Abstract
Classification of pathogenic species according to the distinct host transcriptional responses that they elicit may become a relevant tool for microarray-based diagnosis of infection. Individual strains of Mycobacterium avium, an opportunistic pathogen in humans, have previously been shown to differ in terms of growth and persistence. In order to cover a wide spectrum of virulence, we selected four M. avium isolates (2151SmO, 2151SmT, SE01, TMC724) that have distinct intramacrophage replication characteristics and cause differential activation in human macrophages. Following infection with each of these strains, the expression of 12,558 genes in human macrophages was systematically analyzed by microarray technology. Fifty genes (including genes encoding proinflammatory cytokines, chemokines, signaling, and adhesion molecules) were differentially expressed more than twofold in response to all of the M. avium isolates investigated and therefore constitute a common macrophage signature in response to M. avium. The magnitude of regulation of most of these genes was directly correlated with the host cell-activating capacity of the particular M. avium strain. The regulation of a number of genes not previously associated with mycobacterial infections was apparent; these genes included genes encoding lymphocyte antigen 64 and myosin X. In addition, individual response patterns typical for some M. avium isolates could be defined by the pronounced upregulation of interleukin-12p40 (IL-12p40) (in the case of 2151SmO) or the specific upregulation of SOCS-1 and IL-10 (in the case of SE01) in macrophages. TMC724, a strain of avian origin, could not be classified by any one of these schemes, possibly indicating the limits of pathogen categorization solely by immune response signatures.
The outcome of infection is the net consequence of the immune defenses of the host and a pathogen's capacity to subvert them. Macrophages play a central role in regulating innate and acquired immune responses against pathogens, but several mycobacterial species have developed strategies to persist in these cells even in the face of fully developed T-cell immunity (reviewed in references 40 and 51). Mycobacterium avium is an environmental microorganism and an opportunistic pathogen in humans (33). M. avium causes lymphadenopathies in children, pneumonia in the context of predisposing lung conditions, and disseminated infections in advanced-stage AIDS patients (31, 34). M. avium has proven to be useful in mouse models of infection to define key requirements in mycobacterial infections (3, 6, 8, 15, 21, 29).
Of particular interest when host-pathogen relationships at the species level are compared, M. avium strains having different origins or morphotypes have been shown to differ widely in terms of replication and persistence, both in vitro and in vivo in a mouse model of infection (5, 9, 15, 29, 43, 48, 52), although the molecular basis for this variability in virulence remains undetermined. Previous studies demonstrated an inverse correlation between the virulence of some M. avium isolates and their ability to induce tumor necrosis factor alpha (TNF-α) in murine macrophages, suggesting that virulent strains elicit only very limited activation of macrophage effector mechanisms, thereby escaping elimination (24, 49). Using primary human monocyte-derived macrophages, we recently demonstrated that intracellular growth control of some M. avium strains was critically dependent on the extent of mitogen-activated protein (MAP) kinase phosphorylation, one essential signaling pathway of macrophage activation (9). However, our studies also demonstrated that there is no direct and simple correlation between MAP kinase phosphorylation, TNF-α secretion, and the capacity of different M. avium strains to replicate inside macrophages.
The mechanisms underlying virulence and persistence may therefore be more complex and may involve the coregulation of specific signaling and transcriptional machinery in response to each individual M. avium isolate. In the current study we performed a systematic, comparative analysis addressing the transcriptional response of macrophages after infection with M. avium strains with different origins and virulence characteristics. We carefully selected four strains which reflect a wide repertoire of M. avium infectivity and persistence.
M. avium 2151 was initially isolated from an AIDS patient in the United States and was classified as a serovar 2 strain (7). Its stably subcultivated morphotypes (smooth opaque [SmO] and smooth transparent [SmT]) have become an intensively studied and well-established model for colony morphotypes in M. avium (5, 15, 20, 41). 2151SmO is known to strongly activate macrophages of mouse and human origin and fails to multiply intracellularly. SmT only weakly activates human and mouse macrophages and shows progressive intracellular growth (9, 54). A second isolate from a European AIDS patient (SE01) was included in the study because it represents a different, yet common serovar (serovar 4), is virulent in mice, and has been shown to readily replicate within human macrophages in the face of massive host cell activation (9). The fourth strain, TMC724 (= ATCC 25291), has been used by various laboratories to study macrophage activation and granuloma formation in the mouse model, in which it is the most virulent strain known to date (5, 21, 29). In contrast to the clinical isolates of M. avium, TMC724 was originally cultured from fowl. Since nothing is known about the virulence of avian isolates in human macrophages, analysis of the gene expression pattern induced by this strain afforded a unique opportunity to compare evolutionarily distant prototypes of M. avium.
When they encounter microbes and microbial structures, macrophages undergo complex functional reprogramming, as recently shown by comprehensive gene expression analyses (10, 14, 22, 42). Our study corroborates that there is an overlapping pattern of regulated genes in human macrophages representing a general inflammatory response to all isolates of M. avium. The quantity of differentially expressed genes and the magnitude of gene regulation are directly correlated with the overall macrophage-activating capacity of the individual M. avium strain used for infection, as determined by MAP kinase phosphorylation and TNF-α production. Additionally, some M. avium strains preferentially induce regulation of a private set of genes that may strain specifically perturb the macrophage response to infection or that may counteract antimycobacterial effector mechanisms. Importantly, however, one strain (TMC724) could not be classified by gene response signatures, most likely due to a lack of overall host cell activation.
MATERIALS AND METHODS
Bacteria.
M. avium strain TMC724 (serovar 2), originally isolated from fowl, causes a progressive infection with a high bacterial burden in all infected organs and accelerated death of infected mice (21, 29). The two morphotypes of M. avium strain 2151 (serovar 2), smooth transparent (SmT) and smooth opaque (SmO), are subclones of an isolate obtained from an AIDS patient (7). While the SmT morphotype causes a progressive infection in mice comparable to that caused by strain TMC724, the SmO morphotype of strain 2151 is eradicated and therefore classified as nonvirulent (4, 5, 9). M. avium strain SE01 (serovar 4), initially cultivated from the blood of an AIDS patient, causes a chronic, nonprogressive infection in the mouse model and can therefore be classified as intermediately virulent (29). M. avium strains were passaged twice in C57BL/6 mice, and the colony phenotype and serotype were confirmed. All strains were then cultured in Middlebrook 7H9 medium (Difco, Detroit, Mich.) containing 10% oleic acid-albumin-dextrose-catalase (Becton Dickinson, Heidelberg, Germany) and 0.05% Tween 80 (Sigma, Deisenhofen, Germany) until the mid-log phase. The absence of contaminating microorganisms was verified by plating culture material on brain heart infusion agar (Difco) and Ziehl-Neelsen staining. The suspension was frozen in aliquots at −70°C until use. For calculation of CFU, aliquots were serially diluted in sterile, distilled water containing 0.05% Tween 80 and plated on 7H10 agar containing 0.075% pyruvate. After incubation for 3 weeks at 37°C CFU were determined. For infection bacterial aliquots were thawed and centrifuged for 10 min at 835 × g. Bacteria were resuspended in phosphate-buffered saline and sonicated (3 to 5 min, 35 kHz; Bandelin, Berlin, Germany) to disrupt aggregates. To rule out the presence of lipopolysaccharide (LPS) in the assays, all strains were tested with a Limulus amebocyte lysate assay (Biowhittaker, Walkersville, MD). The effective LPS concentration in the experiments at inoculum ratios of 10:1 was below 2 pg/ml.
Isolation and cultivation of human monocyte-derived macrophages.
Mononuclear cells were isolated from the peripheral blood of healthy volunteers by density gradient centrifugation (11). Lymphocytes and monocytes were separated by counterflow elutriation as previously described (27). Highly purified (consistently >95%) monocytes were cultivated for 7 days in Teflon culture bags (CellGenix, Freiburg, Germany) in RPMI 1640 (Biochrom, Berlin, Germany) in the presence of 2% (vol/vol) heat-inactivated human AB serum, 2 ng/ml human macrophage colony-stimulating factor (R&D Systems, Wiesbaden, Germany), 2 mmol/liter l-glutamine (Biochrom), 100 U/ml penicillin G (Biochrom), and 100 μg/ml streptomycin (Biochrom). The viability of cells was always >95%, as determined by trypan blue staining. Macrophages were phenotyped by determining the cell surface marker expression profile (CD14, macrophage mannose receptor, carboxypeptidase M, HLA-DR) as described previously (46). Macrophages were cultivated in RPMI 1640 containing 10% (vol/vol) heat-inactivated fetal calf serum (Biochrom) and 2 mmol/liter l-glutamine. Cultures were incubated at 37°C in a humidified atmosphere containing 5% CO2.
Infection of human macrophages with M. avium: in vitro infection and MAP kinase phosphorylation.
A total of 0.4 × 106 human monocyte-derived macrophages (MDM) (at a concentration of 0.8 × 106 cells per ml) were inoculated with 1.2 × 106 CFU of M. avium per well. A lower initial infection rate than the infection rates used in all other experimental procedures was chosen to prevent macrophage death during the 7-day culture, especially with highly replicative M. avium isolates. After 4 h cells were washed vigorously with warm Hanks balanced salt solution to remove extracellular bacteria. To determine bacterial uptake, macrophages were lysed by addition of 0.1% saponin. Lysates were serially diluted in sterile, distilled water containing 0.05% Tween 80 and plated on 7H10 agar containing 0.075% pyruvate. For investigation of bacterial growth, parallel cultures were maintained for 3 and 7 days in 500 μl medium (after 3 days 500 μl fresh medium was added to 7-day cultures). Subsequently, cells were washed, lysed, diluted, and plated for CFU determination as described above. For analysis of MAP kinase phosphorylation, macrophages were incubated with M. avium (4.0 × 106 CFU per well) for 30 min as described above. Cell lysates were analyzed as described previously (46).
RNA isolation, reverse transcription PCR (RT-PCR), and ELISA.
For microarray analyses 4.5 × 106 human macrophages were cultivated in tissue culture plates (Nunc, Roskilde, Denmark) and infected with M. avium at a ratio of 10 mycobacteria per macrophage for 4 h. Culture supernatants of stimulated macrophages were harvested and stored at −20°C until analysis. A sandwich enzyme-linked immunosorbent assay (ELISA) was used for detecting TNF-α (H. Gallati, Intex, Muttenz, Switzerland) (25). Assays were performed as recommended by the manufacturer.
Macrophages were lysed by addition of TriFast FL (PeqLab, Erlangen, Germany). Total RNA was isolated by phase separation with 1-bromo-3-chloropropane (Sigma).
For validation of gene expression, equal amounts of RNA were reverse transcribed (Superscript II RNase H− reverse transcriptase; Invitrogen, Karlsruhe, Germany) and used for quantitative RT-PCR (LightCycler technology; Roche) using the following gene-specific primer pairs: for beta-2-microglobulin, forward primer 5′ GCTGTGCTCGCGCTACTCTC 3′ and reverse primer (5′ GCGGCATCTTCAAACCTCCAT 3′); for TNF-α, forward primer 5′ GGCTCCAGGCGGTGCTTGTTC 3′ and reverse primer 5′ AGACGGCGATGCGGCTGATG 3′; for interleukin-12p40 (IL-12p40), forward primer 5′ TCAGAGGGGACAACAAGGAGTATG 3′ and reverse primer 5′ CTGGGCCCGCACGCTAAT 3′; for lymphocyte antigen 64 (LY64) (CD180), forward primer 5′ AGCTGCTTCTTTTGGGTGGTG 3′ and reverse primer 5′ TTGGGGAACTTAATGGAGGAAATA 3′; and for myosin X, forward primer 5′ CACGCTGCCATCCCACCTCTC 3′ and reverse primer 5′ CCGCGCTGACACCCAACCA 3′. For RT-PCR of RANTES and pentraxin 3 (PTX3) the primer pairs described previously were used (36, 44). Gene expression was expressed as the relative expression normalized to beta-2-microglobulin expression. The specificity of amplified products was confirmed by sequence analyses.
DNA microarray hybridization and analysis.
The quality and integrity of total RNA isolated from human macrophages were controlled by analyzing all samples with an Agilent Technologies 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany). Equal amounts of total RNA of five individual donors were pooled to perform biotin-labeled target synthesis for array hybridization. The entire experimental approach was repeated with a second donor pool that was comparable in terms of the distribution of the ages, genders, and purified protein derivative reactivities of the individuals.
For biotin-labeled target synthesis starting from total RNA, reactions were performed using standard protocols supplied by the manufacturer (Affymetrix, Santa Clara, CA). Briefly, 8 μg total RNA (equal amounts from five different donors per hybridization) were converted to double-stranded DNA using 100 pmol of a T7T23V primer (Eurogentec, Seraing, Belgium) containing a T7 promoter. The double-stranded DNA was then used directly in an in vitro transcription reaction in the presence of biotinylated nucleotides using a T7 RNA polymerase (BioArray high-yield RNA transcript labeling kit; ENZO, New York, NY). The concentration of biotin-labeled cRNA was determined by UV absorbance. In all cases, 12.5 μg of each biotinylated cRNA preparation was fragmented and placed in a hybridization cocktail containing four biotinylated hybridization controls (BioB, BioC, BioD, and Cre) as recommended by the manufacturer. Samples were hybridized to an identical lot of Affymetrix U95Av2 human genome GeneChips for 16 h. Each U95Av2 GeneChip contains oligonucleotides for 12,558 human genes and expressed sequence tags derived from common UniGene clusters. After hybridization the GeneChips were washed, stained with streptavidin-phycoerythrin, and read using an Affymetrix GeneChip fluidic station and scanner.
Fluorescence intensities were normalized to median array intensities for all conditions tested, and the fold changes were calculated relative to unstimulated baseline controls. Analysis of data was done with gene expression software (GeneChip, MicroDB, and Data Mining Tool; all obtained from Affymetrix) with a filter for regulated genes that employed the following stringent criteria to define genes as significantly differentially expressed: (i) a signal log2 ratio of >1 or <−1, signifying changes in the expression level between control conditions (baseline) and stimulated cells; and (ii) a change in the P value of <0.001 or >0.999, describing the likelihood and direction of change of expression for each transcript. P values indicated the level of significance of the difference between the baseline and experimental conditions based on the Wilcoxon signed-rank test. Genes defined as differentially expressed were significantly regulated in both donor pools, each of which consisted of five individual donors. Unless indicated otherwise specifically below, genes represented by more than one transcript on the Affymetrix U95Av2 GeneChip were considered a single gene for the discussion of absolute numbers of differentially expressed individual genes.
Statistical analysis.
For comparison of RT-PCR results, the global hypothesis was tested according to Friedman. For posttesting, data obtained from independent experiments were compared using the Wilcoxon rank test. The α-error was corrected by the Shaffer procedure.
RESULTS
Intramacrophage replication characteristics of M. avium strains used in this study.
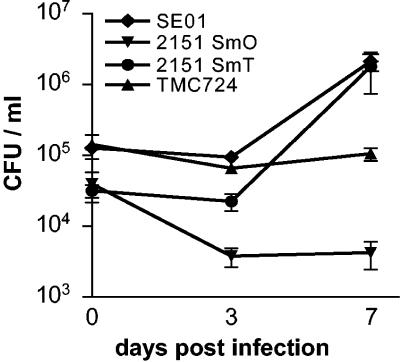
Four different M. avium isolates (TMC724, 2151SmT, 2151SmO, and SE01) that have previously been well characterized in terms of growth and persistence in vitro and in vivo were used in this study (4, 5, 9, 29). Determination of the viable CFU inside macrophages over a 7-day period postinfection revealed that 2151SmO was slowly eliminated, that TMC724 persisted at inoculum levels, and that 2151SmT and SE01 replicated extensively over time (Fig. 1). Overall, these characteristics of replication and persistence are in good accordance with results from murine infection models, with the notable exception of TMC724, which failed to show the supreme virulence exhibited in mice in vivo (21, 29). Nevertheless, with one strain nearly eradicated, one persisting, and two highly replicating, a wide spectrum of virulence was represented even in this small sample of M. avium strains.
FIG. 1.
Uptake and intracellular replication of four M. avium strains in human macrophages. Human MDM were infected with the smooth transparent (SmT) and smooth opaque (SmO) morphotypes of M. avium strain 2151, strain TMC724, and strain SE01 at an MOI of 3. CFU counts in lysed macrophages were determined 4 h and 3 and 7 days after infection. The values are means ± standard deviations for duplicates of one representative experiment of the experiments performed.
TNF-α release and MAP kinase activation of macrophages infected with different strains of M. avium.
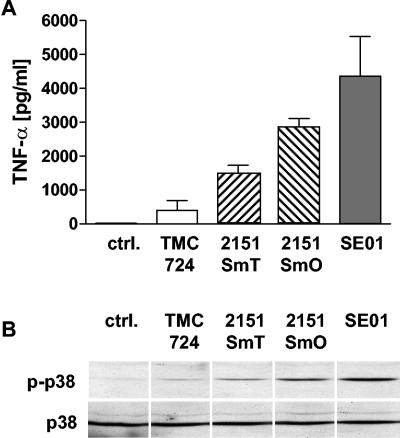
The virulence of M. avium strains has previously been correlated to their capacity to induce cytokine release from infected macrophages (5, 24). We compared the TNF-α production of human monocyte-derived macrophages after infection (10 bacteria per macrophage) with four selected M. avium strains (Fig. 2A). TMC724 and 2151SmT induced only small amounts of TNF-α, and consistently higher TNF-α levels were induced by 2151SmT than by TMC724 when an identical multiplicity of infection (MOI) was used for stimulation. In contrast, 2151SmO and SE01 induced the release of large amounts of TNF-α, and SE01 reproducibly triggered the highest levels of TNF-α.
FIG. 2.
TNF-α release and p38 MAP kinase activation in cultures of human macrophages infected with different M. avium strains. (A) Human MDM were infected with M. avium strains TMC724, 2151SmT, 2151SmO, and SE01 at an MOI of 10. Supernatants were harvested after 4 h. Cytokine concentrations were measured by ELISA. The values are means ± standard deviations for three independent experiments. ctrl., control. (B) Human MDM were incubated with M. avium strains TMC724, 2151SmT, 2151SmO, and SE01 at an MOI of 10 for 30 min. Cells were lysed, and aliquots of cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted onto nitrocellulose membranes. The membranes were incubated with specific anti-phospho-p38 antibodies, followed by a peroxidase-coupled secondary antibody. Visualization was performed by enhanced chemiluminescence. To control equal protein loading, the amounts of total p38 were detected in the same lysates. The results of one representative experiment of three independent experiments are shown.
It appeared that it is possible that the differential capacity of macrophages to secrete TNF-α in response to different M. avium strains might be determined by the differential intensity of pretranscriptional signaling events. We therefore analyzed MAP kinase activation in human macrophages induced by the four M. avium strains after 30 min of stimulation. TMC724 and 2151SmT induced only weak phosphorylation of p38 and ERK1/2 in human macrophages (Fig. 2B and data not shown). However, the signal obtained for 2151SmT was usually stronger than that obtained for TMC724 when a similar MOI (10 bacteria per macrophage) was used for stimulation. In contrast, 2151SmO and SE01 induced marked MAP kinase phosphorylation, and the signal induced by SE01 was consistently stronger than that induced by 2151SmO (Fig. 2B).
These data indicate that the virulence, as measured by the capacity to replicate within macrophages (Fig. 1), of individual M. avium strains is not consistently associated with the absence of strong macrophage activation. Conversely, the lowest levels of TNF-α induction or MAP kinase phosphorylation need not necessarily correlate with a pronounced capacity of a given strain to multiply. Overall, the quantity of macrophage responses, such as the level of TNF-α secretion, after stimulation with individual M. avium isolates is governed by the magnitude of signal events, such as MAP kinase activation, occurring very early after infection.
Gene expression profiles of human macrophages induced by different strains of M. avium.
In order to investigate whether the macrophage response after infection with individual mycobacterial isolates might also differ qualitatively (i.e., in terms of the kind of transcriptional response), we applied a systematic approach using microarray analyses. Macrophages of different donors were infected with each of the four M. avium strains at a ratio of 10 bacteria per macrophage. Total RNA was isolated 4 h postinfection. We and other workers have previously noted the donor-dependent variability of macrophage responses to M. avium (9, 28). To minimize donor-dependent confounding effects, equal amounts of total RNA obtained from cells of five individual donors were pooled to perform biotin-labeled target synthesis for array hybridization. The entire experimental approach was repeated with a second donor pool comparable in terms of the distribution of the ages, genders, and purified protein derivative reactivities of the individuals. Only genes that were differentially expressed compared to unstimulated control macrophages with a change in the expression level of at least twofold (induced or repressed) in both donor pools were considered for further in-depth analysis and are included in the tables if the statistical criteria described in Materials and Methods were met.
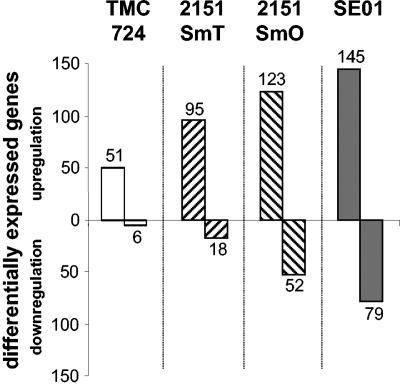
TMC724 induced differential expression of 57 genes (51 genes induced, 6 genes repressed) compared to uninfected macrophages (Fig. 3). 2151SmT induced regulation of 113 macrophage genes (95 genes induced, 18 genes repressed), whereas 175 genes (123 genes induced, 52 genes repressed) were regulated in response to 2151SmO. SE01 induced differential expression of 224 genes (145 genes induced, 79 genes repressed) (Fig. 3). Thus, the numbers of differentially expressed genes in response to these different M. avium isolates reflect the individual capacity of each strain to activate macrophages, as measured by MAP kinase phosphorylation or TNF-α release (Fig. 2 and 3).
FIG. 3.
Numbers of differentially expressed genes in human macrophages after infection with four M. avium strains. Human macrophages were infected with M. avium strains TMC724, 2151SmT, 2151SmO, and SE01 (MOI, 10) for 4 h. Differential gene expression was compared to expression in uninfected control cells using microarray technology. An array analysis was performed with two independent donor pools, each comprising five individuals. The total number of differentially expressed genes (induction or suppression) found in both donor pools is indicated.
When the infection-induced macrophage transcriptomes were compared, the gene expression patterns in response to the M. avium strains were found to overlap. Fifty different genes (47 genes induced, 3 genes repressed) were identified as differentially expressed in response to all strains (Table 1). At this early time (4 h after infection) more than one-third of these differentially expressed genes are involved in a proinflammatory macrophage response, and the molecules involved include cytokines (IL-1β, IL-6, TNF-α, lymphotoxin α), chemokines (RANTES, IL-8, monocyte chemoattractant protein 1, macrophage inflammatory protein 1α [MIP-1α], MIP-1β, MIP-2α, MIP-2β, MIP-3α, etc.), cytokine and chemokine receptors (IL-7 receptor, CCR2, CXCR4), and adhesion molecules (ICAM-1) (Table 1). The changes in the expression levels of genes in these functional categories were more pronounced than the changes in the expression levels of genes having a different functional background. Additionally, a number of genes whose products are involved in signaling cascades, transcriptional events, the cell cycle, and antiapoptosis, as well as genes encoding metabolic enzymes, were differentially expressed in response to infection with all M. avium strains (Table 1). In general, for the majority of genes, the magnitude of regulation in response to the individual M. avium strains could be directly correlated with the TNF-α-inducing capacity of each isolate (Fig. 2).
TABLE 1.
Shared pattern of differentially expressed macrophage genes in response to M. avium strains TMC724, 2151SmT, 2151SmO, and SE01
| Category | Gene name (UniGene) | Designation | Fold regulation ina
|
|||
|---|---|---|---|---|---|---|
| TMC724 | 2151SmT | 2151SmO | SE01 | |||
| Cytokines | Interleukin 1, beta | IL-1b | 7.41 | 15.51 | 19.87 | 16.02 |
| Interleukin 6 (interferon, beta 2) | IL-6 | 68.83 | 264.55 | 489.99 | 306.51 | |
| Tumor necrosis factor | TNF | 4.58 | 3.76 | 3.89 | 10.02 | |
| TNF superfamily, member 2 | 9.08 | 10.07 | 13.32 | 17.24 | ||
| Lymphotoxin alpha TNF superfamily member 1 | LTA | 4.58 | 3.76 | 3.89 | 10.02 | |
| Chemokines | Interleukin 8 | IL-8 | 2.91 | 4.21 | 4.50 | 4.07 |
| Small inducible cytokine A5 | RANTES | 4.36 | 19.02 | 21.86 | 20.20 | |
| GRO1 oncogene | MGSA | 36.55 | 63.10 | 81.27 | 75.78 | |
| GRO2 oncogene | MIP-2A | 11.32 | 15.57 | 19.93 | 18.96 | |
| GRO3 oncogene | MIP-2B | 11.61 | 15.00 | 17.36 | 21.64 | |
| Small inducible cytokine A2 | MCP-1 | 2.55 | 4.21 | 5.09 | 3.69 | |
| Monocyte chemotactic protein 1 | 2.85 | 4.53 | 5.12 | 3.98 | ||
| Small inducible cytokine A3 | MIP-1A | 3.08 | 4.19 | 4.71 | 4.76 | |
| Small inducible cytokine A4 | MIP-1B | 8.16 | 11.10 | 12.31 | 11.63 | |
| Small inducible cytokine subfamily A (Cys-Cys), member 20 | MIP-3A | 24.43 | 70.86 | 114.24 | 132.60 | |
| Receptors | Adenosine A2a receptor | ADORA2A | 12.37 | 9.44 | 12.80 | 13.95 |
| Interleukin 7 receptor | IL-7R | 3.20 | 4.09 | 5.37 | 5.29 | |
| Tumor necrosis factor receptor superfamily, member 4 | TNFRSF4 | 2.81 | 4.58 | 5.01 | 5.58 | |
| Chemokine (C-C motif) receptor 2 | CCR2 | −5.01 | −6.84 | −6.56 | −16.20 | |
| Chemokine (C-X-C motif), receptor 4 (fusin) | CXCR4 | −3.94 | −4.35 | −4.79 | −11.98 | |
| Signal transduction | Nef-associated factor 1 | NAF1 | 2.41 | 2.50 | 3.02 | 2.72 |
| TNF receptor-associated factor 1 | TRAF1 | 4.83 | 6.86 | 8.15 | 8.40 | |
| Jagged 1 (Alagille syndrome) | JAG1 | 3.59 | 4.51 | 5.03 | 6.76 | |
| Phosphodiesterase 4B, cAMP specific | PDE4B | 2.96 | 4.29 | 4.32 | 3.40 | |
| Thyroid hormone receptor interactor 10 | TRIP10 | 2.96 | 4.06 | 5.60 | 7.17 | |
| Seven in absentia (Drosophila) homolog 2 | SIAH2 | 2.41 | 2.88 | 2.66 | 3.95 | |
| Cytoskeleton | Singed (Drosophila)-like (sea urchin fascin homolog-like) | SNL | 3.65 | 8.06 | 11.90 | 12.74 |
| Transcription | Nuclear factor of kappa light polypeptide | LYT-10 | 2.19 | 2.48 | 3.02 | 3.44 |
| Gene enhancer in B-cells 2 (p49/p100) | LOC125387 | 3.46 | 2.91 | 4.83 | 5.03 | |
| Butyrate response factor 2 (EGF-resp. factor 2) | BRF2 | −2.41 | −4.23 | −6.44 | −8.55 | |
| Enzymes | Adenosine deaminase | ADA | 2.31 | 3.76 | 5.78 | 6.12 |
| Prostaglandin-endoperoxide synthase 2 | PTGS2 | 27.29 | 27.68 | 54.59 | 153.21 | |
| GTP cyclohydrolase 1 (dopa-responsive dystonia) | GCH1 | 4.32 | 7.68 | 10.21 | 9.51 | |
| Carbohydrate (chondroitin 6/keratan) | C6ST | 2.46 | 2.52 | 3.02 | 3.28 | |
| Sulfotransferase 2 | ||||||
| Antiapoptosis | Tumor necrosis factor, alpha-induced protein 3 | TNFAIP3 | 3.06 | 3.20 | 3.65 | 4.09 |
| Immediate early response 3 | IER3 | 3.38 | 3.98 | 4.34 | 4.66 | |
| Adhesion | Intercellular adhesion molecule 1 (CD54), human rhinovirus receptor | ICAM1 | 2.99 | 3.64 | 4.06 | 4.25 |
| Tumor necrosis factor, alpha-induced protein 6 | TNFAIP6 | 5.62 | 11.80 | 16.53 | 17.93 | |
| Proliferation/ cell cycle | Tumor necrosis factor receptor superfamily, member 9 | TNFRSF9 | 2.87 | 3.79 | 4.42 | 3.48 |
| Tumor necrosis factor (ligand) superfamily, member 9 | TNFSF9 | 13.92 | 13.84 | 27.99 | 97.96 | |
| Putative lymphocyte G0/G1 switch gene | GOS2 | 3.07 | 3.04 | 3.20 | 3.95 | |
| Pim-2 oncogene | PIM2 | 2.62 | 3.55 | 4.44 | 4.51 | |
| Clotting | Glycoprotein 1b (platelet), alpha polypeptide | GP1B | 3.06 | 4.00 | 3.67 | 3.99 |
| Miscellaneous | Tumor necrosis factor alpha-induced protein 2 | TNFAIP2 | 3.68 | 5.02 | 5.90 | 6.61 |
| Tumor suppressing subtransferable candidate 3 | TSSC3 | 2.29 | 3.11 | 4.22 | 4.59 | |
| Myristoylated alanine-rich protein kinase C substrate (MARCKS, 80K-L) | MACS | 2.39 | 2.87 | 2.91 | 3.24 | |
| Adrenomedullin | ADM | 3.81 | 3.74 | 4.78 | 8.82 | |
| Pentaxin-related, rapidly induced by IL-1beta | PTX3 | 12.78 | 39.59 | 106.07 | 120.67 | |
| EH domain containing 1 | EHD1 | 3.31 | 4.27 | 5.76 | 6.14 | |
| KIAA0854 protein | KIAA0854 | 2.23 | 2.71 | 3.22 | 4.49 | |
| Wilms' tumor 1-associating protein | KIAA0105 | 2.81 | 3.55 | 4.45 | 4.62 | |
| Hypothetical protein | FLJ20500 | 2.83 | 6.62 | 6.51 | 5.05 | |
| Human DNA sequence from clone RP5-1174N9 on chromosome 1p34.1-35.3 | 3.44 | 3.03 | 2.93 | 4.25 | ||
Positive values indicate fold induction, and negative values indicate fold suppression.
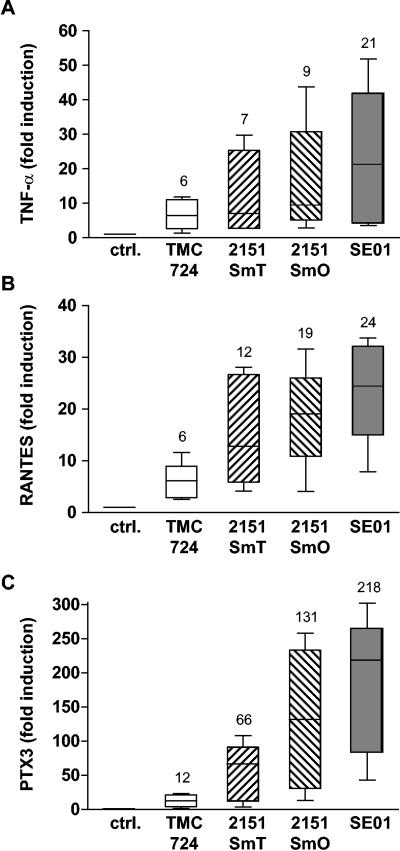
To independently confirm the microarray results for gene induction by M. avium, three genes induced by all M. avium strains were selected from different categories, and their expression was analyzed by quantitative PCR using the same RNA samples with which microarrays had been hybridized. Figure 4 shows that the expression levels of the cytokine TNF-α (Fig. 4A), the chemokine RANTES (Fig. 4B), and the ancestral pattern recognition receptor PTX3 (Fig. 4C) induced by each strain indeed were the same order of magnitude as the expression levels observed in the microarray analyses (Fig. 4 and Table 1).
FIG. 4.
Expression of TNF-α, RANTES, and PTX3 mRNA in human monocyte-derived macrophages after infection with M. avium. Human MDM were incubated with M. avium isolates TMC724, 2151SmT, 2151SmO, and SE01 (MOI, 10) for 4 h. Total RNA was isolated, reverse transcribed, and used for quantitative RT-PCR of TNF-α (A), RANTES (B), and PTX3 (C) mRNA normalized to beta-2-microglobulin mRNA levels. The values indicate the fold induction compared to untreated cells for duplicate determinations for five different donors. The boxes indicate the 25th and 75th percentiles. The whiskers above and below the boxes indicate the highest and lowest values. The medians are marked and indicated above the boxes. ctrl., control.
Search for a “virulence response signature”: M. avium strain-specific gene expression profiles of human macrophages.
All events examined thus far (MAP kinase phosphorylation, TNF-α secretion, quantity of genes regulated) pointed to a macrophage-activating capacity that was intrinsic to each M. avium strain but partially disparate from its virulence properties. We therefore sought to determine whether there are any transcriptional signatures that allow categorization of isolates according to their intracellular replicating characteristics.
The two morphotypes of M. avium strain 2151 have a common genotype, yet they differ profoundly in intracellular growth. Therefore, they provided an excellent starting point to study the macrophage responses to virulent and nonvirulent M. avium isolates in a comparison of the host cell gene expression profiles induced by them. In addition to the general macrophage response signature for infection with M. avium as described above, direct comparison indeed identified genes that were individually regulated following infection with these two morphotypes (Table 2). The individual responses to 2151SmT and 2151SmO covered 9% (10 genes; 5 genes induced and 5 genes repressed) and 39% (68 genes; 31 genes induced and 37 genes repressed), respectively, of all genes differentially expressed after infection with the individual morphotypes (Table 2).
TABLE 2.
Individual patterns of differentially expressed macrophage genes in response to two morphotypes of M. avium strain 2151
| Morphotype | Gene name (UniGene) | Designation | Fold regulationa |
|---|---|---|---|
| 2151SmT | Myosin X | MYO10 | 20.82 |
| RAS guanyl releasing protein 1 calcium and DAG regulated | RASGRP1 | 4.47 | |
| Neutrophil cytosolic factor 1 | NCF1 | 2.62 | |
| DEAD/H Asp-Glu-Ala-Asp/His box polypeptide Y chromosome | DBY | 2.22 | |
| Hypothetical protein | HSPC111 | 2.08 | |
| BAI1-associated protein 2 | BAIAP2 | −2.17 | |
| Ecotropic viral integration site 2A | EVI2A | −2.36 | |
| Histidyl-tRNA synthetase-like | HARSL | −2.39 | |
| Dual-specificity tyrosine-Y phosphorylation-regulated kinase 2 | DYRK2 | −4.96 | |
| i-Beta-1,3-N-acetylglucosaminyltransferase | BETA3GNTI | −5.76 | |
| 2151SmO | Interleukin 12B/p40 | IL12B | 47.19 |
| CGI-142 | CGI-142 | 13.51 | |
| Mevalonate diphosphodecarboxylase | MVD | 8.71 | |
| Basic leucine zipper transcription factor ATF-like | BATF | 5.86 | |
| RAB33A member RAS oncogene family | RAB33A | 5.80 | |
| Dual-specificity phosphatase 2 | DUSP2 | 4.15 | |
| Syntaxin 11 | STX11 | 3.95 | |
| Phosphatidic acid phosphatase type 2B | PPAP2B | 3.40 | |
| Ubiquitin-specific protease 12 | USP12 | 3.21 | |
| Chemokine (C-C motif) receptor 7 | CCR7 | 3.16 | |
| Regulator of G-protein signaling 16 | RGS16 | 2.94 | |
| v-rel avian reticuloendotheliosis viral oncogene homolog | REL | 2.90 | |
| Serine or cysteine proteinase inhibitor clade B ovalbumin member 9 | SERPINB9 | 2.81 | |
| CASP8 and FADD-like apoptosis regulator | CFLAR | 2.70 | |
| Musculin-activated B-cell factor-1 | MSC | 2.66 | |
| Translation factor sui1 homolog | GC20 | 2.64 | |
| Interferon regulatory factor 1 | IRF1 | 2.63 | |
| Chromosome X open reading frame 6 | CXORF6 | 2.55 | |
| KIAA0286 protein | KIAA0286 | 2.43 | |
| Stannin | SNN | 2.42 | |
| Uridine phosphorylase | UP | 2.40 | |
| Nuclear receptor subfamily 4 group A member 3 | NR4A3 | 2.40 | |
| Serine or cysteine proteinase inhibitor clade B ovalbumin member 8 | SERPINB8 | 2.37 | |
| Tight junction protein 2 zona occludens 2 | TJP2 | 2.35 | |
| LPS-induced TNF-alpha factor | PIG7 | 2.33 | |
| KIAA0084 protein | KIAA0084 | 2.28 | |
| Ninjurin 1 | NINJ1 | 2.23 | |
| Poliovirus receptor | PVR | 2.21 | |
| SRY sex-determining region Y box 4 | SOX4 | 2.16 | |
| v-ets avian erythroblastosis virus E26 oncogene homolog 2 | ETS2 | 2.10 | |
| Nucleolar phosphoprotein p130 | P130 | 2.07 | |
| Chromosome 21 open reading frame 25 | C21ORF25 | −2.01 | |
| Hypothetical protein FLJ13052 | FLJ13052 | −2.04 | |
| p8 protein candidate of metastasis 1 | P8 | −2.04 | |
| Sialyltransferase 1-beta-galactoside alpha-2,6-sialytransferase | SIAT1 | −2.08 | |
| Murine leukemia viral bmi-1 oncogene homolog | BMI1 | −2.10 | |
| Zinc finger protein 148 pHZ-52 | ZNF148 | −2.11 | |
| Sorting nexin 2 | SNX2 | −2.12 | |
| LIM domain only 2 rhombotin-like 1 | LMO2 | −2.13 | |
| Adipose differentiation-related protein | ADFP | −2.14 | |
| Coronin actin-binding protein 1A | CORO1A | −2.16 | |
| Interleukin 17 receptor | IL17R | −2.16 | |
| KIAA1224 protein | KIAA1224 | −2.25 | |
| ATP-binding cassette subfamily C CFTR/MRP member 5 | ABCC5 | −2.25 | |
| Conserved gene amplified in osteosarcoma | OS4 | −2.27 | |
| Three prime repair exonuclease 1 | DKFZp434J0310 | −2.33 | |
| LCAT-like lysophospholipase | LLPL | −2.36 | |
| KIAA1093 protein | KIAA1093 | −2.37 | |
| Proline-serine-threonine phosphatase interacting protein 1 | PSTPIP1 | −2.38 | |
| FYN-binding protein FYB-120/130 transcription factor 3 E2A immunoglobulin enhancer binding factors | FYB | −2.39 | |
| E12/E47 | TCF3 | −2.42 | |
| Regulator of G-protein signaling 2.24kD | RGS2 | −2.44 | |
| Protein predicted by clone 23627 | HSU79266 | −2.48 | |
| Inositol 1,4,5-trisphosphate 3-kinase B | ITPKB | −2.56 | |
| Glutamate-cysteine ligase catalytic subunit | GCLC | −2.61 | |
| Disabled Drosophila homolog 2 mitogen-responsive phosphoprotein | DAB2 | −2.69 | |
| Transforming growth factor beta receptor II 70-80kD | TGFBR2 | −2.80 | |
| Mad4 homolog myeloid/lymphoid or mixed-lineage leukemia trithorax Drosophila | MAD4 | −2.92 | |
| Homolog translocated to 2 | MLLT2 | −2.98 | |
| Ecotropic viral integration site 2B | EVI2B | −3.05 | |
| Endocrine regulator | HRIHFB2436 | −3.27 | |
| Peroxisome proliferative activated receptor gamma | PPARG | −3.63 | |
| Interferon gamma-inducible protein 16 | IFI16 | −3.64 | |
| Protein tyrosine phosphatase nonreceptor type 22 lymphoid | PTPN22 | −3.72 | |
| B cell linker protein | SLP65 | −4.70 | |
| KIAA0500 protein | KIAA0500 | −4.94 | |
| KIAA0846 protein | KIAA0846 | −6.28 | |
| Lymphocyte antigen 64 mouse homolog radioprotective 105kD | LY64/CD180 | −23.10 |
Positive values indicate fold induction, and negative values indicate fold suppression.
In contrast to the overall amount of differentially expressed genes that are induced by all four M. avium strains, the individual response patterns show equal numbers of induced and suppressed genes for 2151SmT and 2151SmO. While the majority of genes were regulated only between two- and fourfold (close to the cutoff for twofold regulation), three genes were found to be regulated more than 20-fold, the genes encoding IL-12p40 (47-fold induction) and LY64 (23-fold repression) in response to 2151SmO and the gene encoding myosin X (20-fold induction) after infection with 2151SmT (Table 2).
IL-12 is well known to be a critical factor in controlling mycobacterial infections (reviewed in references 13, 19, and 53). In contrast, there have been no reports on the role of either LY64 or myosin X for modulating mycobacterial growth. To independently confirm the differential expression of the genes encoding these three molecules, human macrophages of five individuals were infected with M. avium 2151SmT or 2151SmO for 4 h, and total RNA was isolated and used for quantitative RT-PCR.
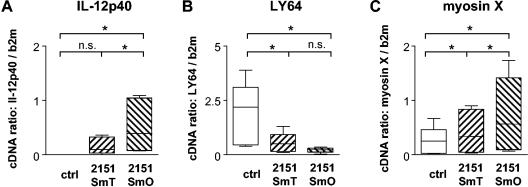
2151SmO significantly induced expression of IL-12p40 mRNA in macrophages of all five donors compared to unstimulated control cultures (Fig. 5A). The closely related SmT morphotype of strain 2151 also induced IL-12p40 mRNA expression in four of five donors (Fig. 5A), albeit to a significantly lower extent. Stimulation of human macrophages with 2151SmO as well as 2151SmT significantly reduced LY64 mRNA expression in all individuals (Fig. 5B), and the inhibition was more pronounced in response to SmO than in response to SmT (Fig. 5B). Expression of myosin X mRNA was significantly enhanced in response to 2151SmT and, in contrast to the microarray results, also after stimulation with 2151SmO (Fig. 5C). Furthermore, 2151SmO consistently induced myosin X mRNA expression to a significantly higher extent than 2151SmT.
FIG. 5.
Expression of IL-12p40, LY64, and myosin X mRNA in human monocyte-derived macrophages after infection with M. avium. Human MDM were incubated with M. avium isolates 2151SmT and 2151SmO (MOI, 10) for 4 h. Total RNA was isolated, reverse transcribed, and used for quantitative RT-PCR of IL-12p40 (A), LY64 (B), and myosin X (C) mRNA normalized to beta-2-microglobulin mRNA levels. The values are the means for duplicate determinations for five different donors. The boxes indicate the 25th and 75th percentiles. The whiskers above and below the boxes indicate the highest and lowest values. The medians are marked. Significant differences are indicated; an asterisk indicates that the P value is <0.05. n.s., not significant; ctrl, control; b2m, beta-2-microglobulin.
To address this discrepancy between the results from microarray analysis and RT-PCR, reanalysis of the raw, array-based gene expression data was performed. Expression of myosin X mRNA was induced 33- and 2.8-fold in response to 2151SmT in the two replicate microarray experiments. In response to 2151SmO, the induction was 12- and 3.6-fold, but the difference did not reach statistical significance. Thus, the raw gene expression values were actually well in accordance with the RT-PCR data. This is even more remarkable since the array-based myosin X mRNA expression values were generally very low (signal, <50). The fact that changes in gene expression even at the very low signal end were confirmed by an independent method again underscores the robustness of our microarray-based gene expression analysis.
Finally, we scrutinized the macrophage gene expression profiles induced by TMC724 and SE01, two genetically completely unrelated M. avium strains with highly discrepant intracellular growth properties. The private macrophage expression signature evoked by TMC724 (compared to all other isolates) contained only two genes, the proto-oncogene c-Myc (twofold induction) and the vaccinia-related kinase 2 gene (twofold repression), once more reflecting the overall weak potential of this M. avium strain to activate macrophages. In contrast, the individual signature of differentially expressed macrophage genes in response to SE01 consisted of 87 genes (38 genes induced, 49 genes suppressed) (Table 3), the highest number of privately regulated genes in this series.
TABLE 3.
Individual pattern of differentially expressed macrophage genes in response to M. avium strain SE01
| Gene name (UniGene) | Designation | Fold regulationa |
|---|---|---|
| JAK binding protein | SSI-1 | 15.77 |
| Plasminogen activator, tissue | PLAT | 10.26 |
| Hairy (Drosophila) homolog | HRY | 8.51 |
| Platelet-derived growth factor beta polypeptide | PDGFB | 7.96 |
| Solute carrier family 1, glial high-affinity glutamate transport, membrane 2 | SLC1A2 | 7.33 |
| BCL2-like 1 | BCL2L1 | 7.18 |
| Signaling lymphocytic activation molecule | SLAM | 6.92 |
| Colony-stimulating factor 3 (granulocyte) | CSF3 | 6.69 |
| Colony-stimulating factor 2 (granulocyte-macrophage) | CSF2 | 6.66 |
| Interleukin 10 | IL10 | 5.95 |
| Dual-specificity phosphatase 8 | DUSP8 | 5.58 |
| Aldehyde dehydrogenase 5 | ALDH5 | 4.65 |
| Dual-specificity tyrosine-phosphorylation-regulated kinase 3 | DYRK3 | 3.56 |
| v-src avian sarcoma (Schmidt-Ruppin A-2) | SRC | 3.49 |
| Homo sapiens C2H2-type zinc finger protein mRNA, complete cds | 3.37 | |
| Thymopoietin | TMPO | 3.33 |
| CD83 antigen | CD83 | 3.31 |
| Growth arrest and DNA damage-inducible, alpha | GADD45A | 3.21 |
| Coagulation factor III (thromboplastin, tissue factor) | F3 | 3.20 |
| EST, moderately similar to POL2 human retrovirus-related POL polyprotein | 2.90 | |
| Dishevelled 3 (homologous to Drosophila dsh) | DVL3 | 2.78 |
| NCK adaptor protein 2 | NCK2 | 2.69 |
| Cytokine-inducible kinase | CNK | 2.69 |
| Acetyl LDL receptor; SREC | SREC | 2.69 |
| Mitogen-activated protein kinase-activated protein kinase 2 | MAPKAPK2 | 2.67 |
| Prostaglandin I2 (prostacyclin) receptor (IP) | PTGIR | 2.60 |
| Dual-specificity phosphatase 1 | DUSP1 | 2.60 |
| Glycophorin C (Gerbich blood group) | GYPC | 2.60 |
| Creatine kinase, brain | CKB | 2.53 |
| Glutaredoxin (thioltransferase) | GLRX | 2.38 |
| X-box binding protein 1 | XBP1 | 2.36 |
| Prostaglandin E receptor 4 (subtype EP4) | PTGER4 | 2.31 |
| Signal transducer and activator of transcription 5A | STAT5A | 2.28 |
| Pleiomorphic adenoma gene-like 2 | PLAGL2 | 2.24 |
| CCAAT/enhancer binding protein (C/EBP), delta | CEBPD | 2.20 |
| Guanylate binding protein 2, interferon inducible | GBP2 | 2.18 |
| BTG family, member 3 | BTG3 | 2.13 |
| ras homolog gene family, member G (rho G) | ARHG | 2.09 |
| KIAA0759 protein | KIAA0759 | −2.07 |
| cAMP-responsive element modulator | CREM | −2.09 |
| Diacylglycerol kinase, zeta (104kD) | DGKZ | −2.11 |
| Myeloid cell nuclear differentiation antigen | MNDA | −2.12 |
| Homo sapiens cDNA: FLJ21449 fis, clone COL04483, highly similar to AF010235 H. sapiens mRNA from chromosome 5q31-33 region | −2.15 | |
| Putative human HLA class II associated protein I | PHAP1 | −2.17 |
| Interferon gamma receptor 1 | IFNGR1 | −2.19 |
| Src-like-adapter | SLA | −2.19 |
| RAS p21 protein activator (GTPase activating protein) 1 | RASA1 | −2.20 |
| Homo sapiens mRNA; cDNA (from clone DKFZp564N1272); complete cds | −2.22 | |
| DKFZP566O1646 protein | −2.24 | |
| Proliferating cell nuclear antigen | PCNA | −2.26 |
| KIAA0229 protein | KIAA0229 | −2.28 |
| Nucleoporin 214kD (CAIN) | NUP214 | −2.30 |
| Human clone 23801 mRNA sequence | −2.30 | |
| MKP-1 like protein tyrosine phosphatase | MKP-L | −2.31 |
| Speckle-type POZ protein | SPOP | −2.33 |
Positive values indicate fold induction, and negative values indicate fold suppression.
The SE01-specific induced macrophage transcriptome was remarkable in that a number of genes involved in the downregulation of a proinflammatory immune response were highly upregulated. Thus, profound upregulation (15-fold) of suppressor of cytokine signaling 1 (SOCS-1/SSI-1), a negative regulator of gamma interferon (IFN-γ) signaling (38), was accompanied by repression of IFN-γ receptor 1 (twofold). In addition, the anti-inflammatory cytokine IL-10 was significantly induced (sixfold) only by SE01 (Table 3).
DISCUSSION
This study shows that the extent of early macrophage signaling events induced by individual strains of M. avium is directly correlated with the quantity of genes and the overall magnitude of gene regulation elicited by each individual strain. While this type of classification of M. avium strains proved to be robust, in that three different methods of evaluation (MAP kinase phosphorylation, TNF-α secretion, and size of infection-induced transcriptome) showed consistent results with four different isolates, we found no straightforward association of a specific macrophage response signature with the pathogenicities of the infecting strains, as measured by their intracellular replication rates in vitro. Therefore, the virulence of the species M. avium is not mirrored by a stereotypic, species-specific macrophage response, as previously suggested (28). Nor is virulence of M. avium invariably a unique failure to activate macrophages, as hypothesized previously (24, 49). Rather, our data imply that virulence must be explained at the level of the individual isolate of M. avium; while some isolates may modulate host defenses by manipulating specific host defense genes (such as the gene encoding IL-12p40, LY64, SOCS-1, or IL-10, as identified in this study), other isolates may simply rely on their intrinsic resistance to antimycobacterial effector mechanisms.
Our data corroborate previous findings from microarray analyses performed with macrophages infected with mycobacteria or stimulated with mycobacterial components (10, 14, 42). In particular, they support the concept that following interaction with mycobacteria, a prewired infection-related gene expression program is induced in macrophages, consisting of signaling components, transcriptional machinery, and mediators and receptors necessary to enhance myelomonocytic cell recruitment and activation. This program differs from that elicited by protozoan or helminth parasites in that, for example, many more interferon-inducible genes are triggered (14).
A previous study detailing the expression signature of macrophages in response to M. avium described the upregulation of approximately 150 genes involved in signal transduction, transcription, adhesion, apoptosis, protein turnover, homeostasis, detoxification, and general metabolism or coding for receptor-associated proteins, cytokines, and chemokines (28). While many of the genes described in that report are also listed in our M. avium-induced common signature (28) (Table 1), there are individual differences in both the number and the type of genes that were found to be regulated. These differences are most likely due to donor variability and different times of analysis, but they probably also reflect the use of yet another M. avium strain in that study (only described as a smooth transparent isolate). The fact that IL-12p40 and IL-10 were not appreciably induced but that a number of ras-associated signaling molecules were expressly regulated by that strain put it in a category not dissimilar to that shown here for 2151SmT (28).
The results of this study and a recent study (9) provide evidence that within minutes following exposure, the macrophage responses to M. avium strains already differ at the level of signal transduction. Virulent or persistent strains, such as M. avium 2151SmT or TMC724, induce much less phosphorylation of MAP kinases than avirulent isolates, such as 2151SmO, induce. Virulence may therefore be an intrinsic inability of certain isolates to activate macrophages (e.g., due to a lack of stimulating cell wall-associated structures). TMC724 and both morphotypes of strain 2151 contain the ser2 gene cluster, which is involved in glycosylation of glycopeptidolipids of the cell wall. Genetic characterization of this region revealed strain-specific variations in this region, indicating structural differences in the individual cell wall composition (7, 20). The quantity of receptor-ligating structures in individual strains of M. avium has, however, not been accurately determined thus far.
Alternatively, virulence may be the consequence of active suppression of early signaling events within the host cell (e.g., by activating phosphatases) (37). In this respect, it is noteworthy that the virulent SE01 strain significantly induced two dual-specificity phosphatases (phosphatases 1 and 8).
Highly stringent statistical evaluation of the microarray data initially showed strain-specific transcriptional activation of several macrophage genes, e.g., the genes encoding IL-12p40, LY64, and myosin X. Independent confirmation of the expression levels of these genes by quantitative RT-PCR revealed that rather than representing “exclusively” regulated genes, the genes were regulated in a more pronounced fashion by infection with 2151SmO than by infection with 2151SmT. Further analysis of this discrepancy showed that in the microarray-based screening these genes did in fact also show regulation following infection with other strains but that the magnitude was not sufficient to meet our statistical criteria.
The control of mycobacterial infections is known to depend on IL-12, which is essential for inducing an appropriate IFN-γ response (2, 17, 39). Impaired and/or delayed expression of IL-12 may therefore be interpreted as a corollary of virulence in individual M. avium strains (such as 2151SmT or TMC724). Conversely, the high induction of IL-12 formation by the avirulent 2151SmO strain may be termed suicidal. However, it should be mentioned that the biology of the heterodimeric IL-12 family of cytokines is more complex and that excess IL-12p40 dimers can also antagonize IL-12p70 functions at the IL-12 receptor (12, 30).
The individual response to M. avium 2151SmO indicated profound downregulation of LY64 (CD180, RP105), a member of the Toll-like receptor (TLR) family, which mediates the LPS response in B lymphocytes (reviewed in reference 35). LY64 has previously been found to be differentially expressed in tumor-associated macrophages (26) but has not been linked to mycobacterial infections thus far. Interestingly, LY64 has functional similarities with TLR4. While there is a report that TLR4-defective mice are as susceptible to infection with M. avium as with wild-type controls, the role of TLR4 in the control of mycobacterial infections is controversial (1, 23, 47, 50). Elucidating the role of LY64 in M. avium pathogenesis might shed new light on the contribution of different members of the TLR family in the innate response against mycobacterial infection.
Collectively, our data show that for some strains, such as 2151SmT and 2151SmO, the overall magnitude of host cell activation, as determined by MAP kinase phosphorylation, TNF-α production, and the size of the induced transcriptome, is inversely correlated with the capacity to persist and grow inside macrophages. While there is no specific “virulence-associated” signature of the virulent 2151SmT strain, the strong regulation of some genes, such as the IL-12p40, myosin X, and LY64 genes, by 2151SmO may be a good corollary of this strain's lack of virulence. For TMC724, there is also no “persistence-associated,” strain-specific macrophage gene expression signature. In fact, TMC724 induces only very little gene regulation, and therefore activation, in human host cells. This may reflect its avian origin, which endowed it with a heightened imperviousness to constitutively expressed effector molecules in human macrophages.
The macrophage transcriptome induced by M. avium SE01 is particularly enlightening, as it shows that the analysis of a strain-specific signature may help us understand this isolate's behavior inside the macrophage. Obviously, despite very strong activation of the innate immune response, this strain is able to persist in macrophages. Here, SE01 induces upregulation of SOCS-1/SSI-1, a negative feedback regulator of signaling events via janus kinases induced by cytokines (38). SOCS-1-mediated negative regulation of IFN-γ-induced signals can be hypothesized as one mechanism by which SE01 promotes its survival inside macrophages. Moreover, infection with SE01 induces the transcriptional downregulation of IFN-γ receptor 1, again indicating effective counteraction of IFN-γ-mediated eradication (16, 18, 32). In addition, SE01 was the only strain found to induce very high IL-10 gene transcription, which may interfere with antimycobacterial effector mechanisms (45, 55). If the results are taken together, SE01 seems to be capable of tipping the scale of macrophage activation toward a net “anti-inflammatory” score which favors persistence.
In conclusion, microarray analyses of gene expression patterns of human macrophages in response to M. avium strains with different levels of virulence yielded a number of genes associated with a common antimicrobial macrophage response, consistent with previous reports. Overall, quantitative rather than qualitative differences of gene regulation were associated with the capacity of M. avium strains to grow inside macrophages. In particular, our data provide no evidence that the virulence and pathogenicity of M. avium as a species can be unequivocally deduced from the macrophage gene response signatures that individual isolates elicit. Instead, there are at least three distinct explanations for virulence at the level of the individual strain. First, isolates such as 2151SmT or TMC724 elicit only a low or delayed type of host response, which contributes to their escaping antimycobacterial effector mechanisms. Second, isolates such as SE01 induce a strong macrophage response in which the net balance of pro- and anti-inflammatory activities appears to be skewed towards suppressing antimycobacterial protection. Third, isolates such as TMC724 may be largely impervious to antimycobacterial effector molecules in human macrophages irrespective of the host response that they elicit.
Acknowledgments
This work was supported in part by the Deutsche Forschungsgemeinschaft (SFB 415, project C7) and the BMBF (NGFN-1).
We thank Svenja Kröger for expert technical assistance and acknowledge Robert Geffers for helpful discussions.
Editor: J. D. Clements
REFERENCES
- 1.Abel, B., N. Thieblemont, V. J. Quesniaux, N. Brown, J. Mpagi, K. Miyake, F. Bihl, and B. Ryffel. 2002. Toll-like receptor 4 expression is required to control chronic Mycobacterium tuberculosis infection in mice. J. Immunol. 169:3155-3162. [DOI] [PubMed] [Google Scholar]
- 2.Altare, F., A. Durandy, D. Lammas, J. F. Emile, S. Lamhamedi, F. Le Deist, P. Drysdale, E. Jouanguy, R. Doffinger, F. Bernaudin, O. Jeppsson, J. A. Gollob, E. Meinl, A. W. Segal, A. Fischer, D. Kumararatne, and J. L. Casanova. 1998. Impairment of mycobacterial immunity in human interleukin-12 receptor deficiency. Science 280:1432-1435. [DOI] [PubMed] [Google Scholar]
- 3.Appelberg, R., A. G. Castro, J. Pedrosa, R. A. Silva, I. M. Orme, and P. Minoprio. 1994. Role of gamma interferon and tumor necrosis factor alpha during T-cell-independent and -dependent phases of Mycobacterium avium infection. Infect. Immun. 62:3962-3971. (Erratum, 63:1145, 1995.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appelberg, R., and I. M. Orme. 1993. Effector mechanisms involved in cytokine-mediated bacteriostasis of Mycobacterium avium infections in murine macrophages. Immunology 80:352-359. [PMC free article] [PubMed] [Google Scholar]
- 5.Appelberg, R., A. Sarmento, and A. G. Castro. 1995. Tumour necrosis factor-alpha (TNF-alpha) in the host resistance to mycobacteria of distinct virulence. Clin. Exp. Immunol. 101:308-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Appelberg, R., and A. M. Sarmento. 1990. The role of macrophage activation and of Bcg-encoded macrophage function(s) in the control of Mycobacterium avium infection in mice. Clin. Exp. Immunol. 80:324-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belisle, J. T., K. Klaczkiewicz, P. J. Brennan, W. R. Jacobs, Jr., and J. M. Inamine. 1993. Rough morphological variants of Mycobacterium avium. Characterization of genomic deletions resulting in the loss of glycopeptidolipid expression. J. Biol. Chem. 268:10517-10523. [PubMed] [Google Scholar]
- 8.Benini, J., E. M. Ehlers, and S. Ehlers. 1999. Different types of pulmonary granuloma necrosis in immunocompetent vs. TNFRp55-gene-deficient mice aerogenically infected with highly virulent Mycobacterium avium. J. Pathol. 189:127-137. [DOI] [PubMed] [Google Scholar]
- 9.Blumenthal, A., S. Ehlers, M. Ernst, H. D. Flad, and N. Reiling. 2002. Control of mycobacterial replication in human macrophages: roles of extracellular signal-regulated kinases 1 and 2 and p38 mitogen-activated protein kinase pathways. Infect. Immun. 70:4961-4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boldrick, J. C., A. A. Alizadeh, M. Diehn, S. Dudoit, C. L. Liu, C. E. Belcher, D. Botstein, L. M. Staudt, P. O. Brown, and D. A. Relman. 2002. Stereotyped and specific gene expression programs in human innate immune responses to bacteria. Proc. Natl. Acad. Sci. USA 99:972-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyum, A.. 1968. Isolation of mononuclear cells and granulocytes from human blood. Isolation of monuclear cells by one centrifugation, and of granulocytes by combining centrifugation and sedimentation at 1 g. Scand. J. Clin. Lab. Investig. Suppl. 97:77-89. [PubMed] [Google Scholar]
- 12.Brombacher, F., R. A. Kastelein, and G. Alber. 2003. Novel IL-12 family members shed light on the orchestration of Th1 responses. Trends Immunol. 24:207-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casanova, J. L., and L. Abel. 2002. Genetic dissection of immunity to mycobacteria: the human model. Annu. Rev. Immunol. 20:581-620. [DOI] [PubMed] [Google Scholar]
- 14.Chaussabel, D., R. T. Semnani, M. A. McDowell, D. Sacks, A. Sher, and T. B. Nutman. 2003. Unique gene expression profiles of human macrophages and dendritic cells to phylogenetically distinct parasites. Blood 102:672-681. [DOI] [PubMed] [Google Scholar]
- 15.Cooper, A. M., R. Appelberg, and I. M. Orme. 1998. Immunopathogenesis of Mycobacterium avium infection. Front. Biosci. 3:e141-e148. [DOI] [PubMed] [Google Scholar]
- 16.Cooper, A. M., D. K. Dalton, T. A. Stewart, J. P. Griffin, D. G. Russell, and I. M. Orme. 1993. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J. Exp. Med. 178:2243-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Jong, R., F. Altare, I. A. Haagen, D. G. Elferink, T. Boer, P. J. Breda Vriesman, P. J. Kabel, J. M. Draaisma, J. T. van Dissel, F. P. Kroon, J. L. Casanova, and T. H. Ottenhoff. 1998. Severe mycobacterial and Salmonella infections in interleukin-12 receptor-deficient patients. Science 280:1435-1438. [DOI] [PubMed] [Google Scholar]
- 18.Dorman, S. E., and S. M. Holland. 1998. Mutation in the signal-transducing chain of the interferon-gamma receptor and susceptibility to mycobacterial infection. J. Clin. Investig. 101:2364-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dorman, S. E., and S. M. Holland. 2000. Interferon-gamma and interleukin-12 pathway defects and human disease. Cytokine Growth Factor Rev. 11:321-333. [DOI] [PubMed] [Google Scholar]
- 20.Eckstein, T. M., J. T. Belisle, and J. M. Inamine. 2003. Proposed pathway for the biosynthesis of serovar-specific glycopeptidolipids in Mycobacterium avium serovar 2. Microbiology 149:2797-2807. [DOI] [PubMed] [Google Scholar]
- 21.Ehlers, S., J. Benini, H.-D. Held, C. Roeck, G. Alber, and S. Uhlig. 2001. Alphabeta T cell receptor-positive cells and interferon-gamma, but not inducible nitric oxide synthase, are critical for granuloma necrosis in a mouse model of mycobacteria-induced pulmonary immunopathology. J. Exp. Med. 194:1847-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ehrt, S., D. Schnappinger, S. Bekiranov, J. Drenkow, S. Shi, T. R. Gingeras, T. Gaasterland, G. Schoolnik, and C. Nathan. 2001. Reprogramming of the macrophage transcriptome in response to interferon-gamma and Mycobacterium tuberculosis: signaling roles of nitric oxide synthase-2 and phagocyte oxidase. J. Exp. Med. 194:1123-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng, C. G., C. A. Scanga, C. M. Collazo-Custodio, A. W. Cheever, S. Hieny, P. Caspar, and A. Sher. 2003. Mice lacking myeloid differentiation factor 88 display profound defects in host resistance and immune responses to Mycobacterium avium infection not exhibited by Toll-like receptor 2 (TLR2)- and TLR4-deficient animals. J. Immunol. 171:4758-4764. [DOI] [PubMed] [Google Scholar]
- 24.Furney, S. K., P. S. Skinner, A. D. Roberts, R. Appelberg, and I. M. Orme. 1992. Capacity of Mycobacterium avium isolates to grow well or poorly in murine macrophages resides in their ability to induce secretion of tumor necrosis factor. Infect. Immun. 60:4410-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallati, H., and I. Pracht. 1985. Horseradish peroxidase: kinetic studies and optimization of peroxidase activity determination using the substrates H2O2 and 3,3′,5,5′-tetramethylbenzidine. J. Clin. Chem. Clin. Biochem. 23:453-460. [PubMed] [Google Scholar]
- 26.Gottfried, E., S. Faust, J. Fritsche, L. A. Kunz-Schughart, R. Andreesen, K. Miyake, and M. Kreutz. 2003. Identification of genes expressed in tumor-associated macrophages. Immunobiology 207:351-359. [DOI] [PubMed] [Google Scholar]
- 27.Grage-Griebenow, E., D. Lorenzen, R. Fetting, H.-D. Flad, and M. Ernst. 1993. Phenotypical and functional characterization of Fcgamma receptor I (CD64)-negative monocytes, a minor human monocyte subpopulation with high accessory and antiviral activity. Eur. J. Immunol. 23:3126-3135. [DOI] [PubMed] [Google Scholar]
- 28.Greenwell-Wild, T., N. Vazquez, D. Sim, M. Schito, D. Chatterjee, J. M. Orenstein, and S. M. Wahl. 2002. Mycobacterium avium infection and modulation of human macrophage gene expression. J. Immunol. 169:6286-6297. [DOI] [PubMed] [Google Scholar]
- 29.Hänsch, H. C., D. A. Smith, M. E. Mielke, H. Hahn, G. J. Bancroft, and S. Ehlers. 1996. Mechanisms of granuloma formation in murine Mycobacterium avium infection: the contribution of CD4+ T cells. Int. Immunol. 8:1299-1310. [DOI] [PubMed] [Google Scholar]
- 30.Hölscher, C.. 2004. The power of combinatorial immunology: IL-12 and IL-12-related dimeric cytokines in infectious diseases. Med. Microbiol. Immunol. (Berl.) 193:1-17. [DOI] [PubMed] [Google Scholar]
- 31.Horsburgh, C. R., Jr. 1991. Mycobacterium avium complex infection in the acquired immunodeficiency syndrome. N. Engl. J. Med. 324:1332-1338. [DOI] [PubMed] [Google Scholar]
- 32.Hussain, S., B. S. Zwilling, and W. P. Lafuse. 1999. Mycobacterium avium infection of mouse macrophages inhibits IFN-gamma Janus kinase-STAT signaling and gene induction by down-regulation of the IFN-gamma receptor. J. Immunol. 163:2041-2048. [PubMed] [Google Scholar]
- 33.Inderlied, C. B., C. A. Kemper, and L. E. Bermudez. 1993. The Mycobacterium avium complex. Clin. Microbiol. Rev. 6:266-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iseman, M. D. 1989. Mycobacterium avium complex and the normal host: the other side of the coin. N. Engl. J. Med. 321:896-898. [DOI] [PubMed] [Google Scholar]
- 35.Kimoto, M., K. Nagasawa, and K. Miyake. 2003. Role of TLR4/MD-2 and RP105/MD-1 in innate recognition of lipopolysaccharide. Scand. J. Infect. Dis. 35:568-572. [DOI] [PubMed] [Google Scholar]
- 36.Klouche, M., N. Brockmeyer, C. Knabbe, and S. Rose-John. 2002. Human herpesvirus 8-derived viral IL-6 induces PTX3 expression in Kaposi's sarcoma cells. AIDS 16:F9-F18. [DOI] [PubMed] [Google Scholar]
- 37.Knutson, K. L., Z. Hmama, P. Herrera-Velit, R. Rochford, and N. E. Reiner. 1998. Lipoarabinomannan of Mycobacterium tuberculosis promotes protein tyrosine dephosphorylation and inhibition of mitogen-activated protein kinase in human mononuclear phagocytes. Role of the Src homology 2 containing tyrosine phosphatase 1. J. Biol. Chem. 273:645-652. [DOI] [PubMed] [Google Scholar]
- 38.Kubo, M., T. Hanada, and A. Yoshimura. 2003. Suppressors of cytokine signaling and immunity. Nat. Immunol. 4:1169-1176. [DOI] [PubMed] [Google Scholar]
- 39.Magram, J., S. E. Connaughton, R. R. Warrier, D. M. Carvajal, C. Y. Wu, J. Ferrante, C. Stewart, U. Sarmiento, D. A. Faherty, and M. K. Gately. 1996. IL-12-deficient mice are defective in IFN gamma production and type 1 cytokine responses. Immunity 4:471-481. [DOI] [PubMed] [Google Scholar]
- 40.Manabe, Y. C., and W. R. Bishai. 2000. Latent Mycobacterium tuberculosis—persistence, patience, and winning by waiting. Nat. Med. 6:1327-1329. [DOI] [PubMed] [Google Scholar]
- 41.Mills, J. A., M. R. McNeil, J. T. Belisle, W. R. J. Jacobs, and P. J. Brennan. 1994. Loci of Mycobacterium avium ser2 gene cluster and their functions. J. Bacteriol. 176:4803-4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nau, G. J., J. F. Richmond, A. Schlesinger, E. G. Jennings, E. S. Lander, and R. A. Young. 2002. Human macrophage activation programs induced by bacterial pathogens. Proc. Natl. Acad. Sci. USA 99:1503-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pedrosa, J., M. Florido, Z. M. Kunze, A. G. Castro, F. Portaels, J. McFadden, M. T. Silva, and R. Appelberg. 1994. Characterization of the virulence of Mycobacterium avium complex (MAC) isolates in mice. Clin. Exp. Immunol. 98:210-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pierer, M., J. Rethage, R. Seibl, R. Lauener, F. Brentano, U. Wagner, H. Hantzschel, B. A. Michel, R. E. Gay, S. Gay, and D. Kyburz. 2004. Chemokine secretion of rheumatoid arthritis synovial fibroblasts stimulated by Toll-like receptor 2 ligands. J. Immunol. 172:1256-1265. [DOI] [PubMed] [Google Scholar]
- 45.Redpath, S., P. Ghazal, and N. R. Gascoigne. 2001. Hijacking and exploitation of IL-10 by intracellular pathogens. Trends Microbiol. 9:86-92. [DOI] [PubMed] [Google Scholar]
- 46.Reiling, N., A. Blumenthal, H.-D. Flad, M. Ernst, and S. Ehlers. 2001. Mycobacteria-induced TNF-alpha and IL-10 formation by human macrophages is differentially regulated at the level of mitogen-activated protein kinase activity. J. Immunol. 167:3339-3345. [DOI] [PubMed] [Google Scholar]
- 47.Reiling, N., C. Hölscher, A. Fehrenbach, S. Kröger, C. J. Kirschning, S. Goyert, and S. Ehlers. 2002. Cutting edge: Toll-like receptor (TLR)2- and TLR4-mediated pathogen recognition in resistance to airborne infection with Mycobacterium tuberculosis. J. Immunol. 169:3480-3484. [DOI] [PubMed] [Google Scholar]
- 48.Sarmento, A., and R. Appelberg. 1996. Involvement of reactive oxygen intermediates in tumor necrosis factor alpha-dependent bacteriostasis of Mycobacterium avium. Infect. Immun. 64:3224-3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarmento, A. M., and R. Appelberg. 1995. Relationship between virulence of Mycobacterium avium strains and induction of tumor necrosis factor alpha production in infected mice and in in vitro-cultured mouse macrophages. Infect. Immun. 63:3759-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shim, T. S., O. C. Turner, and I. M. Orme. 2003. Toll-like receptor 4 plays no role in susceptibility of mice to Mycobacterium tuberculosis infection. Tuberculosis (Edinburgh) 83:367-371. [DOI] [PubMed] [Google Scholar]
- 51.Stewart, G. R., B. D. Robertson, and D. B. Young. 2003. Tuberculosis: a problem with persistence. Nat. Rev. Microbiol. 1:97-105. [DOI] [PubMed] [Google Scholar]
- 52.Torrelles, J. B., D. Ellis, T. Osborne, A. Hoefer, I. M. Orme, D. Chatterjee, P. J. Brennan, and A. M. Cooper. 2002. Characterization of virulence, colony morphotype and the glycopeptidolipid of Mycobacterium avium strain 104. Tuberculosis (Edinburgh) 82:293-300. [DOI] [PubMed] [Google Scholar]
- 53.Trinchieri, G., S. Pflanz, and R. A. Kastelein. 2003. The IL-12 family of heterodimeric cytokines: new players in the regulation of T cell responses. Immunity 19:641-644. [DOI] [PubMed] [Google Scholar]
- 54.Tse, H. M., S. I. Josephy, E. D. Chan, D. Fouts, and A. M. Cooper. 2002. Activation of the mitogen-activated protein kinase signaling pathway is instrumental in determining the ability of Mycobacterium avium to grow in murine macrophages. J. Immunol. 168:825-833. [DOI] [PubMed] [Google Scholar]
- 55.Turner, J., M. Gonzalez-Juarrero, D. L. Ellis, R. J. Basaraba, A. Kipnis, I. M. Orme, and A. M. Cooper. 2002. In vivo IL-10 production reactivates chronic pulmonary tuberculosis in C57BL/6 mice. J. Immunol. 169:6343-6351. [DOI] [PubMed] [Google Scholar]