Abstract
Using a promoter trap, we have identified 56 Mycobacterium tuberculosis genes preferentially expressed in the mouse lung. Quantitative real-time PCR showed that RNA levels of several genes were higher from bacteria growing in mouse lungs than from broth cultures. These results support the current hypothesis that Mycobacterium tuberculosis utilizes fatty acids as a carbon source in the mouse lung.
In one of the earliest experiments comparing the physiology of Mycobacterium tuberculosis growing in broth culture with that of bacteria isolated from the mouse lung, Segal and Bloch (16) showed that bacteria isolated from lungs only responded to substrates containing fatty acids, whereas broth-grown bacilli responded to a variety of substrates, including carbohydrates. This was the first indication that the physiological state of this important pathogen was altered during infection of a mammalian host and that M. tuberculosis growing in the host obtains its energy mainly from degradation of fatty acids rather than from carbohydrates. Consistent with this idea, the annotated DNA sequence (http://genolist.pasteur.fr/TubercuList/) of M. tuberculosis (2) contains more than 250 genes involved in fatty acid metabolism, compared with an estimated 50 in Escherichia coli. Genetic experiments have shown that icl, coding for isocitrate lyase, is required for persistence of M. tuberculosis in the mouse lung (9). This enzyme is part of the glyoxylate cycle, an anapleurotic pathway required for growth on acetate, the end product of fatty acid oxidation. Results from several laboratories indicate that numerous genes probably involved in lipid metabolism are transcriptionally upregulated during growth in the mammalian macrophage (4, 15) and the mouse lung (15, 20). Genes required for iron uptake, various stress responses, and other processes are also upregulated during growth in the host (4, 15, 17, 20). This regulation may reflect the requirements of M. tuberculosis for survival and/or growth during infection, and such information may be useful in the design of new antibiotics or vaccines.
Classic promoter trap technology, which predates the modern use of DNA microarrays for genomic transcriptional analyses, is still useful under conditions where transcriptional profiling is impractical. We have described a promoter trap for M. tuberculosis, based upon overexpression of inhA, coding for an enoyl-ACP-reductase, a protein required for mycolic acid biosynthesis and the major target of isoniazid (INH) (1). Since overexpression of inhA confers resistance to INH (1), a promoter trap using this INH resistance was used to identify M. tuberculosis genes expressed during infection of human macrophages but not during growth in broth (4). We have now applied this technology to identify genes specifically expressed during mouse infection and have identified 56 promoters that drive inhA expression at higher levels during infection in the mouse lung than during growth in laboratory medium. The differential expression of seven of these genes has been validated by quantitative reverse transcription-PCR (RT-PCR) comparing M. tuberculosis RNA prepared from broth cultures with RNA from infected mouse lungs.
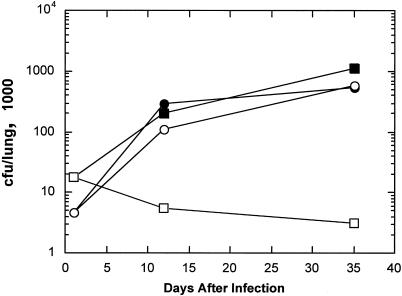
An M. tuberculosis strain expressing inhA from the hsp60 promoter confers resistance to INH in mouse lungs.
We infected mice intravenously with 105 to 106 CFU of M. tuberculosis H37Rv carrying plasmid pJD32 (the promoter trap vector with no promoter driving inhA) or plasmid pJD33 (same as pJD32, with the hsp60 promoter driving inhA) (4). One group of mice was treated with INH (0.1 g/liter of drinking water) (14) 24 h postinfection, and the other group was not given the antibiotic, and at various times after infection, the bacterial load was measured in the lung. As expected, the strain carrying pJD32 was sensitive to INH and the one carrying pJD33 was resistant (Fig. 1). This result indicated that the promoter trap selection could work in mice, since a difference of approximately 100-fold was evident between treated and untreated INH-sensitive (INH-s) strains after 35 days. We then infected mice with a library of strains carrying DNA fragments of M. tuberculosis cloned upstream of inhA in the pJD32 vector (4). The mice were provided INH in the drinking water and sacrificed after 5 weeks or 3 months. Whereas only 0.01% of the original library of M. tuberculosis clones were INH resistant (0.5 μg/ml INH) in vitro, after 5 weeks of selection in INH-treated mice, 5% surviving in the lungs were INH resistant in vitro (108 clones were tested), and after 3 months of selection in INH-treated mice, 55% surviving in the lungs were resistant when 1,122 clones were picked and tested by streaking them on plates containing 0.5 μg/ml INH. These results show that the selection worked in the mice since INH-sensitive clones were eliminated with increased selection time. The clones resistant on plates carry constitutively active cloned promoters, whereas the clones remaining sensitive on plates presumably carry promoters active in the mouse lung, but not in vitro. One hundred fifty-five clones isolated from the 3-month sample that were INH sensitive on plates were picked for determination of the DNA sequence cloned upstream of inhA in pJD32. Such clones are presumed to carry promoter sequences driving inhA expression specifically in the mouse lungs but not on laboratory medium.
FIG. 1.
INH sensitivity of strains in mouse lungs. M. tuberculosis H37Rv(pJD32) or M. tuberculosis H37Rv(pJD33) was used to infect C57BL/6 mice by tail vein injection. One group of mice was treated with INH, and the other group was not treated. Lungs were removed at various times after the infection, and the CFU were measured. pJD32 − INH, ▪; pJD32 + INH, □; pJD33 − INH, •; pJD33 + INH, ○.
Sequencing DNA inserts in selected clones.
Colonies of M. tuberculosis obtained from mouse lungs which were INH-s on plates were selected, the inserts were amplified by PCR, and the amplicons were sequenced as described previously (4). A total of 75/155 clones contained sequences corresponding to DNA regions 5′ to a known open reading frame in the M. tuberculosis H37Rv genome database (2) in the correct orientation to drive inhA expression, whereas the others carried internal fragments of open reading frames or fragments oriented in the wrong orientation. These inserts could contain sequences which function artifactually as promoters in the host. A total of 56/75 sequences were unique, and the genes driven by these promoters are shown in Table 1. Three (Rv0036c, Rv0406c, and Rv1323) had been identified as upregulated in THP-1 macrophages using the same promoter library (4).
TABLE 1.
Genes identified by promoter trap in mouse lungsa
| Rv no. | Gene annotation listing and possible function |
|---|---|
| Rv0036c | Unknown |
| Rv0079 | Unknown |
| Rv0158 | Transcriptional regulator |
| Rv0243 | fadA2; fatty acid oxidation |
| Rv0270 | fadD2; fatty acid oxidation |
| Rv0276 | Unknown |
| Rv0288 | esxH; ESAT-6 family of antigens |
| Rv0315 | Unknown |
| Rv0406c | Unknown |
| Rv0471c | Unknown |
| Rv0486 | mshA; mycothiol biosynthesis |
| Rv0628c | Unknown |
| Rv0812 | pabC; amino acid transferase |
| Rv0896 | gltA2; citrate synthase |
| Rv0945 | Short-chain dehydrogenase/reductase |
| Rv1056 | Unknown |
| Rv1151c | Transcriptional regulator |
| Rv1323 | fadA4; fatty acid oxidation |
| Rv1419 | Unknown |
| Rv1508c | Membrane protein |
| Rv1541c | lprl; lipoprotein |
| Rv1731 | gabD2 succinate semiald̈ehyde dehydrogenase |
| Rv1779c | Membrane protein |
| Rv1931c | Transcriptional regulatory protein |
| Rv1963c | mce3R transcriptional regulator of mce3 operon |
| Rv1997 | ctpF; metal cation transport ATPase, membrane protein |
| Rv2011c | Unknown |
| Rv2024c | Unknown |
| Rv2141c | Unknown |
| Rv2144c | Transmembrane protein |
| Rv2266 | cyp124: cytochrome P450 family, oxidation of fatty acids |
| Rv2336 | Unknown |
| Rv2346c | esXO; ESAT-6 family of antigens |
| Rv2376c | cfp2; low-molecular-weight antigen |
| Rv2405 | Unknown |
| Rv2510c | Unknown |
| Rv2673 | Membrane protein |
| Rv2713 | sthA; soluble pyridine nucleotide transhydrogenase |
| Rv2738c | Unknown |
| Rv2799 | Membrane protein |
| Rv2854 | Unknown |
| Rv2989 | Transcriptional regulator |
| Rv3206c | moeB1; molybdopterin biosynthesis |
| Rv3230c | Oxido-reductase |
| Rv3248c | sahH; adenosylhomocysteinase |
| Rv3260c | whiB2; transcriptional regulator |
| Rv3266c | rmlD; dTDP l-rhamnose biosynthesis |
| Rv3323c | moaX; molybdopterin biosynthesis |
| Rv3371 | Unknown |
| Rv3404c | Methionyl-tRNA formyl transferase |
| Rv3619c | esxV; esat-6 family of antigens |
| Rv3683 | Unknown |
| Rv3732 | Unknown |
| Rv3801c | fadD32; fatty acid oxidation |
| Rv3831 | Unknown |
| Rv3883c | mycP1; membrane-anchored serine protease |
| ValV | tRNA valine |
The genes were identified by sequencing the DNA upstream of inhA in M. tuberculosis clones surviving INH treatment in mice which were INH-s on plates.
Differential expression of selected genes measured by RT-PCR.
Mice were infected with the wild-type M. tuberculosis H37Rv by tail vein injection, and RNA was prepared from lungs 3 weeks after infection as well as from exponentially growing broth cultures. It should be emphasized that these measurements of RNA levels were done with a plasmid-free wild-type strain. Infected mouse lungs were removed from mice, snap-frozen in liquid nitrogen, and then maintained at −80°C. A frozen lung was placed in a petri dish and quickly cut into four pieces with a scalpel. Each piece was placed into a bead beater tube containing 0.5 ml of an equal mixture of 1- and 0.1-mm-diameter glass beads (Biospec products) and 1 ml Trizol. The samples were disrupted using the bead beater set at maximum speed for 1 min and then cooled on ice for 2 min. This procedure was repeated seven times, and the RNA was purified as previously described (4). RNA was also prepared from 40-ml broth cultures growing exponentially (optical density, 0.3 to 0.6) in Middlebrook 7H9 medium. Reverse transcription and PCR using molecular beacons were done as described previously (4). RNA was prepared from lungs of three infected mice, as well as from two different exponentially growing broth cultures. The values obtained for each gene are normalized to 16S RNA levels, which do not change during mouse lung infection (17). RNA levels of seven genes (esxH, whiB2, moeB1, sthA, Rv2854, Rv3230c, and fadA4) identified by the promoter trap selection in this paper and two (echA19 and fadA5^) identified previously in THP-1 cells (4) were all induced in mouse lungs (Table 2). echA19 and several other genes involved in fatty acid metabolism have been shown to be upregulated in murine bone marrow macrophages and mice (15). Three genes identified by the promoter trap described in this paper were probably not induced in mouse lungs (fadD2, cyp124, and lprl), although they may be induced at later times of infection. The differential expression of a few genes not identified by the promoter trap but predicted to code for proteins involved in lipid metabolism was also tested. As previously reported (9), iclA was upregulated. choD coding for cholesterol oxidase, a protein known to be required for virulence in Rhodococcus equi (10), was upregulated, but desA3, coding for a desaturase, was not. bfrB, coding for a bacteriferritin, which is induced under conditions of high iron (13), was not expressed at higher levels in the mouse lung. The bfrB results are consistent with the finding that mbtB, induced by low iron (5, 13), is upregulated in mouse lungs and that bfrA, also requiring high iron for its expression, was not induced in mouse lungs (20). These results support the hypothesis that M. tuberculosis faces a low-iron environment in mouse lungs.
TABLE 2.
Ratios of expression of selected genes in mouse lungs versus brotha
| Gene | Rv no. | Lung/broth gene expression ratio
|
Avg | ||
|---|---|---|---|---|---|
| Lung 1 | Lung 2 | Lung 3 | |||
| esxHb | Rv0288 | 49 | 25.3 | 20.6 | 31.6 |
| whiB2b | Rv3260c | 5.5 | 1.8 | 2.5 | 3.3 |
| moeB1b | Rv3206c | 2.38 | 0.76 | 5 | 2.7 |
| sthAb | Rv2713 | 7.1 | 17.9 | 15.7 | 13.6 |
| Unknownb | Rv3230c | 0.29 | 4.9 | 4.9 | 3.4 |
| Unknownb | Rv2854 | NDg | 8.7 | 11.8 | 10.25 |
| fadA4bc | Rv1323 | 6.6 | 11.9 | ND | 9.25 |
| echA19c | Rv3516 | 56 | 69.4 | 27.6 | 51 |
| fadA5c | Rv3546 | 6.1 | 2.4 | ND | 4.25 |
| iclAd | Rv0467 | 57.5 | ND | ND | 57.5 |
| choDe | Rv03409c | 7.3 | 10.7 | 11.3 | 9.8 |
| cyp124b | Rv2266 | 0.25 | 0.73 | 0.51 | 0.5 |
| lprlb | Rv1541c | 0.51 | 2 | ND | 1.26 |
| fadD2b | Rv0270 | 0.128 | 0.318 | 2.9 | 1.1 |
| bfrBf | Rv3841 | 1.2 | 0.35 | 0.36 | 0.64 |
| desA3e | Rv3229c | 0.165 | 0.8 | ND | 0.48 |
Mice were infected with M. tuberculosis H37Rv by tail vein injection, and the lungs were harvested 3 weeks after the infection when the bacteria are still growing. Gene ratios (lung/broth) are expressed as the number of cDNA copies for a particular gene/0.1 μl RNA isolated from infected mouse lungs, determined by mbRT-PCR, normalized to the number of cDNA copies for 16S RNA, compared with the same number determined for RNA isolated from broth cultures. The results shown are from three separate mice.
Genes identified by promoter trap in mice.
Genes identified by promoter trap in macrophages (4).
iclA was published as upregulated in mice (20); therefore, the analysis was only done with one lung.
Genes annotated as involved in lipid metabolism (2).
Induced by high iron (13).
ND, not determined.
Although we identified fadA4 in this screen as well as the one in macrophages, (4), there was no other overlap in the genes identified by the two screens. We should emphasize that neither screen should be considered as saturated for all genes upregulated during infection. The promoter trap technology can result in artifacts because we used a multicopy plasmid in which gene expression does not necessarily reflect normal gene expression from the chromosome. Also, the readout for gene expression is resistance to INH in the host, and the dose received in the lung may be variable in different areas of the lung and at different times. In addition, we selected clones which were completely sensitive to INH in vitro. This selection would therefore eliminate many genes which are differentially expressed but which have some level of expression in vitro.
We identified three members of the ESAT-6 family, esxH, esXO, and esxV, as upregulated during growth in mouse lungs. ESAT-6 and TB10.4 are members of a large family of small secreted proteins. These two proteins are also immunodominant antigens in tuberculosis patients (12, 18). TB10.4 is coded for by Rv0288, one of the genes identified by our promoter trap and validated by reverse transcription-PCR using molecular beacons. ESAT-6 was recognized as of possible importance for virulence since it is one of the genes present in the RD1 region of the M. tuberculosis chromosome, which is deleted in the avirulent vaccine strain, Mycobacterium bovis BCG (8). Since the host responds vigorously to this group of proteins, there are several promising strategies for vaccine production involving the expression of ESAT-6 either as a recombinant protein fused to other antigens (11) or as a DNA vaccine (6, 7).
Molybdopterin is a cofactor required for nitrate reductase and other enzymes involved in anaerobic metabolism. M. tuberculosis dedicates 21 genes to the biosynthesis of this cofactor (2), and we identified two of them (moaX and moeB1) as upregulated in mouse lungs. Seven orthologues of the Streptomyces whi genes are found in the M. tuberculosis chromosome (2), and we identified whiB2 as upregulated in mouse lungs. In Streptomyces, they code for small transcriptional regulators, but, with the exception of whiB3, their function is unknown in M. tuberculosis. whiB3 codes for a small DNA binding protein which interacts with SigA, the housekeeping sigma factor (19). Specific mutations in sigA cause attenuation of M. bovis (3) due to the inability of the mutant SigA to interact with WhiB3 (19).
Four new genes involved in lipid metabolism were shown to be upregulated during growth in mouse lungs, lending further support to the idea that M. tuberculosis utilizes fatty acids during infection of the host. Some of the genes may be indispensable for growth and/or persistence in the host, and we are currently constructing strains carrying disruptions in the genes identified in this report. Preliminary experiments have shown that a mutation in fadA5 causes attenuation (Fontan et al., unpublished observations).
Acknowledgments
We thank Lidya Sanchez for designing most of the RT primers, Patricia Fontán for help with some of the experiments and useful discussions, and Dave Dubnau for helpful discussions and critical reading of the manuscript.
This work was supported by National Institutes of Health grant HL 64544 (awarded to I.S.).
Editor: J. L. Flynn
REFERENCES
- 1.Banerjee, A., E. Dubnau, A. Quemard, V. Balasubramanian, K. S. Um, T. Wilson, D. Collins, G. de Lisle, and W. R. Jacobs, Jr. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227-230. [DOI] [PubMed] [Google Scholar]
- 2.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 3.Collins, D. M., R. P. Kawakami, G. W. de Lisle, L. Pascopella, B. R. Bloom, and W. R. Jacobs, Jr. 1995. Mutation of the principal sigma factor causes loss of virulence in a strain of the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 92:8036-8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubnau, E., P. Fontán, R. Manganelli, S. Soares-Appel, and I. Smith. 2002. Mycobacterium tuberculosis genes induced during infection of human macrophages. Infect. Immun. 70:2787-2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gold, B., G. M. Rodriguez, S. A. E. Marras, M. Pentecost, and I. Smith. 2001. The Mycobacterium tuberculosis IdeR is a dual functional regulator that controls transcription of genes involved in iron acquisition, iron storage and survival in macrophages. Mol. Microbiol. 42:851-865. [DOI] [PubMed] [Google Scholar]
- 6.Kamath, A. T., C. G. Feng, M. Macdonald, H. Briscoe, and W. J. Britton. 1999. Differential protective efficacy of DNA vaccines expressing secreted proteins of Mycobacterium tuberculosis. Infect. Immun. 67:1702-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li, Z., A. Howard, C. Kelley, G. Delogu, F. Collins, and S. Morris. 1999. Immunogenicity of DNA vaccines expressing tuberculosis proteins fused to tissue plasminogen activator signal sequences. Infect. Immun. 67:4780-4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 178:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKinney, J. D., K. Honer zu Bentrup, E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 10.Navas, J., B. Gonzalez-Zorn, N. Ladrón, P. Garrido, and J. A. Vázquez-Boland. 2001. Identification and mutagenesis by allelic exchange of choE, encoding a cholesterol oxidase from the intracellular pathogen Rhodococcus equi. J. Bacteriol. 183:4796-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olsen, A. W., L. A. H. van Pinxteren, L. M. Okkels, P. B. Rasmussen, and P. Andersen. 2001. Protection of mice with a tuberculosis subunit vaccine based on a fusion protein of antigen 85B and ESAT-6. Infect. Immun. 69:2773-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ravn, P., A. Demissie, T. Eguale, H. Wondwosson, D. Lein, H. A. Amoudy, A. S. Mustafa, A. K. Jensen, A. Holm, I. Rosenkrands, F. Oftung, J. Olobo, F. von Reyn, and P. Andersen. 1999. Human T cell responses to the ESAT-6 antigen from Mycobacterium tuberculosis. J. Infect. Dis. 179:637-645. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez, G. M., M. I. Voskuil, B. Gold, G. K. Schoolnik, and I. Smith. 2002. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70:3371-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scanga, C. A., V. P. Mohan, H. Joseph, K. Yu, J. Chan, and J. L. Flynn. 1999. Reactivation of latent tuberculosis: variations on the Cornell murine model. Infect. Immun. 67:4531-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segal, W., and H. Bloch. 1956. Biochemical differentiation of Mycobacterium tuberculosis grown in vivo and in vitro. J. Bacteriol. 72:132-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi, L., Y.-J. Jung, S. Tyagi, M. L. Gennaro, and R. J. North. 2002. Expression of Th1-mediated immunity in mouse lungs induces a Mycobacterium tuberculosis transcription pattern characteristic of nonreplicating persistence. Proc. Natl. Acad. Sci. USA 100:241-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skjøt, R. L. V., T. Oettinger, I. Rosenkrands, P. Ravn, I. Brock, S. Jacobsen, and P. Andersen. 2000. Comparative evaluation of low-molecular-mass proteins from Mycobacterium tuberculosis identifies members of the ESAT-6 family as immunodominant T-cell antigens. Infect. Immun. 68:214-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steyn, A. J. C., D. M. Collins, M. K. Hondalus, W. R. J. Jacobs, R. P. Kawakami, and B. R. Bloom. 2002. Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc. Natl. Acad. Sci. USA 99:3147-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Timm, J., F. A. Post, L.-G. Bekker, G. B. Walther, H. C. Wainwright, R. Manganelli, W.-T. Chan, L. Tsenova, B. Gold, I. Smith, G. Kaplan, and J. D. McKinney. 2003. Differential expression of iron-, carbon-, and oxygen-responsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc. Natl. Acad. Sci. USA 24:14321-14326. [DOI] [PMC free article] [PubMed] [Google Scholar]