Abstract
Doxorubicin (DOX)-induced cardiomyopathy constitutes dose-dependent cardiac toxicity, culminating in fatal heart failure progression. Cardiac toxicity limits effective and subsequent use of DOX in chemotherapy regimens in pediatric, adult, and recurrent cancer patients. DOX-induced profound alterations in mitochondrial morphology, dynamics, and defects in mitochondrial energy metabolism in the heart comprise key stressors in DOX-induced cardiotoxicity. Hence, the discovery of novel molecular targets and therapeutics to mitigate DOX-induced mitochondrial dysfunctions are imperative. Herein, we provided two laboratory protocols to monitor DOX-induced alterations in mitochondrial morphology and respiration in isolated primary neonatal rat cardiomyocytes. Neonatal rat cardiomyocytes are extensively used to monitor signaling mechanisms regulating cardiomyopathy in vitro. Therefore, these protocols will help researchers study the effects of novel pharmacological and genetic manipulations against DOX-induced alterations in mitochondrial morphology and energy metabolism in cardiomyocytes.
Keywords: Doxorubicin-induced cardiomyopathy, Mitochondrial morphology, Mitochondrial respiration, Oxygen consumption rates
1. Introduction
Doxorubicin (Dox) is an efficacious anthracycline-based chemotherapeutic agent widely used against a variety of solid tumors, i.e., breast, gynecological, urogenital, endocrine, brain tumors, and also in leukemia [1–3]. Despite being an effective anti-cancer drug and first-line choice of chemotherapeutic agent, DOX’s use is limited due to its dose-dependent cardiotoxicity. DOX-induced cardiotoxicity causes pathological changes in the heart termed as “Doxorubicin-induced Cardiomyopathy” that underlies fatal heart failure development in adolescent and adult cancer surviving DOX recipient patients [4–6]. Nearly 26% of the patients suffer heart failure who received a lifetime cumulative dose of DOX greater than 500 mg/m2 while heart failure prevalence reaches 36% in patients receiving 600 mg/m2 of cumulative dose of DOX [1, 6–8]. Extensive studies confirmed DOX profoundly affects cardiac mitochondrial ultrastructure and respiratory functions, including mitochondrial swelling and abnormal cristae structure [9, 10], mitochondrial DNA (mtDNA) damage [11, 12], excessive reactive oxygen species generation through redox cycling at respiratory Complex I [13–15], suppression of mitochondrial oxidative phosphorylation involved respiration [16, 17], defects in fatty acid β-oxidation resulting switch in substrate metabolism [18], reduced mitochondrial membrane potential (Ψm) culminating to the opening of mitochondrial permeability transition pore (mPTP) [9, 19]. This leads to apoptotic mediators release from mitochondria intermembrane and matrix that eventually potentiate cell demise, constituting heart failure development [9, 19, 20].
Cardiomyocytes require a continuous supply of energy in the form of ATP (adenosine triphosphate) to maintain their contraction–relaxation cycles [21, 22]. Oxidative phosphorylation at mitochondrial ETS accounts for nearly 95% of ATP supply in the heart [23, 24]. To this point, accumulating evidence underscores a homeostatic balance between mitochondrial fission and fusion processes, which is essential in maintaining mitochondrial quality control, ultrastructure, and respiratory functions [16, 25–31]. In the light of well-documented DOX-induced defects in cardiac mitochondrial fission–fusion signaling, ultrastructure, ETS functions, and respiration, there is an urgent need to discover novel molecular effectors and therapeutic agents to protect against DOX-induced mitochondrial structural and bioenergetics defect to prevent heart failure initiation and progression.
Isolated primary neonatal rat cardiomyocytes (NRCs) are post-mitotic, terminally differentiated cells with intrinsic beating rhythms, can be cultured for an extended period, responsive to pharmacological stimuli, and amenable for genetic manipulations for gene gain-of-function and loss-of-function studies [32–34]. Hence, the NRCs culture technique provides an excellent preclinical research platform to study molecular underpinnings and to discover novel molecular effectors in cardiomyocytes in vitro exclusive of confounding effects, including in vivo neuro-humoral regulation, extracellular matrix, autocrine, endocrine, and paracrine effects [33, 35].
Our current protocols describe two step-by-step methods to visualize mitochondrial network morphology and mitochondrial oxygen consumption rates (OCR) in Doxorubicin-treated isolated primary ventricular cardiomyocytes from neonatal rat pups [16, 28, 30, 31]. The first protocol will allow researchers to visualize the effects of pharmacological and genetic manipulations in preserving DOX-induced altered mitochondrial network morphology. The second protocol utilizes the Seahorse XFe24 Extracellular Flux analyzer, which offers microplate, live cell-based oxygen consumption rates (OCRs) measurement enabling offline mitochondrial bioenergetic parameters analysis and assessment. Together, these protocols will provide guidelines to the investigators to study the effects of novel genetic manipulations and pharmacological interventions to protect against DOX-induced pathological alterations in mitochondrial morphology and respirations.
2. Materials
2.1. Equipment
Agilent Seahorse XFe24 Extracellular Flux Analyzer.
Nikon A1R high-speed confocal microscope.
Olympus CKX53 inverted cell culture microscope.
Thermo Scientific™ Heracell™ VIOS 160i CO2 incubator equipped with HEPA Filters.
Thermo Scientific™ 1300 Series Class II Biological Safety Cabinet.
VWR Non-CO2 Isotemp incubator.
Eppendorf 5810R centrifuge.
Two-key cell counter (202C, BAL Supply, LLC).
Bright-Line Hemacytometer (Z359629, Sigma-Aldrich).
DeNovix DS-11 FX+ Spectrophotometer/Fluorometer.
2.2. Cell Line
Primary Neonatal Rat Ventricular Cardiomyocytes (NRCs).
2.3. Reagents and Cell Culture Supplies
Doxorubicin Hydrochloride (D1515–10mg, Sigma-Aldrich).
DMEM High Glucose with GlutaMAX™ supplement Cell Culture Media (10566–016, Gibco).
Seahorse XF DMEM medium, pH 7.4 (103575–100, Agilent Technologies Inc.).
MEM alpha (1×) (12571048, Gibco).
Fetal Bovine Serum (FBS) (16000044, Gibco).
Antibiotic-Antimycotic (100×) (15240–062, Gibco).
0.1% Gelatin in Water (07903, STEMCELL Technologies).
MitoTracker™ Green FM (M7514, Invitrogen).
Quick Start™ Bradford 1× Dye Reagent (Cat# 500–0205; Bio-Rad).
Nunc™ Lab-Tek™ II Two-well Chamber slides with cover (154461; Thermo Fisher).
Phosphate Buffered Saline, pH 7.4 (1× PBS) (10010031, Gibco).
Dimethyl sulfoxide (DMSO) (D2650, Sigma-Aldrich).
VECTASHIELD® Hardset™ Antifade Mounting Medium (H-1400–10; Vector Laboratories).
Fisherfinest™ Premium Cover Glasses (24×60–1) (125485P, Fisher Scientific).
Seahorse XFe24 FluxPak (102340–100, Agilent Technologies, Inc.).
Seahorse XF Cell Mito Stress Test Kit includes Oligomycin, Carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP), Rotenone/Antimycin A (103015–100, Agilent Technologies Inc.).
Seahorse XF Glucose solution (1 M) (103577–100, Agilent Technologies Inc.).
Seahorse XF Pyruvate solution (100 mM) (103578–100, Agilent Technologies Inc.).
3. Methods
3.1. Mitochondrial Network Morphology Visualization in Dox-Treated Cardiomyocytes Through Confocal Fluorescence Microscopy
Isolate neonatal rat ventricular cardiomyocytes (NRCs) from 1 to 2 days old rat pups from timed-pregnant Sprague-Dawley rats as detailed previously [33].
Count the cells (see Note 1) and seed 1 × 105 cells per well in gelatin-coated (see Note 2) Nunc™ Lab-Tek™ II Two-well Chamber slides with 800 μL of MEM α cell culture media supplemented with 10% v/v FBS and 1% v/v antibiotic-antimycotic overnight in a humidified cell culture incubator supplied with 5% CO2 and 95% air at 37 °C (see Note 3).
Next morning, add an additional 1200 μL of MEM α media containing 10% v/v FBS and 1% v/v antibiotic-antimycotic per well of chamber slides.
After 24 hours of initial cell seeding, change the media to 2 mL of DMEM High Glucose GlutaMAX™ medium containing 2% v/v FBS and 1% v/v antibiotic-antimycotic per well of chamber slides (see Note 4).
After 72 hours of culture, treat NRCs for 24 h with DOX (prepare stock in DMSO at 50 mM) at 1 μM and 10 μM doses dissolved 1 mL per well of DMEM High Glucose GlutaMAX™ medium containing 2% v/v FBS and 1% v/v antibiotic-antimycotic. Treat vehicle group wells with the same DMSO volume as with DOX treated wells in 1 mL of DMEM High Glucose GlutaMAX™ medium containing 2% v/v FBS and 1% v/v antibiotic-antimycotic.
After 24 hours of treatment, load each well with 200 nM of mitochondrial membrane potential independent Mito-Tracker™ Green FM dye dissolved in 1 mL of DMEM High Glucose GlutaMAX™ medium containing 2% v/v FBS and 1% v/v antibiotic-antimycotic (see Note 5).
Incubate the chamber slides in an incubator supplied with 5% CO2 and 95% air at 37 °C for 40 minutes.
Discard the media, add 1 mL of 3.7% paraformaldehyde in 1× PBS per well for 10 minutes at room temperature to fix the cardiomyocytes.
Discard the fixation solution gently using a micropipette and wash the cells twice with 1 mL of 1× PBS each time for 5 minutes.
Mount the cardiomyocytes with VECTASHIELD® Hardset™ Antifade Mounting Medium.
Observe and image the cardiomyocytes at high magnification to visualize MitoTracker Green stained mitochondrial networks using a confocal fluorescence microscope. We use Nikon A1R high-speed confocal microscope using a 60× oil objective (NA = 1.4) to observe Nikon NIS-Elements C software to image the cardiomyocytes (see Note 6) (Fig. 1).
Mitochondrial networks length and width can be measured on NIH ImageJ software (v1.6.0) using “Freehand Line” drawing on captured digital micrographs [16].
Fig. 1.
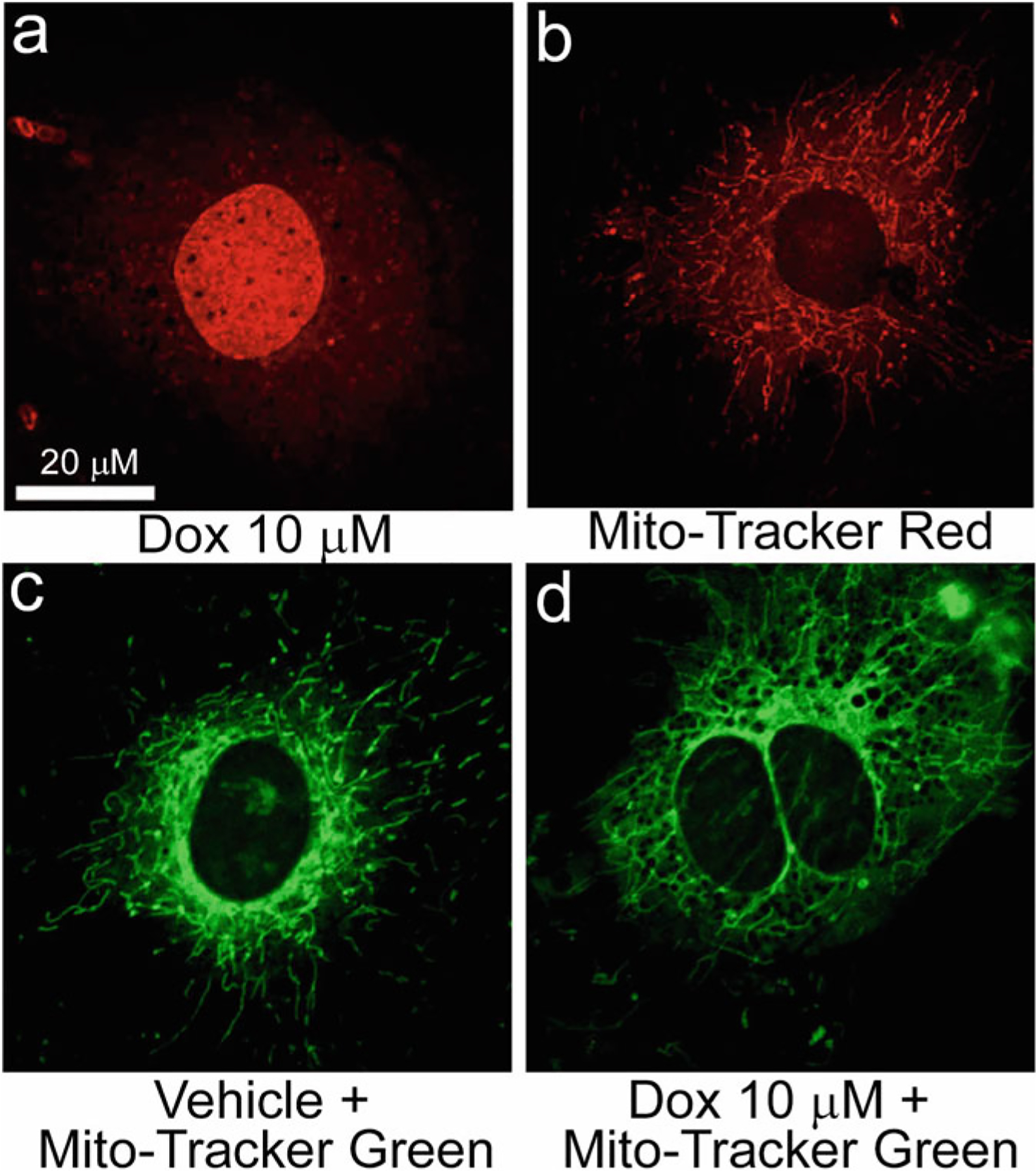
Monitoring mitochondrial network organization and morphology in cardiomyocytes. Representative confocal fluorescence microscope images demonstrating Dox alone (a), MitoTracker® Red alone (b), and MitoTracker® Green (c, d) labeled neonatal rat cardiomyocytes. (a) DOX (red) accumulates at subcellular locations as appeared as fluorescent dots in the cytosol and diffuse accumulation in the nucleus of cardiomyocytes following DOX-treatment (10 μM, 24 hours). (b) MitoTracker® Red staining reveals mitochondrial network (red) as appeared in a blend of fragmented, short tubular, and long tubular forms. Hence, we do not recommend the use of MitoTracker Red dyes to use in DOX-treated cells to avoid potential overlap between them in captured fluorescence images which may lead to erroneous interpretation of the data (please see Note 5 for explanation). (c, d) MitoTracker® Green staining reveals a mixture of a fragmented, short, and long tubular network of mitochondria (green) in vehicle (c) treated cardiomyocytes while DOX treatment (10 μM, 24 hours) resulted in hyperfused tubular mitochondrial network in cardiomyocytes
3.2. Mitochondrial Oxygen Consumption Rates Measurement and Bioenergetic Parameters Calculation in Cardiomyocytes
3.2.1. Cardiomyocytes Seeding in XF24 Cell Culture Plates and Doxorubicin Treatment
Isolate neonatal rat cardiomyocytes (NRCs) from 1 to 2 days old rat pups heart ventricles from timed-pregnant Sprague-Dawley rats as detailed.
Count the cardiomyocytes using a hemacytometer (see Note 1).
Seed 8 × 104 cells per well in gelatin-coated (see Note 2) XF24 cell culture plates initially in 200 μL of MEM α cell culture media containing 10% v/v FBS and 1% v/v antibiotic-antimycotic for 3–4 h in a humidified cell culture incubator supplied with 5% CO2 and 95% air at 37 °C (see Notes 3 and 7).
Add only MEM α cell culture media in four wells (one well per row) in XF24 cell culture plates to use temperature and background control in Seahorse assay.
Add an additional 400 μL of MEM α media containing 10% v/v FBS and 1% v/v antibiotic-antimycotic per well of cardiomyocytes after initial 3–4 hours of seeding.
After 24 hours, change the media to 650 μL of DMEM High Glucose GlutaMAX™ medium supplemented with 2% v/v FBS and 1% v/v antibiotic-antimycotic per well of XF24 cell culture plates (see Note 8).
After 72 hours of culture, treat NRCs for 24 h with Doxorubicin (DOX) (prepare stock in DMSO at 100 mg/mL) at 1 μM and 10 μM concentrations dissolved in DMEM High Glucose GlutaMAX™ medium containing 2% v/v FBS and 1% v/v antibiotic-antimycotic. Treat vehicle group wells with the same DMSO volume as with DOX treated wells in DMEM High Glucose GlutaMAX™ medium containing 2% v/v FBS and 1% v/v antibiotic-antimycotic.
3.2.2. Sensor Cartridge Hydration and Inhibitors Loading into the Sensor Cartridge
The day before the Seahorse assay, separate cartridge lid, Sensor cartridge, Hydro booster, and Utility plate provided in XFe24 FluxPak.
Add 1 mL of XF Calibrant to each well of the Utility plate.
Place the Hydro booster on top of the Utility plate.
Carefully place the Sensor cartridge through the Hydro Booster plate’s openings into the Utility plate, submerging the sensors in XF calibrant.
Cover the Sensor cartridge with the cartridge lid and place the setup in a non-CO2 humidified incubator at 37 °C overnight (see Note 9).
The next day, remove the Cartridge lid from the top of the Sensor cartridge and Hydro booster from between the Sensor cartridge and Utility plate.
Prepare XF assay media by supplementing XF DMEM media with 10 mM Glucose and 2 mM Pyruvate. Check and adjust (if necessary) the pH to 7.4 and sterile filter under a biological safety cabinet.
Prepare Oligomycin (100 μM), FCCP (100 μM), Rotenone, and Antimycin A (50 μM) stock solutions in XF assay media according to the manufacturer’s instructions (Agilent).
Load Oligomycin, FCCP, Rotenone, and Antimycin A diluted in XF assay medium (at 10× concentration) in three ports of Sensor cartridge to deliver sequentially 1 μM Oligomycin, 4 μM FCCP, 0.5 μM Rotenone, and 0.5 μM Antimycin A per well according to the Table 1 (see Note 10).
Table 1.
Inhibitors preparation for loading to XFe24 sensor cartridges
| Inhibitors | Injection port | DMEM media (μL) | The volume of stock compound (μL) | Concentration at ports (10×) (μM) | Loading on sensor cartridge port (μL) |
|---|---|---|---|---|---|
| Oligomycin (100 μM stock) | A | 1350 | 150 | 10 | 56 |
| FCCP (100 μM stock) | B | 900 | 600 | 40 | 62 |
| Rotenone and Antimycin A (50 μM stock) | C | 1350 | 150 | 5 | 69 |
3.2.3. Sensor Cartridge Calibration and Running the Assay
Set up assay protocol in “Wave” software to measure mitochondrial Oxygen Consumption Rates (OCR). An example of the protocol is described in Table 2.
Place the Sensor cartridge on top of the Utility plate into the Seahorse XFe24 Analyzer for calibration.
When the calibration is running, remove the XF24 cell culture plate from the incubator. Gently aspirate the cell culture medium leaving 50 μL of the medium in each well.
Gently add 600 μL of warmed XF assay media (no glucose, pyruvate) into each well.
Gently aspirate the media while leaving 50 μL of the media in each well.
Add 450 μL of warmed XF assay media supplemented with 10 mM Glucose and 2 mM Pyruvate into each well of XF24 cell culture plates.
Place the plate in a humidified non-CO2 incubator at 37 °C until the Sensor cartridge calibration is complete.
Once the calibration is completed, remove and discard the Utility plate while leaving the instrument’s Sensor cartridge.
Place XF24 cell culture pate on the platform and run the already set up protocol to measure mitochondrial oxygen consumption rates.
Following the assay, carefully discard the assay medium, homogenize the cardiomyocytes by adding 100 μL of Cell Lytic M buffer per well on a rocker at room temperature for 15 minutes.
Measure protein concentrations in cell lysates using Bradford assay (Bio-Rad) according to manufacturer’s instructions to normalize OCR values per μg of protein.
Table 2.
Seahorse XF24 analyzer OCR measurements protocol
| Steps | Measurement loops |
|---|---|
| Sensor cartridge calibration | – |
| Equilibration | – |
| Baseline measurement | 2–3 cycles: Mix 3 min, Wait 2 min, Measure 3 min |
| Inject port A—oligomycin | – |
| Measure | 2–3 cycles: Mix 3 min, Wait 2 min, Measure 3 min |
| Inject port B—FCCP | – |
| Measure | 2–3 cycles: Mix 3 min, Wait 2 min, Measure 3 min |
| Inject port C—rotenone and antimycin A | – |
| Measure | 2–3 cycles: Mix 3 min, Wait 2 min, Measure 3 min |
3.2.4. Mitochondrial Respiratory Parameters Analysis
Once the assay protocol is finished, transfer all the wells’ OCR values for each time point of measurement into a spreadsheet for further calculations and data analysis (Fig. 2).
Subtract four empty wells’ average OCR values from treatment wells’ OCR values at each time point to account for background well readings.
Herein, the described protocol utilizes sequential addition of Oligomycin, FCCP, Rotenone, and Antimycin A to assess mitochondrial respirations in real-time in Seahorse XFe24 Flux Analyzer.
Following the measurements of baseline OCRs (sum of all physiological oxygen consumptions), Oligomycin, an ATP synthase (Complex V) inhibitor, is injected to measure ATP-linked OCRs.
Next, carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP), an uncoupling agent to collapse the proton gradient and uncouples the mitochondrial respiration from oxidative phosphorylation, is injected to measure maximal mitochondrial OCRs.
The last injection delivers a mixture of Rotenone, a Complex I inhibitor, and Antimycin A, a complex III inhibitor. This injection halts cellular mitochondrial respiration and allows to measure of nonmitochondrial respiration driven by processes outside the mitochondria.
Real-time OCR values normalized to μg of protein can be utilized to determine key mitochondrial respiratory parameters related to mitochondrial oxidative phosphorylation, as outlined in Table 3 based on the OCR plots in Fig. 2.
Fig. 2.
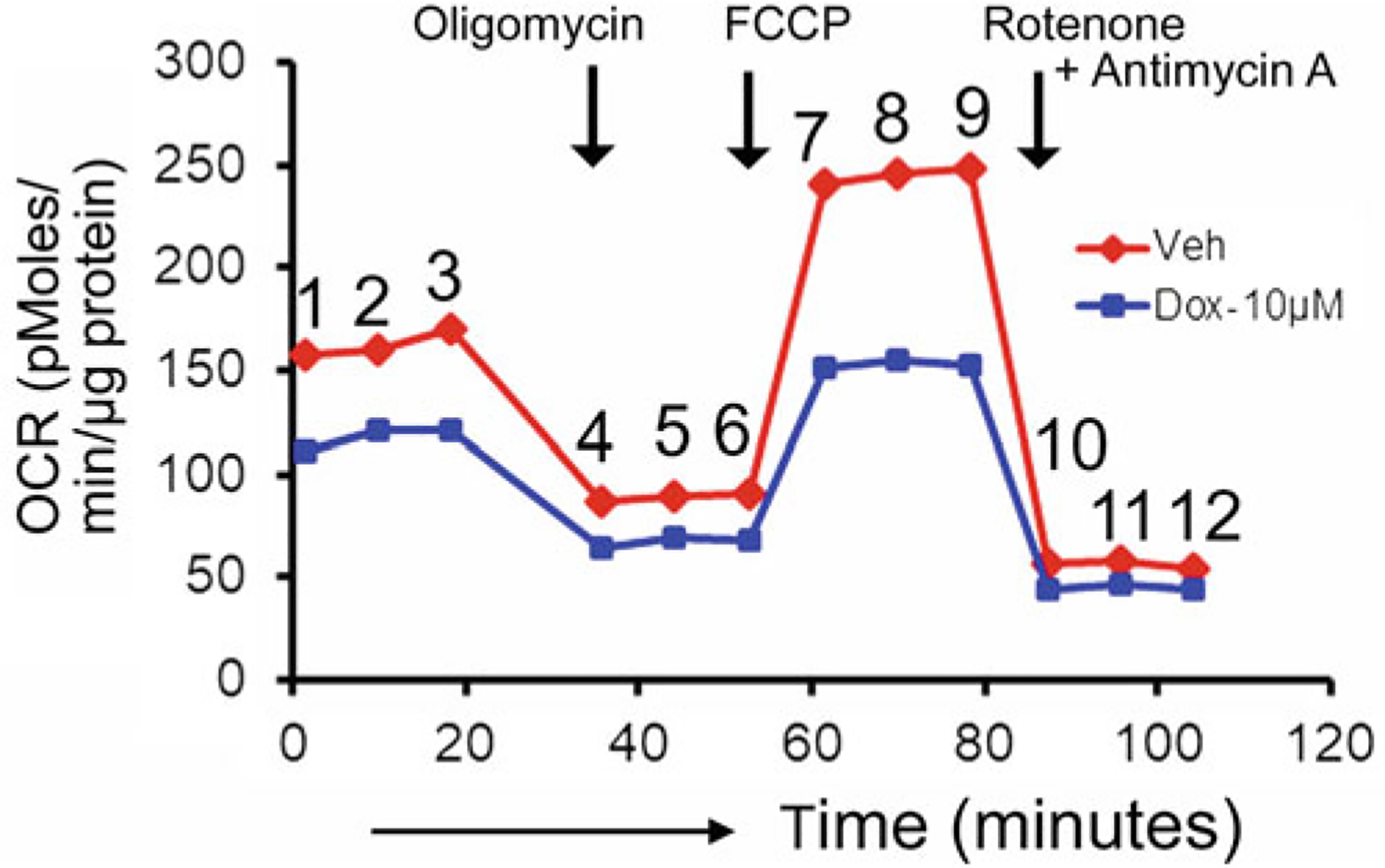
Measurement of mitochondrial respiration in DOX-treated neonatal rat cardiomyocytes. Examples of oxygen consumption rates (OCR) tracing in vehicle and DOX-treated (10 μM, 24 h) cardiomyocytes. Oligomycin (1 μM), FCCP (4 μM), Rotenone (0.5 μM) and Antimycin A (0.5 μM) were sequentially injected at indicated time points (black arrows). OCR values are represented as pmol/min/μg of protein. Numerical values indicate data points where average values from replicate wells at particular time points are analyzed and plotted for OCR profiles in vehicle and DOX-treated cardiomyocyte wells
Table 3.
Key mitochondrial respiratory parameters calculation
| Parameters | Equations |
|---|---|
| ATP Turnover | Basal OCR (1,2,3)—OCR with Oligomycin (4,5,6) |
| Maximal Respiration | OCR with FCCP (7,8,9)—OCR with Rotenone/Antimycin A (10,11,12) |
| Reserve Capacity | OCR with FCCP (7,8,9)—Basal OCR (1,2,3) |
| State Apparent | 4-[{Basal OCR (1,2,3)—Oligomycin OCR (4,5,6)}/{FCCP OCR (7,8,9)—Oligomycin OCR (4,5,6)}] |
| Coupling Efficiency | OCR with Oligomycin (4,5,6)/Basal OCR (1,2,3) |
4. Notes
Incorporate Trypan Blue (T8154, Sigma-Aldrich) during cell counting to count viable cells for subsequent seeding into the Chamber slides and XF24 cell culture plates.
Please coat Nunc™ Lab-Tek™ II Two-well Chamber slides (1 mL per well) and XF24 cell culture plates (500 μL per well) with 0.1% Gelatin (STEMCELL Technologies) in a biological safety cabinet at room temperature overnight before seeding cardiomyocytes into them. This step is essential to improve homogenous cardiomyocytes settling and seeding into the wells.
To improve cardiomyocytes settling and seeding into the wells of two-well chamber slides and XF24 cell culture plates, please follow sequential addition of cell culture media as described (initially in a low volume of cell culture media, then the addition of a remaining volume of cell culture media). The initial culture period in a low volume of cell culture media allows cardiomyocytes to settle rapidly and homogenously in the wells’ bottom.
During aspiration of cell culture media, in all steps, please avoid using a high vacuum suction technique to aspirate cell culture media from the wells as this leads to cell dislodging and inadvertent drying of the cardiomyocytes. Instead, use a micropipette to aspirate the media or buffer and add relevant cell culture media or buffer immediately.
Doxorubicin (DOX) possesses intrinsic fluorescence properties with fluorescence emission maximum (λemission, max) near 600 nm, which are readily detectable with photomultipliers and CCD cameras fluorescence microscopes [36–40]. Hence, we recommend using MitoTracker® Green dye (excitation maximum: 490 nm, emission maximum: 516 nm) over MitoTracker® dyes that emission maximum lies near 600 nm (e.g., MitoTracker® Red CMXRos, λemission, max 599 nm, Catalogue Number: M7512, Invitrogen) (Fig. 1).
We do not recommend storing MitoTracker™ Green labeled cardiomyocyte slides long term as we observed fading of MitoTracker™ Green dye’s fluorescence intensity over time. The stained cardiomyocyte slides should be kept in the dark slide box at 4 °C and imaged within 24–48 hours of staining (Fig. 1).
During loading the XF24 cell culture plates with cell suspension in cell culture media, do not place the pipette tip directly to the bottom of the wells. Place the pipette tip at the side of the wall of wells at a slanted angle and slowly add the cell suspension for loading.
During change and replenishment of cell culture media, do not entirely aspirate the media to prevent cells’ drying. Leave about 50 μL of medium to avoid cells being dried out. Be cautious and gentle to avoid inadvertent cell dislodging and damage.
Seahorse Sensor Cartridge hydration is a required step for its proper functioning. We recommend Seahorse Sensor Cartridge hydration in a humidified non-CO2 incubator at 37 °C overnight before using in Seahorse XF Test assays.
Mitochondrial electron transport complex inhibitors are required to be loaded into the ports prior to calibrating the Sensor Cartridge.
Acknowledgment
This work was supported by the National Institutes of Health grants: HL122354 and HL145753 to M.S.B; HL141998 and HL141998-01S1 to SM; AA025744, AA026708, and AA025744-02S1 to MP; LSUHSC-S CCDS Finish Line Award, COVID-19 Research Award, LARC Research Award, and Feist-Weiller Cancer Center IDEA Grant to M.S.B.; LSUHSC-S Malcolm Feist Cardiovascular and AHA Postdoctoral Fellowship to C.S.A. (20POST35210789) and LSUHSC-S Malcolm Feist Pre-doctoral Fellowship to R.A. We would also like to thank the COBRE research core facility.
References
- 1.Bonadonna G, Monfardini S, De Lena M, Fossati-Bellani F (1969) Clinical evaluation of adriamycin, a new antitumour antibiotic. Br Med J 3(5669):503–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonadonna G, Monfardini S, De Lena M, Fossati-Bellani F, Beretta G (1970) Phase I and preliminary phase II evaluation of adriamycin (NSC 123127). Cancer Res 30(10): 2572–2582 [PubMed] [Google Scholar]
- 3.Sledge GW, Neuberg D, Bernardo P, Ingle JN, Martino S, Rowinsky EK, Wood WC (2003) Phase III trial of doxorubicin, paclitaxel, and the combination of doxorubicin and paclitaxel as front-line chemotherapy for metastatic breast cancer: an intergroup trial (E1193). J Clin Oncol 21(4):588–592 [DOI] [PubMed] [Google Scholar]
- 4.Kremer LC, van der Pal HJ, Offringa M, van Dalen EC, Voute PA (2002) Frequency and risk factors of subclinical cardiotoxicity after anthracycline therapy in children: a systematic review. Ann Oncol 13(6):819–829 [DOI] [PubMed] [Google Scholar]
- 5.Steinherz L, Steinherz P (1991) Delayed cardiac toxicity from anthracycline therapy. Pediatrician 18(1):49–52 [PubMed] [Google Scholar]
- 6.Kajihara H, Yokozaki H, Yamahara M, Kadomoto Y, Tahara E (1986) Anthracycline induced myocardial damage. An analysis of 16 autopsy cases. Pathol Res Pract 181(4): 434–441 [DOI] [PubMed] [Google Scholar]
- 7.Steinherz LJ, Steinherz PG, Tan CT, Heller G, Murphy ML (1991) Cardiac toxicity 4 to 20 years after completing anthracycline therapy. JAMA 266(12):1672–1677 [PubMed] [Google Scholar]
- 8.Swain SM, Whaley FS, Ewer MS (2003) Congestive heart failure in patients treated with doxorubicin: a retrospective analysis of three trials. Cancer 97(11):2869–2879 [DOI] [PubMed] [Google Scholar]
- 9.Dhingra R, Margulets V, Chowdhury SR, Thliveris J, Jassal D, Fernyhough P, Dorn GW II, Kirshenbaum LA (2014) Bnip3 mediates doxorubicin-induced cardiac myocyte necrosis and mortality through changes in mitochondrial signaling. Proc Natl Acad Sci U S A 111(51):E5537–E5544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoshino A, Mita Y, Okawa Y, Ariyoshi M, Iwai-Kanai E, Ueyama T, Ikeda K, Ogata T, Matoba S (2013) Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat Commun 4:2308. [DOI] [PubMed] [Google Scholar]
- 11.Palmeira CM, Serrano J, Kuehl DW, Wallace KB (1997) Preferential oxidation of cardiac mitochondrial DNA following acute intoxication with doxorubicin. Biochim Biophys Acta 1321(2):101–106 [DOI] [PubMed] [Google Scholar]
- 12.Serrano J, Palmeira CM, Kuehl DW, Wallace KB (1999) Cardioselective and cumulative oxidation of mitochondrial DNA following sub-chronic doxorubicin administration. Biochim Biophys Acta 1411(1):201–205 [DOI] [PubMed] [Google Scholar]
- 13.Davies KJ, Doroshow JH (1986) Redox cycling of anthracyclines by cardiac mitochondria. I. Anthracycline radical formation by NADH dehydrogenase. J Biol Chem 261(7):3060–3067 [PubMed] [Google Scholar]
- 14.Davies KJ, Doroshow JH, Hochstein P (1983) Mitochondrial NADH dehydrogenase-catalyzed oxygen radical production by adriamycin, and the relative inactivity of 5-iminodaunorubicin. FEBS Lett 153(1): 227–230 [DOI] [PubMed] [Google Scholar]
- 15.Doroshow JH, Davies KJ (1986) Redox cycling of anthracyclines by cardiac mitochondria. II. Formation of superoxide anion, hydrogen peroxide, and hydroxyl radical. J Biol Chem 261(7):3068–3074 [PubMed] [Google Scholar]
- 16.Abdullah CS, Alam S, Aishwarya R, Miriyala S, Bhuiyan MAN, Panchatcharam M, Pattillo CB, Orr AW, Sadoshima J, Hill JA, Bhuiyan MS (2019) Doxorubicin-induced cardiomyopathy associated with inhibition of autophagic degradation process and defects in mitochondrial respiration. Sci Rep 9(1):2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berthiaume JM, Wallace KB (2007) Adriamycin-induced oxidative mitochondrial cardiotoxicity. Cell Biol Toxicol 23(1):15–25 [DOI] [PubMed] [Google Scholar]
- 18.Berthiaume JM, Wallace KB (2007) Persistent alterations to the gene expression profile of the heart subsequent to chronic Doxorubicin treatment. Cardiovasc Toxicol 7(3):178–191 [DOI] [PubMed] [Google Scholar]
- 19.Amgalan D, Garner TP, Pekson R, Jia XF, Yanamandala M, Paulino V, Liang FG, Corbalan JJ, Lee J, Chen Y, Karagiannis GS, Sanchez LR, Liang H, Narayanagari SR, Mitchell K, Lopez A, Margulets V, Scarlata M, Santulli G, Asnani A, Peterson RT, Hazan RB, Condeelis JS, Oktay MH, Steidl U, Kirshenbaum LA, Gavathiotis E, Kitsis RN (2020) A small-molecule allosteric inhibitor of BAX protects against doxorubicin-induced cardiomyopathy. Nat Can 1(3):315–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vercesi AE, Castilho RF, Kowaltowski AJ, de Oliveira HCF, de Souza-Pinto NC, Figueira TR, Busanello ENB (2018) Mitochondrial calcium transport and the redox nature of the calcium-induced membrane permeability transition. Free Radic Biol Med 129:1–24 [DOI] [PubMed] [Google Scholar]
- 21.Stanley WC, Recchia FA, Lopaschuk GD (2005) Myocardial substrate metabolism in the normal and failing heart. Physiol Rev 85(3):1093–1129 [DOI] [PubMed] [Google Scholar]
- 22.Kolwicz SC Jr, Purohit S, Tian R (2013) Cardiac metabolism and its interactions with contraction, growth, and survival of cardiomyocytes. Circ Res 113(5):603–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doenst T, Nguyen TD, Abel ED (2013) Cardiac metabolism in heart failure: implications beyond ATP production. Circ Res 113(6): 709–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosca MG, Tandler B, Hoppel CL (2013) Mitochondria in cardiac hypertrophy and heart failure. J Mol Cell Cardiol 55:31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song M, Mihara K, Chen Y, Scorrano L, Dorn GW II (2015) Mitochondrial fission and fusion factors reciprocally orchestrate mitophagic culling in mouse hearts and cultured fibroblasts. Cell Metab 21(2):273–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song M, Franco A, Fleischer JA, Zhang L, Dorn GW II (2017) Abrogating mitochondrial dynamics in mouse hearts accelerates mitochondrial senescence. Cell Metab 26(6): 872–883.e875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wai T, Garcia-Prieto J, Baker MJ, Merkwirth C, Benit P, Rustin P, Ruperez FJ, Barbas C, Ibanez B, Langer T (2015) Imbalanced OPA1 processing and mitochondrial fragmentation cause heart failure in mice. Science 350(6265):aad0116. [DOI] [PubMed] [Google Scholar]
- 28.Abdullah CS, Aishwarya R, Alam S, Morshed M, Remex NS, Nitu S, Kolluru GK, Traylor J, Miriyala S, Panchatcharam M, Hartman B, King J, Bhuiyan MAN, Chandran S, Woolard MD, Yu X, Goeders NE, Dominic P, Arnold CL, Stokes K, Kevil CG, Orr AW, Bhuiyan MS (2020) Methamphetamine induces cardiomyopathy by Sigmar1 inhibition-dependent impairment of mitochondrial dynamics and function. Commun Biol 3(1):682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abdullah CS, Alam S, Aishwarya R, Miriyala S, Panchatcharam M, Bhuiyan MAN, Peretik JM, Orr AW, James J, Osinska H, Robbins J, Lorenz JN, Bhuiyan MS (2018) Cardiac dysfunction in the sigma 1 receptor knockout mouse associated with impaired mitochondrial dynamics and bioenergetics. J Am Heart Assoc 7(20):e009775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alam S, Abdullah CS, Aishwarya R, Miriyala S, Panchatcharam M, Peretik JM, Orr AW, James J, Robbins J, Bhuiyan MS (2018) Aberrant mitochondrial fission is maladaptive in desmin mutation-induced cardiac proteotoxi-city. J Am Heart Assoc 7(14):e009289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alam S, Abdullah CS, Aishwarya R, Morshed M, Nitu SS, Miriyala S, Panchatcharam M, Kevil CG, Orr AW, Bhuiyan MS (2020) Dysfunctional mitochondrial dynamic and oxidative phosphorylation precedes cardiac dysfunction in R120G-alphaB-crystallin-induced desmin-related cardiomyopathy. J Am Heart Assoc 9(23):e017195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutherland FJ, Hearse DJ (2000) The isolated blood and perfusion fluid perfused heart. Pharmacol Res 41(6):613–627 [DOI] [PubMed] [Google Scholar]
- 33.Razzaque MA, Robbins J (2014) Isolation of neonatal and adult rat cardiomyocytes. In: Manual of research techniques in cardiovascular medicine. John Wiley & Sons, Ltd, New York, NY, pp 117–124 [Google Scholar]
- 34.Parameswaran S, Kumar S, Verma RS, Sharma RK (2013) Cardiomyocyte culture - an update on the in vitro cardiovascular model and future challenges. Can J Physiol Pharmacol 91(12): 985–998 [DOI] [PubMed] [Google Scholar]
- 35.Tachibana S, Chen C, Zhang OR, Schurr SV, Hill C, Li R, Manso AM, Zhang J, Andreyev A, Murphy AN, Ross RS, Cho Y (2019) Analyzing oxygen consumption rate in primary cultured mouse neonatal cardiomyocytes using an extracellular flux analyzer. J Vis Exp (144):59052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdullah CS, Ray P, Alam S, Kale N, Aishwarya R, Morshed M, Dutta D, Hudziak C, Banerjee SK, Mallik S, Banerjee S, Bhuiyan MS, Quadir M (2020) Chemical architecture of block copolymers differentially abrogate cardiotoxicity and maintain the anti-cancer efficacy of doxorubicin. Mol Pharm 17(12):4676–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duray PH, Cuono CB, Madri JA (1986) Demonstration of cutaneous doxorubicin extravasation by rhodamine-filtered fluorescence microscopy. J Surg Oncol 31(1):21–25 [DOI] [PubMed] [Google Scholar]
- 38.Karukstis KK, Thompson EH, Whiles JA, Rosenfeld RJ (1998) Deciphering the fluorescence signature of daunomycin and doxorubicin. Biophys Chem 73(3):249–263 [DOI] [PubMed] [Google Scholar]
- 39.Lankelma J, Dekker H, Luque FR, Luykx S, Hoekman K, van der Valk P, van Diest PJ, Pinedo HM (1999) Doxorubicin gradients in human breast cancer. Clin Cancer Res 5(7): 1703–1707 [PubMed] [Google Scholar]
- 40.Shah S, Chandra A, Kaur A, Sabnis N, Lacko A, Gryczynski Z, Fudala R, Gryczynski I (2017) Fluorescence properties of doxorubicin in PBS buffer and PVA films. J Photochem Photobiol B 170:65–69 [DOI] [PMC free article] [PubMed] [Google Scholar]


