Abstract
The persistent increase in the worldwide burden of type 2 diabetes (T2D) and the accompanying rise of its complications, including cardiovascular disease, necessitates our understanding of the metabolic disturbances that cause diabetes. Metabolomics and proteomics, facilitated by recent advances in high-throughput technologies, have given us unprecedented insight into circulating biomarkers of T2D even over a decade prior to overt disease. These markers may be effective tools for diabetes screening, diagnosis, and prognosis. As participants of metabolic pathways, metabolite and protein markers may also highlight pathways involved in T2D development. The integration of metabolomics and proteomics with genomics in “multi-omics” strategies provide an analytical method that can begin to decipher causal associations. These methods are without their limitations, however, but with careful study design and sample handling, these methods represent powerful scientific tools that can be leveraged for the study of T2D. In this paper, we aim to give a timely overview of circulating metabolomics and proteomics findings with type 2 diabetes observed in large human population studies to provide the reader with a snapshot into these emerging fields of research.
Keywords: Metabolomics, Proteomics, Type 2 Diabetes Mellitus, Obesity, Diabetes, Type 2, Metabolism
Challenges in the Care of Individuals with Diabetes
It is projected that in 2040, 642 million adults worldwide will have diabetes—the vast majority of which will be type 2 diabetes (T2D)1. The unprecedented increase in the global burden of this “lifestyle disease” will be accompanied by rising mortality and disability rates, especially among adults during their most productive years. A multinational observational study that included countries in South America, North Africa, South and East Asia, the Middle East, and Russia, estimated ~50% prevalence of microvascular complications (e.g. retinopathy, nephropathy, and neuropathy) and ~30% prevalence of macrovascular complications, including coronary heart disease and peripheral vascular disease among individuals with T2D2. In regards to atherosclerotic cardiovascular disease, T2D confers a 2–4 fold increased risk of cardiovascular events and death3, a similar increase in risk for lower extremity amputations4, and is the leading modifiable risk factor for heart failure5. In turn, cardiovascular disease is estimated to cause two-thirds of deaths in individuals with T2D6. Together, this translates to a 2–3 fold increase in medical expenditures7.
The complications, of diabetes, and associated health care costs, can be avoided to a significant degree by effective prevention and treatment. Changes in diet and increasing physical activity have been found to be more effective than pharmacotherapy (e.g., metformin) to delay and potentially prevent diabetes8. It is difficult, however, to identify individuals at risk since the metabolic dysfunction associated with diabetes development begins decades before increases in blood glucose. Also, not all individuals with elevated blood glucose will progress to diabetes. Population approaches to increase diabetes awareness, physical activity, and healthy food access while decreasing added sugar intake—all considered to be risk factors for diabetes—may be effective. Diabetes incidence in the U.S. consistently rose from 1990 to a peak incidence of 8.2 per 100 adults in 2009, but from 2011 to 2017 has remained stable with a reduction in incidence rate of 35%9, possibly due to these population approaches. However, the casual role of these risk factors have yet to be confirmed. Furthermore, obesity, a traditional risk factor for T2D, continues to rise10 and the prevalence of pre-diabetes remains high11 despite the decline in frank diabetes. These findings suggest there is still much we do not understand about the pathophysiology of T2D development.
There is also evidence of significant heterogeneity of diabetes clinical presentation and course. Leveraging unsupervised clustering analyses of clinical data as well as genetics and outcomes data, Ahllqvist et al. recently identified 5 new diabetes “subgroups” among individuals traditionally considered to have T2D.12 They identified 2 clusters of individuals, labeled as severe autoimmune diabetes (SAID) and severe insulin-deficient diabetes (SIDD), with characteristics more similar to type 1 diabetes including lower BMI, higher rates of diabetic ketoacidosis, and faster progression to insulin therapy despite one cluster having no antibody positivity. Another group that was identified, labeled as having severe insulin-resistant diabetes (SIRD), had higher risk of progression to chronic kidney disease and trended toward an increased risk for coronary events. Improving our ability to identify individuals at the highest risk for developing specific complications will help guide clinical care. Also, with the recent availability of cardiovascular and renal outcomes data for medications previously used only for glycemic control13,14, there is now an unprecedented number of therapeutic options for specific subsets of patients with type 2 diabetes including those with established cardiovascular disease, heart failure, or kidney disease.
A Need for New Diabetes Biomarkers
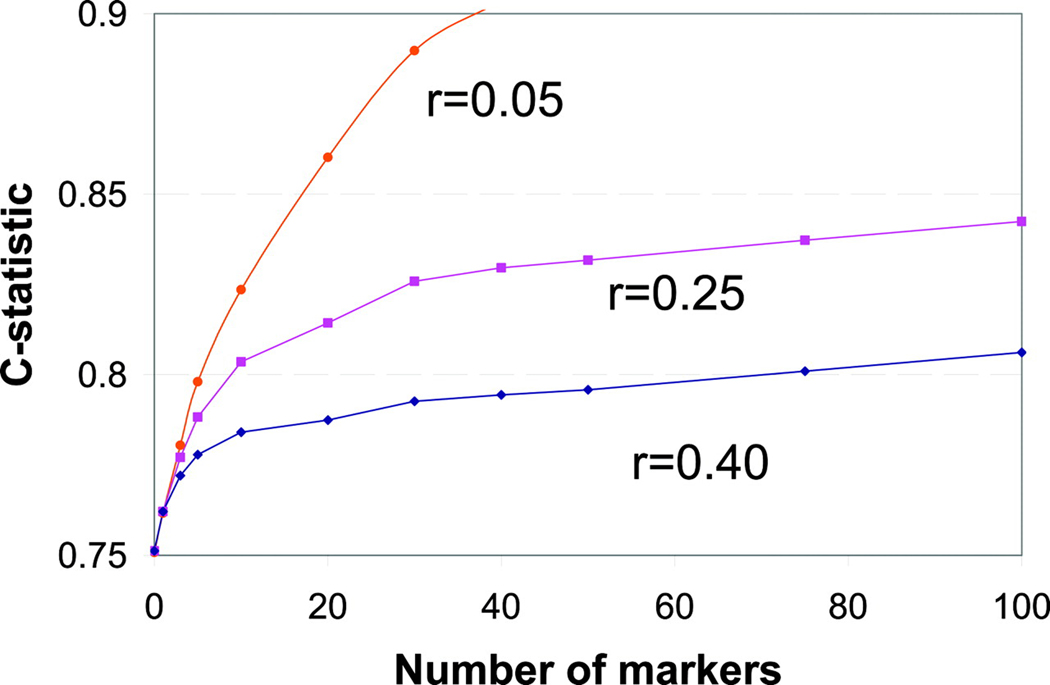
Biomarkers can be effective tools for disease screening, diagnosis, and prognosis. If these biomarkers also participate in disease pathways, which is frequently the case for metabolites and proteins, they can highlight mechanisms of disease development or therapeutic effect. Many current biomarkers for diabetes, however, are strongly correlated with dysglycemia, limiting their predictive and diagnostic value beyond a fasting blood glucose or hemoglobin A1c (HbA1c) level. This point was illustrated by Wang et al. when they simulated the stepwise addition of 100 hypothetical biomarkers to a traditional risk model for cardiovascular disease (Figure 1)15. The degree of correlation between biomarkers included in the clinical model was a key determinant of how much additional information each biomarker provided and how many biomarkers were needed to meaningfully change the predictive power of the risk model16,17. For example, more than 50 biomarkers were needed to improve the c statistic of the model by 0.05 for a set of biomarkers with mean inter-marker correlation of r = 0.4 (i.e., moderately correlated). By contrast, less than 10 biomarkers were needed if their mean inter-marker correlation was r = 0.05 (i.e., weakly correlated)17. High throughput technologies that allow the unbiased quantification of all circulating metabolites and proteins can help facilitate the identification of biomarkers that are from orthogonal pathways and are therefore weakly correlated.
Figure 1.
Incremental improvements in discrimination of hypothetical biomarkers based on a simulation of the predicted hazards ratio per 1 SD increase in a variable number of biomarkers with different marker-marker correlation (r). This figure was generated by Thomas Wang M.D., and Michael Pencina Ph.D. (Wang, Circulation. 2011.)
Metabolites and Proteins are the Product of Genetic, Physiologic, and Environmental Stimuli
The aim of metabolomics is to measure metabolite concentrations in cells, tissues, organs, and biological systems to study the chemical processes involved in metabolism in a systematic fashion. Similarly, proteomics aims to quantify and characterize all proteins that participate in the biological processes of an organism. These should not be considered mutually exclusive fields of study, but rather a continuum of biochemical profiling that focuses—when included with peptidomics—on characterizing the molecular products of genetic transcription. These products could be a single compound (i.e., an amino acid such as aspartic acid), a structure composed of several constituents (i.e., a dipeptide such as aspartame), or a protein that is made up of multiple peptides (i.e., an enzyme such as aspartate transaminase); but due to differences in molecular sizes, require different technologies to identify and quantify. Metabolites and proteins are of particular interest because they are influenced by physiologic changes and environmental stimuli as well as genomic inputs. T2D is specifically suited for metabolomics and proteomics methods since it is generally a polygenic metabolic disease influenced by physiologic changes such as diet and physical exercise. Recent technological advances have allowed scientists to profile circulating metabolites and proteins rapidly and on an increasingly larger scale, several which are briefly highlighted in Table 1. This has facilitated the mapping of the complete human metabolome and proteome, analogous to how genomic advances have allowed the mapping of the human genome.
Table 1.
Comparison of metabolomics and proteomics technologies for circulating biomarker discovery
| Metabolomics | Proteomics | |
|---|---|---|
| Analytes Measured | Circulating small molecules | Circulating proteins |
| Technologies | 1. NMR 2. MS a. GC-MS b. LC-MS |
1. MS 2. Multiplex-nucleic acid affinity reagents (aptamers) 3. PEA with nucleotide-labeled antibodies |
| Strengths | 1. Technologies are high-throughput 2. High sensitivity and specificity 3. NMR is sample non-destructive 4. Small sample amount needed 5. Unbiased measurement of unknown compounds are possible |
1. Most technologies are high-throughput 2. Relatively good sensitivity and specificity 3. Small sample amount needed 4. Proteins are direct products of transcription, facilitating integration with genomic data |
| Limitations | 1. Identification of unknown compounds is labor intensive 2. Metabolite pathway analysis is complex |
1. High-throughput methods may have slightly lower specificity 2. Limited ability to identify unknown proteins |
NMR: nuclear magnetic resonance, MS: mass spectrometry, GC: gas chromatography, LC: liquid chromatography, PEA: proximity extension assay
A percentage of the inter-individual variability of circulating metabolites, or proteins, can be attributed to genetics. Twin studies were initially used to study the heritability of circulating concentrations of specific blood factors18,19. The heritability of a wide range of metabolite concentrations in large populations can now be estimated with the availability of genome wide association study (GWAS) data. A single nucleotide polymorphism (SNP) can explain up to 16–36% of a metabolite’s variance20,21. Metabolites, and proteins, can also be influenced by multiple genes simultaneously. General heritability estimates in a Finnish study suggested genetics explained 23–55% of the variability in amino acid and small molecule levels. The percentages were higher, 48–76%, for lipids and lipoproteins22. While genetic influences are substantial, there remains a percentage that is not explained by genetics. GWAS in the Framingham Offspring cohort showed less than 20% of inter-individual variation in 34% of 217 measured plasma metabolites was attributable to genetics23. Some of these non-genetic contributors could be known clinical or environmental factors, but some remain unidentified.
Genetics also influence circulating protein concentrations. In a comprehensive GWAS conducted on human plasma proteins in the European INTERVAL study, genetic variants explained more than 20% of variation in only approximately 10% of the proteins measured. A higher percentage of heritability, approximately 90%, was found in the Framingham Offspring cohort but significantly fewer proteins were measured24. The mean heritability, however, was only 49%, suggesting again that there is a percentage of circulating protein variation that is unexplained by genetics in humans that warrants further study.
Biomarker Integration with Genomics Could Uncover Causal Relationships with Disease
Metabolite and protein biomarkers can illuminate disease pathophysiology, however, a limitation of biomarker studies are that causality is difficult to establish. Mechanistic studies in cell lines and model organ systems are usually required that are time and resource intensive. With the increasing availability of genomic data, however, advances in statistical methods allows the determination of if biomarkers are possibly causal to a disease and allows for the prioritization of specific biomarkers for further mechanistic studies.
Instrumental variable analysis exploits Mendelian randomization, or the random inheritance of genes, to make causal inferences about genetic variants and intermediate phenotypes with an outcome of interest. For example, randomized control trials have shown that the inhibition of proprotein convertase subtilisin-kexin type 9 (PCSK-9), a protein involved in low density lipoprotein (LDL) receptor degradation, successfully reduces LDL levels and likely reduces the risk for major cardiovascular events25,26. Ference et al. demonstrated with Mendelian randomization in a cohort of over 110,000 individuals that loss of function genetic variants of PCSK-9 were associated with lower clinical LDL cholesterol levels (e.g., the intermediate phenotype)27. These same variants were also associated with lower risk of cardiovascular events (e.g., the outcome of interest). This suggested that reductions in cardiovascular risk associated with these specific PCSK-9 variants were mediated by their effect on LDL levels. Interestingly, this study also found these same genetic variants, associated with the same LDL lowering effect, were also associated with increased risk for T2D in individuals with impaired fasting glucose. This is supported by clinical findings that statins, another class of LDL cholesterol lowering medications, are also associated with a small but significant increase in T2D risk. Utilizing this framework, exposure to either high or low levels of a biomarker of interest can now be substituted as the intermediate phenotype to interrogate if the biomarker is in the causal pathway for a disease of interest.
Analytical Methods in Metabolomics
Nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS)—usually coupled to gas chromatography (GC-MS) or liquid chromatography (LC-MS)—are the most common profiling technologies used. An understanding of the strengths and weakness of the different technologies as well as appropriate sample preparation methods are imperative to generate usable data.
Nuclear Magnetic Resonance Spectroscopy (NMR)
NMR has been used for the simultaneous detection of several metabolites in different bodily fluids for decades28,29. It utilizes the predictable behavior of atomic nuclei (e.g. 1H is most commonly measured in metabolomics) when they are exposed to strong magnetic fields based on neighboring atoms and the frequency of the electromagnetic radiation exposure to identify molecular compound structures. The presence of protons in the vast majority of biological compounds makes this identification technique almost universally applicable. NMR is also a nondestructive analytical method that allows for in vivo compound identification. Also, when coupled with imaging techniques such as MRI, in vivo localization of metabolic activity is now possible within an ogransim30. A limitation of NMR, however, is that the peaks generated are often an integration of signals from several different compounds, especially when compounds of similar species are present. Also, MS is better suited for the identification of metabolites that are in higher abundance.
Mass Spectrometry (MS)
MS utilizes chromatographic separation, analyte ionization, and ionic separation by mass to quantify and identify compounds from biological samples with high resolution and sensitivity. Chromatography is not a required step, but this technique reduces ion suppression and improves the quantitative accuracy of MS results especially for lower abundant analytes. It also allows the identification of isomers (compounds that have the same molecular formula and mass but different structural arrangements) which commonly occur in biological metabolites. In chromatography, there is a mobile phase that the analytes are dissolved into that interacts in a predefined manner with a stationary phase that the mobile phase is passed through, allowing for physical separation of the analytes. With GC, analytes are vaporized into a mobile gas phase and passed through a liquid layer in the chromatograph column that serves as the stationary phase. GC has superior resolution to LC and is well suited for nonpolar, low molecular weight, volatile analytes. With LC, analytes are dissolved in a liquid mobile phase that is then passed through a column filled with beads coated with different compounds with predefined chemical properties that serves as the stationary phase. LC is better suited for polar, high-molecular-mass compounds that are heat-labile. There are a variety of LC columns and techniques that facilitate the separation of a large spectrum of metabolites. Hydrophilic interaction chromatography columns (HILIC) contain hydrophilic beads with an organic solvent to water gradient are effective in separating small, polar analytes such as amino acids, nucleotides, and organic acids. Columns that contain hydrophobic beads with a gradient from water to organic solvent are effective in separating polar molecules and lipids.
There are different methods used to ionize analytes with MS, the most common being electrospray ionization (ESI). ESI is typically coupled with LC and utilizes a highly charged needle tip to create charged analyte droplets from a liquid that can be heated and ionized. There are also several different methods to achieve ionic separation in MS using different analyzers including time-of flight (TOF), quadrupole, ion trap mass, and orbitrap. The TOF instruments utilize a fixed electric field to accelerate ions through a voltage drop to impart kinetic energy and then down a flight tube of known length. The time an ion requires to reach the end detector is dependent on its charge and mass and therefore a mass to charge ratio (m/z) can be calculated with high resolution and mass accuracy. A quadrupole mass spectrometer uses oscillating electrical fields to selectively stabilize the flight path of an ion with a specific m/z of interest, serving as a mass filter. A triple quadrupole measure metabolites with high sensitivity, but can only measure a smaller subset of preselected metabolites. An ion trap mass spectrometer traps ions of a specific m/z using an electrical field and calculates the m/z based on the radio frequency required to retain the ions. The ion trap has increased sensitivity but is less accurate with quantification. An orbitrap mass analyzer monitors the frequency of oscillations of an ion once it is trapped around a central spindle-shaped electrode and calculates the m/z based on the ion oscillations around a specific axis. This analyzer provides exceptional resolution and mass accuracy. These different analyzers can also be coupled together in a single method to provide increased mass resolution and accuracy.
Metabolomics in Diabetes
Over the past decade there has been a growing body of literature describing metabolomic profiles associated with type 2 diabetes. Both targeted methods, that measure a defined group of known chemical compounds, and untargeted methods, that measure all chemicals present including those that have never been previously annotated, have been used.
Amino Acids
One of the strongest associations with diabetes and glycemic traits that has emerged from metabolomics studies is the positive association of branch chain amino acids (BCAAs)—e.g., leucine, isoleucine, and valine. Evidence relating amino acid metabolism with insulin resistance and obesity in humans was initially described decades ago31. Newgard et al. confirmed these associations in a cross-sectional metabolomics analysis of obese and lean individuals. BCAAs in particular were higher in individuals that were obese and, furthermore, in those that had higher insulin resistance (defined by the homeostatic model assessment of insulin resistance or HOMA-IR) even after adjustments for adiposity32. Wang et al. then demonstrated in a prospective analysis in the Framingham Heart Study (FHS) that individuals with BCAA concentrations in the highest quarter had a 2–3.5 fold higher odds of developing type 2 diabetes up to 12 years later compared to those in the lowest quarter33. These associations remained after adjustments for clinical risk factors including body mass index (BMI) and have subsequently been replicated in multiple other cohorts34–39. Growing experimental evidence have posited potential mechanisms40,41 including increased BCAAs, either due to increased dietary contribution and/or defective metabolism, activating mammalian target of rapamycin (mTOR) kinase activity that uncouples insulin signaling32 or leading to a buildup of cytoxic metabolites that could adversely affect the pancreatic islet β-cells42–44 or adipocytes45.
It is still unclear, however, if BCAAs have a causal relationship with diabetes or if these associations are due to reverse causality or confounders such as obesity. Work integrating these findings with human genetics has been completed to begin to clarify these issues. Lotta et al. completed a GWAS of BCAA levels in 16,596 individuals that identified several top SNPs, some of which also imparted an increased risk for diabetes among 47,877 cases and 267,694 controls across several European cohorts46. Analyses in a smaller cohort utilizing Mendelian Randomization suggested a causal association of insulin resistance with BCAA levels47, but not the reveres, which was further supported by a second analysis that included more SNPs and individuals48. Taken together, these findings point to elevated BCAAs as a downstream effect of adiposity and insulin resistance but that temporally precedes the development of clinical diabetes, suggesting they may be mediators to some degree in disease development.
Several studies have also found positive associations of aromatic amino acids (AAAs), including tyrosine and phenylalanine, with future development of diabetes34–37,48–50. AAAs in solution with BCAA also alter cellular insulin signaling through the mTOR pathway51 and they compete with BCAA for the same intracellular transporter52 but further data on AAA causing diabetes or insulin resistance is limited. Glutamate and glutamine, two amino acids central to both nitrogen and carbon cycling and linked with BCAA metabolism, have also been associated with the development of diabetes in several cohorts.
Glutamate, synthesized from the citric acid cycle product α-ketogluteric acid and an intermediate in the generation of the antioxidant glutathione53, has been consistently found to be positively associated with diabetes39,54. Glutamine, a transamination product of glutamate55, has been found to be inversely associated with the development of diabetes as well as the ratio of glutamine/glutamate35,37,49,54,56,57. Glycine58, an amino acid synthesized from serine, has also been consistently found to be inversely associated with development of T2D34–37,57 and impaired glucose tolerance34. The roles these metabolites could potentially have in diabetes development have yet to be clarified; however, each have central roles in several cellular metabolism pathways. An interesting proposed common pathway could be through their interaction with NMDA glutamate receptors that may regulate insulin secretion in the β-cell59. Serine, which has also been found to have an inverse association with incident diabetes35,36,57, could also participate in this pathway.
Higher levels of 2-aminoadipic acid (2-AAA), a lysine degradation product, was also found to be associated with increased risk for incident diabetes in FHS and MDC60. Individuals with concentrations in the highest quarter had more than a 4-fold increased odds of developing diabetes over 12 years. These results mirrored findings of increased concentrations of 2-AAA found in obese mice, hyperinsulinemic mice fed a high fat diet, and diabetic rats. Also, augmented insulin secretion was demonstrated in both murine and human islet cells that were acutely or chronically exposed to 2-AAA. While it remains unclear if 2-AAA levels rise prior to the development of insulin resistance, it may have a role in compensatory mechanisms afterward.
Organic Acids
Alpha-hydroxybutyrate, a product of amino acid catabolism that is derived from α-ketobutyrate, a participant in the glutathione production pathway, has been positively associated with incident diabetes39,57. In a group of individuals free of diabetes, α-hydroxybutyrate levels have also been found to be inversely associated with insulin sensitivity61. Acetoacetate, a ketone body synthesized from fatty acids as an energy source when glucose is low, has also been positively associated with the risk for diabetes in a group of Finnish men62 and in a smaller Bavarian study63. Alpha-keto acids, specifically branched chain α-keto acids, are of interest because they are formed in the first irreversible step of BCAA catabolism64.
Bile Acids
In a European study that utilized a non-targeted approach, three bile acids—e.g., deoxycholic, glycocholic, and glycodeoxycholic acid—were positively associated with incident disease in age- and sex-adjusted models50. Only the association of deoxycholic acid remained significant after additional adjustments for clinical risk factors50. This study also identified a SNP in the CYP7A1 coding region associated with deoxycholic acid levels that was also associated with type 2 diabetes in published GWAS meta-analyses. While the genetic findings were not a formal Mendelian randomization analysis and thus cannot prove causality, these findings support emerging experimental evidence of both receptor-mediated and non-receptor-mediated mechanisms (that involve incretin stimulation) for circulating bile acids to effect glycemia65.
Carbohydrates
Hexose sugars—typically measured as a composite of multiple different isomers of 6 carbon monosaccharides including glucose and fructose—are the most frequently analyzed carbohydrate in metabolomics studies of incident diabetes35,39,66,67. These composite measures consistently have a positive association with disease even after adjustments for clinical measures of glucose. This reflects the high degree of sensitivity of the analytical technologies to detect the hexose sugars present in the samples that are not measured by clinical glucose assays. Circulating levels of trehalose—a non-endogenous sugar obtained from the diet in humans—has also been positively associated with diabetes39,66. Using untargeted methods, a species of mannitol and several deoxy-hexose sugars were found to be inversely associated with diabetes risk in a nested case-control study of the EPIC-Potsdam Germany cohort68.
Lipids and Acylcarnitines
Lipids are an integral part of cellular energy homeostasis, serving in multiple roles including as metabolic substrates, signaling hormones, or cellular membrane building blocks. Elevated clinical measures of lipids, specifically of bulk triglycerides, is considered a tradition risk factor for T2D. In the 1960s, Randle et al. described the competitive relationship of fatty acids and glucose for oxidative cycling and proposed that excessive fatty acid oxidation contributed to impaired glucose homeostasis and insulin resistance69. The intracellular accumulation of fatty acid oxidation products such as diacylglycerols (DAGs), triacylglycerols (TAGs), and ceramides have also been linked with insulin resistance70. Extensive experimental work is being conducted to understand the mechanisms causing these associations and whether these oxidation products—as well as other lipid species—are causative or consequences of insulin resistance67.
Traditional clinical measurements of lipids often lacked specificity, but with GC and LC-MS techniques, unique lipid species can now be identified by total acyl chain carbon number and double bond content. In FHS, individuals with higher levels of TAGs with shorter acyl carbon chains and fewer double bonds were at increased risk for the development of diabetes even after adjustments for clinical risk factors71. A similar trend was also observed for cholesterol esters (CEs) and specific phospholipids including lysophosphatidylcholines (LPCs), phosphatidylcholines (PCs), and lysophosphatidylethanolamines (LPEs). A study in Finnish men found a positive association of the ratio of monounsaturated fatty acids (FAs) to total FAs with increased risk for future T2D. The ratio of saturated and n-7 and n-9 FAs to total FAs along with glycerol, total FAs, and total TAGs levels were also found to be positively associated with T2D risk while the ratios of docosahexaenoic acid (DHA), an omega-3 FA, and linoleic acid, a n-6 FA, with total FAs were inversely associated72. Lipoprotein lipid subclasses can also be profiled with NMR techniques, and in the Metabolic Syndrome in Men (METSIM) study of Finnish men, the ratio of apolipoprotein A1 to HLD was the strongest predictor of future T2D risk73. In a more recent study of young Fins, lipid subfractions of lipoproteins were measured and higher cholesterol concentrations in very large LDL particles was positively associated with T2D risk while higher concentrations in very large and large HDL particles—especially of non-esterified cholesterols—were inversely associated74. Higher relative TAG content in all lipoprotein subclasses was positively associated with T2D risk.
Specific phospholipid species have also been studied. Wang-Sattler et al. found an inverse association of C18:2 (denoting acyl chain carbon length:number of double bonds) LPC, along with glycine, with future development of both impaired glucose tolerance and incident T2D in the Cooperative Health Research in the Region of Augsburg (KORA) cohort and in the European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam cohort34. In a separate analysis that used an EPIC-Potsdam subcohort for discovery and KORA for validation, specific diacyl-phosphatidylcholines (C32:1, C36:1, C38:3, and C40:5) were positively associated with T2D risk. They also confirmed the inverse association of C18:2 LPC and found C16:1 sphingomyelin, and specific acyl-alkyl-phosphatidylcholines or plasmalogen PCs (C34:3, C40:6, C42:5, C44:4, and C44:5) were inversely associated with T2D risk35. Further untargeted work in the EPIC-Potsdam cohort has confirmed the inverse association of specific LPCs and PCs with T2D68, while work in a small American Indian cohort has also confirmed the findings in PCs, albeit with different subspecies75. Phospholipid linoleoyl-glycerophosphocholine (L-GPC)57 has also been inversely associated with future diabetes.
The transport of fatty acids into the mitochondria for cellular β-oxidation is facilitated by the formation of acylcarnitines, especially medium-length acylcarnitines. BCAA catabolism also leads to the production of specific acylcarnitine species. This class has been studied in several metabolomics cohorts given their position at the intersection of BCAA and fatty acid metabolism. Elevated C3 and C5 acylcarnitines, products of BCAA catabolism, were found by Newgard et al. to be strongly associated with insulin resistance32. C2 acetylcarnitine was also found to be positively associated with incident diabetes in KORA34.
Proteomics
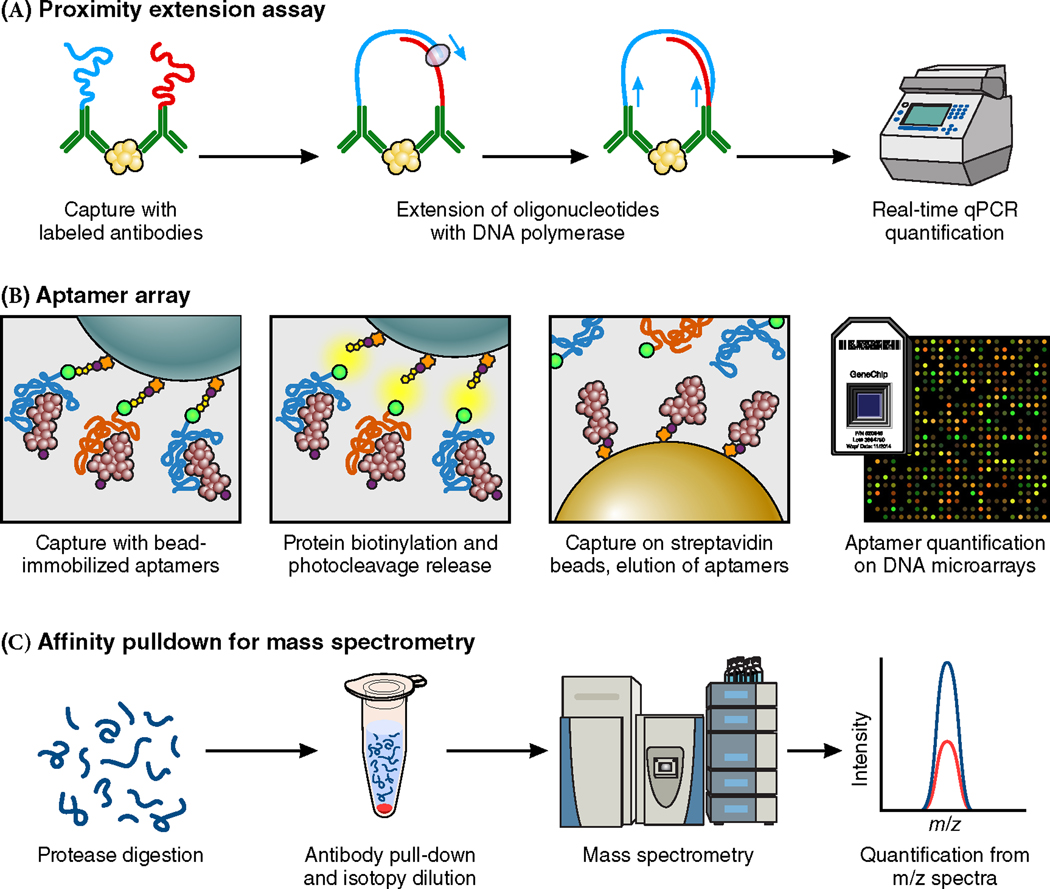
The study of proteins, as the final product of genetic transcription and post-transcriptional modifications, has also played a pivotal role in the understanding of disease. Mass spectrometry and immuno-assays of single proteins were utilized to identify and uncover the association of circulating levels of adiponectin76, leptin77, sex-hormone binding globulin78, and the vitamin E binding, afamin79, with type 2 diabetes risk. Recent developments, however, in protein profiling techniques have increased the efficiency and numbers of circulating proteins that can now be measured. While MS remains a powerful tool for the detection and quantification of proteins, the process is labor and time-intensive and this technique remains limited in the number of proteins it can measure simultaneously. The development of affinity-based methods utilizing multiplexing antibodies and/or novel affinity reagents has drastically expanded the number of proteins that can be quantified. Two high-throughput technologies now commonly used include the use of nucleic acid affinity reagents (aptamers) or nucleotide-labeled antibodies. With aptamers, the diverse structural confirmations that can be achieved with oligonucleotides are utilized to bind to target protein epitopes to facilitate protein detection and quantification. Nucleic acid labeling of antibodies has also allowed the use of polymerase chain reaction (PCR) technology to amplify, detect, and quantify proteins. The natural tendency of complimentary DNA oligonucleotide sequences to anneal and serve as PCR templates has been leveraged in proximity extension assays (PEAs) to improve the specificity of antibody mediated protein identification, especially in high-throughput methods80,81. Binding specificity of either the aptamer or nucleic acid labeled antibodies remains one of the greatest limitations of these techniques. Confirmation with traditional immunoassays, MS, and integrative genomics, however, can help confirm the specificity of protein identification82,83. Figure 2 depicts the workflow for these methods.
Figure 2.
Workflow for different high-throughput proteomics technologies.
DNA: deoxyribonucleic acid. m/z: mass to charge ratio. qPCR: quantitative polymerase chain reaction. Adapted from a figure by J. Gustav Smith, M.D., Ph.D., and Robert Gerszten, M.D. (Smith and Gerszten. Circulation. 2017).
A Swedish study utilizing nucleic acid labeled antibodies and PEA identified 7 circulating proteins associated with HOMA-IR—including the novel association of cathepsin D as well as previously reported proteins leptin, renin, interleukin-1 receptor antagonist (IL-1ra), hepatocyte growth factor, fatty acid-binding protein 4 (FABP4), and tissue plasminogen activator (t-PA). Of these, IL-1ra and t-PA were also positively associated with incident diabetes, however, these associations were completely attenuated after adjustments for fasting glucose84. Mendelian randomization analyses also suggested insulin resistance had a casual effect on t-PA antigen levels. In a more recent, and larger, cross-sectional Swedish study, 29 proteins were found to be associated with prevalent diabetes at a false discovery rate < 5%. Of these, 14 were reportedly novel associations85. However, none of these were found to be causally associated with diabetes in Mendelian randomization analyses. A recent example of MS proteomics analysis, paired with 2-dimentional gel electrophoresis, was conducted in a small cross-sectional cohort of normoglycemic lean, normoglycemic abdominally obese, prediabetic, and diabetic Koreans showed higher levels of serpin peptidase inhibitor A1 (AAT/SERPINA1), haptoglobin protein (HP), zinc-alpha2-glycoprotein (ZAG), apolipoprotein A-1 (APOA1), and retinol binding protein 4 (RBP4) and lower levels of growth-inhibiting protein 25 (GIG25/AACT/SERPINA3), albumin (ALB), and transthyretin (TTR) in those with abdominal adiposity or insulin resistance compared to normal individuals86.
Associations with Cardiovascular Disease
Metabolomics and proteomics have also been leveraged to study biomarkers of cardiovascular disease. These findings have revealed some potential common metabolic pathways involved in insulin resistance, T2D, and cardiovascular disease. To list a few, BCAA and BCAA related metabolites have been positively associated with coronary artery disease (CAD)87–89. Levels of glutamate/glutamine and several acylcarnitines were also shown to differentiate between individual with CAD and controls even after adjustments for traditional clinical factors including BMI and diabetes87. For lipids, specific LPC and sphingomyelin species have been associated with incident CAD90 while higher levels of LPC and LPC plasmalogens containing unsaturated fatty acids—as well as PCs containing DAGs, sphingomyelins, and ceramides—and decreased levels of LPC and LPC plasmalogens containing saturated fatty acids—were associated with increased prevalent CAD91. For proteins, in a study conducted in Americans from the San Francisco Bay Area with stable coronary heart disease, 200 proteins were associated with cardiovascular events including several families also associated with T2D (e.g., interleukins, cathepsins, and fatty acid-binding proteins). The protease SERPINA3 was one of nine proteins included in a clinical risk prediction model that improved on previously established clinical risk factors92. These findings could begin to highlight important metabolic pathways that may be used to untangle the mechanism for T2D associated cardiovascular disease.
Future Directions
Metabolomic and proteomic studies provide a wealth of information, especially when combined with genomic, transcriptomic, epigenomic, and microbiome information. A current challenge, however, is how to organize this data into meaningful information and successfully prioritize relevant associations for further scientific discovery. A common approach, which has been described extensively above, is to leverage genomic data to identify biomarkers that are disease causal46,47,85. Pathway analysis is also an emerging statistical methodology used to cluster disparate biomarkers together into hypothetical pathways. Further work including statistical innovation, however, is needed to overcome these hurdles.
The vast majority of metabolomic and proteomic analyses have also been focused on known metabolites or proteins included on high-throughput platforms due to a priori knowledge of their associations with specific biological pathways. Still, there are hundreds to potentially thousands of circulating low abundance biomarkers that have yet to be described. Untargeted metabolomics can be utilized to study these molecules in an unbiased fashion. Dimethylguanidino valeric acid (DMGV), a novel circulating biomarker of nonalcoholic fatty liver disease also associated with the future development of T2D93 and reduced cardiometabolic fitness94, was identified through the integration of untargeted MS data with genomic and phenotypic data in large human cohorts. The availability of a growing amount of genomic data will be instrumental in facilitating the identification of these unknown compounds.
The majority of studies described thus far in this review have been population studies focused on assessing biomarker associations with T2D and glycemic traits. These technologies can also be leveraged to study treatment effect. Individual responses to treatment and prevention of T2D can be markedly different8,95. Genetic variants in enzymes involved in drug metabolism (e.g. SLC22A1 genetic variant effects on metformin pharmacokinetics96) as well as differences in clinical factors8,95 can explain some of these differences. In the Diabetes Prevention Program, metabolomics has demonstrated that the baseline concentration of specific metabolites are associated with difference in lifestyle modification verses metformin effects on the prevention of T2D97. Changes in metabolites including betaine, due to changes in physical activity and diet98, and arginine and arginine metabolites, due to changes in diet alone99, have also been associated with decreased risk for development of T2D with these interventions. For proteomics, Williams et al. utilized the previously generated nine protein risk score for cardiovascular events in individuals with stable coronary heart disease92 to demonstrate they could predict that Torcetrapib, a novel cholesterol medicine found in clinical trials to be associated with increased cardiovascular events, would be associated with adverse cardiovascular effects within 3 months of clinical trial initiation compared to the median 550 days of follow up that occurred before trial termination100. These findings, taken in total, suggest that metabolomics and proteomic biomarkers could have a role in therapeutic prognosis as well as elucidate therapeutic mechanisms.
Conclusion
In conclusion, metabolomics and proteomics are powerful technologies that can be leveraged to study biomarkers of T2D. The integration of data from these platforms with genomics and other omics information could help elucidate pathways of disease development as well as therapeutic response. Overlapping findings with cardiovascular disease also highlight common pathways that may explain T2D associated cardiovascular morbidity and mortality. Significant care, however, must be put into technology choice, study design, sample preparation, and data analysis to obtain informative results. In the future, untargeted methods can drastically expand the pool of circulating biomarkers that can be studied while use of these technologies in therapeutic trials could also identify markers of individual response to therapies. In this way, these technologies can further the clinical treatment as well as scientific understanding of T2D.
Sources of funding
This work was supported by grant funding from the National Heart, Lung and Blood Institute including T32 HL007374 for Z.-Z.C. and R01HL132320, R01HL133870, and R01HL144483 - 01A1 for R.E.G who was also supported by R01DK081572, R01DK108159, and U24DK112340 from the National Institute of Diabetes and Digestive and Kidney Disease.
Nonstandard Abbreviations and Acronyms:
- T2D
type 2 diabetes
- ASCVD
atherosclerotic cardiovascular disease
- HbA1c
hemoglobin a1C
- NMR
nuclear magnetic resonance
- MS
mass spectrometry
- GC
gas chromatography
- LC
liquid chromatography
- PEA
proximit extension assay
- ESI
Electrospray ionization
- TOF
time-of flight
- VLDL
very low density liopoprotein cholesterol
- IDL
intermediate density lipoprotein cholesterol
- HDL
high density lipoprotein cholesterol
- BCAA
branch chain amino acids
- mTOR
mammalian target of rapamycin
- GWAS
genome wide association study
- SNP
single nucleotide polymorphism
- AAA
aromatic amino acid
- 2-AAA
2-aminoadipic acid
- DAG
diacylglycerol
- TAG
triacylglycerol
- LPC
lysophosphatidylcholines
- PC
phosphatidylcholine
- LPE
lysophosphatidylethanolamine
- FA
fatty acid
- DHA
docosahexaenoic acid
- L-GPC
linoleoyl-glycerophosphocholine
- p,p’-DDE
or trans-nonachlo, or ordichlorodiphenyltrichloroethane
- IL-1ra
interleukin-1 receptor antagonist
- FADBP4
fatty acid-binding protein 4
- t-PA
tissue plaminogen activator
- AAT/SERPINA1
serpin peptidase inhibitor A1
- HP
haptoglobin protein
- ZAG
zinc-alpha2-glycoprotein (ZAG)
- APOA1
apolipoprotein A-1
- RBP4
retinol binding protein 4
- GIG25/AACT/SERPINA3
growth-inhibiting protein 25
- ALB
albumin
- TTR
transthyretin
- CAD
coronary artery disease
- PCSK-9
proprotein convertase subtilisin-kexin type 9
- DMGV
dimethylguanidino valeric acid
- NAFLD
nonalcoholic fatty liver disease
Footnotes
Disclosures.
The authors have no conflict of interests to report.
References
- 1.Zheng Y, Ley SH, Hu FB. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat Rev Endocrinol. 2018. Feb;14(2):88–98. [DOI] [PubMed] [Google Scholar]
- 2.Litwak L, Goh S-Y, Hussein Z, Malek R, Prusty V, Khamseh ME. Prevalence of diabetes complications in people with type 2 diabetes mellitus and its association with baseline characteristics in the multinational A1chieve study. Diabetol Metab Syndr [Internet]. 2013. Dec [cited 2019 Dec 2];5(1). Available from: https://dmsjournal.biomedcentral.com/articles/10.1186/1758-5996-5-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rawshani A, Rawshani A, Franzén S, Eliasson B, Svensson A-M, Miftaraj M, McGuire DK, Sattar N, Rosengren A, Gudbjörnsdottir S. Mortality and Cardiovascular Disease in Type 1 and Type 2 Diabetes. N Engl J Med. 2017. Apr 13;376(15):1407–1418. PMID: 28402770 [DOI] [PubMed] [Google Scholar]
- 4.Vamos EP, Bottle A, Edmonds ME, Valabhji J, Majeed A, Millett C. Changes in the Incidence of Lower Extremity Amputations in Individuals With and Without Diabetes in England Between 2004 and 2008. Diabetes Care. 2010. Dec;33(12):2592–2597. PMCID: PMC2992196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Packer M Heart Failure: The Most Important, Preventable, and Treatable Cardiovascular Complication of Type 2 Diabetes. Diabetes Care. 2018. Jan 1;41(1):11–13. PMID: 29263193 [DOI] [PubMed] [Google Scholar]
- 6.Low Wang Cecilia C., Hess Connie N., Hiatt William R., Goldfine Allison B. Clinical Update: Cardiovascular Disease in Diabetes Mellitus. Circulation. 2016. Jun 14;133(24):2459–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang P, Zhang X, Brown J, Vistisen D, Sicree R, Shaw J, Nichols G. Global healthcare expenditure on diabetes for 2010 and 2030. Diabetes Res Clin Pract. 2010. Mar;87(3):293–301. [DOI] [PubMed] [Google Scholar]
- 8.The Diabetes Prevention Program. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med. 2002. Feb 7;346(6):393–403. PMCID: PMC1370926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benoit SR, Hora I, Albright AL, Gregg EW. New directions in incidence and prevalence of diagnosed diabetes in the USA. BMJ Open Diabetes Res Care [Internet]. 2019. May 1 [cited 2019 Dec 2];7(1). Available from: https://drc.bmj.com/content/7/1/e000657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adult Obesity Facts | Overweight & Obesity | CDC [Internet]. 2019. [cited 2019 Dec 2]. Available from: https://www.cdc.gov/obesity/data/adult.html
- 11.National Diabetes Statistics Report | Data & Statistics | Diabetes | CDC [Internet]. 2018. [cited 2018 Dec 15]. Available from: https://www.cdc.gov/diabetes/data/statistics/statistics-report.html
- 12.Ahlqvist E, Storm P, Käräjämäki A, Martinell M, Dorkhan M, Carlsson A, Vikman P, Prasad RB, Aly DM, Almgren P, Wessman Y, Shaat N, Spégel P, Mulder H, Lindholm E, Melander O, Hansson O, Malmqvist U, Lernmark Å, Lahti K, Forsén T, Tuomi T, Rosengren AH, Groop L. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol. 2018. May 1;6(5):361–369. PMID: 29503172 [DOI] [PubMed] [Google Scholar]
- 13.Marso SP, Daniels GH, Brown-Frandsen K, Kristensen P, Mann JFE, Nauck MA, Nissen SE, Pocock S, Poulter NR, Ravn LS, Steinberg WM, Stockner M, Zinman B, Bergenstal RM, Buse JB. Liraglutide and Cardiovascular Outcomes in Type 2 Diabetes. N Engl J Med. 2016. Jul 28;375(4):311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neal B, Perkovic V, Mahaffey KW, de Zeeuw D, Fulcher G, Erondu N, Shaw W, Law G, Desai M, Matthews DR. Canagliflozin and Cardiovascular and Renal Events in Type 2 Diabetes. N Engl J Med. 2017. Aug 17;377(7):644–657. [DOI] [PubMed] [Google Scholar]
- 15.Wang TJ, Gona P, Larson MG, Tofler GH, Levy D, Newton-Cheh C, Jacques PF, Rifai N, Selhub J, Robins SJ, Benjamin EJ, D’Agostino RB, Vasan RS. Multiple biomarkers for the prediction of first major cardiovascular events and death. N Engl J Med. 2006. Dec 21;355(25):2631–2639. PMID: 17182988 [DOI] [PubMed] [Google Scholar]
- 16.Wang TJ. Assessing the Role of Circulating, Genetic, and Imaging Biomarkers in Cardiovascular Risk Prediction. Circulation. 2011. Feb 8;123(5):551–565. PMCID: PMC3059807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts LD, Gerszten RE. Toward New Biomarkers of Cardiometabolic Diseases. Cell Metab. 2013. Jul 2;18(1):43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osborne RH, Adlersberg D, Degeorge FV, Wang C. Serum lipids, heredity and environment: A study of adult twins. Am J Med. 1959. Jan 1;26(1):54–59. [DOI] [PubMed] [Google Scholar]
- 19.Christian JC, Feinleib M, Hulley SB, Castelli WP, Fabsitz RR, Garrison RJ, Borhani NO, Rosenman RH, Wagner J. Genetics of Plasma Cholesterol and Triglycerides: A Study of Adult Male Twins*. Acta Genet Medicae Gemellol Twin Res. 1976. Jan;25(1):145–149. [DOI] [PubMed] [Google Scholar]
- 20.Gieger C, Geistlinger L, Altmaier E, Angelis MH de, Kronenberg F, Meitinger T, Mewes H-W, Wichmann H-E, Weinberger KM, Adamski J, Illig T, Suhre K. Genetics Meets Metabolomics: A Genome-Wide Association Study of Metabolite Profiles in Human Serum. PLOS Genet. 2008. Nov 28;4(11):e1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Illig T, Gieger C, Zhai G, Römisch-Margl W, Wang-Sattler R, Prehn C, Altmaier E, Kastenmüller G, Kato BS, Mewes H-W, Meitinger T, de Angelis MH, Kronenberg F, Soranzo N, Wichmann H-E, Spector TD, Adamski J, Suhre K. A genomewide perspective of genetic variation in human metabolism. Nat Genet. 2010. Feb;42(2):137–141. PMCID: PMC3773904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kettunen J, Tukiainen T, Sarin A-P, Ortega-Alonso A, Tikkanen E, Lyytikäinen L-P, Kangas AJ, Soininen P, Würtz P, Silander K, Dick DM, Rose RJ, Savolainen MJ, Viikari J, Kähönen M, Lehtimäki T, Pietiläinen KH, Inouye M, McCarthy MI, Jula A, Eriksson J, Raitakari OT, Salomaa V, Kaprio J, Järvelin M-R, Peltonen L, Perola M, Freimer NB, Ala-Korpela M, Palotie A, Ripatti S. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat Genet. 2012. Mar;44(3):269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhee EP, Ho JE, Chen M-H, Shen D, Cheng S, Larson MG, Ghorbani A, Shi X, Helenius IT, O’Donnell CJ, Souza AL, Deik A, Pierce KA, Bullock K, Walford GA, Vasan RS, Florez JC, Clish C, Yeh J-RJ, Wang TJ, Gerszten RE. A Genome-Wide Association Study of the Human Metabolome in a Community-Based Cohort. Cell Metab. 2013. Jul 2;18(1):130–143. PMCID: PMC3973158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benson MD, Yang Q, Ngo D, Zhu Y, Shen D, Farrell LA, Sinha S, Keyes MJ, Vasan RS, Larson MG, Smith JG, Wang TJ, Gerszten RE. The Genetic Architecture of the Cardiovascular Risk Proteome. Circulation. 2018. Mar 13;137(11):1158–1172. PMCID: PMC5849518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sabatine MS, Giugliano RP, Wiviott SD, Raal FJ, Blom DJ, Robinson J, Ballantyne CM, Somaratne R, Legg J, Wasserman SM, Scott R, Koren MJ, Stein EA. Efficacy and Safety of Evolocumab in Reducing Lipids and Cardiovascular Events. N Engl J Med. 2015. Apr 16;372(16):1500–1509. PMID: 25773607 [DOI] [PubMed] [Google Scholar]
- 26.Robinson JG, Farnier M, Krempf M, Bergeron J, Luc G, Averna M, Stroes ES, Langslet G, Raal FJ, El Shahawy M, Koren MJ, Lepor NE, Lorenzato C, Pordy R, Chaudhari U, Kastelein JJP. Efficacy and Safety of Alirocumab in Reducing Lipids and Cardiovascular Events. N Engl J Med. 2015. Apr 16;372(16):1489–1499. PMID: 25773378 [DOI] [PubMed] [Google Scholar]
- 27.Ference BA, Robinson JG, Brook RD, Catapano AL, Chapman MJ, Neff DR, Voros S, Giugliano RP, Davey Smith G, Fazio S, Sabatine MS. Variation in PCSK9 and HMGCR and Risk of Cardiovascular Disease and Diabetes. N Engl J Med. 2016. Dec 1;375(22):2144–2153. PMID: 27959767 [DOI] [PubMed] [Google Scholar]
- 28.Nicholson JK, Buckingham MJ, Sadler PJ. High resolution 1H n.m.r. studies of vertebrate blood and plasma. Biochem J. 1983. Jun 1;211(3):605–615. PMCID: PMC1154405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bales JR, Higham DP, Howe I, Nicholson JK, Sadler PJ. Use of high-resolution proton nuclear magnetic resonance spectroscopy for rapid multi-component analysis of urine. Clin Chem. 1984. Mar;30(3):426–432. PMID: 6321058 [PubMed] [Google Scholar]
- 30.Salamanca-Cardona L, Shah H, Poot AJ, Correa FM, Di Gialleonardo V, Lui H, Miloushev VZ, Granlund KL, Tee SS, Cross JR, Thompson CB, Keshari KR. In vivo imaging of glutamine metabolism to the oncometabolite 2-hydroxyglutarate in IDH1/2 mutant tumors. Cell Metab. 2017. Dec 5;26(6):830–841.e3. PMCID: PMC5718944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Felig P, Marliss E, Cahill GF. Plasma Amino Acid Levels and Insulin Secretion in Obesity. N Engl J Med. 1969. Oct 9;281(15):811–816. PMID: 5809519 [DOI] [PubMed] [Google Scholar]
- 32.Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, Rochon J, Gallup D, Ilkayeva O, Wenner BR, Yancy WS, Eisenson H, Musante G, Surwit RS, Millington DS, Butler MD, Svetkey LP. A Branched-Chain Amino Acid-Related Metabolic Signature that Differentiates Obese and Lean Humans and Contributes to Insulin Resistance. Cell Metab. 2009. Apr 8;9(4):311–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, O’Donnell CJ, Carr SA, Mootha VK, Florez JC, Souza A, Melander O, Clish CB, Gerszten RE. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011. Apr;17(4):448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang-Sattler R, Yu Z, Herder C, Messias AC, Floegel A, He Y, Heim K, Campillos M, Holzapfel C, Thorand B, Grallert H, Xu T, Bader E, Huth C, Mittelstrass K, Döring A, Meisinger C, Gieger C, Prehn C, Roemisch-Margl W, Carstensen M, Xie L, Yamanaka-Okumura H, Xing G, Ceglarek U, Thiery J, Giani G, Lickert H, Lin X, Li Y, Boeing H, Joost H-G, de Angelis MH, Rathmann W, Suhre K, Prokisch H, Peters A, Meitinger T, Roden M, Wichmann H-E, Pischon T, Adamski J, Illig T. Novel biomarkers for pre-diabetes identified by metabolomics. Mol Syst Biol. 2012. Sep 25;8:615. PMCID: PMC3472689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Floegel A, Stefan N, Yu Z, Mühlenbruch K, Drogan D, Joost H-G, Fritsche A, Häring H-U, Angelis MH de, Peters A, Roden M, Prehn C, Wang-Sattler R, Illig T, Schulze MB, Adamski J, Boeing H, Pischon T. Identification of Serum Metabolites Associated With Risk of Type 2 Diabetes Using a Targeted Metabolomic Approach. Diabetes. 2013. Feb 1;62(2):639–648. PMID: 23043162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmer ND, Stevens RD, Antinozzi PA, Anderson A, Bergman RN, Wagenknecht LE, Newgard CB, Bowden DW. Metabolomic Profile Associated With Insulin Resistance and Conversion to Diabetes in the Insulin Resistance Atherosclerosis Study. J Clin Endocrinol Metab. 2015. Mar 1;100(3):E463–E468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tillin T, Hughes AD, Wang Q, Würtz P, Ala-Korpela M, Sattar N, Forouhi NG, Godsland IF, Eastwood SV, McKeigue PM, Chaturvedi N. Diabetes risk and amino acid profiles: cross-sectional and prospective analyses of ethnicity, amino acids and diabetes in a South Asian and European cohort from the SABRE (Southall And Brent REvisited) Study. Diabetologia. 2015;58(5):968–979. PMCID: PMC4392114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Semiz S, van der Lee SJ, van der Spek A, Verhoeven A, van Klinken JB, Sijbrands E, Harms AC, Hankemeier T, van Dijk KW, van Duijn CM, Demirkan A. Metabolomics based markers predict type 2 diabetes in a 14-year follow-up study. Metabolomics. 2017. Jul 28;13(9):104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peddinti G, Cobb J, Yengo L, Froguel P, Kravić J, Balkau B, Tuomi T, Aittokallio T, Groop L. Early metabolic markers identify potential targets for the prevention of type 2 diabetes. Diabetologia. 2017;60(9):1740–1750. PMCID: PMC5552834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lynch CJ, Adams SH. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol. 2014. Dec;10(12):723–736. PMCID: PMC4424797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoon M-S. The Emerging Role of Branched-Chain Amino Acids in Insulin Resistance and Metabolism. Nutrients [Internet]. 2016. Jul 1 [cited 2019 Dec 5];8(7). Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4963881/ PMCID: PMC4963881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olson KC, Chen G, Xu Y, Hajnal A, Lynch CJ. Alloisoleucine differentiates the branched-chain aminoacidemia of obese Zucker and diet-induced obesity (DIO) rats. Obes Silver Spring Md. 2014. May;22(5):1212–1215. PMCID: PMC4008669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jouvet P, Kozma M, Mehmet H. Primary human fibroblasts from a maple syrup urine disease patient undergo apoptosis following exposure to physiological concentrations of branched chain amino acids. Ann N Y Acad Sci. 2000;926:116–121. PMID: 11193026 [DOI] [PubMed] [Google Scholar]
- 44.Amaral AU, Leipnitz G, Fernandes CG, Seminotti B, Schuck PF, Wajner M. Alpha-ketoisocaproic acid and leucine provoke mitochondrial bioenergetic dysfunction in rat brain. Brain Res. 2010. Apr 9;1324:75–84. PMID: 20153737 [DOI] [PubMed] [Google Scholar]
- 45.Green CR, Wallace M, Divakaruni AS, Phillips SA, Murphy AN, Ciaraldi TP, Metallo CM. Branched-chain amino acid catabolism fuels adipocyte differentiation and lipogenesis. Nat Chem Biol. 2016. Jan;12(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lotta LA, Scott RA, Sharp SJ, Burgess S, Luan J, Tillin T, Schmidt AF, Imamura F, Stewart ID, Perry JRB, Marney L, Koulman A, Karoly ED, Forouhi NG, Sjögren RJO, Näslund E, Zierath JR, Krook A, Savage DB, Griffin JL, Chaturvedi N, Hingorani AD, Khaw K-T, Barroso I, McCarthy MI, O’Rahilly S, Wareham NJ, Langenberg C. Genetic Predisposition to an Impaired Metabolism of the Branched-Chain Amino Acids and Risk of Type 2 Diabetes: A Mendelian Randomisation Analysis. PLoS Med [Internet]. 2016. Nov 29 [cited 2019 Sep 13];13(11). Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5127513/ PMCID: PMC5127513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahendran Y, Jonsson A, Have CT, Allin KH, Witte DR, Jørgensen ME, Grarup N, Pedersen O, Kilpeläinen TO, Hansen T. Genetic evidence of a causal effect of insulin resistance on branched-chain amino acid levels. Diabetologia. 2017;60(5):873–878. PMID: 28184960 [DOI] [PubMed] [Google Scholar]
- 48.Wang Q, Holmes MV, Davey Smith G, Ala-Korpela M. Genetic Support for a Causal Role of Insulin Resistance on Circulating Branched-Chain Amino Acids and Inflammation. Diabetes Care. 2017. Dec;40(12):1779–1786. PMCID: PMC5701741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stančáková A, Civelek M, Saleem NK, Soininen P, Kangas AJ, Cederberg H, Paananen J, Pihlajamäki J, Bonnycastle LL, Morken MA, Boehnke M, Pajukanta P, Lusis AJ, Collins FS, Kuusisto J, Ala-Korpela M, Laakso M. Hyperglycemia and a Common Variant of GCKR Are Associated With the Levels of Eight Amino Acids in 9,369 Finnish Men. Diabetes. 2012. Jul;61(7):1895–1902. PMCID: PMC3379649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fall T, Salihovic S, Brandmaier S, Nowak C, Ganna A, Gustafsson S, Broeckling CD, Prenni JE, Kastenmüller G, Peters A, Magnusson PK, Wang-Sattler R, Giedraitis V, Berne C, Gieger C, Pedersen NL, Ingelsson E, Lind L. Non-targeted metabolomics combined with genetic analyses identifies bile acid synthesis and phospholipid metabolism as being associated with incident type 2 diabetes. Diabetologia. 2016. Oct 1;59(10):2114–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tremblay F, Marette A. Amino Acid and Insulin Signaling via the mTOR/p70 S6 Kinase Pathway A NEGATIVE FEEDBACK MECHANISM LEADING TO INSULIN RESISTANCE IN SKELETAL MUSCLE CELLS. J Biol Chem. 2001. Oct 12;276(41):38052–38060. [DOI] [PubMed] [Google Scholar]
- 52.Fernstrom JD. Branched-Chain Amino Acids and Brain Function. J Nutr. 2005. Jun 1;135(6):1539S–1546S. [DOI] [PubMed] [Google Scholar]
- 53.Meister A, Tate SS. Glutathione and Related γ-Glutamyl Compounds: Biosynthesis and Utilization. Annu Rev Biochem. 1976;45(1):559–604. PMID: 9027 [DOI] [PubMed] [Google Scholar]
- 54.Cheng S, Rhee EP, Larson MG, Lewis GD, McCabe EL, Shen D, Palma MJ, Roberts LD, Dejam A, Souza AL, Deik AA, Magnusson M, Fox CS, O’Donnell CJ, Vasan RS, Melander O, Clish CB, Gerszten RE, Wang TJ. Metabolite Profiling Identifies Pathways Associated with Metabolic Risk in Humans. Circulation. 2012. May 8;125(18):2222–2231. PMCID: PMC3376658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krebs HA. Metabolism of amino-acids. Biochem J. 1935. Aug;29(8):1951–1969. PMCID: PMC1266709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu X, Zheng Y, Guasch-Ferré M, Ruiz-Canela M, Toledo E, Clish C, Liang L, Razquin C, Corella D, Estruch R, Fito M, Gómez-Gracia E, Arós F, Ros E, Lapetra J, Fiol M, Serra-Majem L, Papandreou C, Martínez-González MA, Hu FB, Salas-Salvadó J. High plasma glutamate and low glutamine-to-glutamate ratio are associated with type 2 diabetes: Case-cohort study within the PREDIMED trial. Nutr Metab Cardiovasc Dis. 2019. Oct;29(10):1040–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferrannini E, Natali A, Camastra S, Nannipieri M, Mari A, Adam K-P, Milburn MV, Kastenmüller G, Adamski J, Tuomi T, Lyssenko V, Groop L, Gall WE. Early Metabolic Markers of the Development of Dysglycemia and Type 2 Diabetes and Their Physiological Significance. Diabetes. 2013. May;62(5):1730–1737. PMCID: PMC3636608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan-Do R, MacDonald PE. Impaired “Glycine”-mia in Type 2 Diabetes and Potential Mechanisms Contributing to Glucose Homeostasis. Endocrinology. 2017. May 1;158(5):1064–1073. [DOI] [PubMed] [Google Scholar]
- 59.Marquard J, Otter S, Welters A, Stirban A, Fischer A, Eglinger J, Herebian D, Kletke O, Klemen MS, Stožer A, Wnendt S, Piemonti L, Köhler M, Ferrer J, Thorens B, Schliess F, Rupnik MS, Heise T, Berggren P-O, Klöcker N, Meissner T, Mayatepek E, Eberhard D, Kragl M, Lammert E. Characterization of pancreatic NMDA receptors as possible drug targets for diabetes treatment. Nat Med. 2015. Apr;21(4):363–372. PMID: 25774850 [DOI] [PubMed] [Google Scholar]
- 60.Wang TJ, Ngo D, Psychogios N, Dejam A, Larson MG, Vasan RS, Ghorbani A, O’Sullivan J, Cheng S, Rhee EP, Sinha S, McCabe E, Fox CS, O’Donnell CJ, Ho JE, Florez JC, Magnusson M, Pierce KA, Souza AL, Yu Y, Carter C, Light PE, Melander O, Clish CB, Gerszten RE. 2-Aminoadipic acid is a biomarker for diabetes risk. J Clin Invest. 2013. Oct 1;123(10):4309–4317. PMCID: PMC3784523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gall WE, Beebe K, Lawton KA, Adam K-P, Mitchell MW, Nakhle PJ, Ryals JA, Milburn MV, Nannipieri M, Camastra S, Natali A, Ferrannini E, Group for the the RS. α-Hydroxybutyrate Is an Early Biomarker of Insulin Resistance and Glucose Intolerance in a Nondiabetic Population. PLOS ONE. 2010. May 28;5(5):e10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mahendran Y, Vangipurapu J, Cederberg H, Stančáková A, Pihlajamäki J, Soininen P, Kangas AJ, Paananen J, Civelek M, Saleem NK, Pajukanta P, Lusis AJ, Bonnycastle LL, Morken MA, Collins FS, Mohlke KL, Boehnke M, Ala-Korpela M, Kuusisto J, Laakso M. Association of Ketone Body Levels With Hyperglycemia and Type 2 Diabetes in 9,398 Finnish Men. Diabetes. 2013. Oct;62(10):3618. PMID: 23557707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Padberg I, Peter E, González-Maldonado S, Witt H, Mueller M, Weis T, Bethan B, Liebenberg V, Wiemer J, Katus HA, Rein D, Schatz P. A New Metabolomic Signature in Type-2 Diabetes Mellitus and Its Pathophysiology. PLOS ONE. 2014. Jan 17;9(1):e85082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Menni C, Fauman E, Erte I, Perry JRB, Kastenmüller G, Shin S-Y, Petersen A-K, Hyde C, Psatha M, Ward KJ, Yuan W, Milburn M, Palmer CNA, Frayling TM, Trimmer J, Bell JT, Gieger C, Mohney RP, Brosnan MJ, Suhre K, Soranzo N, Spector TD. Biomarkers for Type 2 Diabetes and Impaired Fasting Glucose Using a Nontargeted Metabolomics Approach. Diabetes. 2013. Dec 1;62(12):4270–4276. PMID: 23884885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ahmad TR, Haeusler RA. Bile acids in glucose metabolism and insulin signalling — mechanisms and research needs. Nat Rev Endocrinol. 2019. Dec;15(12):701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rebholz CM, Yu B, Zheng Z, Chang P, Tin A, Köttgen A, Wagenknecht LE, Coresh J, Boerwinkle E, Selvin E. Serum metabolomic profile of incident diabetes. Diabetologia [Internet]. 2018. Mar 20 [cited 2018 Apr 12]; Available from: http://link.springer.com/10.1007/s00125-018-4573-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang SJ, Kwak S-Y, Jo G, Song T-J, Shin M-J. Serum metabolite profile associated with incident type 2 diabetes in Koreans: findings from the Korean Genome and Epidemiology Study. Sci Rep. 2018. May 29;8(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Drogan D, Dunn WB, Lin W, Buijsse B, Schulze MB, Langenberg C, Brown M, Floegel A, Dietrich S, Rolandsson O, Wedge DC, Goodacre R, Forouhi NG, Sharp SJ, Spranger J, Wareham NJ, Boeing H. Untargeted Metabolic Profiling Identifies Altered Serum Metabolites of Type 2 Diabetes Mellitus in a Prospective, Nested Case Control Study. Clin Chem. 2015. Mar 1;61(3):487–497. PMID: 25524438 [DOI] [PubMed] [Google Scholar]
- 69.Randle PJ, Garland PB, Hales CN, Newsholme EA. THE GLUCOSE FATTY-ACID CYCLE ITS ROLE IN INSULIN SENSITIVITY AND THE METABOLIC DISTURBANCES OF DIABETES MELLITUS. The Lancet. 1963. Apr 13;281(7285):785–789. [DOI] [PubMed] [Google Scholar]
- 70.Shulman GI. Ectopic Fat in Insulin Resistance, Dyslipidemia, and Cardiometabolic Disease. N Engl J Med. 2014. Sep 18;371(12):1131–1141. PMID: 25229917 [DOI] [PubMed] [Google Scholar]
- 71.Rhee EP, Cheng S, Larson MG, Walford GA, Lewis GD, McCabe E, Yang E, Farrell L, Fox CS, O’Donnell CJ, Carr SA, Vasan RS, Florez JC, Clish CB, Wang TJ, Gerszten RE. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J Clin Invest. 2011. Apr;121(4):1402–1411. PMCID: PMC3069773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mahendran Y, Cederberg H, Vangipurapu J, Kangas AJ, Soininen P, Kuusisto J, Uusitupa M, Ala-Korpela M, Laakso M. Glycerol and Fatty Acids in Serum Predict the Development of Hyperglycemia and Type 2 Diabetes in Finnish Men. Diabetes Care. 2013. Nov;36(11):3732–3738. PMCID: PMC3816902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fizelova M, Miilunpohja M, Kangas AJ, Soininen P, Kuusisto J, Ala-Korpela M, Laakso M, Stančáková A. Associations of multiple lipoprotein and apolipoprotein measures with worsening of glycemia and incident type 2 diabetes in 6607 non-diabetic Finnish men. Atherosclerosis. 2015. May 1;240(1):272–277. [DOI] [PubMed] [Google Scholar]
- 74.Ahola-Olli AV, Mustelin L, Kalimeri M, Kettunen J, Jokelainen J, Auvinen J, Puukka K, Havulinna AS, Lehtimäki T, Kähönen M, Juonala M, Keinänen-Kiukaanniemi S, Salomaa V, Perola M, Järvelin M-R, Ala-Korpela M, Raitakari O, Würtz P. Circulating metabolites and the risk of type 2 diabetes: a prospective study of 11,896 young adults from four Finnish cohorts. Diabetologia. 2019;62(12):2298–2309. PMCID: PMC6861432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao J, Zhu Y, Hyun N, Zeng D, Uppal K, Tran VT, Yu T, Jones D, He J, Lee ET, Howard BV. Novel Metabolic Markers for the Risk of Diabetes Development in American Indians. Diabetes Care. 2015. Feb;38(2):220–227. PMCID: PMC4302260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Spranger J, Kroke A, Möhlig M, Bergmann MM, Ristow M, Boeing H, Pfeiffer AFH. Adiponectin and protection against type 2 diabetes mellitus. Lancet Lond Engl. 2003. Jan 18;361(9353):226–228. PMID: 12547549 [DOI] [PubMed] [Google Scholar]
- 77.Sun Q, Dam RM van, Meigs JB, Franco OH, Mantzoros CS, Hu FB. Leptin and Soluble Leptin Receptor Levels in Plasma and Risk of Type 2 Diabetes in U.S. Women: A Prospective Study. Diabetes. 2010. Mar 1;59(3):611–618. PMID: 19959759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ding EL, Song Y, Manson JE, Hunter DJ, Lee CC, Rifai N, Buring JE, Gaziano JM, Liu S. Sex hormone-binding globulin and risk of type 2 diabetes in women and men. N Engl J Med. 2009. Sep 17;361(12):1152–1163. PMCID: PMC2774225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kollerits B, Lamina C, Huth C, Marques-Vidal P, Kiechl S, Seppälä I, Cooper J, Hunt SC, Meisinger C, Herder C, Kedenko L, Willeit J, Thorand B, Dähnhardt D, Stöckl D, Willeit K, Roden M, Rathmann W, Paulweber B, Peters A, Kähönen M, Lehtimäki T, Raitakari OT, Humphries SE, Vollenweider P, Dieplinger H, Kronenberg F. Plasma Concentrations of Afamin Are Associated With Prevalent and Incident Type 2 Diabetes: A Pooled Analysis in More Than 20,000 Individuals. Diabetes Care [Internet]. 2017. Aug 10 [cited 2020 Jan 17]; Available from: https://care.diabetesjournals.org/content/early/2017/08/09/dc17-0201 PMID: 28877915 [DOI] [PubMed] [Google Scholar]
- 80.Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gústafsdóttir SM, Östman A, Landegren U. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002. May;20(5):473–477. [DOI] [PubMed] [Google Scholar]
- 81.Lundberg M, Eriksson A, Tran B, Assarsson E, Fredriksson S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 2011. Aug;39(15):e102. PMCID: PMC3159481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ngo D, Sinha S, Shen D, Kuhn EW, Keyes MJ, Shi X, Benson MD, O’ Sullivan JF, Keshishian H, Farrell LA, Fifer MA, Vasan RS, Sabatine MS, Larson MG, Carr SA, Wang TJ, Gerszten RE. Aptamer-Based Proteomic Profiling Reveals Novel Candidate Biomarkers and Pathways in Cardiovascular Disease. Circulation. 2016. Jul 26;134(4):270–285. PMCID: PMC4963294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, Burgess S, Jiang T, Paige E, Surendran P, Oliver-Williams C, Kamat MA, Prins BP, Wilcox SK, Zimmerman ES, Chi A, Bansal N, Spain SL, Wood AM, Morrell NW, Bradley JR, Janjic N, Roberts DJ, Ouwehand WH, Todd JA, Soranzo N, Suhre K, Paul DS, Fox CS, Plenge RM, Danesh J, Runz H, Butterworth AS. Genomic atlas of the human plasma proteome. Nature. 2018. Jun 1;558(7708):73–79. PMCID: PMC6697541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nowak C, Sundström J, Gustafsson S, Giedraitis V, Lind L, Ingelsson E, Fall T. Protein Biomarkers for Insulin Resistance and Type 2 Diabetes Risk in Two Large Community Cohorts. Diabetes. 2016. Jan;65(1):276–284. PMCID: PMC5860375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Beijer K, Nowak C, Sundström J, Ärnlöv J, Fall T, Lind L. In search of causal pathways in diabetes: a study using proteomics and genotyping data from a cross-sectional study. Diabetologia. 2019;62(11):1998–2006. PMCID: PMC6805963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim SW, Choi J-W, Yun JW, Chung I-S, Cho HC, Song S-E, Im S-S, Song D-K. Proteomics approach to identify serum biomarkers associated with the progression of diabetes in Korean patients with abdominal obesity. PLoS ONE [Internet]. 2019. Sep 10 [cited 2019 Dec 16];14(9). Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6736247/ PMCID: PMC6736247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shah Svati H., Bain James R., Muehlbauer Michael J., Stevens Robert D., Crosslin David R., Haynes Carol, Dungan Jennifer, Kristin Newby L., Hauser Elizabeth R., Ginsburg Geoffrey S., Newgard Christopher B., Kraus William E. Association of a Peripheral Blood Metabolic Profile With Coronary Artery Disease and Risk of Subsequent Cardiovascular Events. Circ Cardiovasc Genet. 2010. Apr 1;3(2):207–214. [DOI] [PubMed] [Google Scholar]
- 88.Bhattacharya S, Granger CB, Craig D, Haynes C, Bain J, Stevens RD, Hauser ER, Newgard CB, Kraus WE, Newby LK, Shah SH. Validation of the association between a branched chain amino acid metabolite profile and extremes of coronary artery disease in patients referred for cardiac catheterization. Atherosclerosis. 2014. Jan 1;232(1):191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang R, Dong J, Zhao H, Li H, Guo H, Wang S, Zhang C, Wang S, Wang M, Yu S, Chen W. Association of Branched-Chain Amino Acids with Carotid Intima-Media Thickness and Coronary Artery Disease Risk Factors. PLOS ONE. 2014. Jun 9;9(6):e99598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ganna A, Salihovic S, Sundström J, Broeckling CD, Hedman ÅK, Magnusson PKE, Pedersen NL, Larsson A, Siegbahn A, Zilmer M, Prenni J, Ärnlöv J, Lind L, Fall T, Ingelsson E. Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart Disease. PLOS Genet. 2014. Dec 11;10(12):e1004801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Park JY, Lee S-H, Shin M-J, Hwang G-S. Alteration in Metabolic Signature and Lipid Metabolism in Patients with Angina Pectoris and Myocardial Infarction. PLOS ONE. 2015. Aug 10;10(8):e0135228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ganz P, Heidecker B, Hveem K, Jonasson C, Kato S, Segal MR, Sterling DG, Williams SA. Development and Validation of a Protein-Based Risk Score for Cardiovascular Outcomes Among Patients With Stable Coronary Heart Disease. JAMA. 2016. Jun 21;315(23):2532–2541. [DOI] [PubMed] [Google Scholar]
- 93.O’Sullivan JF, Morningstar JE, Yang Q, Zheng B, Gao Y, Jeanfavre S, Scott J, Fernandez C, Zheng H, O’Connor S, Cohen P, Vasan RS, Long MT, Wilson JG, Melander O, Wang TJ, Fox C, Peterson RT, Clish CB, Corey KE, Gerszten RE. Dimethylguanidino valeric acid is a marker of liver fat and predicts diabetes. J Clin Invest. 2017. Dec 1;127(12):4394–4402. PMCID: PMC5707166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Robbins JM, Herzig M, Morningstar J, Sarzynski MA, Cruz DE, Wang TJ, Gao Y, Wilson JG, Bouchard C, Rankinen T, Gerszten RE. Association of Dimethylguanidino Valeric Acid With Partial Resistance to Metabolic Health Benefits of Regular Exercise. JAMA Cardiol. 2019. Jul 1;4(7):636–643. PMCID: PMC6551586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Donnelly LA, Doney ASF, Hattersley AT, Morris AD, Pearson ER. The effect of obesity on glycaemic response to metformin or sulphonylureas in Type 2 diabetes. Diabet Med J Br Diabet Assoc. 2006. Feb;23(2):128–133. PMID: 16433709 [DOI] [PubMed] [Google Scholar]
- 96.Shu Y, Sheardown SA, Brown C, Owen RP, Zhang S, Castro RA, Ianculescu AG, Yue L, Lo JC, Burchard EG, Brett CM, Giacomini KM. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J Clin Invest. 2007. May 1;117(5):1422–1431. PMCID: PMC1857259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen Z-Z, Liu J, Morningstar J, Heckman-Stoddard BM, Lee CG, Dagogo-Jack S, Ferguson JF, Hamman RF, Knowler WC, Mather KJ, Perreault L, Florez JC, Wang TJ, Clish C, Temprosa M, Gerszten RE, Group the DPPR. Metabolite Profiles of Incident Diabetes and Heterogeneity of Treatment Effect in the Diabetes Prevention Program. Diabetes. 2019. Dec 1;68(12):2337–2349. PMID: 31582408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Walford GA, Ma Y, Clish C, Florez JC, Wang TJ, Gerszten RE. Metabolite Profiles of Diabetes Incidence and Intervention Response in the Diabetes Prevention Program. Diabetes. 2016. May;65(5):1424–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yu E, Ruiz‐Canela M, Razquin C, Guasch‐Ferré M, Toledo E, Wang DD, Papandreou C, Dennis C, Clish C, Liang L, Bullo M, Corella D, Fitó M, Gutiérrez‐Bedmar M, Lapetra J, Estruch R, Ros E, Cofán M, Arós F, Romaguera D, Serra‐Majem L, Sorlí JV, Salas‐Salvadó J, Hu FB, Martínez‐González MA. Changes in arginine are inversely associated with type 2 diabetes: A case-cohort study in the PREDIMED trial. Diabetes Obes Metab. 2019;21(2):397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Williams Stephen A., Murthy Ashwin C., DeLisle Robert K., Hyde Craig, Malarstig Anders, Ostroff Rachel, Weiss Sophie J., Segal Mark R., Ganz Peter. Improving Assessment of Drug Safety Through Proteomics. Circulation. 2018. Mar 6;137(10):999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]