 1989;40:1–11. [Google Scholar]
- Fry SC. Polysaccharide-modifying enzymes in the plant cell wall. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:497–520. [Google Scholar]
- Gilmartin PM, Chua NH. Spacing between GT-1 binding sites within a light-responsive element is critical for transcriptional activity. Plant Cell. 1990;2:447–455. doi: 10.1105/tpc.2.5.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén R, York WS, Pauly M, An G, Albersheim P, Darvill AG. Metabolism of xyloglucan generates xylose-deficient oligosaccharide subunits of this polysaccharide in etiolated peas. Carbohydr Res. 1995;277:291–311. doi: 10.1016/0008-6215(95)00220-n. [DOI] [PubMed] [Google Scholar]
- Hayashi T. Xyloglucans in the primary cell wall. Annu Rev Plant Physiol Mol Biol. 1989;40:139–168. [Google Scholar]
- Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoson T. Regulation of polysaccharide breakdown during auxin-induced cell wall loosening. J Plant Res. 1993;106:369–381. [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Koyama T, Hayashi T, Kato Y, Matsuda K. Degradation of xyloglucan by wall-bound enzymes from soybean tissue II: degradation of the fragment heptasaccharide from xyloglucan and the characteristic action pattern of the α-d-xylosidase in the enzyme system. Plant Cell Physiol. 1983;24:155–162. [Google Scholar]
- Lorences EP, Fry SC. Xyloglucan oligosaccharides with at least two α-d-xylose residues act as acceptor substrates for xyloglucan endotransglycosylase and promote the depolymerisation of xyloglucan. Physiol Plant. 1993;88:105–112. [Google Scholar]
- Lorences EP, McDougall GJ, Fry SC. Xyloglucan- and cello-oligosaccharides: antagonists of the growth promoting effect of H+ Physiol Plant. 1990;80:109–113. [Google Scholar]
- Marra MA, Kucaba TA, Dietrich NL, Green ED, Brownstein B, Wilson RK, McDonald KM, Hillier LW, McPherson JD, Waterston RH. High throughput fingerprint analysis of large-insert clones. Genome Res. 1997;11:1072–1084. doi: 10.1101/gr.7.11.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita J, Kato Y, Matsuda K. Purification and properties of an α-d-xylosidase from Aspergillus niger. J Biochem. 1985;98:825–832. doi: 10.1093/oxfordjournals.jbchem.a135341. [DOI] [PubMed] [Google Scholar]
- McDougall G, Fry SC. Inhibition of auxin-stimulated growth of pea stem segments by a specific nonasaccharide of xyloglucan. Planta. 1988;175:412–416. doi: 10.1007/BF00396348. [DOI] [PubMed] [Google Scholar]
- McDougall G, Fry SC. Xyloglucan oligosaccharides promote growth and activate cellulase: evidence for a role of cellulase in cell expansion. Plant Physiol. 1990;93:1042–1048. doi: 10.1104/pp.93.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe JD, Gough CM, Chandler LE, Loch CM, Ferrante JE, Wright PW. Structure, properties, and tissue localization of apoplastic α-glucosidase in crucifers. Plant Physiol. 1999;119:385–397. doi: 10.1104/pp.119.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moracci M, Ponzano BC, Trincone A, Fusco S, De Rosa M, van der Oost J, Sensen CW, Charlebois RL, Rossi M. Identification and molecular characterization of the first α-xylosidase from an Archaeon. J Biol Chem. 2000;275:22082–22089. doi: 10.1074/jbc.M910392199. [DOI] [PubMed] [Google Scholar]
- Mozo T, Fischer S, Meier-Ewert S, Lehrach H, Altmann T. Use of the IGF BAC library for physical mapping of the Arabidopsis thalianagenome. Plant J. 1998a;16:377–384. doi: 10.1046/j.1365-313x.1998.00299.x. [DOI] [PubMed] [Google Scholar]
- Mozo T, Fischer S, Shizusa H, Altmann T. Construction and characterization of the IGF Arabidopsis BAC library. Mol Gen Genet. 1998b;258:562–570. doi: 10.1007/s004380050769. [DOI] [PubMed] [Google Scholar]
- Newman T, deBruijn FJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M. Genes galore: a summary of methods for accessing results from large-scale partial sequencing of anonymous ArabidopsiscDNA clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani K, Masuda Y. Auxin-induced changes in the cell wall xyloglucans: effects of auxin on the two different subfractions of xyloglucans in the epicotyl cell wall of Vigna angularis. Plant Cell Physiol. 1983;24:345–355. [Google Scholar]
- O'Neill R, Albersheim P, Darvill AG. Purification and characterization of a xyloglucan oligosaccharide-specific xylosidase from pea seedlings. J Biol Chem. 1989;260:20430–20437. [PubMed] [Google Scholar]
- Roe JH, Rice EW. A photometric method for the determination of free pentoses in animal tissues. J Biol Chem. 1948;173:507–512. [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sánchez M, Queijeiro E, Revilla G, Zarra I. Changes in ascorbic acid levels in apoplastic fluid during growth of pine hypocotyls: effect on peroxidase activities associated with cell walls. Physiol Plant. 1997;101:815–820. [Google Scholar]
- Takahama U. Regulation of peroxidase dependent oxidation of phenolics by ascorbic acid: different effects of ascorbic acid on the oxidation of coniferyl alcohol by the apoplastic soluble and cell wall-bound peroxidases from epicotyls of Vigna angularis. Plant Cell Physiol. 1993;39:809–817. [Google Scholar]
- Villain P, Clabault G, Mache R, Zhou DX. S1F binding site is related to but different from the light–responsive GT-1 binding site and differentially represses the spinach rps1 promoter in transgenic tobacco. J Biol Chem. 1994;269:16626–16630. [PubMed] [Google Scholar]
- Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Pruss M, Reuter I, Schacherer F. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisselaar HA, Kroos MA, Hermans MMP, Van Beeument J, Reuser AJJ. Structural and functional changes of lysosomal acid α-glucosidase during intracellular transport and maturation. J Biol Chem. 1993;268:2223–2231. [PubMed] [Google Scholar]
- Zong N, Kamiyama Y, Yasui T. Substrate specificity of Bacillus α-d-xylosidase. Agric Biol Chem. 1989;53:2129–2139. [Google Scholar]
- Zong N, Yasui T. Purification and some properties of an α-d-xylosidase from Bacillussp. no. 693-1. Agric Biol Chem. 1989;53:187–195. [Google Scholar]