Abstract
Background:
Aortic structure impacts cardiovascular health through multiple mechanisms. Aortic structural degeneration occurs with aging, increasing left ventricular afterload and promoting increased arterial pulsatility and target organ damage. Despite the impact of aortic structure on cardiovascular health, three-dimensional (3D) aortic geometry has not been comprehensively characterized in large populations.
Methods:
We segmented the complete thoracic aorta using a deep learning architecture and used morphological image operations to extract aortic geometric phenotypes (AGPs, including diameter, length, curvature, and tortuosity) across multiple subsegments of the thoracic aorta. We deployed our segmentation approach on imaging scans from 54,241 participants in the UK Biobank and 8,456 participants in the Penn Medicine Biobank. Age-related structural remodeling was quantified on a reference cohort of normative participants. The genetic architecture of three-dimensional aortic geometry was quantified using genome-wide association studies, followed by gene-level analysis and drug-gene interactions.
Results:
Aging was associated with various 3D-geometric changes, reflecting structural aortic degeneration, including decreased arch unfolding, descending aortic lengthening and luminal dilation across multiple subsegments of the thoracic aorta. Male aortas exhibited increased length and diameters compared to female aortas across all age ranges, whereas female aortas exhibited increased curvature compared with males. We identified 209 novel genetic loci associated with various 3D-AGPs. 357 significant gene-level associations were uncovered, with Fibrillin-2 gene polymorphisms being identified as key determinants of aortic arch structure. Drug-gene interaction analysis identified 25 cardiovascular drugs potentially interacting with aortic geometric loci.
Conclusion:
Our analysis identified key patterns of aortic structural degeneration linked to aging. We present the first GWAS results for multiple 3D-structural parameters of the aorta, including length, curvature, and tortuosity. Additionally, we confirm various loci associated with aortic diameter. These results expand the genetic loci associated aortic structure and will provide crucial insights into the joint interplay between aging, genetics and cardiovascular structure.
Keywords: Deep learning, 3D aortic structure, aortic aging, GWAS
1. Introduction
The aorta is the largest conduit artery in the human body1. In addition to its conduit function, the aorta plays a key role in modulating pulsatile arterial hemodynamics, mediated by its cushioning function of the intermittent left ventricular ejection. Aortic structural parameters (including geometry and wall stiffness) have been shown to be key determinants of aortic hemodynamic function1,2. Despite its prominent age-associated changes and its key hemodynamic role, studies related to structural properties of aortic geometric phenotypes (AGPs) are lacking3,4. Moreover, previous studies have been limited to two-dimensional cross-sectional geometric parameters of the aorta, neglecting important three-dimensional (3D) aspects of its geometry, including elongation, tortuosity/unfolding, and curvature, all of which influence aortic function. Understanding 3D aortic structure would help to identify patterns of normative and accelerated vascular aging, uncover the genetic architecture of aortic geometry, and identify possible therapeutic targets to delay aortic aging and degeneration.
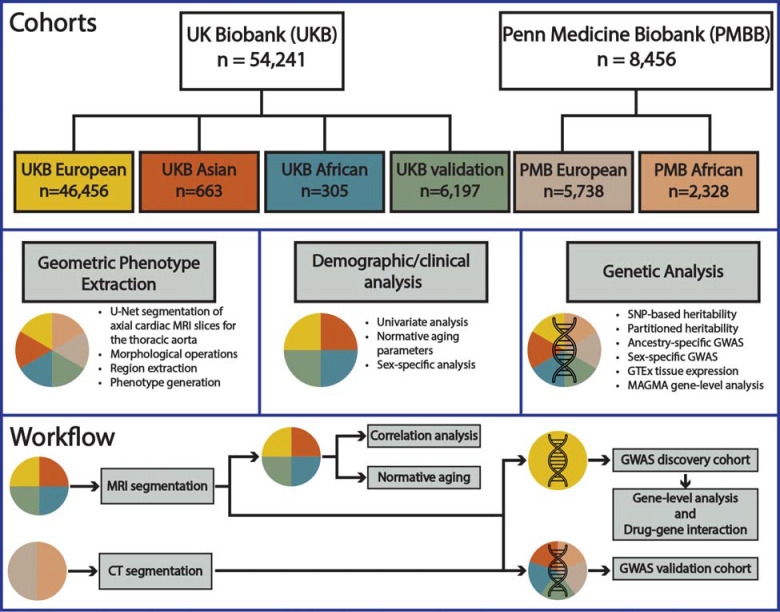
Cardiovascular magnetic resonance (CMR) imaging has become a valuable resource for quantifying structural properties of the heart. Previous analyses of image-derived cardiac phenotypes acquired through the segmentation of CMR imaging have been used to uncover the genetic basis and prognostic value for cardiac structure and function5,6. However, analyses of 3D aortic geometry have not been performed to date. To comprehensively characterize the three-dimensional structural parameters of the thoracic aorta, we leveraged imaging data from 54,241 participants enrolled in the UK Biobank7 (UKB) and 8,456 individuals enrolled in the Penn Medicine Biobank8 (PMBB). We aimed to (1) develop a deep learning approach to segment and comprehensively characterize the 3D geometry of the thoracic aorta, and measure key phenotypes (diameter, length, curvature, tortuosity) across various thoracic aortic subsegments, (2) examine normative aging patterns for the thoracic aorta, and (3) characterize the genetic architecture of the thoracic aorta. Figure 1 provides a summary of our study.
Figure 1.
Overview of (1) complete study cohorts for the UK Biobank (UKB) and Penn Medicine Biobank (PMBB) as well as the ancestry specific subgroups for genetic analysis, (2) phenotype extraction overview, demographic and clinical correlation analysis, and genetic analysis, and (3) analysis workflow.
2. Methods
An overview of the cohorts and methodologies used in this study are presented in Figure 1. For a comprehensive description of the methodology please refer to the Supplemental Material. The PMBB is approved under IRB protocol# 813913. This research has been conducted using the UK Biobank Resource under Application Number 81032.
Data Availability Statement
UK Biobank data is available to researchers following UKB application approval and IRB approval. Researchers can apply at https://www.ukbiobank.ac.uk/. Access to Penn Medicine BioBank data is provided to investigators at the University of Pennsylvania. External access can be provided through collaboration with an investigator at the university. All GWAS summary statistics will be made available upon publication.
Code availability
All code for aortic geometric phenotype (AGP) extraction can be accessed at https://github.com/cams2b/ukb_aorta/. The provided repository contains pretrained weights for U-Net segmentation as well as the required morphological pipeline for extraction AGPs.
3. Results
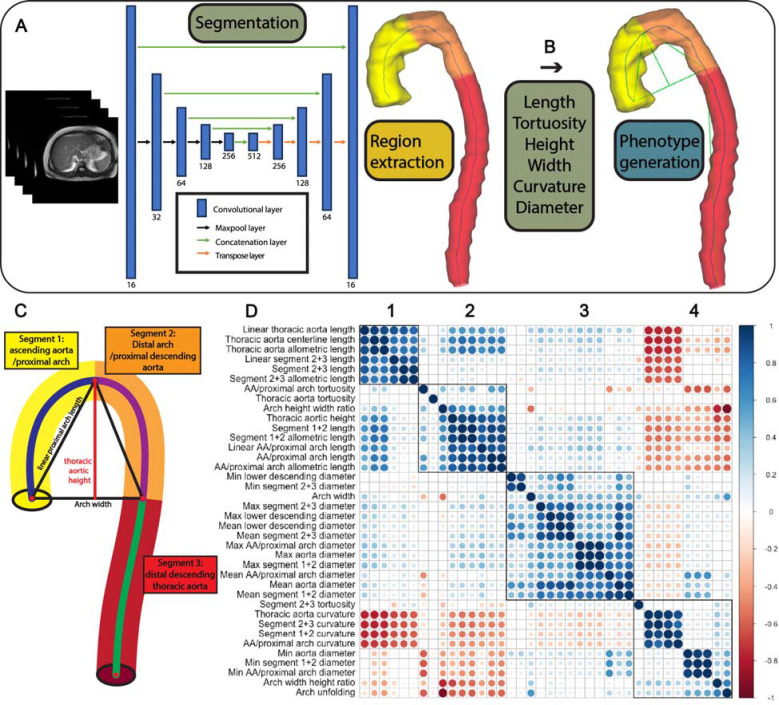
We automatically segmented the entire thoracic aorta and derived over 35 unique 3D-aortic geometric phenotypes (AGPs) (Fig. 2A–C) from 54,241 UKB participants via U-Net segmentation combined with morphological operations (Figs. 2A–2C). The identification of each segment and specific landmarks used to define them are detailed in the methods section and in Figures 2B and 2C. We note that these segments do not necessarily correspond to standard anatomical segmental definitions, but were rather derived based on objective unequivocal landmarks in the three-dimensionally segmented thoracic aorta. In particular, the segment extending from the root to the apex of the aorta (segment 1 in Figure 2C), comprising the proximal transverse aortic arch, was identified as a single segment. Similarly, the segment from the apex to the plane intersecting the aortic root (segment 2 in Figure 2C) was identified as a single segment, whereas the sum of the 2 segments above were used to calculate the height and width of segments 1+2, as well as the width-to-height ratio. Given that this height computation incorporates the ascending aorta and the proximal descending aorta, we note that these height and width indices from our study are not directly comparable to previous studies that measured aortic arch height and width starting from an arbitrarily defined plane intersecting some point of the middle of the anatomic ascending and descending aorta9.
Figure 2.
(A) Overview of the U-Net segmentation architecture. (B) Thoracic aorta segmentation and phenotype generation. After segmenting the entire thoracic aorta and extracting its centerline, we identified the apex of the aorta (i.e., maximum centerline point on the vertical axis), and the centerline point on the descending aorta with the smallest Euclidean distance on the vertical axis with the aortic root. The segment extending from the root to the apex of the aorta (ascending aorta/proximal arch) was identified as segment 1. The segment from the aortic apex to the point on the descending aorta vertically corresponding to the level of the aortic root (distal arch/proximal descending aorta) was identified as a segment 2. Segment 3 comprised the remainder of the descending aorta (i.e., distal descending aorta). (C) Legend for each AGP region. (D) Correlation matrix of select aortic geometric phenotypes (AGPS), showing the 4 variable clusters (see text for details).
Figure 2D shows the correlation matrix of various 3D-AGPs. There were four well-defined AGP clusters, which related to: aortic length and specifically distal arch/descending aortic length (cluster 1), aortic arch 3D geometric phenotypes (cluster 2), aortic diameter phenotypes (cluster 3) and aortic curvature phenotypes (cluster 4). Interestingly, the length of the proximal arch/ascending aorta segregated separately (cluster 2) from overall thoracic aortic length and distal arch/descending aorta, which clustered with metrics of aortic length (cluster 1).
Aortic aging patterns
Normative aging patterns were computed on a healthy subset of UKB participants (n=4,725) who met the following criteria: (1) BMI >18 and <25 kg/m2, (2) low-density lipoprotein (LDL) cholesterol <130 mg/dL, (3) high-density lipoprotein (HDL) cholesterol >50 (female) or >40 (male) mg/dL, (4) triglycerides <200 mg/dL, (5) hemoglobin A1c <6% (<42 mmol/mol), (6) never a smoker, and (7) no history of cardiovascular disease. Participants with AGP measurements that fell outside of 3 standard deviations of the mean were removed to account for phenotype stochasticity (i.e., model error). Participants were classified according to the following age range categories: <50, 50–60, 60–70, and >70 years (Supplemental Table 1, Supplemental Figs. 2–5) and geometric patterns were compared across age strata. The aorta demonstrated a clear pattern of increased elongation, dilation, and tortuosity with age (Supplemental Figs. 2–4). In contrast, there was a clear pattern of decreased unfolding and curvature across the full thoracic aorta as well as various subsegments (Supplemental Fig. 5).
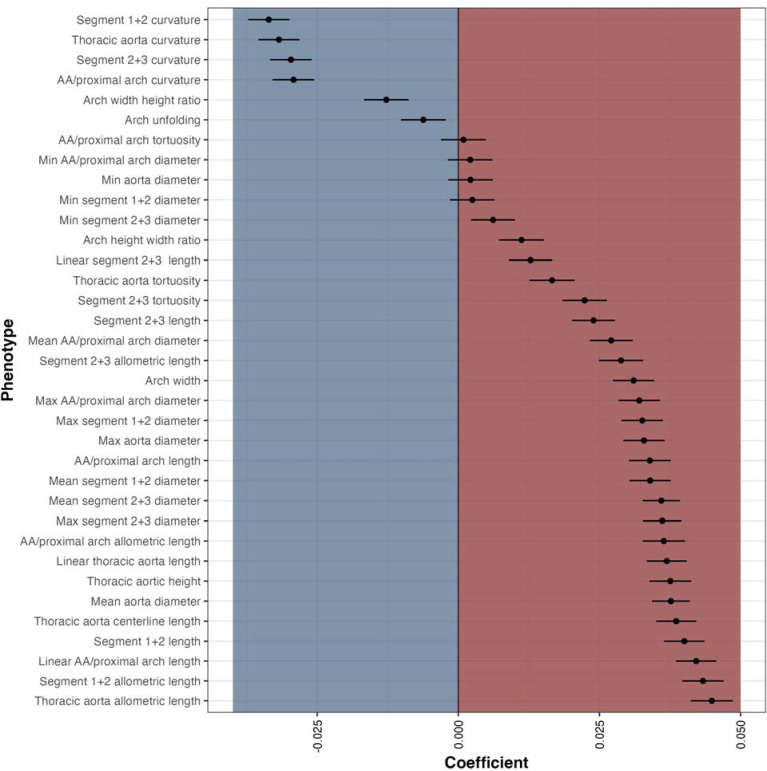
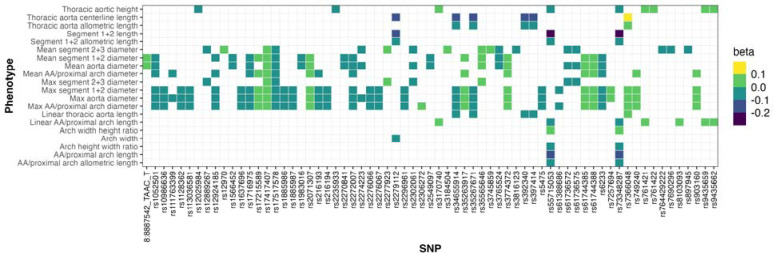
Sex-adjusted linear models were generated to assess the relationship between AGPs and aging (Supplemental Table 2). Ascending/proximal arch length (segment 1; β=0.6 mm/year; p=2.79×10−72) and distal arch/descending aortic length (segments 2+3; β=0.4 mm/year; p=3.96×10−36) were directly related to age. The aortic arch length (segments 1+2) exhibited the steepest association with age (segments 1+2; β =1.1 mm/year; p=4.76×10−105). The mean diameter of the ascending aorta/proximal arch (segment 1) and the distal arch/descending aorta (segments 2+3) exhibited comparable slopes with age, of 0.077 mm/year (p=1.21×10−45) and 0.070 mm/year (p=4.81×10−100) respectively. Thoracic arch unfolding (segments 1+2) was inversely related to age (β=−0.0035; p=0.001), demonstrating that the centerline length (segments 1+2; β=1.1 mm/year; p=4.76×10−105) elongates more than the width increases (β=0.3 mm/year; p=1.49×10−62). The normalized β coefficients for the sex-adjusted linear models for assessing the relationship between age and AGPs are depicted in Figure 3 (Supplemental Table 3).
Figure 3.
Sex-adjusted, normalized beta coefficients for the relationship between AGPs and age in the UK Biobank “healthy” cohort with normative lab measurements and no history of cardiovascular disease (n=4,725).
Aging patterns were largely similar in the sex-specific analyses, with a β coefficient correlation of 0.98 between the male and female cohorts (Supplemental Table 4). Male aortas exhibited increased length and diameters compared to female aortas across all age ranges; however, female aortas exhibited increased curvature compared with males.
SNP-based heritability estimates of AGPs
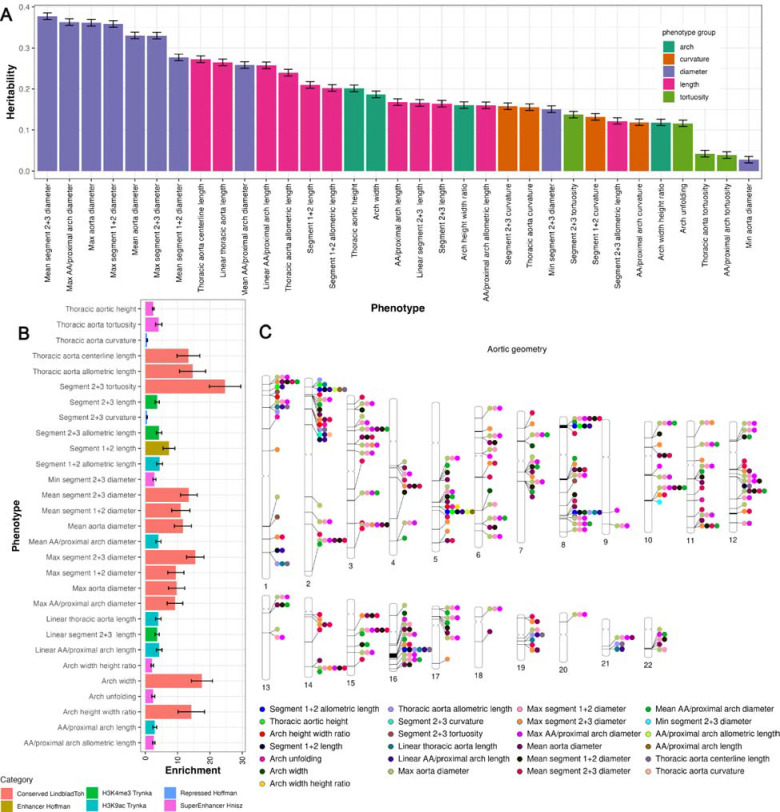
We estimated the SNP-based heritability (h2) for 35 AGPs representative of the complete thoracic aorta (Supplemental Table 5). After Bonferroni correction, 33 AGPs were significantly heritable, with an average heritability of 0.20 (±0.09). The AGP exhibiting the highest heritability was notably the diameter of the distal arch/descending aorta (segments 2+3; h2=0.38±0.01). Thoracic aortic length (allometrically indexed and non-indexed) showed h2 of 0.24 and 0.27, respectively. Thoracic aortic width heritability was 0.19 (±0.01). Figure 4A shows estimates of the SNP-based heritability (h2) for 33 AGPs using all UKB imaging participants with European ancestry (n=52,665). We additionally computed sex-specific heritability for males (n=25,209) and females (n=27,456) in the European ancestry cohort (Supplemental Fig. 6). After Bonferroni correction, 31 AGPs displayed significant heritability for males and females (Supplemental Tables 6–7). Males had an average heritability of 0.24 (±0.13), whereas females had an average heritability of 0.24(±0.12). Interestingly, there was a noticeable increase in the heritability of the distal arch/descending aorta (segments 2+3) in the sex-specific analysis, with males exhibiting an h2=0.54(±0.01) and females exhibiting an h2=0.49 (±0.01).
Figure 4.
(A) SNP-based heritability of all significant (p<1e-4) aortic geometric phenotypes. (B) partitioned heritability of the strongest significant enrichment category (p<0.05/53). (C) visualization of significant genetic loci (p<5×10−8/31).
Partitioned heritability analysis
We performed partitioned heritability analysis to explore the enrichment patterns in 53 overlapping functional genomic categories. After Bonferroni correction, 29 AGPs displayed significant enrichment (p<0.05/53) of at least 1 functional region, with 12 AGPs having primary enrichment in conserved functional regions of the genome10,11. Measurements of thoracic aortic length across several subsegments as well as the diameter of the proximal aorta (segment 1) exhibited primary enrichment in gene promoter regions, specifically among SNPs surrounding the chromatin marks acetylation of histone 3 at lysine 9 (H3K9ac)12. A subset of histone 3 Lysine 27 acetylation (H3K27ac) super-enhancer regions had primary enrichment in thoracic aortic height, indexed thoracic aortic length of segments 1+2, and tortuosity of the thoracic aorta13. Descending aortic length (segments 2+3) had primary enrichment among histone promoter regions involving trimethylation of histone 3 at lysine 4 (H3K4me3). A complete list of significant enrichment regions can be found at Supplemental Table 8.
Genetic architecture of 3D aortic geometry
We performed GWAS on 35 AGPs representative of the complete thoracic aorta as well as three key subsegments, namely the proximal aorta (segment 1), the descending/distal aorta (segments 2+3), and the sum of segments 1+2 in the UKB European ancestry sub cohort (n=46,456). Four AGPs displayed no significant SNPs at genome wide significance (p<5×10−8) and were discarded from future analysis.
All GWAS results are available in Supplemental Datasets (will be made available upon publication). LD and genomic inflation (λ) were tested to verify the robustness of each GWAS (LD mean=1.01±0.01, λ mean=1.10±0.06). LD scores, genomic inflation and GWAS-based heritability is shown in Supplemental Table 9. After adjusting for Bonferroni correction at the genome-wide significance level (p<5×10−8/31) we identified 209 unique loci (Fig. 4C). More specifically, we identified 25 loci associated with aortic length, 7 loci associated with aortic tortuosity, 1 locus associated with aortic curvature, 172 loci associated with aortic diameter, and 12 aortic arch loci (arch width, thoracic-aortic height) (Supplemental Table 10). In the sex-stratified analysis for males (n=25,209) and females (n=27,456) of European ancestry the GWAS results were strongly correlated (LDSC correlation = 0.944±0.09).
Validation of GWAS results
In the UKB validation cohort (Supplemental Table 11, Supplemental Fig. 7), we replicated 4 aortic length loci, 1 tortuosity loci, 39 aortic diameter loci at Bonferroni significance (p<0.05/209). In total we validated 44 unique loci at Bonferroni significance. Furthermore, at nominal significance (p<0.05) we verified 7 arch loci (arch width, thoracic-aortic height). In our PMBB external validation cohort (Supplemental Table 12, Supplemental Fig. 8) among participants with European ancestry we replicated 25 aortic diameter loci at Bonferroni significance. At nominal significance we validated 7 length loci, 1 tortuosity loci, 1 curvature loci and 6 arch loci (arch width, thoracic-aortic height). All loci were concordant in the same direction.
Assessment in participants of different ancestries
We assessed whether the genetic associations demonstrated among participants of European ancestry in the UKB were present among participants with Asian (n=663) and African (n=305) genetic ancestry in the UKB, as well as participants in the PMBB with African ancestry (n=2,328). In our PMBB African cohort (Supplemental Table 13, Supplemental Fig. 9), we replicated 8 diameter loci Bonferroni significance. At nominal significance we verified 10 aortic length loci, 1 curvature loci, 1 tortuosity loci, and 5 aortic arch loci (arch width, thoracic-aortic height). In the UKB African cohort (Supplemental Table 14, Supplemental Fig. 10), we replicated 3 diameter loci at Bonferroni significance. At nominal significance we verified 11 aortic length loci, 3 tortuosity loci, and 5 arch loci (arch width, thoracic-aortic height). In the UKB Asian cohort (Supplemental Table 15, Supplemental Fig. 11), we replicated 4 diameter loci at Bonferroni significance. At nominal significance we verified 6 aortic length loci, 1 tortuosity loci, and 4 related to the aortic arch (arch width, thoracic-aortic height).
Concordance of results with previous GWAS findings
To assess the concordance of our results with previous findings from GWAS, we queried independent SNPs among AGPs to previously reported GWAS summary data (NHGRI-EBI GWAS Catalog). We identified shared variants between our measurements for the aorta/proximal arch (segment 1) and the distal arch/descending aorta (segments 2+3) diameter measurements with previous in-plane measurements of the diameter of the ascending and descending thoracic aorta14. More specifically, we validated 29 loci previously associated with the ascending and descending aortic diameter14. We highlight genetic concordance between thoracic aortic length and aortic root diameter15. Furthermore, we identified shared variants between the distal arch/descending aortic (segments 2+3) diameter and left and right ventricular end-systolic and end-diastolic volume, whereas the diameter of segments 1+2 shared variants with left ventricular wall thickness5,16. Specifically, the genes, SH2B3, ATXN2, and CKDN1A were identified in GWAS of the aorta and the left ventricle. Refer to Supplemental Table 16 for a complete list of concordant results.
Gene and gene-set analysis
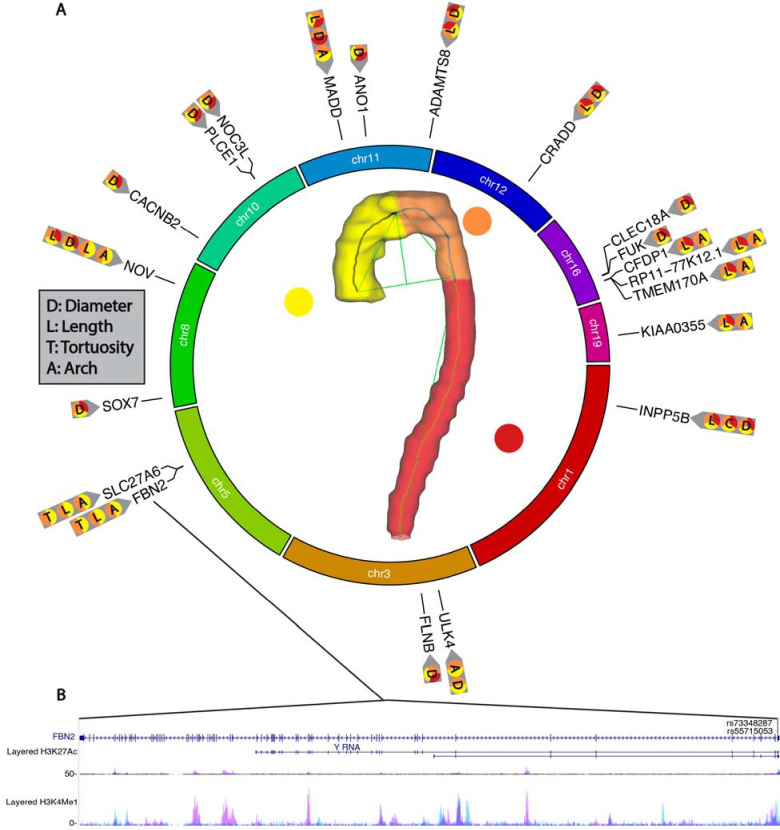
We identified candidate gene-level and gene-set associations for their relationship with aortic geometric phenotypes using MAGMA and identified expression quantitative trait loci (eQTL) in aortic tissue using GTEx v8. Additionally, we queried our GWAS results to identify any significant exonic variants using FUMA. Next, we mapped significant SNPS (p<5×10−8/31) within each identified gene region and identified overlapping histone modifier tracks (Supplemental Figs. 12–24)17. Firstly, we identified 67 unique SNP variants in exon regions of the genome (Fig. 5, Supplemental Table 17). Furthermore, several exonic SNPs were localized to specific AGP categories; specifically, rs55715053 and rs73348287 were identified in GWAS results of thoracic aortic length and segment 1+2 length. Interestingly, some SNPs had cross categorical localization between aortic length and diameter (rs34655914, rs35267671). Next, MAGMA identified 357 significant gene-level associations (p<0.05/19,809) (Supplemental Table 18). Some genes were repeatedly identified across multiple AGPs, as well as being localized to specific subsegments of the aorta (Fig. 6A). Notably, fibrillin-2 (FBN2) was identified across 18 AGPs with primary associations being localized to the aortic arch (Fig. 6A–B, Supplemental Fig. 12). There were 91 significant variants (p<5×10−8/31) within FBN2 and two exonic variants (rs73348287 and rs55715053) overlapping enhancer histone modification tracks. FBN2 has previously been associated with CVD, coronary artery disease, as well as calcium-channel blocker and beta-blocker use18–21. 18 SNP variants were identified across 11 AGPs in the gene a disintegrin and metalloproteinase with thrombospondin motifs 8 (ADAMTS8) which has been implicated in the structural remodeling of vessels and its upregulation has been associated with pulmonary arterial hypertension22. Four significant SNP variants overlapped histone modifier tracks in ADAMTS8 (Supplemental Fig. 13). While ADAMTS7 has been previously documented in GWAS of the aorta, ADAMTS8 was thought to be primarily associated with heart and lung morphology. Cellular communication network factor 3 (NOV) was identified with aortic length, arch width, and aortic diameter and has previously been associated with hypertension23. 36 significant variants were identified in NOV with three SNPs, (rs56013326, rs2279112, rs7834596) existing within histone modifier regions (Supplemental Fig. 14). 710 unique SNP variants were identified in Unc-51 Like Kinase 4 (ULK4) and were only associated with diameter AGPs (Supplemental Fig. 15). ULK4 has previously been associated with neuropsychiatric disorders and neurogenesis24. In the GTEx tissue specific analysis, we identified enriched gene-expression (p<5×10−8) among aortic artery, coronary artery, tibial artery, as well as the uterus, cervix, and fallopian tube tissues.
Figure 5.
All significant exonic SNP variants (p<5×10−8/31).
Figure 6.
(A) Gene-level associations (P<0.05/19,809) mapped to their corresponding genome location and their corresponding AGP. (B) FBN2 genetic locus, with corresponding histone modification tracks (H3K4Me3 and H3K27Ac) and corresponding exonic SNP variants (rs73348287 and rs55715053).
Identification of candidate genes for drug target prioritization
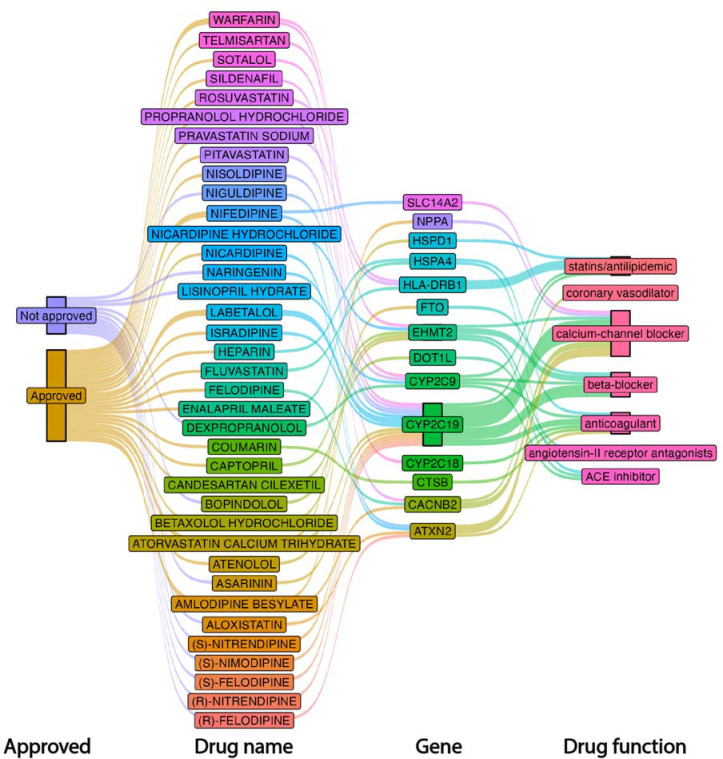
Using the AGP-associated genes identified via MAGMA, we queried our results to identify potential therapeutic drugs using the Drug-Gene interaction database (DGIB)25. The 357 significant genes identified in our analyses yielded 2,741 potential drug-gene interactions, composed of 966 approved drugs, a more restricted 762 drugs that are not cancer drugs (anti neoplastic) and 8 immunotherapy drugs (Supplemental Table 19). Among approved drugs we identified 25 cardiovascular drugs mapped to 14 genes. More specifically, seven drugs were calcium-channel blockers which targeted six unique genes (ATXN2, CACNB2, CYP2C19, EHMT2, NPPA, SLC14A2). There were two angiotensin-converting enzyme (ACE) inhibitors that mapped to EHMT2 and HSPA4. Two drugs were angiotensin-II receptor antagonists mapping to DOT1L, EHMT2. There were five beta-blockers that mapped to three genes, namely, CYP2C19, EHMT2, FTO. Five antilipidemic drugs (statins) mapped to four genes (CYP2C19, CYP2C9, HLA-DRB1, HSPD1). Three anticoagulants, namely, warfarin, coumarin, and heparin mapped to four genes (CTSB, CYP2C18, CYP2C9, HSPA4). Examining the unapproved drugs, we identified an additional 12 cardiovascular drugs that mapped to four unique genes. More specifically, six calcium-channel blockers were mapped to two genes, namely CYP2C19 and ATXN2. Two beta-blockers mapped to three unique genes (CYP2C9, EHMT2). One ACE inhibitor mapped to CYP2C9. Three anticoagulants, aloxistatin, naringin, and asarinin mapped to CYP2C19 and CYP2C19. Figure 7 provides an overview of cardiovascular drugs and their associated genetic interactions.
Figure 7.
Visualization of approved and not approved drugs, their corresponding gene interactions and their functional category.
4. Discussion
This study leveraged aortic imaging data and genetics from two large independent cohorts, the UKB and the PMBB, to comprehensively characterize 3D structural parameters of the thoracic aorta, their aging patterns, and their underlying genetic architecture. We developed, for the first time, a phenotype extraction protocol consisting of (1) a deep learning segmentation architecture that accurately segments the thoracic aorta, and (2) morphological operations that operate directly on three-dimensional mesh representations to compute aortic length, diameter, tortuosity, curvature and arch measurements, as well as identify subsegments of the aorta. We then deployed our segmentation approach on 54,241 UKB imaging participants and 8,456 PMBB participants. Next, we computed normative aging profiles for all AGPs that were extracted from our pipeline and comprehensively characterized their genetic architecture.
Using a highly selective subset of healthy UKB participants (n=4,725) we identified a clear pattern of normal aortic aging involving elongation, widening, reduced unfolding and dilation. Our results extend a previous investigation of 210 patients undergoing aortic imaging, which reported that the aorta elongates with age3, as well as a previous study of 100 participants which found changes in diameter, arch height, and width with aging9. In contrast to previous studies, we studied large samples of participants with a wide age range from 2 large biobanks, comprising >60,000 participants. Our approach was fully automated, removing observer bias that may arise from manual segmentation of the aorta. Moreover, our utilization of deep neural networks facilitated the segmentation of far more imaging studies, which in turn allowed us to study a highly selected subgroup of participants to assess the relationships between aortic 3D-geometry and normal aging. This further allowed us to characterize normal allometric relationships to properly account for the influence of body size and differences in body size between males and females. This is important, because various conditions (such as diabetes, hypertension and various other cardiovascular risk factors) that increase in prevalence with age or that vary in prevalence between males and females, can impact the thoracic aortic wall. We identified remodeling patterns characteristic of normal aortic aging, including decreased thoracic aorta unfolding, reduced curvature and reduced tortuosity of the aorta distal to the apex (segments 2+3). In addition, aortic curvature displayed a strong negative relationship with age1,26. We also found that the proximal aorta (segment 1) displays faster elongation and dilation when compared with the distal arch/descending aorta (segments 2+3). The geometric changes were characterized by an overall reduced unfolding of the thoracic aorta. We note that previous smaller studies reported increased unfolding of the aortic arch with age9. This is not inconsistent with our results, since we found that all thoracic aortic segments increase in length with aging, resulting in widening and unfolding of the transverse arch. However, the increase in length of the thoracic aorta (segments 1+2) exceeded the widening of the arch, resulting in decreased overall unfolding indices of the thoracic aorta with increasing age.
To our knowledge, our study is the first to uncover the genetic architecture of the 3D-geometry of the thoracic aorta. Our AGPs exhibited moderate to high heritability, with h2 ranging from 23%, for length AGPs (namely, thoracic aortic length) to 38% for diameter phenotypes of the distal aorta. Some heritability metrics of aortic diameter have been previously reported in the literature, which are comparable to our findings27. We additionally highlight an increased heritability of the distal arch/descending aortic diameter in our sex-specific heritability analysis. Using a GWAS discovery cohort of 46,456 participants with European ancestry in the UK Biobank, we report 209 unique genetic loci for across all AGPs. Next, we validated our results on three separate validation cohorts of the UKB and two external cohorts from the PMBB. UKB validation results replicated 39 unique genomic loci associated with AGPs at Bonferroni significance. We additionally validate 29 aortic diameter loci that were previously identified by Pirruccello et. al14. This is the first study to examine the genetic architecture of several 3D-aortic geometric phenotypes, including aortic length, tortuosity, curvature, as well as arch-specific geometric indices, namely, arch width, as well as aortic diameter across the entire thoracic aorta, including validation in independent cohorts. Previous aortic GWAS have been conducted only on cross sectional measurements of the aorta, which largely discards the heterogenous differences between proximal and distal regions28 and neglects the complex 3D structure of the aorta.
Despite the recognized importance of the aorta in aging, hypertension and hypertensive target organ damage, data regarding the molecular determinants of aortic structural degeneration are scarce1 and derived mostly from animal studies or non-genetic observational studies. Genetic determinants of aortic geometry can provide important clues regarding its pathogenesis. We identified several genes that were significantly associated with multiple AGPs across several subsegments of the aorta (Fig. 6). Furthermore, we identify several SNP variants that overlapped histone modifier tracks. NOV was identified across all AGP categories, as well as several subsegments of the aorta. Furthermore, NOV has been linked to increased inflammation, as well as degradation of cartilage29. Waldman et al. found that inhibition of NOV expression restores the function of anti-inflammatory, thermogenic, and mitochondrial gene expression, resulting in improved vascular function in mice30. FBN2 is a protein-encoder gene responsible for the production of fibrillin-2 and is essential for maintaining elastin deposits and the elastic fiber system.31 FBN2 was primarily associated with AGP structures in segments 1+2. It has also been documented that FBN2 loss of function mutations result in a reduction of fibrillin-2 and disorganized smooth muscle cells in epithelial tube formation, as well as a shortened tracheal tube length32. To our knowledge, FBN2 has not previously been identified in aortic GWAS results, suggesting that it is primarily associated with 3-dimensional geometric properties of the aorta.
We highlight 14 genes that have the potential to be targeted by drugs, including currently approved cardiovascular drugs. Specifically, calcium voltage-gated channel auxiliary subunit beta 2 (CACBN2) was implicated in multiple drug-gene interactions. CACBN2 has been previously associated with cardiac ion channel function. Cytochrome P450 2C19 (CYP2C19) was identified in multiple drug-gene interactions and loss of function mutations have been identified with adverse cardiovascular events33. Additionally, the fat mass and obesity-associated (FTO) gene was identified to interact with beta-blockers. While primarily associated with obesity, FTO SNP variants have increasingly been recognized with the risk of adverse cardiovascular events34. Asarinin was identified among unapproved drugs and has previously demonstrated potential as a therapeutic to target pulmonary fibrosis35. Further research is required to assess the potential therapeutic value of these agents to impact aortic remodeling and function with aging.
Our work also has limitations. Firstly, the imaging modality varied between the UKB (CMR) and the PMBB (CT). However, given the large dimensions of the aorta, its 3D-segmentation is less sensitive to the imaging modality and resolution. Additionally, the participants in the PMBB were undergoing imaging for medical evaluation, whereas UKB participants were randomly selected. Nevertheless, the replication of findings across these two cohorts with different inclusion and exclusion criteria and different imaging modalities indicates robust associations, adding confidence to our results. We note that our anatomic segmentation was based on objectively identifiable three-dimensional landmarks in the aorta (such as the aortic apex) and may not necessarily correspond to standard anatomic definitions or aortic subsegments with different embryologic origins. Finally, our genetic analysis was largely limited to participants with European ancestry; however, we were able to replicate genetic loci in participants with African and Asian ancestries.
In conclusion, we present a novel segmentation approach that accurately quantifies the three-dimensional geometry of the thoracic aorta, key subsegments, and provides comprehensive phenotyping of key 3D aortic geometric properties. We leverage our segmentation approach to provide normative aging patterns for all AGPs, and we present the first GWAS results for many key aortic structural parameters. Future work should investigate the prognostic value of AGPs as well as the causal relationships of AGP GWAS results.
Supplementary Material
Acknowledgements
This research has been conducted using the UK Biobank Resource under Application Number 81032.
We acknowledge the Penn Medicine BioBank (PMBB) for providing data and thank the patient-participants of Penn Medicine who consented to participate in this research program. We would also like to thank the Penn Medicine BioBank team and Regeneron Genetics Center for providing genetic variant data for analysis. The PMBB is approved under IRB protocol# 813913 and supported by Perelman School of Medicine at University of Pennsylvania, a gift from the Smilow family, and the National Center for Advancing Translational Sciences of the National Institutes of Health under CTSA award number UL1TR001878.
Funding:
J.A.C. is supported by NIH grants R01-HL 121510, R33-HL-146390, R01HL153646, R01-AG058969, 1R01-HL104106, P01-HL094307, R03-HL146874, K24-AG070459 and R56-HL136730. W.R.W. is supported by NIH grants P41-EB029460, R01 HL169378, R01 HL137984, UL1 TR001878. J.G is supported by R01EB031722, R01HL133889.
Disclosures:
Dr. Chirinos is supported by NIH grants U01-TR003734, U01-TR003734–01S1, UO1-HL160277, R33-HL-146390, R01-HL153646, K24-AG070459, R01-AG058969, R01-HL157108, R01-HL155599, R01-HL104106 and R01HL155764. He has recently consulted for Bayer, Fukuda-Denshi, Bristol-Myers Squibb, Biohaven Pharmaceuticals, Johnson & Johnson, Edwards Life Sciences, Merck, and NGM Biopharmaceuticals. He received University of Pennsylvania research grants from National Institutes of Health, Fukuda-Denshi, Bristol-Myers Squibb, Microsoft and Abbott. He is named as inventor in a University of Pennsylvania patent for the use of inorganic nitrates/nitrites for the treatment of Heart Failure and Preserved Ejection Fraction and for the use of biomarkers in heart failure with preserved ejection fraction. He has received payments for editorial roles from the American Heart Association, the American College of Cardiology, Elsevier and Wiley, and payments for academic roles from the University of Texas, Boston University, and Virginia Commonwealth University. He has received research device loans from Atcor Medical, Fukuda-Denshi, Unex, Uscom, NDD Medical Technologies, Microsoft and MicroVision Medical. The remaining authors have nothing to disclose.
Footnotes
Supplemental Note
Penn Medicine BioBank Banner Author List and Contribution Statements
PMBB Leadership Team
Daniel J. Rader, M.D., Marylyn D. Ritchie, Ph.D.
Contribution: All authors contributed to securing funding, study design and oversight. All authors reviewed the final version of the manuscript.
Patient Recruitment and Regulatory Oversight
JoEllen Weaver, Nawar Naseer, Ph.D., M.P.H., Giorgio Sirugo, M.D., P.h.D., Afiya Poindexter, Yi-An Ko, Ph.D., Kyle P. Nerz
Contributions: JW manages patient recruitment and regulatory oversight of study. NN manages participant engagement, assists with regulatory oversight, and researcher access. GS assists with researcher access. AP, YK, KPN perform recruitment and enrollment of study participants.
Lab Operations
JoEllen Weaver, Meghan Livingstone, Fred Vadivieso, Stephanie DerOhannessian, Teo Tran, Julia Stephanowski, Salma Santos, Ned Haubein, P.h.D., Joseph Dunn
Contribution: JW, ML, FV, SD conduct oversight of lab operations. ML, FV, AK, SD, TT, JS, SS perform sample processing. NH, JD are responsible for sample tracking and the laboratory information management system.
Clinical Informatics
Anurag Verma, Ph.D., Colleen Morse Kripke, M.S. DPT, MSA, Marjorie Risman, M.S., Renae Judy, B.S., Colin Wollack, M.S.
Contribution: All authors contributed to the development and validation of clinical phenotypes used to identify study subjects and (when applicable) controls.
Genome Informatics
Anurag Verma Ph.D., Shefali S. Verma, Ph.D., Scott Damrauer, M.D., Yuki Bradford, M.S., Scott Dudek, M.S., Theodore Drivas, M.D., Ph.D.
Contribution: AV, SSV, and SD are responsible for the analysis, design, and infrastructure needed to quality control genotype and exome data. YB performs the analysis. TD and AV provides variant and gene annotations and their functional interpretation of variants.
References
- 1.Chirinos J.A., Segers P., Hughes T. & Townsend R. Large-Artery Stiffness in Health and Disease: JACC State-of-the-Art Review. Journal of the American College of Cardiology 74, 1237–1263 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vasan R.S., Song R.J., Xanthakis V. & Mitchell G.F. Aortic Root Diameter and Arterial Stiffness: Conjoint Relations to the Incidence of Cardiovascular Disease in the Framingham Heart Study. Hypertension 78, 1278–1286 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouke P.A. et al. Aortic elongation part I: the normal aortic ageing process. Heart 104, 1772 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Samuel H. et al. Aortic elongation part II: the risk of acute type A aortic dissection. Heart 104, 1778 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Aung N. et al. Genome-wide association analysis reveals insights into the genetic architecture of right ventricular structure and function. Nature Genetics 54, 783–791 (2022). [DOI] [PubMed] [Google Scholar]
- 6.Gomes B. et al. Genetic architecture of cardiac dynamic flow volumes. Nature Genetics (2023). [DOI] [PubMed] [Google Scholar]
- 7.Littlejohns T.J. et al. The UK Biobank imaging enhancement of 100,000 participants: rationale, data collection, management and future directions. Nature Communications 11, 2624 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verma A. et al. The Penn Medicine BioBank: Towards a Genomics-Enabled Learning Healthcare System to Accelerate Precision Medicine in a Diverse Population. Journal of Personalized Medicine 12, 1974 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Redheuil A. et al. Age-Related Changes in Aortic Arch Geometry: Relationship With Proximal Aortic Function and Left Ventricular Mass and Remodeling. Journal of the American College of Cardiology 58, 1262–1270 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindblad-Toh K. et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476–482 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward L.D. & Kellis M. Evidence of Abundant Purifying Selection in Humans for Recently Acquired Regulatory Functions. Science 337, 1675–1678 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trynka G. et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nature Genetics 45, 124–130 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hnisz D. et al. Super-Enhancers in the Control of Cell Identity and Disease. Cell 155, 934–947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pirruccello J.P. et al. Deep learning enables genetic analysis of the human thoracic aorta. Nature Genetics 54, 40–51 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vasan R.S. et al. Genetic Variants Associated With Cardiac Structure and Function: A Meta-analysis and Replication of Genome-wide Association Data. JAMA 302, 168–178 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tadros R. et al. Shared genetic pathways contribute to risk of hypertrophic and dilated cardiomyopathies with opposite directions of effect. Nature Genetics 53, 128–134 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kent W.J. et al. The Human Genome Browser at UCSC. Genome Research 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kichaev G. et al. Leveraging Polygenic Functional Enrichment to Improve GWAS Power. The American Journal of Human Genetics 104, 65–75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aragam K.G. et al. Discovery and systematic characterization of risk variants and genes for coronary artery disease in over a million participants. Nature Genetics 54, 1803–1815 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y. et al. Genome-wide association study of medication-use and associated disease in the UK Biobank. Nature Communications 10, 1891 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakaue S. et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nature Genetics 53, 1415–1424 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Omura J. et al. ADAMTS8 Promotes the Development of Pulmonary Arterial Hypertension and Right Ventricular Failure. Circulation Research 125, 884–906 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Slavin T.P., Feng T., Schnell A., Zhu X. & Elston R.C. Two-marker association tests yield new disease associations for coronary artery disease and hypertension. Human Genetics 130, 725–733 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo S., Zheng N. & Lang B. ULK4 in Neurodevelopmental and Neuropsychiatric Disorders. Frontiers in Cell and Developmental Biology 10(2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freshour S.L. et al. Integration of the Drug–Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Research 49, D1144–D1151 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohyama Y., Redheuil A., Kachenoura N., Ambale Venkatesh B. & Lima J.A.C. Imaging Insights on the Aorta in Aging. Circulation: Cardiovascular Imaging 11, e005617 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao B. et al. Heart-brain connections: Phenotypic and genetic insights from magnetic resonance images. Science 380, abn6598. [DOI] [PubMed] [Google Scholar]
- 28.Francis C.M. et al. Genome-wide associations of aortic distensibility suggest causality for aortic aneurysms and brain white matter hyperintensities. Nature Communications 13, 4505 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuwahara M. et al. CCN3 (NOV) Drives Degradative Changes in Aging Articular Cartilage. in International Journal of Molecular Sciences Vol. 21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waldman M. et al. Silencing the Adipocytokine NOV: A Novel Approach to Reversing Oxidative Stress-Induced Cardiometabolic Dysfunction. in Cells Vol. 11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Putnam E.A., Zhang H., Ramirez F. & Milewicz D.M. Fibrillin–2 (FBN2) mutations result in the Marfan–like disorder, congenital contractural arachnodactyly. Nature Genetics 11, 456–458 (1995). [DOI] [PubMed] [Google Scholar]
- 32.Wenguang Y. et al. Fibrillin-2 is a key mediator of smooth muscle extracellular matrix homeostasis during mouse tracheal tubulogenesis. European Respiratory Journal 53, 1800840 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Tunehag K.R. et al. CYP2C19 Genotype Is Associated With Adverse Cardiovascular Outcomes in Black Patients Treated With Clopidogrel Undergoing Percutaneous Coronary Intervention. Journal of the American Heart Association 13, e033791 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Z.-Y., Jing X. & Xiong X.-D. Emerging Role and Mechanism of the FTO Gene in Cardiovascular Diseases. Biomolecules 13, 850 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng Q. et al. Asarinin attenuates bleomycin-induced pulmonary fibrosis by activating PPARγ. Scientific Reports 13, 14706 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
UK Biobank data is available to researchers following UKB application approval and IRB approval. Researchers can apply at https://www.ukbiobank.ac.uk/. Access to Penn Medicine BioBank data is provided to investigators at the University of Pennsylvania. External access can be provided through collaboration with an investigator at the university. All GWAS summary statistics will be made available upon publication.