Abstract
We hypothesize that Haemophilus influenzae, as a species, possesses a much greater number of genes than that found in any single H. influenzae genome. This supragenome is distributed throughout naturally occurring infectious populations, and new strains arise through autocompetence and autotransformation systems. The effect is that H. influenzae populations can readily adapt to environmental stressors. The supragenome hypothesis predicts that significant differences exist between and among the genomes of individual infectious strains of nontypeable H. influenzae (NTHi). To test this prediction, we obtained 10 low-passage NTHi clinical isolates from the middle ear effusions of patients with chronic otitis media. DNA sequencing was performed with 771 clones chosen at random from a pooled genomic library. Homology searching demonstrated that ∼10% of these clones were novel compared to the H. influenzae Rd KW20 genome, and most of them did not match any DNA sequence in GenBank. Amino acid homology searches using hypothetical translations of the open reading frames revealed homologies to a variety of proteins, including bacterial virulence factors not previously identified in the NTHi isolates. The distribution and expression of 53 of these genes among the 10 strains were determined by PCR- and reverse transcription PCR-based analyses. These unique genes were nonuniformly distributed among the 10 isolates, and transcription of these genes in planktonic cultures was detected in 50% (177 of 352) of the occurrences. All of the novel sequences were transcribed in one or more of the NTHi isolates. Seventeen percent (9 of 53) of the novel genes were identified in all 10 NTHi strains, with each of the remaining 44 being present in only a subset of the strains. These genic distribution analyses were more effective as a strain discrimination tool than either multilocus sequence typing or 23S ribosomal gene typing methods.
Haemophilus influenzae is a small, naturally transformable, gram-negative coccobacillus that is associated etiologically with acute and chronic infections of the upper and lower human respiratory tracts. H. influenzae strains can be divided into two groups, according to the presence or lack of a polysaccharide capsule. The encapsulated strains are typed as a to f, depending on the antigenicity of their capsule, while the unencapsulated strains are referred to as nontypeable (nontypeable H. influenzae; NTHi). H. influenzae enters the body through respiration and establishes either an asymptomatic colonization or a frank infectious process within the host respiratory mucosa. It is estimated that up to 80% of the population are chronic H. influenzae carriers. Infections may be local, contributing to diseases such as otitis media, sinusitis, bronchitis, pneumonia, and chronic obstructive pulmonary disease (COPD), or they can be systemic when bacteria spread through the bloodstream to distant organs, causing other infections, such as meningitis (46, 70). NTHi strains are associated with chronic infections of the respiratory tree, including otitis media with effusion (OME) and COPD, both of which have been hypothesized to be biofilm illnesses (17, 27); however, definitive findings regarding the phenotypic nature of the NTHi strains in natural infections are still forthcoming.
NTHi is the predominant colonizer of the nasopharyngeal mucosa in patients with otitis media (38, 61). Characterization of NTHi populations in the nasopharynx has suggested the possibility that children and adults sometimes harbor multiple distinct strains concurrently (27, 57-59). In other cases, it appears that children will carry a given strain for several weeks to months, lose the first strain, and then acquire a new strain (19, 34). The number of separate times a child is colonized with a strain of NTHi correlates directly with the frequency of H. influenzae-associated otitis media. After an episode of otitis media, immunity against a specific NTHi isolate does not prevent recurrence caused by a different strain (9). Smith-Vaughan et al. (57-59) described simultaneous carriage and horizontal gene transfer among multiple NTHi strains in Australian Aboriginals. They documented numerous cases of horizontal transfer among individual strains within infectious isolates of the gene encoding the major outer membrane protein P2. Similarly, Hiltke et al. demonstrated in situ transfer of the P2 gene between NTHi strains during cocolonization of the respiratory tract of a patient with COPD (27). In another series of studies, by Murphy et al. (49), it was reported that multiple strains of NTHi were present simultaneously in the sputum of 26.3% of adults with COPD. Moreover, the authors suggested that these numbers likely underestimated the true frequency of the presence of multiple strains of NTHi, as they had sampled an average of only 6.3 colonies per isolate. These data, along with numerous specific reports of genic differences among the NTHi strains, support the concept that there is significant heterogeneity among the NTHi strains (12, 39, 51, 54, 65), which we have postulated may be important for chronic persistence in the host (11, 15, 52, 55). Thus, a better understanding of the genomic engine that drives pathogenicity of NTHi is important.
A significant number of genomic sequences identified in clinical strains of NTHi are absent from the genome of H. influenzae Rd KW20 (Rd), a laboratory strain with reduced virulence (22, 23). These sequences include genes encoding virulence-associated autotransporters (14, 48), hemagglutinating pili (66), novel NTHi sequences expressed during interaction with human epithelial cell lines (67), genes that are upregulated in the middle ear in a chinchilla model of acute otitis media (42), and uncharacterized sequences predicted to encode hypothetical or phage-like proteins (47). Such sequences have come to be recognized as components of a set of contingency genes that are available to H. influenzae at a population level and are distributed among the clinical strains; however, no single strain has the full complement of genes present in the population-based supragenome (15, 21, 24).
The ability to take up DNA provides a means for the exchange of these novel sequences among the H. influenzae. Competent H. influenzae preferentially take up DNA from highly related organisms via the recognition of an overrepresented uptake signal sequence (USS) (13, 56). The highly efficient exchange of genes among competent strains is important; e.g., the Rd genome contains 1,465 USSs (56). Evidence that bacteria take up DNA more efficiently while growing as a biofilm (45), accompanied by the preliminary observation that H. influenzae grows as a biofilm in an experimental model of OME (17) supports, but does not prove, the hypothesis that horizontal gene transfer is an important pathogenic process for persistence during the course of a chronic infection. Such an increase in gene transfer would theoretically contribute to additional genetic heterogeneity (allelic differences) and genomic plasticity (genic differences) among the strains in a polyclonal infection, thereby further increasing the fitness of the bacterial population as a whole through the continual generation of novel recombinant strains. These results underscore the importance of elucidating the full repertoire of virulence genes available to NTHi at a population level.
MATERIALS AND METHODS
Isolation of NTHi clinical strains.
Ten H. influenzae strains were obtained as first-plate isolates on chocolate agar from the middle ear effusions of 10 children undergoing myringotomy for the treatment of otitis media at Children's Hospital of Pittsburgh. Each of these strains was streaked for single colonies and then grown in broth for the preparation of aliquots for cryopreservation. The strains, designated PittAA through PittJJ, were all determined to be nontypeable (NTHi) by slide agglutination assay-based typing at the Pittsburgh Public Health Laboratory, with the exception of AA, which was initially reported to be weakly reactive when tested with polyclonal sera for all capsular types, a to f. This specimen was nonreactive with individual anti-A and anti-B sera. This specimen was sent for typing to the New York State Department of Health's Wadsworth Laboratory, where it was determined to be nontypeable. In addition, a PCR-based capsular typing method (20), which has previously been used to resolve discrepancies in slide agglutination serotyping results (36), demonstrated that all 10 strains were nontypeable.
Bacterial culture conditions.
All NTHi strains were cultured in brain heart infusion broth (Becton Dickinson, Sparks, MD) supplemented with 10 μg/ml hemin (Fisher Scientific, Pittsburgh, PA), 2 μg/ml NAD (Sigma, St. Louis, MO), and 20 μg/ml thiamine HCl (Sigma) and grown at 37°C in a humidified 5% CO2 atmosphere. Escherichia coli TOP10 cells were grown in LB broth (Becton Dickinson) supplemented with 50 μg/ml kanamycin, when necessary, at 37°C.
Extraction of bacterial genomic DNA.
Bacterial genomic DNA was extracted using a modification of the method described by Ausubel et al. (2). The DNA was analyzed by UV spectrophotometry and agarose gel electrophoresis.
Construction of a pooled genomic library.
A pooled genomic library DNA was constructed (18) to maximize efficiency and to minimize strain-specific biases which could result from the construction of multiple independent libraries. Individual genomic DNAs from each of the 10 clinical strains were isolated and hydrodynamically sheared to an average fragment length of 1.5 kb using a HydroShear (GeneMachines, San Carlos, CA) according to the manufacturer's instructions. Ten-microgram aliquots of each of the sheared DNAs were pooled, end repaired using T4 and Klenow polymerases (Invitrogen), ligated into the plasmid pCR4Blunt-TOPO, and transformed into E. coli TOP10 cells according to the manufacturer's protocol (Invitrogen) (18). A Q-bot multitasking robot (Genetix Limited, United Kingdom) was used to construct an addressable array containing 76,800 transformants, which were replica plated and stored in 10% glycerol at −80°C. Clones in the library were chosen randomly for further analysis.
DNA sequencing.
Plasmid DNA templates were prepared for sequencing using QIAprep Miniprep kits (QIAGEN, Inc., Valencia, CA) with a Beckman FX robot (Beckman Instruments). Prior to sequencing, plasmid preparations were digested with EcoRI (Invitrogen) and analyzed on ethidium bromide-stained 1% agarose gels in Tris-acetate-EDTA buffer. Only constructs containing inserts larger than 0.5 kb were used as templates. Dideoxy sequencing was performed according to standard protocols for both IR2 Gene ReadIR instruments (LiCor, Inc., Lincoln, Nebraska) and Beckman CEQ 2000 XL automated capillary electrophoresis sequencing instruments (Beckman Coulter, Inc., Fullerton, CA). Additional sequencing was performed using an ABI 3730 DNA analyzer in which 3-μl reactions were prepared, using a Parallab 350-nanoliter genomic workstation (Brooks Automation, Inc., Chelmsford, MA), consisting of (i) 1.4 μl of plasmid template (approximately 100 ng DNA), (ii) 0.5 μl of primer (10 pmol/μl), and (iii) 1.1 μl of a BigDye Terminator v.3.1 cycle sequencing kit (Applied Biosystems, Inc.) (63). Reaction aliquots of 500 nl were then thermal cycled and purified within the Nano-Pipetter of the Parallab. Cycling conditions were 35 cycles with a 0-s denaturation step at 96°C, a 0-s annealing step at 50°C, and a 60°C extension step for 45 sec. The purified samples were then run on the ABI 3730 Analyzer and analyzed using ABI analysis software v.5.0.
DNA sequence analysis.
Sequences were analyzed and contigs formed using Sequencher (v. 4.1.4) (Gene Codes Corporation, Ann Arbor, MI). DNA sequence similarity searches using the BLASTn and BLASTx algorithms (1) were performed using the Center for Genomic Sciences high-speed BLAST cluster (G. Erdos, unpublished results) that is connected to the NCBI website (http://www.ncbi.nlm.nih.gov/). This system, including a custom-designed software package, automatically performs vector trimming, sequence quality checks, and BLAST homology searches, permitting fast and accurate analyses of thousands of clones daily. Details of these programs and the BLASTn and BLASTx searches will be described elsewhere (J. Gladitz et al., unpublished). The novel nucleotide sequences reported in this paper have been deposited with GenBank.
PCR-based sequence distribution studies.
Primers for distribution and expression studies were designed based on the most likely open reading frame (ORF) in each clone or contig. The primer sequences are posted at http://www.centerforgenomicsciences.org (go to Public Documents, Supplementary Table 1). Primers specific for the H. influenzae glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene served as positive controls for all strains. PCR was performed using an Eppendorf MasterTaq kit (Brinkman Instruments, Inc., Westbury, NY) in a 25-μl reaction mixture containing 0.6 units of Taq DNA polymerase, 50 ng of template DNA, 20 pmol of each primer, 1.5 mM MgCl2, and 0.2 mM deoxynucleoside triphosphates. PCR was carried out by using Perkin Elmer 9600 thermal cyclers beginning with a 10-min denaturation step at 95°C and followed by 35 cycles of 30 s at 94°C, 1 min at 55°C, and 1 min at 72°C. Cycling was followed by a final extension step of 7 min at 72°C and then a 4°C hold. Reactions were analyzed on 1.7% ethidium bromide-stained agarose gels.
RT-PCR detection of transcripts encoded by non-Rd sequences.
Gene expression assays were designed for each of the non-Rd clones solely to determine if the novel sequences were part of functional transcriptional units. Total RNA for each of the 10 NTHi isolates was extracted using the hot phenol method (Invitrogen) from bacterial cells grown in supplemented brain heart infusion broth that had reached mid-exponential phase. Samples were treated with Turbo DNase (Ambion, Inc., Austin, TX) and then used as templates for PCR and reverse transcription (RT)-PCR. RNA quality was checked on an Agilent 2100 Bioanalyzer using an RNA 6000 Nano Assay kit (Agilent Technologies, Palo Alto, CA). All RNA preparations were assayed in RT-PCR-based assays, and sham (−RT) PCR-based assays, the latter to test the efficacy of the DNase treatment. For RT-PCR, random hexamers were mixed with 4 μg of total RNA from each strain, heat denatured, chilled on ice, and then divided equally into two tubes. Two master mixes were prepared for the reverse transcription step, one containing all of the RT components, including Moloney murine leukemia virus (Invitrogen) (+RT) and the other one lacking the RT enzyme (−RT). Each pair of RNA specimens received an aliquot of the +RT mixture in one tube and the -RT mixture in the other. Following the RT reaction, 2.5 μl of the first-strand cDNA from each reaction was used as a PCR template. For all strains, GAPDH mRNA was amplified as a positive control. Negative controls consisted of reaction mixtures prepared with no template nucleic acid.
Southern blot analysis.
High-molecular-weight bacterial genomic DNAs were isolated from each of the 10 NTHi strains and Rd as described previously (18), digested to completion with EcoRI, and electrophoresed into a 1% agarose gel. Each gel also contained as a positive control one lane with a mixture of the unique plasmid clones that we were probing for. The DNAs were then transferred to positively charged nylon membranes (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) by capillary blotting according to the method of Southern (60a) using 0.4 M NaOH. Probe production was accomplished by PCR of the plasmid inserts corresponding to the unique genes under study, followed by purification using QIAquick PCR purification kits (QIAGEN). Labeling of the probe consisted of denaturing 25 ng of each amplimer at 95°C for 5 min; quenching on ice; the addition of random primers buffer mixture (Random Primers DNA labeling system; Invitrogen), 6.0 μl of a dATP-dGTP-dTTP mixture, 50 μCi of 32P dCTP, and 3 U of Klenow fragment; and incubation at 25°C for 1 h, after which the reaction was stopped with 5 μl stop buffer. Purification of the radiolabeled probe was performed using gel exclusion chromatography (G-50 Sephadex columns, Roche Diagnostics, Indianapolis, IN), and the specific activity was measured with a QC4000XER counter (Bioscan, Washington, DC). Membranes were prehybridized at 42°C for 30 min in hybridization tubes using formamide prehybridization solution (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 5× Denhardt solution, 50% (wt/vol) formamide, 1% (wt/vol) sodium dodecyl sulfate [SDS]), with the addition of heat-denatured sheared salmon sperm DNA (Sigma) immediately before incubation in the hybridization oven (Biometra, Goettingen, Germany). After prehybridization, 2 × 107 dpm of heat-denatured probe was added, and hybridization was carried out at 42°C overnight. The membranes were washed with 2× SSC-0.1% (wt/vol) SDS for 5 min at room temperature three times, 0.2× SSC-0.1% (wt/vol) SDS for 15 min at room temperature, 0.2× SSC-0.1% (wt/vol) SDS for 15 min at 42°C, and 0.1× SSC-0.1% SDS for 15 min at 68°C, followed by autoradiography.
Multilocus sequence typing and construction of phylogenetic trees.
DNA sequencing was performed as described previously (http://www.mlst.net) for 7 H. influenzae housekeeping genes, adk, atpG, frdB, fucK, mdh, pgi, and recA, from each of the 10 NTHi clinical strains to determine their multilocus sequence types (MLSTs) (44). PCRs for each gene were performed using the published primers in a 96-well microtiter plate format using Perkin Elmer 9600 thermal cyclers. The PCR conditions were as follows: initial denaturation at 95°C for 4 min; 30 cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 60 sec; and 72°C for 10 min followed by a 4°C hold. All long-term sample storage was at −20°C. The PCR products were purified using the QIAquick PCR purification kit (QIAGEN) according to the manufacturer's protocol. DNA fragments were sequenced bidirectionally on the Beckman capillary DNA analysis system and the sequences were analyzed as described above. The alignment of multiple sequences was made using Clustal X. The sequence of each locus in each strain was then compared with the MLST database to obtain an allelic profile. If a locus sequence had no identical allele in the database, the sequence trace files were sent to the MLST curators for a final determination as to whether it represented a new allele. All confirmed new alleles were assigned a unique allele number for that gene locus. The combined allelic profiles of the seven gene loci for each strain resulted in a sequence type (ST) that was then compared with the existing ST profiles in the MLST database. A new ST number was assigned to a strain with no match in the database. Phylogenetic trees were generated using the unweighted-pair group method with arithmetic mean program MEGA v.2.1 software (35). Trees based on the average distance method were generated using the on-line java script available at the MLST website (http://www.mlst.net).
Characterization of 23S ribosomal RNAs.
Total bacterial RNA from each of the NTHi strains was analyzed for 23S fragmentation patterns using an RNA 6000 Nano Assay chip and an Agilent 2100 Bioanalyzer. The sizes of the bands were measured by the software with the aid of an RNA marker ladder. Primers for amplifying fragments containing intervening sequences (IVSs) were used to detect the existence of the IVS1 and IVS2, according to the method of Song et al. (60). The PCR conditions were as follows: 3 min of denaturation at 94°C, followed by 30 cycles of 1 min at 94°C, 1 min at 55°C and 1 min at 72°C, with a final extension at 72°C for 7 min. The PCR products were run on 1% agarose gels.
Nucleotide sequence accession numbers.
The novel nucleotide sequences reported in this paper have been deposited in GenBank under accession numbers AY599423 to AY599486.
RESULTS AND DISCUSSION
DNA sequence comparisons with the Rd genome.
DNA sequencing was performed for 771 clones from a pooled genomic DNA library prepared from 10 NTHi clinical isolates obtained from children with OME (18). All unique sequences (non-Rd) were evaluated for their potential to encode virulence factors and to determine their distribution and expression among the strains that make up the library. The average insert size was 1.5 kb, and the average read length from each end was 650 bases, providing ∼87% coverage from the initial read. The remaining sequence for each clone was determined by primer walking.
Clones that revealed a minimum of 350 bp of contiguous homology (at least 75% nucleotide identity) to Rd at each end and that produced a DNA fragment size in accordance with that predicted by the Rd genome were classified as being Rd-like. This method identified 699 of 771 clones (90.7%) as being Rd-like. Many of these Rd-like clones were demonstrated to contain small insertions and deletions and numerous point mutations, but in general, they represent allelic variations of known genes.
Identification of novel DNA sequences.
BLASTn analyses of 72 of 771 clones (9.3%) indicated that they contained inserts that were unique with respect to the Rd genome. Each of these clones was then compared to all of the other novel clones to identify overlaps. A total of five contigs were constructed from 12 clones. These contigs, together with the 60 nonoverlapping clones, made up the 65 sequences that were analyzed. Twenty-one of the 65 novel sequences contained a contiguous Rd-like sequence at one end, indicating a chromosomal origin (1). The novel sequences that could be anchored with known H. influenzae genes were demonstrated to be distributed evenly around the Rd genome. This distribution of novel sequences throughout the genome suggests that strain evolution in H. influenzae is mostly incremental and is not associated with the acquisition of large pathogenicity islands, such as has been documented for the enteric pathogens (43). It further suggests that the pooled genomic library was unbiased in its coverage. This was a concern because of reports that genes closer to the origin of replication tend to be overrepresented in libraries prepared from rapidly doubling bacteria (23). The remaining 44 sequences shared no nucleotide-level homology with the Rd genome, consistent with a chromosomal insertion size greater than that of the clone, or of episomal or phage origin.
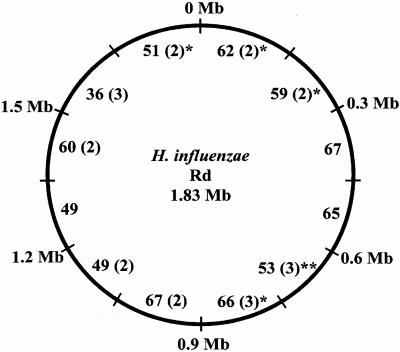
FIG. 1.
Distribution of randomly selected library clones with respect to the H. influenzae Rd KW20 genome. A map of the Rd chromosome was divided into sections, each representing 0.15 Mb. The number of clones demonstrating homology to each Rd section is noted, for a total of 684 Rd-like clones. The locations of 21 clones that contained both Rd and contiguous non-Rd sequences are shown in parentheses. Asterisks represent the positions of the six rRNA loci on the Rd genome (22). The 15 clones that contained rRNA sequence were not included in the figure, as it was not possible to determine which of the six regions was represented in each of these clones.
BLASTx searches were carried out to identify proteins with similarity to the conceptual protein translations of these novel DNA sequences. These novel DNA sequences and their cognate translations have been deposited in GenBank (accession numbers AY599423 to AY599486). Eighteen of the 65 novel sequences are most likely restriction/modification genes (n = 7) or phage genes (n = 11) and are not considered further here. The results of the remaining protein similarity searches produced a number of candidate virulence genes (Table 1).
TABLE 1.
Results of protein similarity searches and RT-PCR analyses for select nonRd H. influenzae sequences
| Clonea | Similar to: | Organism | Database reference | % Identity, % similarity | No. of USSsb | No. strains RT-PCR positivec |
|---|---|---|---|---|---|---|
| 13_I7/135_C22 ORF1 | Chromosome segregation ATPase | H. influenzae R2866 | ZP_00156560 | 90, 94 | 1 | 1 |
| 13_I7/135_C22 ORF2′ | Uncharacterized conserved protein | H. influenzae R2866 | ZP_00156561 | 96, 97 | ||
| 17_D20′ | Hypothetical protein | Burkholderia cepacia R1808 | ZP_00220700 | 47, 65 | 1 | 1 |
| 32_B2′ | Hemoglobin-haptoglobin binding protein (HhuA) | H. influenzae TN106 | AAB36696 | 57, 73 | 3 | |
| 32_F13 ORF1 | ABC-type dipeptide transport system, periplasmic component | H. somnus 2336 | ZP_00204638 | 29, 56 | ||
| 32_F13 ORF2′ | Unknown protein | P. multocida Pm70 | NP_246721 | 38, 58 | 1 | 1 |
| 43_I10′ | Putative glucosidase | Yersinia pestis C092 | NP_404469 | 73, 84 | 6 | |
| 55_M14′ | Predicted hydrolase (metallo-beta-lactamase superfamily) | H. influenzae R2846 | ZP_00155137 | 30, 48 | 2 | |
| 59_C2 ORF1 | Predicted transcriptional regulator | H. somnus 129Pt | ZP_00122724 | 82, 88 | 1 | |
| 59_C2 ORF2 | Hypothetical protein | H. somnus 129Pt | ZP_00123422 | 43, 56 | 1 | |
| 67_D11 ORF1′ | Hypothetical protein | H. influenzae R2866 | ZP_00156558 | 99, 100 | ||
| 67_D11 ORF2 | Hypothetical protein | H. influenzae R2866 | ZP_00156559 | 97, 97 | 4 | |
| 90_A18 ORF1′ | Soluble lytic murein transglycosylase | H. influenzae R2866 | ZP_00157614 | 100, 100 | 1 | 2 |
| 90_A18 ORF2′ | Hypothetical protein | H. influenzae R2866 | ZP_00157615 | 100, 100 | ||
| 93_G12/117_A22 ORF1′ | Hypothetical protein | H. somnus 129Pt | ZP_00122695 | 75, 87 | ||
| 93_G12/117_A22 ORF2 | Hypothetical protein | H. somnus 129Pt | ZP_00122696 | 26, 47 | 1 | 2 |
| 97_H3′ | 2-Methylthioadenine synthetase | H. influenzae R2866 | ZP_00156447 | 98, 99 | 1 | 3 |
| 101_H6 ORF1′ | DNA repair protein | H. influenzae R2866 | ZP_00157610 | 96, 98 | ||
| 101_H6 ORF2 | Malate/l-lactate dehydrogenase | H. influenzae R2866 | ZP_00157611 | 95, 97 | 3 | |
| 101_H6 ORF3 | Permease (major facilitator superfamily) | H. influenzae R2866 | ZP_00203230 | 97, 98 | ||
| 101_H6 ORF4′ | Hypothetical protein | H. influenzae R2866 | ZP_00157612 | 98, 98 | 3 | |
| 104_E15′ | Hypothetical protein | H. somnus 129Pt | ZP_00122672 | 61, 76 | 5 | |
| 110_E11 ORF1′ | Hypothetical protein | Haemophilus ducreyi 35000HP | NP_873930 | 68, 81 | 1 | |
| 110_E11 ORF2′ | Hypothetical protein | H. ducreyi 35000HP | NP_873929 | 76, 86 | 1 | |
| 112_A12 ORF1′ | ADP-heptose: lipopolysaccharide heptosyltransferase | H. influenzae R2866 | ZP_00156725 | 100, 100 | 1 | 3 |
| 112_A12 ORF2′ | Formamidopyrimidine-DNA glycosylase | H. influenzae R2866 | ZP_00156724 | 100, 100 | ||
| 120_O6 ORF1′ | Hypothetical protein | Actinobacillus pleuropneumoniae 4074 | ZP_00135133 | 73, 82 | 2 | 4 |
| 121_L20 | S-adenosylmethionine-dependent methyltransferase | Nostoc punctiforme | ZP_00110044 | 39, 54 | 5 | |
| 122_N17 ORF1′ | ATPase (AAA+ class) | H. influenzae R2866 | ZP_00157321 | 99, 99 | 1 | 9 |
| 122_N17 ORF2 | Leucyl-tRNA synthetase | H. influenzae R2846 | ZP_00155057 | 100, 100 | ||
| 125_L2 ORF1′ | Putative transcriptional regulator (LysR family) | Shigella flexneri 2a | AAL08432 | 68, 77 | ||
| 125_L2 ORF2 | Unknown protein (putative aspartate racemase) | S. flexneri 2a | AAL08433 | 58, 77 | 2 | |
| 125_L2 ORF3′ | Putative anaerobic decarboxylate transporter | S. flexneri 2a | AAL08434 | 71, 83 | ||
| 126_N4 ORF1′ | Predicted ATPase (AAA + superfamily) | H. influenzae R2866 | ZP_00156451 | 86, 88 | 2 | 4 |
| 126_N4 ORF2′ | Hypothetical protein | Enterococcus faecalis V583 | NP_815563 | 43, 59 | ||
| 128_C1 | Hypothetical protein (Lin 1719) | Listeria innocua | NP_471055 | 25, 44 | 1 | 2 |
| 132_O3 ORF1′ | Conserved hypothetical protein | Chromobacterium violaceum ATCC 12472 | NP_901809 | 52, 71 | 1 | 1 |
| 132_O3 ORF2′ | Probable portal protein | C. violaceum ATCC 12472 | NP_901810 | 59, 76 | ||
| 134_O6 | Formamidopyrimidine-DNA glycosylase | H. influenzae R2866 | ZP_00156724 | 98, 99 | 1 | 4 |
| 135_110′ | Las (autotransporter protein) | H. influenzae biotype aegyptius | AAK76426 | 53, 63 | 1 | 3 |
| 151_B3′ | AhpA | P. multocida VP161 | AAF71219 | 43, 64 | 1 | 2 |
| 151_O4′ | YhbX/YhjW/YijP/YjdB family protein | N. meningitidis MC58 | NP_275002 | 48, 69 | 2 | |
| 159_B20 ORF1 | Conserved hypothetical protein (HD0114) | H. ducreyi 35000HP | NP_872728 | 74, 81 | 1 | |
| 159_B20 ORF2′ | Conserved hypothetical protein (HD0115) | H. ducreyi 35000HP | NP_872729 | 40, 69 | 1 | |
| 166_G6 ORF1′ | Soluble lytic murein transglycosylase | H. influenzae R2866 | ZP_00157614 | 94, 97 | ||
| 166_G6 ORF2 | Hypothetical protein Hflu2121901 | H. influenzae R2866 | ZP_00157615 | 98, 99 | ||
| 166_G6 ORF3′ | Type IV secretory pathway, VirD4 components | H. influenzae R2866 | ZP_00157616 | 99, 100 | 3 | |
| 167_A16 ORF1′ | Predicted hydrolase (metallo-beta-lactamase superfamily) | H. influenzae R2846 | ZP_00155137 | 100, 100 | ||
| 167_A16 ORF2 | TPR repeat, SEL-1 subfamily protein | H. influenzae R2846 | ZP_00155136 | 100, 100 | 1 | 2 |
| 167_A16 ORF3′ | TPR repeat, SEL-1 subfamily protein | Psychrobacter 273-4 | ZP_00146612 | 26, 42 | ||
| 168_P21 ORF1 | Conserved hypothetical protein | C. violaceum ATCC 12472 | NP_901800 | 33, 55 | 1 | 4 |
| 168_P21 ORF2′ | Hypothetical protein | Photorhabdus luminescens subsp. Laumondii TTO1 | NP_930650 | 54, 72 | ||
| 173_G10′ | Hypothetical protein | H. influenzae R2866 | ZP_00156458 | 98, 98 | 5 | |
| 179_D14′ | Flp pilus assembly protein, ATPase CpaF | A. vinelandii | ZP_00091413 | 91, 93 | 1 | |
| hgp_contig′ | Hemoglobin binding protein A | A. actinomycetemcomitans | AAK49998 | 49, 67 | 3 | |
| GI_contig | Glucuronate isomerase | H. somnus 129Pt | ZP_00123198 | 78, 89 | 9 |
Partial ORFs are designated with a prime.
Number of USSs (Haemophilus uptake signal sequences) per ORF. Additional USSs were identified in the above clones in Rd-like sequences (data not shown).
Indicates the number of H. influenzae clinical isolates (out of the 10 isolates used to construct the library) that tested positive for a transcript using primers specific to the ORF in RT-PCR analysis. When no number is given, RT-PCR analysis was not performed. None of the transcripts were detected in Rd.
Distribution of USSs among the non-Rd clones.
The H. influenzae USS is a conserved 9-bp core sequence within a 29-bp consensus sequence (5′-aAAGTGCGGTnRW5n6RW5-3′, where boldface indicates invariants and lowercase indicates consensus sequences). DNA containing this sequence is preferentially taken up by competent H. influenzae (13, 25, 56). We identified 36 USSs distributed among 30 of the 65 novel sequences, suggesting that this subset of novel sequences has been in the NTHi gene pool for an extended time. The actual number of USSs among these novel loci is likely greater than 36, since the majority of the clones contain only partial ORFs. Table 1 includes the occurrences of USSs among the novel ORFs that are presented in more detail below.
Interspecies exchange of these genes may also occur, given that the H. influenzae USS is also overrepresented in the genomes of other members of the Pasteurellaceae, including Haemophilus somnus, Pasteurella multocida, and Actinobacillus actinomycetemcomitans (3, 68). Moreover, DNA transfer between the upper respiratory pathogens H. influenzae and Neisseria meningitidis has been previously documented (14, 33). Although USS-mediated DNA uptake was not implicated in the exchanges with Neisseria, transformation with heterologous DNA has been suggested to play an important role in establishing chromosomal mosaicism in these organisms (33). Based on these findings, it seems plausible that the novel H. influenzae sequences identified herein, particularly those that harbor a USS, could be readily transferred not only to other strains of H. influenzae but also to other genera of the Pasteurellaceae.
Incidence of H. influenzae virulence genes among the NTHi strains that are absent from Rd.
Twelve of 65 (18%) of the non-Rd sequences showed amino acid (aa) homology to proteins from the hif, hmw, tna, and lex2 operons that have been previously identified for other H. influenzae strains but are absent from Rd. Gene distribution and expression assays were performed among the clinical isolates for these four gene clusters (Table 2). Three clones (0004_E21, 0133_D06, and 0152_N02) contained genes from the hemagglutinating pilus (hif) gene cluster. We detected transcripts for these hif genes in 2 of the 10 isolates (Table 2) and a hicB transcript in 7 isolates, including the 2 isolates that expressed the hif genes. Three clones (0120_C11, 0036_E20, and 0170_J08) displayed aa homology to the hmw gene cluster that encodes the high-molecular-weight adhesions HMW1 and HMW2 (4, 5, 62). Two clones (0009_E14 and 0013_D09) contained genes from the tryptophanase operon (tnaABC), and 9 of 10 NTHi isolates tested positive for a tnaB expression and indole production, which has been associated with virulence (32, 41). Finally, three clones (0083_M12, 0047_C18, and 0093_M17) contained genes from the lex2AB locus, which controls virulence through the variable expression of lipooligosaccharide epitopes (10, 30). These genes possessed tetranucleotide repeats associated with phase variation.
TABLE 2.
Distribution of Non-Rd DNA sequences and RNA transcripts among NTHi clinical strains via PCR and RT-PCR
| Name of clone and contig | Data set | Product length (bp) |
H. influenzae strain
|
No. of strains with non-Rd sequences | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PittAA | PittBB | PittCC | PittDD | PittEE | PittFF | PittGG | PittHH | PittII | PittJJ | Rd KW20 | ||||
| 0004_E21 (hif) | PCR | 257 | − | + | − | − | − | − | − | + | − | + | − | 3 |
| RT-PCR | 257 | − | + | − | − | − | − | − | + | − | − | − | 2 | |
| 0009_E14 (tna) | PCR | 175 | + | + | + | + | + | + | + | + | + | + | − | 10 |
| RT-PCR | 175 | + | + | + | + | + | + | + | + | + | + | − | 9 | |
| 0013_107 | PCR | 183 | + | − | − | + | − | + | − | − | − | − | − | 3 |
| RT-PCR | 183 | − | − | − | + | − | − | − | − | − | − | − | 1 | |
| 0017_D20 | PCR | 359 | + | + | + | + | + | + | − | − | − | − | − | 6 |
| RT-PCR | 359 | + | − | − | − | − | − | − | − | − | − | − | 1 | |
| 0032_B02 | PCR | 130 | + | − | − | + | − | − | − | + | − | + | − | 4 |
| RT-PCR | 130 | − | − | − | + | − | − | − | + | − | + | − | 3 | |
| 0032_F13 | PCR | 208 | + | − | − | − | + | + | + | − | + | − | − | 5 |
| RT-PCR | 208 | − | − | − | − | − | − | + | − | − | − | − | 1 | |
| 0036_E20 | PCR | 435 | + | − | − | + | + | + | + | + | + | − | − | 7 |
| RT-PCR | 435 | + | − | − | − | + | + | − | − | − | − | − | 3 | |
| 0043_110 | PCR | 368 | + | + | + | + | + | + | − | + | + | + | − | 9 |
| RT-PCR | 368 | + | + | + | − | − | + | − | + | − | + | − | 6 | |
| 0047_C03 | PCR | 182 | + | + | + | + | + | + | + | − | + | − | − | 8 |
| RT-PCR | 182 | + | + | − | − | − | − | + | − | + | − | − | 4 | |
| 0049_H03 | PCR | 127 | + | + | + | + | + | + | + | + | + | + | − | 10 |
| RT-PCR | 127 | + | + | + | + | + | + | + | + | − | + | − | 9 | |
| 0056_M14 | PCR | 110 | − | − | − | − | − | − | − | + | − | + | − | 2 |
| RT-PCR | 110 | − | − | − | − | − | − | − | + | − | + | − | 2 | |
| 0059_C02 | PCR | 235 | + | − | − | − | − | + | − | − | − | − | − | 2 |
| RT-PCR | 235 | + | − | − | − | − | − | − | − | − | − | − | 1 | |
| 0090_A18 | PCR | 329 | + | − | + | + | − | + | − | − | + | − | − | 5 |
| RT-PCR | 329 | − | − | + | − | − | + | − | − | − | − | − | 2 | |
| 0093_G12 | PCR | 344 | + | − | − | + | + | − | − | − | − | − | − | 3 |
| RT-PCR | 344 | − | − | − | + | + | − | − | − | − | − | − | 2 | |
| 0096_C16 | PCR | 318 | + | + | + | + | + | + | + | + | + | + | − | 10 |
| RT-PCR | 318 | − | − | + | − | − | + | − | + | − | + | − | 4 | |
| 0097_H03 | PCR | 206 | + | − | + | + | − | + | + | + | + | + | − | 8 |
| RT-PCR | 206 | + | − | − | + | − | − | + | − | − | − | − | 3 | |
| 0101_H06 | PCR | 479 | + | − | + | + | − | + | + | − | + | − | − | 6 |
| RT-PCR | 479 | − | − | + | − | − | + | − | − | + | − | − | 3 | |
| 0104_E15 | PCR | 326 | + | + | + | + | − | + | − | − | + | − | − | 6 |
| RT-PCR | 326 | + | + | + | − | − | + | − | − | + | − | − | 5 | |
| 0110_E11 | PCR | 321 | + | + | + | + | − | − | − | + | − | − | − | 5 |
| RT-PCR | 321 | + | − | − | − | − | − | − | − | − | − | − | 1 | |
| 0112_A12 | PCR | 480 | + | + | + | + | + | − | + | + | + | + | − | 9 |
| RT-PCR | 480 | − | − | − | − | − | − | − | + | + | + | − | 3 | |
| 0120_C11 (hmw) | PCR | 372 | + | − | − | + | + | + | + | + | + | − | − | 7 |
| RT-PCR | 372 | + | − | − | + | + | + | − | − | − | − | − | 4 | |
| 0120_O06 | PCR | 477 | + | + | + | + | + | + | + | + | + | − | − | 9 |
| RT-PCR | 477 | + | + | − | − | − | − | + | − | + | − | − | 4 | |
| 0121_J07 | PCR | 342 | − | − | − | − | − | − | − | + | + | + | − | 3 |
| RT-PCR | 342 | − | − | − | − | − | − | − | + | − | + | − | 2 | |
| 0121_L20 | PCR | 201 | + | + | + | + | + | + | + | + | + | − | − | 9 |
| RT-PCR | 201 | + | + | − | + | + | + | − | − | − | − | − | 5 | |
| 0122_N17 | PCR | 424 | + | + | + | + | + | + | + | + | + | + | − | 10 |
| RT-PCR | 424 | + | − | + | + | + | + | + | + | + | + | − | 9 | |
| 0124_K2 | PCR | 859 | + | + | + | + | + | + | + | + | + | − | − | 9 |
| RT-PCR | 859 | + | − | + | + | + | + | + | − | + | − | − | 7 | |
| 0125_L02 | PCR | 217 | + | + | + | + | − | + | − | − | − | − | − | 5 |
| RT-PCR | 217 | − | − | + | − | − | + | − | − | − | − | − | 2 | |
| 0126_N04 | PCR | 755 | + | − | − | − | − | − | − | + | + | + | − | 4 |
| RT-PCR | 755 | + | − | − | − | − | − | − | + | + | + | − | 4 | |
| 0128_C01 | PCR | 232 | + | + | + | + | + | + | + | + | + | + | − | 10 |
| RT-PCR | 232 | − | − | − | − | − | + | + | − | − | − | − | 2 | |
| 0132_O03 | PCR | 352 | + | + | + | + | + | + | − | − | + | − | − | 7 |
| RT-PCR | 352 | + | − | − | − | − | − | − | − | − | − | − | 1 | |
| 0133_D06 | PCR | 326 | + | + | + | − | − | − | − | − | − | − | − | 3 |
| RT-PCR | 326 | + | − | − | − | − | − | − | − | − | − | − | 1 | |
| 0134_B22/0134_112 contig | PCR | 314 | + | + | + | + | + | − | − | − | − | − | − | 5 |
| RT-PCR | 314 | − | + | − | − | − | − | − | − | − | − | − | 1 | |
| 0134_O06 | PCR | 96 | + | − | − | − | − | − | − | + | + | + | − | 4 |
| RT-PCR | 96 | + | − | − | − | − | − | − | + | + | + | − | 4 | |
| 0135_110 | PCR | 192 | + | − | − | − | + | + | + | + | + | − | − | 6 |
| RT-PCR | 192 | − | − | − | − | − | + | + | − | + | − | − | 3 | |
| 0141_D07 | PCR | 447 | + | + | − | − | + | + | + | + | + | − | − | 7 |
| RT-PCR | 447 | − | − | − | − | − | − | + | − | + | − | − | 2 | |
| o0151_O04 | PCR | 164 | + | − | − | + | + | − | − | − | − | − | − | 3 |
| RT-PCR | 164 | − | − | − | + | + | − | − | − | − | − | − | 2 | |
| 0151_B03 | PCR | 168 | + | − | − | − | − | − | − | + | + | + | − | 4 |
| RT-PCR | 168 | − | − | − | − | − | − | − | + | − | + | − | 2 | |
| 0153_I16 | PCR | 293 | + | + | + | + | + | − | + | + | + | + | − | 9 |
| RT-PCR | 293 | − | − | − | + | − | − | − | + | + | + | − | 4 | |
| 0157_C17 | PCR | 484 | + | − | + | + | + | + | + | − | + | − | − | 7 |
| RT-PCR | 484 | + | − | + | + | + | + | + | − | + | − | − | 7 | |
| 0159_B20 | PCR | 186 | + | + | + | + | + | + | − | − | − | − | − | 6 |
| RT-PCR | 186 | + | − | − | − | − | − | − | − | − | − | − | 1 | |
| 0160_E24 | PCR | 158 | + | + | + | + | + | − | − | − | + | − | − | 6 |
| RT-PCR | 158 | + | + | − | − | − | − | − | − | + | − | − | 3 | |
| 0164_J22 | PCR | 121 | + | + | + | + | + | + | + | + | + | + | − | 10 |
| RT-PCR | 121 | + | + | − | + | + | − | − | + | + | + | − | 7 | |
| 0166_G06 | PCR | 369 | + | + | + | + | + | + | + | + | + | + | − | 10 |
| RT-PCR | 369 | − | − | + | − | − | + | − | − | + | − | − | 3 | |
| 0167_A16 | PCR | 857 | + | + | − | + | + | + | + | + | − | − | − | 7 |
| RT-PCR | 857 | − | − | − | − | + | + | − | − | − | − | − | 2 | |
| 0168_P21 | PCR | 278 | + | + | + | + | + | − | + | + | + | + | − | 9 |
| RT-PCR | 278 | + | + | − | − | − | − | − | + | − | + | − | 4 | |
| 0170_J08 (hmw) | PCR | 426 | + | + | + | + | + | + | + | + | + | + | − | 10 |
| RT-PCR | 426 | + | − | − | + | + | + | − | − | − | − | − | 4 | |
| 0173_G10 | PCR | 387 | + | + | + | + | + | + | + | + | + | − | − | 9 |
| RT-PCR | 387 | − | − | − | + | + | + | + | − | + | − | − | 5 | |
| 0179_D14 | PCR | 199 | + | + | + | + | + | + | + | + | + | + | − | 10 |
| RT-PCR | 199 | − | − | − | − | − | − | + | − | − | − | − | 1 | |
| 0183_E08 | PCR | 331 | + | − | − | + | + | + | + | + | + | − | − | 7 |
| RT-PCR | 331 | − | − | − | − | − | + | + | − | + | − | − | 3 | |
| 0197_N01 | PCR | 603 | + | + | + | + | + | − | − | − | − | + | − | 6 |
| RT-PCR | 603 | − | + | − | − | − | − | − | − | − | − | − | 1 | |
| 0023_P24 | PCR | 253 | + | − | + | + | + | + | + | + | + | − | − | 8 |
| RT-PCR | 253 | − | − | − | − | − | + | + | − | − | − | − | 2 | |
| 0067_D11 | PCR | 455 | + | + | − | + | + | − | − | + | + | + | − | 7 |
| RT-PCR | 455 | + | + | − | + | + | − | − | + | + | + | − | 7 | |
| HgpA contig | PCR | 974 | + | − | − | + | − | − | − | + | + | + | − | 5 |
| RT-PCR | 974 | − | − | − | + | − | − | − | + | − | + | − | 3 | |
| No. of non-Rd DNA sequences | 50 | 32 | 33 | 42 | 36 | 34 | 28 | 35 | 38 | 24 | 0 | 352 | ||
| No. of non-Rd RNA transcripts | 26 | 14 | 12 | 18 | 14 | 22 | 16 | 18 | 20 | 17 | 0 | 177 | ||
| No. of non-Rd DNA sequences found exclusively in this strain | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| No. of non-Rd RNA transcripts found exclusively in this strain | 6 | 2 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 11 | ||
Novel HGBPs.
We identified three novel hemoglobin binding protein (HGBP) genes that displayed high degrees of homology over short distances to known HGBP genes that alternated with regions that contained no identifiable homology. This mosaic pattern of gene evolution is likely reflective of the fact that heme acquisition is vital for NTHi, but that heme acquisition proteins are in continual contact with the host immune response, which puts enormous selective pressure on them. Thus, novel genes arise through domain swapping, making it difficult to establish evolutionary relationships among them. One of these genes (GenBank accession number AY599483) contained a conserved TonB-dependent receptor domain and was most similar to an HGBP gene of A. actinomycetemcomitans (26).
Novel autotransporter protein.
GenBank accession number AY599439 contains a USS and is predicted to encode a class of autotransporter (Las) that mediates its own outer membrane secretion (6, 14, 37). It is interesting that the only homology between this clone and other members of this family resides in the C-terminal region of the protein (96% aa identity), which is responsible for secretion, whereas the upstream sequence was completely unique and had a lower G+C content (35%) than the Las-like domain, indicating chimerism. Davis et al. and Loveless and Sair also observed that Las homologs often contain variant passenger domains acquired by lateral gene transfer that are linked to the conserved β domain (14).
Novel YijP family protein.
GenBank accession number AY599442 contains the 3′ end of the purD allele, and a unique gene with seven GTTT tandem repeats located 5′ to the novel ORF. The tetranucleotide repeats indicate that this gene is likely phase variable through slip strand mispairing during replication. A USS in both purD alleles was identified at the point where identity ends, suggestive of a recombination event. We identified a consensus −10 promoter sequence and a potential PUR box (5′-AgGgcAACGTTTaCGa-3′) 58 nucleotides upstream of the start codon for the novel ORF, suggesting that this gene is also under control of the PUR repressor. The most significant match (47% identity) was with the YhbX protein of N. meningitidis (64), which is a member of a family of proteins predicted to be membrane-associated metal-dependent hydrolases (43) that are associated in E. coli with the ability to penetrate the blood-brain barrier of newborn rats.
Clonal similarity to a Shigella flexneri pathogenicity island.
Clone 0125_L02 contained two regions, each of ∼75 bp, with 83% nucleotide homology to the Shigella resistance locus-pathogenicity island (SRL PAI). The SRL PAI carries genes for antibiotic resistance, iron uptake, and at least 22 prophage-related ORFs (40). The conceptual translation of our clone revealed three proteins that showed limited homologies to the proteins of ORFs 7, 8, and 9 of the SRL PAI, respectively, including a LysR-like transcriptional regulator, an aspartate racemase, and an anaerobic decarboxylate transporter (40). It is likely that this group of genes, which is present in only 50% of the NTHi clinical strains, was only recently transferred as a group into H. influenzae. The G+C content (33%) of these genes is much lower than either H. influenzae (38%) or Shigella (45%).
TPR proteins.
Clone 0167_A16 contained a >2.5-kb insert with two short regions of nucleotide homology to Rd of 96% and 90% and three ORFs. The recently available unfinished sequence for NTHi strain R2846 (A. L. Erwin, A. Smith, M. Kibukawa, Y. Zhou, R. K. Kaul, and M. V. Olson, GenBank accession number NZ_AADO00000000) revealed the presence of a virtually identical locus. However, the start codon that we identified for ORF 2 is 465 nucleotides upstream of the start codon predicted for the corresponding R2846 gene and downstream of a consensus RBS. Our start site prediction was supported by RT-PCR-based analyses. The three ORFs include a putative 260-aa protein product predicted to be a member of the metallo-beta-lactamase superfamily and two others that both encode proteins containing tetratricopeptide repeat (TPR) domains of the SEL-1 family of proteins. TPRs occur in a wide array of organisms, including bacteria, fungi, plants, and humans, where they participate as scaffolds for the mediation of protein-protein interactions associated with numerous cellular processes, including cell-cycle control, transcription, chaperone assistance and protein transport, and DNA uptake and recombination (8). These three gene products appeared to be coordinately expressed, based on RT-PCR studies.
Putative Flp pilus assembly protein (ATPase CpaF).
Clone 0179_D14 showed 88% nucleotide homology to an Azotobacter vinelandii gene and to a gene of the plant pathogen Ralstonia solanacearum. The deduced aa sequence was 91% identical to the A. vinelandii Flp pilus assembly protein CpaF and 92% identical to the R. solanacearum conjugal transfer protein TrbB. CpaF and TrbB belong to a superfamily of proteins that spans both the archaea and the bacteria and that are involved in the formation of surface-associated protein complexes that mediate diverse processes, such as pilus biosynthesis, DNA transport, and the secretion of virulence factors, including the type II and IV secretion system NTPases (28, 31, 53, 69). The high degree of homology between the 0179_D14 sequence and the Azotobacter and Ralstonia genes, combined with its very high G+C content (68%) suggests that this gene has entered the H. influenzae supragenome relatively recently from a genus outside the Pasteurellaceae. This gene contains multiple tandem pentanucleotide (CCCGG) repeats, which in the present clone interrupt the reading frame. We are confident that this is not due to a sequencing error and propose that this repeat region plays a role in regulation through slip strand mispairing at the translational level, because our RT-PCR results identified transcript sequences extending 451 bp 3′ of the premature stop codon. We speculate that this gene encodes part of a virulence secretion system responsible for the transport of macromolecules. It is noteworthy that NTHi strain PittGG was the only isolate to express both this sequence and the above-described autotransporter sequence, as PittGG has been demonstrated to be the most virulent of the 10 NTHi strains in a blinded in vivo pathogenicity study (G. D. Ehrlich et al., unpublished).
We identified an Hib-like sequence (0162_D23) that displayed 96% nucleotide homology to the genetic island (HiGI7) of the Eagan strain (7). The clone included the 3′ end of holA, suggesting that this genetic island was inserted into the tRNAIle gene found between holA and glyS, as in the Eagan strain. A notable difference between the Eagan sequence and ours is that ours contains seven repeats of an 11-bp sequence (GGAATTATTTG) that occurs only once in the Hib strain. Although HiGI7 was previously thought to be specific to type b strains, it is also present in strain R2846.
Distribution pattern of novel DNA sequences.
The distribution and expression patterns for 53 of the novel DNA sequences among the 10 NTHi clinical strains were studied using PCR- and RT-PCR-based assays (Table 2). The restriction/modification and phage clones were not evaluated. In all cases, genomic DNA and RNA from Rd were used as a negative control. The GAPDH gene served as a positive control for both the DNA and RNA assays. All assays were performed in triplicate, and a positive call required at least two out of three results to be positive. One possible source of error with regard to the PCR-based analyses for distribution of the novel sequences is that different strains may possess different alleles of the same gene, thus preventing amplification with the primers designed from the sequenced clone. Thus, we performed Southern blotting for 7 of the 53 clones to confirm the PCR results. Overall the results of the Southern blots corroborated the PCR results; however, there were a few minor differences (online Supplementary Table 2).
Three hundred fifty-two of 530 (66%) DNA assays were positive, with at least two strains harboring each of the 53 novel sequences, indicating that all of the unique clones corresponded to sequences actually present in the NTHi strains under study. It is possible that a larger survey would have identified genes that are present in only a single strain, as our survey covered only ∼1% of the library. Nine (17%) of the novel sequences were present in all 10 strains; the PittAA strain had the most novel sequences (50 of 53), and the PittJJ had the least (24 of 50). The average number of novel sequences across the 10 strains was 35, and the majority of the sequences (35 of 53) were found in the majority (6 or more) of the strains.
In approximately 50% (177 of 352) of the genic occurrences, RNA transcripts were also detected; RNA expression was observed for at least one of the strains harboring the gene for each of the novel sequences under study, indicating that all of these unique genes are transcriptionally active. In contrast to the DNA results, only 10 of 53 (19%) of the genes were expressed in a majority of the isolates under in vitro planktonic conditions, and no single novel transcript was found to be expressed in all of the strains. Only five clones gave positive RNA results for each genic occurrence. As with the DNA results, the PittAA strain demonstrated the highest rate of expression of these novel sequences (26 of 51), the strain expressing the least of its novel genes was PittCC (12 of 33). Expression was limited to one of the strains for 11 of the genes (online Supplementary Table 3).
Pairwise comparisons of the PCR results were performed for the 53 novel DNA sequences among all 10 strains to evaluate the overall level of genomic diversity (online Supplementary Table 4). The greatest degree of difference was observed between strains PittFF and PittJJ, which varied on 36 sequences. The most closely related strains using this metric were PittAA and PittDD which differed at only eight loci; interestingly, in an independent test of pathogenicity, these two strains were also found to be very similar (Ehrlich et al., unpublished). Overall, PittJJ was found to have the greatest number of differences (240) compared to all other strains, while PittEE had the fewest differences (152). A similar comparison was made using the RT-PCR results (online Supplementary Table 5). These distribution studies demonstrate that the novel genes were not uniformly distributed among the 10 clinical strains, nor were they universally expressed under in vitro planktonic growth conditions. The latter observation is not surprising, as many of these genes are predicted to encode virulence factors that would not necessarily be constitutively expressed under laboratory conditions. The fact that all sequences were found to be expressed in at least one of the strains argues that the novel sequences are functional genes and do not represent junk DNA.
Our investigations were designed to test the hypothesis that among natural infecting populations of H. influenzae, there exists a substantial number of genes that are not represented in the genome of the laboratory strain Rd and that no two clinical strains (obtained from different patients) would be found to be identical in terms of their genic content. Our finding that ∼10% of the clones examined were novel with respect to Rd is in concordance with those of Davis et al. (2001). The most parsimonious explanation for the genomic plasticity observed among the 10 NTHi strains is horizontal gene transfer. The consequence of frequent horizontal gene transfers is that the set of genes (not alleles) that any given strain or isolate contains is unique with respect to all other strains. Our observations support the concept that a bacterial species is defined by a minimal gene set possessed by all organisms within that species (29, 50) and that each strain within that species has a unique distribution of contingency genes derived from a population-based supragenome, the latter of which is much larger than the genome of any single organism (11, 21, 55).
MLST and construction of phylogenetic trees.
We applied the MLST method, which is used to classify strains of a bacterial species based on the degree of genetic similarity among a set of housekeeping genes, to our 10 NTHi strains. The sequences of seven housekeeping loci (adk, atpG, frdB, fucK, mdh, pgi, recA) were obtained and used as described previously (44). All sequences were compared with the MLST database (http://www.mlst.net), and new numbers were assigned to unique alleles and STs (online Supplementary Table 6). PittBB was given new allele numbers for adk (allele 40), and frdB (allele 38); PittHH and PittJJ received the same new fucK allele (allele 31); and PittII received a new fucK allele (allele 32). New STs were assigned to PittHH and PittJJ (ST112), PittBB (ST113), and PittII (ST187). Strains PittFF and PittGG were both ST43. Unweighted-pair group method with arithmetic mean-based phylogenetic trees were generated based on each of the individual housekeeping genes, using all of the loci concatenated together. The seven trees based on the individual genes showed that except for the two strain pairs (PittFF/PittGG and PittHH/PittJJ), there was no common set of relationships among the strains; thus, a completely unique tree was constructed for each gene examined (online Supplementary Fig. 1). An average distance phylogenetic tree was generated using the online Java program at http://www.mlst.net. This tree shows the genetic distances, using the MLST data among all 187 known H. influenzae sequence types compared with the locations of our strains as indicated in Fig. 2. Our MLST study suggests that the STs are not, in fact, useful as NTHi haplotypes, because the phylogenetic trees built using the seven genes and our 10 clinical strains each produced a completely different branching pattern. This heterogeneity among the trees is supportive of the concept that the primary means of H. influenzae strain evolution is horizontal gene transfer. We did not observe a correlation between MLST typing and our novel gene distribution data. For example, strains PittGG and PittFF are both ST112 (suggesting they are closely related); however, we observed 12 differences in the distribution patterns of the novel genes and 18 differences in their expression of the novel genes in vitro. Moreover, the pathogenic character of these two strains is vastly different as assessed in a series of blinded OME animal model experiments in which a quantitative trait approach was used to characterize virulence. Therefore, MLST would appear to be of limited value in estimating the phylogenic relationships among strains and for estimating strain virulence.
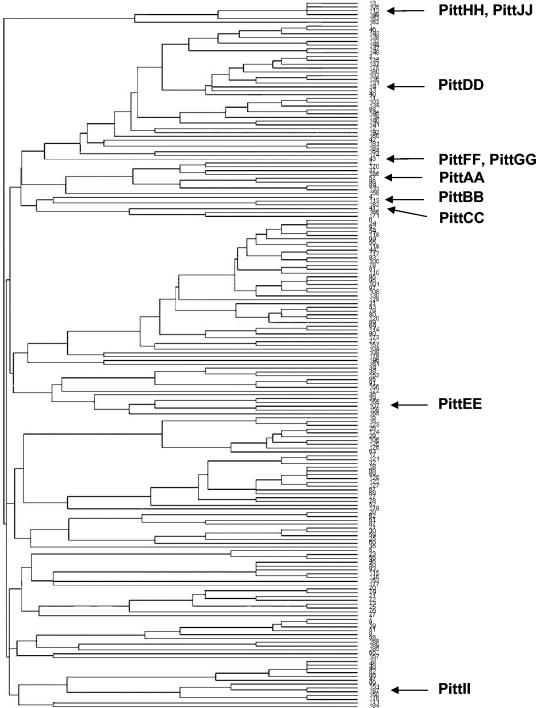
FIG. 2.
Phylogenetic tree generated using sequences from all 187 STs listed in the MLST database and showing the relative distances among the 10 clinical strains of H. influenzae (indicated to the right) characterized in the present study.
23S rRNA fragmentation typing.
The intact 2.9-kb 23S rRNA molecule was observed only in Rd; all 10 of the NTHi clinical strains demonstrated fragmented 23S rRNA patterns (online Supplementary Table 7). Seven of our strains demonstrated the presence of two intervening sequences (IVS1 and IVS2) in both copies of their 23S rDNA. The other strains showed heterogeneous cleavage patterns for the two 23S genes; strains PittAA and PittII had one 23S rRNA gene that contained both IVS1 and IVS2 and one that contained only IVS2. Strain PittEE had one 23S rRNA gene with both IVS1 and IVS2 and one with IVS1 only. It is clear that this methodology has insufficient discriminatory power to accurately type the NTHi strains.
The distributed genome hypothesis states that there exists a population-based supragenome for pathogenic bacteria, which is made up of a set of contingency genes from which each strain has a unique distribution compared with all other component strains of the species (15, 55). The virulence corollary of the distributed genome hypothesis holds that the autocompetence and autotransformation mechanisms of these pathogenic bacteria have been selected for in vivo to provide these pathogens with a rapid means of generating diversity as a way to persist in the face of myriad host defense mechanisms. In other words, the contingency genes, through reassortment during chronic infectious processes, provide for an increased number of genetic characters that enable the population as a whole to adapt rapidly to environmental factors, such as those experienced in the host. The recent understanding that many chronic bacterial infections are caused by biofilms (for reviews, see references 11, 15, 16, and 71) and the fact that bacteria in biofilms have been demonstrated to exchange DNA at rates several orders of magnitude greater than planktonic bacteria (45) provide us with a new rubric for understanding chronic bacterial pathogenesis. This pathogenesis model also helps to explain the difficulty in establishing good animal models of chronic infection, as nearly all such studies start with a single clonal isolate, and we would predict that anagenesis does not produce sufficient genetic heterogeneity for persistence in the face of a concerted host response. The data presented in this paper support the concept that the H. influenzae possesses a population-based supragenome and that no two strains have the same complement of genes. Moreover, it would appear as if the H. influenzae supragenome is necessarily substantially larger in size than the genomes of individual bacteria, but without far more extensive surveys than the one conducted for this study, it will not be possible to accurately estimate the genome space for the H. influenzae on a worldwide pathogen basis.
Acknowledgments
This work was supported by Allegheny General Hospital, Allegheny-Singer Research Institute, and by National Institute on Deafness and Other Communication Disorders grants DC 02148 (G.D.E.) and DC 04173 (G.D.E.).
Editor: J. N. Weiser
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology. Greene Publishing Associates and Wiley Interscience, New York, N.Y.
- 3.Bakkali, M., T. Y. Chen, H. C. Lee, and R. J. Redfield. 2004. Evolutionary stability of DNA uptake signal sequences in the Pasteurellaceae. Proc. Natl. Acad. Sci. USA 101:4513-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barenkamp, S. J., and E. Leininger. 1992. Cloning, expression, and DNA sequence analysis of genes encoding nontypeable Haemophilus influenzae high-molecular-weight surface-exposed proteins related to filamentous hemagglutinin of Bordetella pertussis. Infect. Immun. 60:1302-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barenkamp, S. J., and J. W. St. Geme III. 1994. Genes encoding high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae are part of gene clusters. Infect. Immun. 62:3320-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benz, I., and M. A. Schmidt. 1992. AIDA-I, the adhesin involved in diffuse adherence of the diarrhoeagenic Escherichia coli strain 2787 (O126:H27), is synthesized via a precursor molecule. Mol. Microbiol. 6:1539-1546. [DOI] [PubMed] [Google Scholar]
- 7.Bergman, N. H., and B. J. Akerley. 2003. Position-based scanning for comparative genomics and identification of genetic islands in Haemophilus influenzae type b. Infect. Immun. 71:1098-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blatch, G. L., and M. Lassle. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932-939. [DOI] [PubMed] [Google Scholar]
- 9.Bouchet, V., D. W. Hood, J. Li, J. R. Brisson, G. A. Randle, A. Martin, Z. Li, R. Goldstein, E. K. Schweda, S. I. Pelton, J. C. Richards, and E. R. Moxon. 2003. Host-derived sialic acid is incorporated into Haemophilus influenzae lipopolysaccharide and is a major virulence factor in experimental otitis media. Proc. Natl. Acad. Sci. USA 100:8898-8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cope, L. D., R. Yogev, J. Mertsola, J. L. Latimer, M. S. Hanson, G. H. McCracken, Jr., and E. J. Hansen. 1991. Molecular cloning of a gene involved in lipooligosaccharide biosynthesis and virulence expression by Haemophilus influenzae type B. Mol. Microbiol. 5:1113-1124. [DOI] [PubMed] [Google Scholar]
- 11.Costerton, W., R. Veeh, M. Shirtliff, M. Pasmore, C. Post, and G. Ehrlich. 2003. The application of biofilm science to the study and control of chronic bacterial infections. J. Clin. Investig. 112:1466-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dabernat, H., M. A. Plisson-Saune, C. Delmas, M. Seguy, G. Faucon, R. Pelissier, H. Carsenti, C. Pradier, M. Roussel-Delvallez, J. Leroy, M. J. Dupont, F. De Bels, and P. Dellamonica. 2003. Haemophilus influenzae carriage in children attending French day care centers: a molecular epidemiological study. J. Clin. Microbiol. 41:1664-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danner, D. B., R. A. Deich, K. L. Sisco, and H. O. Smith. 1980. An eleven-base-pair sequence determines the specificity of DNA uptake in Haemophilus transformation. Gene 11:311-318. [DOI] [PubMed] [Google Scholar]
- 14.Davis, J., A. L. Smith, W. R. Hughes, and M. Golomb. 2001. Evolution of an autotransporter: domain shuffling and lateral transfer from pathogenic Haemophilus to Neisseria. J. Bacteriol. 183:4626-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehrlich, G. D., F. Z. Hu, and J. C. Post. 2004. Role for biofilms in infectious disease, p. 332-358. In M. Ghannoum and G. A. O'Toole (ed.), Microbial biofilms. ASM Press, Washington, D.C.
- 16.Ehrlich, G. D., and J. C. Post. 2002. Susceptibility to otitis media: strong evidence that genetics plays a role. JAMA 282:2167-2169. [DOI] [PubMed] [Google Scholar]
- 17.Ehrlich, G. D., R. Veeh, X. Wang, J. W. Costerton, J. D. Hayes, F. Z. Hu, B. J. Daigle, M. D. Ehrlich, and J. C. Post. 2002. Mucosal biofilm formation on middle-ear mucosa in the chinchilla model of otitis media. JAMA 287:1710-1715. [DOI] [PubMed] [Google Scholar]
- 18.Erdos, G., S. Sayeed, P. Antalis, F. Z. Hu, J. Hayes, J. Goodwin, R. Dopico, J. C. Post, and G. D. Ehrlich. 2003. Development and characterization of a pooled Haemophilus influenzae genomic library for the evaluation of gene expression changes associated with mucosal biofilm formation in otitis media. Int. J. Pediatr. Otorhinolaryngol. 67:749-755. [DOI] [PubMed] [Google Scholar]
- 19.Faden, H., L. Duffy, A. Williams, D. A. Krystofik, and J. Wolf. 1995. Epidemiology of nasopharyngeal colonization with nontypeable Haemophilus influenzae in the first 2 years of life. J. Infect. Dis. 172:132-135. [DOI] [PubMed] [Google Scholar]
- 20.Falla, T. J., D. W. Crook, L. N. Brophy, D. Maskell, J. S. Kroll, and E. R. Moxon. 1994. PCR for capsular typing of Haemophilus influenzae. J. Clin. Microbiol. 32:2382-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feil, E. J. 2004. Small change: keeping pace with microevolution. Nat. Rev. Microbiol. 2:483-495. [DOI] [PubMed] [Google Scholar]
- 22.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 23.Frangeul, L., K. E. Nelson, C. Buchrieser, A. Danchin, P. Glaser, and F. Kunst. 1999. Cloning and assembly strategies in microbial genome projects. Microbiology 145:2625-2634. [DOI] [PubMed] [Google Scholar]
- 24.Gilsdorf, J. R., C. F. Marrs, and B. Foxman. 2004. Haemophilus influenzae: genetic variability and natural selection to identify virulence factors. Infect. Immun. 72:2457-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodgal, S. H., and M. A. Mitchell. 1990. Sequence and uptake specificity of cloned sonicated fragments of Haemophilus influenzae DNA. J. Bacteriol. 172:5924-5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashida, H., K. Poulsen, and M. Kilian. 2002. Differences in iron acquisition from human haemoglobin among strains of Actinobacillus actinomycetemcomitans. Microbiology 148:3993-4001. [DOI] [PubMed] [Google Scholar]
- 27.Hiltke, T. J., A. T. Schiffmacher, A. J. Dagonese, S. Sethi, and T. F. Murphy. 2003. Horizontal transfer of the gene encoding outer membrane protein P2 of nontypeable Haemophilus influenzae, in a patient with chronic obstructive pulmonary disease. J. Infect. Dis. 188:114-117. [DOI] [PubMed] [Google Scholar]
- 28.Hobbs, M., and J. S. Mattick. 1993. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 10:233-243. [DOI] [PubMed] [Google Scholar]
- 29.Hutchison, C. A., S. N. Peterson, S. R. Gill, R. T. Cline, O. White, C. M. Fraser, H. O. Smith, and J. C. Venter. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286:2165-2169. [DOI] [PubMed] [Google Scholar]
- 30.Jarosik, G. P., and E. J. Hansen. 1994. Identification of a new locus involved in expression of Haemophilus influenzae type b lipooligosaccharide. Infect. Immun. 62:4861-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kachlany, S. C., P. J. Planet, M. K. Bhattacharjee, E. Kollia, R. DeSalle, D. H. Fine, and D. H. Figurski. 2000. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J. Bacteriol. 182:6169-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kilian, M. 1976. A taxonomic study of the genus Haemophilus, with the proposal of a new species. J. Gen. Microbiol. 93:9-62. [DOI] [PubMed] [Google Scholar]
- 33.Kroll, J. S., K. E. Wilks, J. L. Farrant, and P. R. Langford. 1998. Natural genetic exchange between Haemophilus and Neisseria: intergeneric transfer of chromosomal genes between major human pathogens. Proc. Natl. Acad. Sci. USA 95:12381-12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuklinska, D., and M. Kilian. 1984. Relative proportions of Haemophilus species in the throat of healthy children and adults. Eur. J. Clin. Microbiol. 3:249-252. [DOI] [PubMed] [Google Scholar]
- 35.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 36.LaClaire, L. L., M. L. Tondella, D. S. Beall, C. A. Noble, P. L. Raghunathan, N. E. Rosenstein, and T. Popovic. 2003. Identification of Haemophilus influenzae serotypes by standard slide agglutination serotyping and PCR-based capsule typing. J. Clin. Microbiol. 41:393-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lett, M. C., C. Sasakawa, N. Okada, T. Sakai, S. Makino, M. Yamada, K. Komatsu, and M. Yoshikawa. 1989. virG, a plasmid-coded virulence gene of Shigella flexneri: identification of the virG protein and determination of the complete coding sequence. J. Bacteriol. 171:353-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Long, S. S., F. M. Henretig, M. J. Teter, and K. L. McGowan. 1983. Nasopharyngeal flora and acute otitis media. Infect. Immun. 41:987-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loos, B. G., J. M. Bernstein, D. M. Dryja, T. F. Murphy, and D. P. Dickinson. 1989. Determination of the epidemiology and transmission of nontypable Haemophilus influenzae in children with otitis media by comparison of total genomic DNA restriction fingerprints. Infect. Immun. 57:2751-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luck, S. N., S. A. Turner, K. Rajakumar, H. Sakellaris, and B. Adler. 2001. Ferric dicitrate transport system (Fec) of Shigella flexneri 2a YSH6000 is encoded on a novel pathogenicity island carrying multiple antibiotic resistance genes. Infect. Immun. 69:6012-6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin, K., G. Morlin, A. Smith, A. Nordyke, A. Eisenstark, and M. Golomb. 1998. The tryptophanase gene cluster of Haemophilus influenzae type b: evidence for horizontal gene transfer. J. Bacteriol. 180:107-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mason, K. M., R. S. Munson, Jr., and L. O. Bakaletz. 2003. Nontypeable Haemophilus influenzae gene expression induced in vivo in a chinchilla model of otitis media. Infect. Immun. 71:3454-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meats, E., E. J. Feil, S. Stringer, A. J. Cody, R. Goldstein, J. S. Kroll, T. Popovic, and B. G. Spratt. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J. Clin. Microbiol. 41:1623-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Molin, S., and T. Tolker-Nielsen. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 14:255-261. [DOI] [PubMed] [Google Scholar]
- 46.Moxon, E. R., and R. Wilson. 1991. The role of Haemophilus influenzae in the pathogenesis of pneumonia. Rev. Infect. Dis. 13(Suppl. 6):S518-S527. [DOI] [PubMed] [Google Scholar]
- 47.Munson, R. S., Jr., A. Harrison, A. Gillaspy, W. C. Ray, M. Carson, D. Armbruster, J. Gipson, M. Gipson, L. Johnson, L. Lewis, D. W. Dyer, and L. O. Bakaletz. 2004. Partial analysis of the genomes of two nontypeable Haemophilus influenzae otitis media isolates. Infect. Immun. 72:3002-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Munson, R. S., Jr., H. Zhong, R. Mungur, W. C. Ray, R. J. Shea, G. G. Mahairas, and M. H. Mulks. 2004. Haemophilus ducreyi strain ATCC 27722 contains a genetic element with homology to the vibrio RS1 element that can replicate as a plasmid and confer NAD independence on Haemophilus influenzae. Infect. Immun. 72:1143-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murphy, T. F., S. Sethi, K. L. Klingman, A. B. Brueggemann, and G. V. Doern. 1999. Simultaneous respiratory tract colonization by multiple strains of nontypeable Haemophilus influenzae in chronic obstructive pulmonary disease: implications for antibiotic therapy. J. Infect. Dis. 180:404-409. [DOI] [PubMed] [Google Scholar]
- 50.Mushegian, A. R., and E. V. Koonin. 1996. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. USA 93:10268-10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Musser, J. M., S. J. Barenkamp, D. M. Granoff, and R. K. Selander. 1986. Genetic relationships of serologically nontypable and serotype b strains of Haemophilus influenzae. Infect. Immun. 52:183-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parsek, M. R., and C. Fuqua. 2004. Biofilms 2003: emerging themes and challenges in studies of surface-associated microbial life. J. Bacteriol. 186:4427-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Planet, P. J., S. C. Kachlany, R. DeSalle, and D. H. Figurski. 2001. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl. Acad. Sci. USA 98:2503-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Porras, O., D. A. Caugant, B. Gray, T. Lagergard, B. R. Levin, and C. Svanborg-Eden. 1986. Difference in structure between type b and nontypable Haemophilus influenzae populations. Infect. Immun. 53:79-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen, K., X. Wang, J. C. Post, and G. D. Ehrlich. 2003. Molecular and translational research approaches for the study of bacterial pathogenesis in otitis media, p. 91-119. In R. M. Rosenfeld and C. D. Bluestone (ed.), Evidence-based otitis media, 2nd ed. BC Decker Inc., Hamilton, Ontario, Canada.
- 56.Smith, H. O., J. F. Tomb, B. A. Dougherty, R. D. Fleischmann, and J. C. Venter. 1995. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science 269:538-540. [DOI] [PubMed] [Google Scholar]
- 57.Smith-Vaughan, H. C., A. J. Leach, T. M. Shelby-James, K. Kemp, D. J. Kemp, and J. D. Mathews. 1996. Carriage of multiple ribotypes of non-encapsulated Haemophilus influenzae in Aboriginal infants with otitis media. Epidemiol. Infect. 116:177-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith-Vaughan, H. C., K. S. Sriprakash, J. D. Mathews, and D. J. Kemp. 1995. Long PCR-ribotyping of nontypeable Haemophilus influenzae. J. Clin. Microbiol. 33:1192-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith-Vaughan, H. C., K. S. Sriprakash, J. D. Mathews, and D. J. Kemp. 1997. Nonencapsulated Haemophilus influenzae in Aboriginal infants with otitis media: prolonged carriage of P2 porin variants and evidence for horizontal P2 gene transfer. Infect. Immun. 65:1468-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song, X. M., A. Forsgren, and H. Janson. 1999. Fragmentation heterogeneity of 23S ribosomal RNA in Haemophilus species. Gene 230:287-293. [DOI] [PubMed] [Google Scholar]
- 60a.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 61.St. Geme, J. W., III. 2000. The pathogenesis of nontypable Haemophilus influenzae otitis media. Vaccine 19(Suppl. 1):S41-S50. [DOI] [PubMed] [Google Scholar]
- 62.St. Geme, J. W., III, and S. Grass. 1998. Secretion of the Haemophilus influenzae HMW1 and HMW2 adhesins involves a periplasmic intermediate and requires the HMWB and HMWC proteins. Mol. Microbiol. 27:617-630. [DOI] [PubMed] [Google Scholar]
- 63.Ten Cate, A. R., E. Freeman, and J. B. Dickinson. 1977. Sutural development: structure and its response to rapid expansion. Am. J. Orthod. 71:622-636. [DOI] [PubMed] [Google Scholar]
- 64.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 65.van Alphen, L., D. A. Caugant, B. Duim, M. O'Rourke, and L. D. Bowler. 1997. Differences in genetic diversity of nonencapsulated Haemophilus influenzae from various diseases. Microbiology 143:1423-1431. [DOI] [PubMed] [Google Scholar]
- 66.van Ham, S. M., L. van Alphen, F. R. Mooi, and J. P. van Putten. 1994. The fimbrial gene cluster of Haemophilus influenzae type b. Mol. Microbiol. 13:673-684. [DOI] [PubMed] [Google Scholar]
- 67.van Ulsen, P., M. van Schilfgaarde, J. Dankert, H. Jansen, and L. van Alphen. 2002. Genes of non-typeable Haemophilus influenzae expressed during interaction with human epithelial cell lines. Mol. Microbiol. 45:485-500. [DOI] [PubMed] [Google Scholar]
- 68.Wang, Y., S. D. Goodman, R. J. Redfield, and C. Chen. 2002. Natural transformation and DNA uptake signal sequences in Actinobacillus actinomycetemcomitans. J. Bacteriol. 184:3442-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Whitchurch, C. B., M. Hobbs, S. P. Livingston, V. Krishnapillai, and J. S. Mattick. 1991. Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene 101:33-44. [DOI] [PubMed] [Google Scholar]
- 70.Wilfert, C. M. 1990. Epidemiology of Haemophilus influenzae type b infections. Pediatrics 85:631-635. [PubMed] [Google Scholar]
- 71.Donlan, R. 2002. Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8:881-890. [DOI] [PMC free article] [PubMed] [Google Scholar]