THE GASTROINTESTINAL ECOSYSTEM
The gastrointestinal (GI) tract is the site of the largest and most complex environment in the mammalian host. In the adult human, the total microbial population (ca. 1014 cells) is estimated to exceed the total number of mammalian cells by at least an order of magnitude (5). The GI tract features several diverse “macro” environments, including the oral cavity, the stomach, the small intestine (including the three major regions, the duodenum, jejunum, and ileum), and the large intestine (colon). The density of bacteria along the GI tract can vary greatly, from 103/ml near the gastric outlet to 1010/ml at the ileocecal valve to 1011 to 1012/ml in the colon (6). Within these “macro” environments are several “micro” environments where bacteria can live, such as the lumen of the bowel, the mucus layer overlying the epithelium, mucus within intestinal crypts, and the surface of mucosal epithelial cells. This wide diversity of environments harbors an even wider diversity of bacterial species, and an estimated 500 to 1,000 different species are present in the GI tract, with an aggregate biomass of ca. 1.5 kg (90). This immense microbial population represents enormous genetic diversity. Assuming an average genome the size of that of Escherichia coli for 1,000 species, the number of genes in this “microbiome” may exceed the total number of human genes by a factor of ca. 100 (90).
A sobering fact about the microbial population of the GI tract is that the majority of the estimated 500 to 1,000 species have not yet been cultured in vitro. Molecular techniques, such as microbial community genome sequencing projects (83), are being applied to the microbiome to more fully characterize this population. Of the commensal bacteria that have been cultured from the GI tract, >99.9% are obligate anaerobes. The dominant commensal microbial genera that can be cultured from the GI tract include Bacteroides, Bifidobacterium, Eubacterium, Lactobacillus, Clostridium, Fusobacterium, Peptococcus, Peptostreptococcus, Escherichia, and Veillonella (38). The commensal flora has a number of benefits for the host, including nutritional contributions, protection from infection, maturation of the immune system, and maturation of the intestinal mucosa (5).
Given the enormous number and diversity of bacteria that comprise the GI environment, it would not be surprising if the members of this community were to somehow communicate among themselves to coordinate various processes ranging from maintenance of the commensal population to aiding or resisting infectious diseases. Quorum sensing (QS) is an important mechanism of cell-to-cell communication that involves density-dependent recognition of signaling molecules, resulting in modulation of gene expression. The first report of a potential role for QS in GI infections was published in 1999 (73), and many reports for different GI pathogens have followed. This review discusses recent information concerning QS among bacteria of the GI tract, with a particular emphasis on pathogenic species that cause infection in the GI tract.
QUORUM SENSING
Overview.
QS is a cell-to-cell signaling mechanism that refers to the ability of bacteria to respond to chemical molecules, called autoinducers (AIs), in their environment in a dose-dependent fashion. The autoinducers can be produced from bacteria of the same species or bacteria of different genera. When an autoinducer concentration reaches a critical threshold, the bacteria detect and respond to this signal by altering their gene expression. This phenomenon allows bacteria to act as a collective unit, i.e., a multicellular entity, as opposed to individual cells all performing individual functions.
QS was first characterized in the marine bacterium Vibrio fischeri (reviewed in references 23 and 63). This species lives in symbiotic associations with several different marine animal hosts and can colonize the light organ of the host, in which it grows to high densities. V. fischeri produces a luciferase enzyme complex encoded by the luxCDABEGH genes that is responsible for light production. However, transcription of the lux operon and subsequent light production occur only at high densities of V. fischeri and are repressed at low densities. A protein called LuxI is responsible for production of a signaling molecule, an AI, that diffuses across the membrane into the extracellular environment or back into the cytoplasm, where it binds to a protein called LuxR. The AI of V. fischeri is an acylated homoserine lactone (HSL). When the bacterial density is high, the concentration of AI is high and the LuxR-AI complex becomes an activated transcription factor that induces transcription of the luxCDABEGH genes. When bacterial densities are low, the concentration of AI molecules is below the threshold required to activate transcription and the lux genes encoding luciferase are not expressed.
Since the initial description for V. fischeri, QS has now been recognized to regulate a wide range of activities in diverse bacteria, including plasmid transfer and plant tumor induction by Agrobacterium tumefaciens, antibiotic production in Erwinia carotovora, biofilm production and virulence gene expression in Pseudomonas aeruginosa, competence and sporulation in Bacillus subtilis, competence for DNA uptake in Streptococcus pneumoniae, and virulence gene expression in numerous pathogens, including Staphylococcus aureus, Vibrio cholerae, and diarrheagenic E. coli (reviewed in references 16, 45, and 78). In addition to modulating expression of specific functions that are best achieved by a whole population rather than individual bacteria, QS may be used as a system for bacteria to prevent the population from growing to levels that are unsustainable in their environment. If all the nutrients are depleted and waste products are not removed from their environment, it will be deleterious for the community as a whole. In effect, QS is used to determine the fitness of a population (89).
Three major QS circuits have been described; one is used primarily by gram-negative bacteria, one is used primarily by gram-positive bacteria, and one has been proposed to be universal. The gram-negative QS system involves the use of acyl homoserine lactones (AHLs) as autoinducers, which then bind to response regulators that affect gene expression. The gram-positive bacteria use oligopeptide autoinducers that are detected by two-component systems. The third QS system is proposed to be a universal system that allows interspecies communication and is found in both gram negatives and gram positives. There have been numerous recent reviews of the various QS systems (4, 23, 54, 63, 78, 89), and the reader is referred to one or more of these for additional details. The broad concepts of these three systems are briefly reviewed below, with particular emphasis on the third system since it has been studied in all GI pathogens discussed here.
Gram-negative LuxIR systems.
The paradigm for QS in gram-negative bacteria is the LuxIR system, first described for V. fischeri. The LuxIR system uses the LuxI protein, or a homologue of this protein, to synthesize an autoinducer and LuxR (or a homologue of LuxR) as a regulator that binds to the autoinducer and modulates gene expression (24). This system exhibits great specificity, as the AI produced by one species of bacteria can rarely, if ever, interact with the LuxR-type regulator of another species. More than 70 LuxIR QS systems have been found in gram-negative bacteria (15, 24, 53). Interestingly, the great specificity seen with the AI-LuxR interaction is not seen at the level of binding of the activated LuxR transcriptional factor to DNA in the promoter region of the regulated gene since LuxR proteins from different species all bind to similar DNA sequences called “lux boxes” (78).
The AI molecule produced by the LuxIR systems is an AHL, in which there is a common homoserine lactone moiety but variable acyl side chains. The function of the LuxI protein is to link the side chain group of specific acyl-acyl carrier proteins to the homocysteine moiety of S-adenosylmethionine (SAM). Some species may produce more than one AHL AI and have more than one LuxIR pair. For example, in P. aeruginosa, one pair of LuxIR homologues called LasI/LasR produces and responds to an AHL called 3-oxo-C12-HSL, and in the same strain the RhlI/RhlR proteins produce and respond to an AHL called C4-HSL (15, 24, 53). The LasI/LasR system regulates exotoxin A, LasA, LasB, Xcp, and biofilm formation, while the RhlI/RhlR system regulates LasB, rhamnolipid, RpoS, and secondary metabolites. Interestingly, LasI/LasR regulates RhlI/RhlR, thereby allowing the genes controlled by the former to be expressed prior to genes controlled by the latter in a hierarchy of temporal gene expression.
Gram-positive oligopeptide systems.
Rather than AHLs, the QS system used by gram-positive bacteria utilizes peptides as AI signaling molecules. These autoinducing polypeptides (AIPs) are produced in the cytoplasm as precursor peptides and then cleaved, modified, and exported. The extracellular AIPs are detected via two-component systems in which the external portion of a membrane-bound sensor kinase protein detects the AIP and then phosphorylates and activates a response regulator that binds to DNA and modulates transcription. S. aureus has served as a prototype for the gram-positive AIP systems, and the S. aureus Agr (accessory gene regulator) QS system regulates virulence gene expression and biofilm formation (reviewed in references 40, 50, and 92). The S. aureus Agr system utilizes an oligopeptide produced by AgrD that is modified by AgrB. The resulting AIP is 8 or 9 amino acids long and contains thiolactone rings. Detection of the extracellular AIP and subsequent gene activation is by the two-component system encoded by agrAC. The AIP of S. aureus is even more specific than AHLs, and there are four subgroups of this species defined by the AIP they produce. Not only does an AIP produced by one subgroup of S. aureus not activate gene expression in another subgroup, but it also inhibits the QS system in another subgroup. This was demonstrated in a mouse model of infection in which mice infected with S. aureus from one subgroup were protected from disease if an AIP from another subgroup was added to the inoculum (41).
LuxS/AI-2 system.
The third major QS system present in bacteria is found in a wide variety of bacteria, including both gram-negative and gram-positive species (64). This system, called the LuxS or autoinducer 2 (AI-2) system, has been detected in more than 55 species by sequence analysis or functional assays (47, 89). LuxS was initially characterized in V. harveyi, which also has an AHL QS system. The signal molecule of this system, AI-2, is detected in the environment by a two-component system called LuxP/LuxQ, and the resulting phosphorylation cascade results in modulation of gene transcription. In V. harveyi, AI-2 activates expression of luciferase at high densities, resulting in bioluminescence. V. harveyi mutants deficient in production of AI-2 have been used to test other bacterial species for production of AI-2 by adding culture supernatants of these species to the V. harveyi mutant and testing for bioluminescence (77). LuxS is an enzyme involved in the metabolism of SAM; it converts ribose-homocysteine into homocysteine and 4,5-dihydrody-2,3-pentanedione. 4,5-Dihydrody-2,3-pentanedione is a very unstable compound that reacts with water and cyclizes into several furanones (64, 75, 87), one of which is thought to be the precursor of AI-2 (64). The AI-2 structure has been solved by cocrystallizing this ligand with its receptor, LuxP (a periplasmic protein that resembles the ribose-binding protein RbsB), in V. harveyi, and it has been reported to be a furanosyl-borate diester (8). However, LuxP homologues, as well as homologues from this signaling cascade, have been found only in Vibrio spp. In non-Vibrio species, the only genes shown to be directly regulated by AI-2 encode an ABC transporter in Salmonella enterica serovar Typhimurium named Lsr (LuxS regulated), which is responsible for the AI-2 uptake by this species (80). This ABC transporter is also present in E. coli and exhibits homology with sugar transporters. AI-2 binds to LsrB and is transported inside the cell, where it is phosphorylated by LsrK and proposed to interact with LsrR, which is a SorC-like transcription factor involved in repressing expression of the lsr operon (79, 80) (Fig. 1). Several groups have been unable to detect the furanosyl-borate diester, proposed to be AI-2, in purified fractions containing AI-2 activity from Salmonella and E. coli (as measured using the V. harveyi bioluminescence assay) (64, 75, 87). The results for these fractions yielded only identification of several furanosyl compounds that did not contain boron. These results can be explained now that AI-2 has been cocrystallized with its receptor (the periplasmic protein LsrB) in Salmonella. In these studies the LsrB ligand was not a furanosyl-borate diester but was a furanone [(2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran] (47), consistent with what has been observed in AI-2 fractions of Salmonella and E. coli (64, 75, 87). This scenario is fundamentally different from AI-2 detection in V. harveyi and raises the question of whether all bacteria may actually use AI-2 as a signaling compound or whether it is released as a waste product or used as a metabolite by some bacteria, rather than as a signal (87, 88).
FIG. 1.
(A) Lsr ABC transporter system (LuxS-regulated genes) from S. enterica serovar Typhimurium and EHEC serotype O157:H7. All of the lsr genes from Salmonella are present in EHEC, with the exception of lsrE. The lsrACDBFGE genes are transcribed in an operon, and divergently transcribed from this operon are lsrR, encoding the SorC-like transcription factor that represses expression of the LsrABC transporter, and lsrK, whose product phosphorylates AI-2 upon its entry into the bacterial cell (79, 80). (B) LsrB exhibits homology with the ribose-binding periplasmic protein RbsB, which together with RbsC, RbsA, and RbsK is responsible for transport of ribose into the cell. (C) LsrB is the receptor for AI-2 in Salmonella and E. coli (47). Upon binding to LsrB, AI-2 is transported through the LsrR ABC transporter, which closely resembles the ribose ABC transporter. Once inside the cell AI-2 is phosphorylated by LsrK. Phospho-AI-2 is thought to interact with LsrR to relieve the repression of the lsr operon.
Genes potentially regulated by AI-2 in other species have been identified by constructing a luxS mutant of a test species and comparing gene expression in the wild type and the luxS mutant. Among the phenotypes and functions affected by luxS mutations are type III secretion (TTS) and flagellum expression in enterohemorrhagic E. coli (EHEC) O157:H7 (73, 74), expression of VirB in Shigella flexneri (14), secretion of SpeB cysteine protease in Streptococcus pyogenes (37), TTS in V. harveyi and Vibrio parahaemolyticus (28), and other phenotypes reviewed below. The most comprehensive analysis published so far has been the analysis of E. coli, and two microarray analyses by two different groups have compared genomic differences in gene expression between luxS-negative mutants and their isogenic luxS-positive parent strains. Using an E. coli K-12 microarray and mRNA and cDNA from EHEC (an EHEC array was not available at the time), Sperandio et al. (74) found that approximately 10% of the genes in the K-12 genome were differentially regulated fivefold in a wild-type EHEC strain and its isogenic luxS mutant, with roughly equal numbers of genes being positively regulated and negatively regulated (73, 74). In a separate study, DeLisa and colleagues (17) reported that about 5.6% of the K-12 genome was differentially regulated in a wild-type K-12 strain and its isogenic luxS mutant. The difference in the number of genes regulated in the two reports may have been due to differences in methodology, including differences in growth temperature (37°C in the study of Sperandio et al. versus 30°C in the study of DeLisa et al.), nutrient availability (Dulbecco modified Eagle medium in the study of Sperandio et al. and LB broth in the study of DeLisa et al.), and E. coli strain analyzed (EHEC in the study of Sperandio et al. and E. coli K-12 in the study of DeLisa et al). The observation that up to 10% of the array genes are differentially regulated in an EHEC wild-type strain and its isogenic luxS mutant is not surprising if one considers the pleiotropic nature of a luxS mutation.
LuxS is a metabolic enzyme involved primarily in the conversion of ribosyl-homocysteine into homocysteine and 4,5-dihydroxy-2,3-pentanedione, which is the precursor of AI-2 (64). A luxS mutation interrupts this metabolic pathway, thereby changing the whole metabolism of the bacteria. A luxS mutant accumulates S-ribosyl-homocysteine, because it is unable to catalyze its conversion to homocysteine. This could cause the levels of homocysteine to diminish within the cell. Given that homocysteine is used for the de novo synthesis of methionine, the cell then employs a salvage pathway (e.g., using oxaloacetate to produce homocysteine to synthesize methionine). Since oxaloacetate is necessary together with l-glutamate to synthesize aspartate, the use of this salvage pathway for the de novo synthesis of methionine affects other amino acid synthesis and catabolic pathways within the cell (http://www.ecosal.org/ecosal/index.jsp).
A recent breakthrough in distinguishing the potential cell signaling functions from general metabolic functions was the discovery of a new signaling molecule called AI-3, whose synthesis is dependent on LuxS. Building on their previous studies showing that a luxS mutant of EHEC was deficient in TTS and flagellum production, Sperandio et al. (75) showed that purified and “in vitro” synthesized AI-2 was unable to restore these phenotypes when it was added to the mutant. The autoinducer responsible for this signaling is dependent on the presence of the luxS gene for its synthesis, but it is different from AI-2. AI-2 is a very polar furanone that does not bind to C18 columns. The signaling compound activating the EHEC virulence genes, which was designated AI-3, binds to C18 columns and can be eluted only with methanol (75). Electrospray mass spectrometry analysis of the AI-3 fraction showed a major peak at a mass of 213.1 Da and minor peaks at 109.1, 164.9, 176.1, 196.1, 211.1, 214.1, and 222.9 Da (75). All of these masses are different from that of AI-2 (8), indicating that AI-3 is a novel compound. These results suggest that some of the phenotypes attributed to AI-2 signaling need to be revised in light of the fact that LuxS is not devoted to AI-2 production; it is in fact an important enzyme whose absence affects the metabolism of SAM and various amino acid pathways, as described above. Consequently, altered gene expression due to a luxS mutation includes both genes affected by QS per se and genes differentially expressed because of the interruption of this metabolic pathway. Furthermore, one also has to take into consideration that a knockout of luxS seems to affect the synthesis of at least two autoinducers, AI-2 and AI-3 (75). The activities of the two signals can be uncoupled by utilizing biological tests specific to each signal. For example, AI-3 shows no activity in the V. harveyi bioluminescence assay (75), which is the “gold standard” test for AI-2 production (77). On the other hand, AI-3 activates the transcription of the EHEC TTS genes, while AI-2 has no effect in this assay (75). The only two phenotypes shown to be AI-2 dependent, using either purified or in vitro synthesized AI-2, are bioluminescence in V. harveyi (64) and expression of the lsr operon in S. enterica serovar Typhimurium (80).
Several species of bacteria that are found in the GI tract for either short or long periods of time possess the luxS gene; these bacteria include commensal and pathogenic E. coli, S. enterica serotype Typhimurium, S. enterica serotype Typhi, S. flexneri, Helicobacter pylori, Campylobacter jejuni, V. cholerae, Enterococcus faecalis, S. aureus, Clostridium difficile, Clostridium perfringens, Bacillus species, and Streptococcus species (63). Using the V. harveyi bioluminescence assay, AI-2 has also been shown to be produced by several species of rumen bacteria, including Butyrivibrio fibrisolvens, Eubacterium ruminantium, Ruminococcus flavefaciens, and Succinimonas amylolytica (48). It has recently been shown, using anaerobically cultured stools from healthy human volunteers, that the microbial intestinal flora produces both AI-2 (using the V. harveyi bioluminescence assay) and AI-3 (using the LEE1 transcription AI-3-dependent bioassay) (75). To obtain further information regarding which intestinal commensals and pathogens are able to produce AI-2 and AI-3, freshly isolated strains from patients were tested (M. P. Sircili and V. Sperandio, unpublished results). Using the bioassays described above, AI-2 and AI-3 activity was observed in spent supernatants from enteropathogenic E. coli strains belonging to serogroups O26:H11 and O111ac:H9, Shigella sp., and Salmonella sp. Activity of both autoinducers was also detected in normal flora bacteria, such as a commensal E. coli, Klebsiella pneumoniae, and Enterobacter cloacae (Sircili and Sperandio, unpublished). These results suggest that AI-3 production is not limited to EHEC and that both AI-2 and AI-3 may be involved in interspecies signaling among intestinal bacteria and could play a role in the pathogenesis of disease caused by these other bacteria.
QUORUM SENSING BY GASTRONINTESTINAL PATHOGENS
For bacterial pathogens, QS has been proposed to aid the disease process by allowing bacteria to appropriately time expression of virulence factors that might activate a defensive immune response before the infection has progressed. As described by De Kievit et al., through QS, “bacteria can amass a high cell density before virulence determinants are expressed, and in so doing, the bacteria are able to make a concerted attack and produce ample virulence factors to overwhelm the host defenses” (15). This basic concept may be sufficient to explain the role of QS in extraintestinal infections where the pathogen encounters low levels of competing microflora, but in the complex microbial flora of the GI tract, QS may play additional roles in disease. As described below, QS has been hypothesized to signal transcription of EHEC virulence factors once it arrives in the large intestine, as well as to signal transcription of Vibrio environmental survival factors after expression of virulence factors and prior to environmental release via fecal discharge. QS has been described in numerous bacterial pathogens of the GI tract, and in many cases, the role of QS in the pathogenesis of the diseases caused by these organisms is not so clear-cut. However, many examples of QS in enteric pathogens have been found that have yielded new insights into the disease process (Table 1).
TABLE 1.
Summary of quorum sensing systems described so far in enteric bacteria
| Bacterial species | QS system | Autoinducer(s) | Regulated phenotype(s) |
|---|---|---|---|
| EHEC and EPEC | Qse/LuxS | AI-3 | TTS, flagella, and motility |
| Lsr/LuxS | AI-2 (R-THMF)a | AI-2 uptake by Lsr | |
| SdiA | AHLs (not self-produced) | ? | |
| Salmonella sp. | Qse/LuxS | AI-3 | ? |
| Lsr/LuxS | AI-2 (R-THMF) | AI-2 uptake by Lsr; biofilm formation? | |
| SdiA | AHLs (not self-produced) | Resistance to human complement (Rck) | |
| Vibrio cholerae | System 1 (CqsA/CqsS) | CAI-1 | TCP, CT, HA protease, biofilm |
| System 2 (LuxS) | AI-2 (furanosyl-borate diester?) | TCP, CT, HA protease, biofilm | |
| System 3 | ? | ? | |
| EAEC | Qse/LuxS | AI-3 | ? |
| Lsr/LuxS | AI-2 (R-THMF) | ? | |
| SdiA | AHLs (not self-produced) | ? | |
| Other? | ? | AggR-regulated virulence genes | |
| Enterococcus faecalis | Cyls | CyILs | Cytolysin production |
| FsR | Peptide | Gelatinase, serine protease | |
| LuxS | AI-2 | ? | |
| Yersinia sp. | Qse/LuxS | AI-3 | ? |
| Lsr/LuxS | AI-2 | ? | |
| YenR/I | AHLs | ? | |
| YpsR/I | AHLs | Flagella and motility | |
| YtbR/I | AHLs | Flagella and motility | |
| Shigella flexneri | Qse/LuxS | AI-3 | Expression of VirB? |
| Lsr/LuxS | AI-2 | Expression of VirB? | |
| Campylobacter jejuni | LuxS | AI-2 | Motility |
| Vibrio vulnificus | LuxS | AI-2 (furanosyl-borate diester?) | Protease, hemolysin |
| SmrC | ? | Protease, hemolysin, virulence | |
| Vibrio parahaemolyticus | LuxM, LuxR (OpaR) | AI-1 (AHL?) | TTS |
| LuxP, LuxQ, LuxS | AI-2 (furanosyl-borate diester?) | ? |
R-THMF, (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahyrofuran.
Enteropathogenic and enterohemorrhagic E. coli.
The first evidence that QS could be involved in the regulation of virulence factors of GI pathogens was found with enteropathogenic E. coli (EPEC), a cause of nonbloody diarrhea primarily in infants in developing countries, and EHEC, which causes bloody diarrhea and hemolytic-uremic syndrome. Although an important difference between these two pathogens is the expression of Shiga toxin (Stx) by EHEC but not by EPEC, they both produce a characteristic intestinal histopathology known as attaching and effacing (AE) lesions (33). The genes responsible for the AE phenotype are found in a pathogenicity island known as the locus of enterocyte effacement (LEE) (42). The LEE encodes a TTS system that translocates effector proteins into intestinal epithelial cells, thereby causing the dramatic cytoskeletal changes that lead to the AE lesions. Sperandio et al. (73) found that cell-free supernatants from culture media preconditioned by growth of EHEC or E. coli K-12 strains activated expression of the LEE genes. When the EHEC luxS gene was mutated, this activation was abrogated. Because EHEC is notable for the very low infectious dose needed to cause infection (10 to 100 CFU), it is unlikely that EHEC “talks” to itself via QS, at least in the initial stages of infection. Instead, we proposed a model in which the EHEC LEE genes are induced by signaling molecules produced by large numbers of commensal E. coli and other species present in the colon. In support of this model, a subsequent study showed that fecal specimens from 10 of 12 healthy individuals contained signaling molecules that were capable of inducing light in the V. harveyi bioluminescence assay for AI-2 (75). Thus, it appears that EHEC uses QS to detect signaling molecules produced by high numbers of commensal flora to determine when it is in an appropriate environment to express genes necessary for intestinal colonization and disease.
As noted above, a microarray analysis of a luxS EHEC mutant revealed that ca. 10% of the genes shared by EHEC O157:H7 and K-12 were differentially regulated in the mutant and wild type. Among the QS-regulated genes and phenotypes noted in these studies were the genes encoding flagella and motility (which may also be involved in pathogenesis) (74). Specifically, it was shown that transcription of flhDC (encoding the master regulator of the flagellum regulon) and transcription of the mot operon (encoding motility genes) are decreased in a luxS mutant compared to wild-type and complemented strains. Transcription of these genes, as well as motility, could be restored by addition of signals exogenously, further confirming that regulation of flagellum expression and motility is controlled by a quorum sensing signaling mechanism (74, 75). QS regulation of flhDC expression has far-reaching implications beyond flagellum expression, given that FlhDC has been shown to also regulate bacterial cell division (57, 58) and several metabolic processes (56). QS regulation of the LEE-encoded TTS system and the flagellum regulon in EHEC is dependent on the AI-3 signal; the role of AI-2 signaling in EHEC remains to be established (75). Given the widespread nature of the luxS/AI-3 system in bacteria, an interesting extrapolation is that the AI-3/luxS quorum sensing system might have initially evolved to mediate microflora-host interactions but was subsequently exploited by EHEC to activate its virulence genes. In this manner, the AI-3/luxS system alerts EHEC to when it has reached the large intestine, where large numbers of commensal E. coli, Enterococcus, Clostridium, and Bacteroides, all of which contain the AI-3/luxS quorum sensing system, reside (Sircili and Sperandio, unpublished).
The most recent study in this series demonstrated that an EHEC luxS mutant, which was unable to produce AI-3 and unable to express the LEE-encoded TTS system at normal levels, nonetheless still produced AE lesions on epithelial cells that were indistinguishable from those seen with the wild type (75). The luxS mutant still responded to a eukaryotic cell signal to activate expression of the LEE genes. This signal was identified as the hormone epinephrine, and it was further shown that beta- and alpha-adrenergic antagonists that block the effect of epinephrine can block the bacterial response to this hormone. The luxS mutant also responded similarly to the hormone norepinephrine. Both epinephrine and norepinephrine are present in the GI tract. Norepinephrine is synthesized within the adrenergic neurons present in the enteric nervous system (25). Although epinephrine is not synthesized in the enteric nervous system (it is synthesized in the central nervous system and in the adrenal medulla), it acts in a systemic manner after being released by the adrenal medula into the bloodstream, thereby reaching the intestine (59). These results imply that there is potential cross-communication between the luxS/AI-3 bacterial QS system and the epinephrine-norepinephrine host signaling system. The multiple signals present in the intestine of either bacterial or host origin could allow further fine-tuning for expression of EHEC genes, in which one set of genes (e.g., those encoding flagella) may be expressed at one time while another set of genes (e.g., those encoding the LEE TTS) may be expressed at a slightly different time. Since eukaryotic cell-to-cell signaling typically occurs through hormones and bacterial cell-to-cell signaling occurs through QS, QS might be a “language” by which bacteria and host cells communicate.
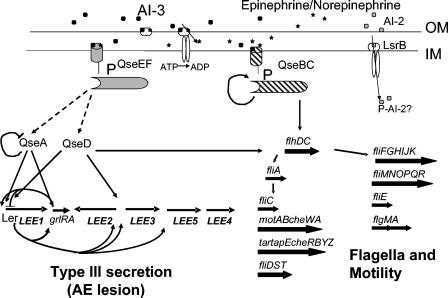
QS regulatory cascades have been extensively studied in organisms such as P. aeruginosa and V. harveyi and have proven to be very complex (16, 63). The regulatory cascade in EHEC and EPEC seems to be similarly complex; so far, at least six regulatory factors have been implicated in this process, and these factors in turn regulate other regulators (Fig. 2). The genes encoding these factors were previously cryptic-hypothetical genes in E. coli K-12 and have been renamed qse (for quorum sensing E. coli regulator) as their role in QS has been established. These factors have been shown to regulate genes involved in TTS and/or flagellum production, primarily by modulating expression of other “master” regulators, such as the LEE-encoded regulator (Ler), which is the activator of all other genes in the LEE (43), or the FlhDC regulator that is the master regulator of the flagellum regulon. Since the qse genes are also present in E. coli K-12 and other Enterobacteriaceae, they presumably regulate many other genes whose expression may be modulated by cell-to-cell signaling.
FIG. 2.
Model of the AI-3 quorum sensing signaling cascade in EHEC. Both AI-3 and epinephrine-norepinephrine appear to be recognized by the same receptor, which is probably in the outer membrane of the bacteria, due to the nonpolar nature of both signals. These signals are probably imported into the periplasmic space, where they interact with two major sensor kinases, QseC and QseE. QseC appears to be the sensor kinase transducing these signals towards activation of the flagellum regulon, whereas QseE appears to be the sensor kinase that transduces these signals to activate transcription of the LEE genes. QseC phosphorylates the QseB response regulator, which binds to the promoter of flhDC (encoding the flagellum master regulators FlhDC) to activate expression of the flagellum regulon. QseB also binds to its own promoter to positively autoregulate its own transcription. The predicted response regulator of QseE is QseF. At what levels QseF regulates transcription of the LEE genes remains to be established. QseA is one of the transcriptional factors involved in the regulation of ler (LEE1) transcription at two levels, by binding and activating transcription of LEE1 and by activating transcription of the grlRA operon, where GrlA and GrlR positively and negatively regulate expression of ler, respectively. Then, in a cascade fashion, Ler activates transcription of the other LEE genes. QseD is a second LysR-like regulator, involved in modulating expression of the LEE and flagellum genes. EHEC also possess an lsr operon involved in recognition and uptake of AI-2; however, the role of AI-2 signaling in EHEC remains to be determined. OM, outer membrane; IM, inner membrane.
The QseA protein is a member of the LysR family of transcriptional regulators (72). Expression of QseA is transcriptionally activated through QS and, in turn, binds to and directly activates transcription of Ler, which is encoded within the LEE1 operon (F. Sharp and Sperandio, unpublished results) (72). Consequently, an EHEC qseA mutant has a striking reduction in TTS but has no defect in flagellation or motility, suggesting that QseA regulates only the LEE genes and plays no role in the flagellum regulon (72). In addition, QseA also activates transcription of the grlRA operon, which is also within the LEE (R. Russell and V. Sperandio, unpublished results). GrlA has been reported to activate transcription of LEE1 (ler), while GrlR seems to repress it (18). These results suggest that QseA regulates transcription of LEE1 at more than one level. A two-component system named QseBC is responsible for the transcriptional activation of the flagellum regulon in response to quorum sensing (76), as well as for autoactivating its own transcription (10). The QseBC system is activated by QS through AI-3 (75, 76). QseC belongs to the sensor kinase family, and a qseC mutant is unable to respond to bacterial autoinducers or epinephrine given exogenously (75, 76). Mutation of qseC reduces motility but does not affect expression of the LEE genes, suggesting that the QseBC system is responsible for sensing AI-3-epinephrine and, in the presence of these signals, activating expression of flhDC. Three additional factors in this signaling cascade have recently been identified. QseD is another regulator of the LysR family and modulates expression of both the LEE and flagellum genes (F. Sharp, M. Walters, and V. Sperandio, unpublished results), and qseE and qseF encode a second two-component system that regulates expression of the LEE genes (N. Reading and V. Sperandio, unpublished results). These data suggest that both AI-3 and epinephrine or norepinephrine are recognized by the same receptor, which is probably in the outer membrane of the bacterium (due to the nonpolar nature of both AI-3 and epinephrine) (75). The current model is that the signaling molecules are imported into the periplasmic space, where they interact with the sensor kinase moieties of two different two-component systems, QseBC and QseEF (Fig. 2). The interaction of AI-3 and epinephrine with more than one sensor kinase could impart a “timing” mechanism to this system to avoid the inefficiency of EHEC producing both the LEE type III secretion system and flagella simultaneously. Therefore, it is hypothesized that EHEC first activates expression of the flagellum regulon through QseBC and then activates the LEE genes at a later time through QseEF. The AI-3-dependent QS signaling cascade is present in all Enterobacteriaceae examined so far (E. coli, Salmonella spp., Shigella spp., and Yersinia spp.). The most striking feature is that the genes encoding the transcriptional factors of this cascade are always in exactly the same context in the chromosomes of all the strains and exhibit high levels of identity in the different species, suggesting that this signaling cascade is functionally conserved in the Enterobacteriaceae.
Although EHEC and EPEC both possess the LEE, there are some differences in QS regulation between the two pathogens (22). Unlike EHEC, EPEC contains a plasmid-encoded regulator (Per) that increases expression of the chromosomal LEE genes (43). Also, EPEC produces the bundle-forming pilus (Bfp) that mediates formation of tight microcolonies of EPEC adhering together on epithelial cells, which allows the accumulation of locally high densities of signaling molecules. Disease due to EPEC also requires a higher infectious dose than that required for disease due to EHEC, and EPEC primarily colonizes the small intestine rather than the colonic location of EHEC (49). These differences suggest a model in which lower numbers of commensal flora in the small intestine require additional regulatory help for EPEC in the form of Per and Bfp-mediated microcolony formation. These compensatory mechanisms then allow the EPEC LEE to be regulated by QS in the small intestine without the extremely high levels of commensal flora present in the colon.
Homologues of the LuxI/LuxR QS system have been sought in E. coli, and although a LuxR homologue known as SdiA has been found, no obvious LuxI homologues that could synthesize an AHL signaling molecule are present in the E. coli genome. Although a cloned sdiA gene on a multicopy plasmid can upregulate expression of ftsQAZ genes, which encode proteins essential for cell division, an sdiA mutant has no apparent cell division defects (85). Kanamaru et al. (32) found that expression of SdiA from a high-copy-number plasmid in EHEC caused abnormal cell division, reduced adherence to cultured epithelial cells, and reduced expression of the intimin adhesin protein and the EspD protein, both of which are encoded on the LEE. However, no sdiA EHEC mutant was constructed and tested, and so the effects seen could have been artifacts due to the abnormally high expression of SdiA. Because no E. coli genes from either EHEC or E. coli K-12 have been demonstrated to be regulated by the single chromosomal copy of sdiA, Ahmer (1) recently concluded that there are no confirmed members of a SdiA regulon in this species.
Salmonella.
Two QS systems have been characterized in S. enterica (reviewed in reference 1). The LuxR homologue SdiA has been characterized in Salmonella, but there does not appear to be a corresponding signal-generating enzyme similar to LuxI in this species. However, Salmonella SdiA can detect AHLs produced by a variety of bacterial species, leading to the suggestion that SdiA appears to be dedicated to detecting signals produced by other species without any role in autoregulation (44). These results suggest that both AHL and AI-2 can be used in interspecies communication within a mixed-species community. SdiA regulates few genes in Salmonella, but one gene potentially involved in resistance to human complement, rck, is regulated by SdiA (2). However, mutation of the sdiA gene had no effect on the virulence of Salmonella in mouse, chicken, and bovine models of disease (1).
Salmonella also produces AI-2, and the only potential virulence phenotype that has been identified so far with a luxS mutant is a failure to form biofilms in an in vitro model of biofilm formation on human gallstones (55). As described above, LuxS regulates expression of an ABC transport system encoded by the lsr (LuxS regulated) operon, which is involved in uptake and internalization of the AI-2 molecule (79). The signaling cascade for AI-3 and epinephrine described above for EHEC is also present in Salmonella, and production of AI-3 by this species has been found (M. P. Sircili and Sperandio, unpublished observations).
V. cholerae.
In contrast to the usual paradigm of QS increasing expression of bacterial virulence factors at high cell densities, QS in V. cholerae appears to act in a way that results in repression of the major virulence factors at high densities and expression at low cell densities. The major virulence factors for V. cholerae are cholera toxin (CT) and the toxin-coregulated pilus (TCP), both of which are regulated as part of the ToxR regulon. There appear to be three parallel QS systems that all converge at the response regulator LuxO, which is a homologue of the LuxO regulator in V. harveyi (27, 46) (Fig. 3). Mutation of luxO in V. cholerae results in severe intestinal colonization defects (94). System 1 has homology with the V. harveyi QS system 1, which is an HSL AI system. The AI synthase for V. cholerae system 1 is called CqsA, and the sensor for this system is CqsS, a homologue of V. harveyi LuxN. The AI for this system is called CAI-1. The second QS system in V. cholerae is the LuxS/AI-2 system, which uses LuxQ and LuxP as sensors of AI-2. Genetic evidence suggests that there is a third system, whose components have not been identified yet. All three systems involve a LuxR homologue called HapR (31) that serves as a repressor of virulence genes and biofilm formation and as an activator of the Hap protease. The LuxO regulator is activated by phosphorylation and in turn activates transcription of small RNAs which together with Hfq mediate the destabilization of the hapR mRNA, repressing expression of hapR posttranscriptionally (36). As if this were not sufficiently complicated, some V. cholerae strains, including the prototypic strain N16961 that is used for human challenge studies, contain a frameshift in hapR, resulting in an inactive HapR protein with no apparent attenuation of disease in human or animal infections (94). The observation that several of the seventh pandemic strains do not contain a functional QS signaling system due to mutations in hapR raises the question of whether QS signaling might be selected against by evolution in V. cholerae. Future epidemiological studies addressing the frequency of mutations in the V. cholerae QS system should be pursued to answer this question.
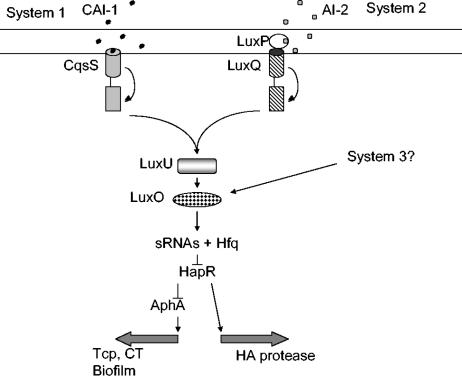
FIG. 3.
Quorum sensing signaling cascade in V. cholerae. There are potentially three quorum sensing systems in V. cholerae (36). System 1 is comprised of the CqsS sensor that responds to the autoinducer CAI-1. System 2 is the AI-2 system similar to the one in V. harveyi, where AI-2 is recognized by the periplasmic protein LuxP, which then interacts with the sensor LuxQ. Both LuxQ and CqsS phosphorylate LuxU, which in turn phosphorylates LuxO. Phosphorylated LuxQ then activates expression of small RNAs (sRNAs) that together with Hfq repress transcription of HapR. HapR activates transcription of the hemagglutinin (HA) protease and represses transcription of virulence genes encoding CT and the TCP via repression of aphA (35). A third system has been proposed, but nothing is known about this potential circuit at this time.
Details of how QS in V. cholerae responds to cell density can be found in the original papers (27, 35, 46, 93, 94), but a broad outline is as follows. At low cell densities, the AI concentrations are low and phosphate is transferred from the unoccupied sensors to the LuxU protein, which in turn transfers phosphate to LuxO. The phosphorylated LuxO activates transcription of small RNAs, which together with Hfq posttranscriptionally repress expression of hapR (36). The low level of HapR allows expression of CT and TCP, as well as vps genes involved in biofilm formation, but does not allow expression of the Hap protease. At high cell densities, the flow of phosphate reverses, resulting in inactivation of LuxO, leading to expression of HapR. HapR then represses CT and TCP indirectly via repression of aphA (35) and directly represses biofilm formation and activates expression of the gene encoding the Hap protease. Hammer and Bassler (27) have proposed a model in which TCP and CT are expressed early in the infection at low densities along with biofilm, which helps intestinal colonization. Later in the infection, when V. cholerae is at high densities, biofilm formation ceases and Hap protease production increases, thereby allowing the organism to exit the host and adapt to an environmental reservoir where expression of CT and TCP are not necessary. Given the strong conservation of QS systems in V. cholerae and the marine organism V. harveyi, it is quite plausible that the QS systems have some common functions that allow the organisms to survive in an environmental reservoir.
Enteroaggregative E. coli.
Enteroaggregative E. coli (EAEC) is an increasingly recognized cause of diarrhea that is often persistent in children and adults in both developing and developed countries. Evidence for QS in the regulation of virulence genes of EAEC was recently discovered using a continuous-flow anaerobic fecal culture system. Using this simulated model of the colonic environment, Ruiz-Perez et al. (61) found that the presence of fecal commensal bacteria increased expression of aggR, which encodes a global transcriptional regulator of EAEC virulence genes. By coculturing EAEC with individual strains of typical commensal bacteria, these investigators found that one or more substances produced by strains of Enterococcus and Clostridium increased expression of aggR, while strains of Lactobacillus and Veillonella downregulated expression of this gene. Although the specific QS systems responsible for modulating expression of aggR by these commensal species have not been identified, these results clearly show that EAEC responds to signaling compounds produced by colonic flora and that these signals modulate expression of a crucial global regulator of virulence.
E. faecalis.
Enterococcus species are normal inhabitants of the human and other mammalian GI tracts, but they are also important causes of nosocomial infections, including surgical site infections, bloodstream infections, and urinary tract infections. A major virulence factor of enterococci is a cytolysin that contributes to the pathogenesis of a variety of infections caused by E. faecalis (11). Not only is this cytolysin lethal for a broad range of eukaryotic cells, but it also is toxic to a number of gram-positive bacteria and serves as an autoinducer for QS induction of the cytolysin operon. The cytolysin, which can be encoded on a plasmid or in the chromosome, is unique among bacterial hemolytic toxins in having both bacteriocin and hemolytic activities in a single system. The cytolysin is made up of two subunits, CylLL and CylLS, that are posttranslationally modified by other proteins encoded in the eight-open-reading-frame cyl operon to produce the active extracellular forms designated CylLL" and CylLS′. Haas et al. (26) demonstrated that expression of the cytolysin is autoregulated by the presence of a threshold concentration of the mature extracellular CylLS" subunit. In this case the cytolysin subunit acts as the autoinducer that activates transcription from the cyl promoter. Recent analysis of conditions for expression of the toxin revealed that cytolysin expression is increased under anaerobic conditions (13). Anaerobic conditions are a major environmental signal in the GI tract, and so sorting out the direct regulation of enterococcal genes via anaerobiasis versus indirect regulation by anaerobiasis through QS is a complicated endeavor.
The cylR1 and cylR2 genes are divergently transcribed from cylL genes. The predicted CylR1 and CylR2 proteins form a two-component regulatory system, but neither protein shows similarity to the superfamily of two-component regulators (26). Mutation in either CylR1 or CylR2 leads to derepression of the cyl operon. CylR2 contains a helix-turn-helix DNA-binding motif, and recent structural and DNA-binding studies of purified CylR2 show that it specifically binds a 22-bp fragment of the cytolysin promoter region that contains an inverted repeat (62). The authors speculated that in the presence of the autoinducer CylLS", CylR1 shifts the DNA-binding specificity of CylR2 to sequences adjacent to the inverted repeat. Recently, Coburn et al. (12) showed that CylLL" preferentially binds to target cell membranes, allowing free CylLS" to accumulate at levels above the induction threshold. They present a model in which enterococci use CylLL" to actively probe the environment for cytolysin targets, and when such targets are detected, the unbound CylLS" activates cyl expression, resulting in high levels of cytolysin production.
The E. faecalis cytolysin not only serves as an autoinducer of enterococcal gene expression, as a lethal toxin active against a variety of eukaryotic cells, and as a “sonarlike” mechanism to detect eukaryotic target cells; it also is a bacteriocin and thereby can kill other bacterial cells in the same environment. Interestingly, a recent study (52) examined 139 healthy subjects for intestinal colonization by C. difficile, an organism that is notorious for causing diarrhea and colitis in hospitalized individuals who have had the normal intestinal flora disrupted by treatment with broad-spectrum antibiotics. This study found that many healthy individuals without a recent history of diarrhea or antibiotic usage were persistently colonized by C. difficile, and the number of fecal enterococci was significantly higher in these individuals than in individuals who were not colonized with C. difficile. It is tempting to speculate that the cytolysin of E. faecalis, which has both AI and bacterocin activities, was responsible for the change in intestinal flora that allowed C. difficile to colonize these healthy individuals, but this hypothesis remains to be proven.
Another QS system in Enterococcus species, called Fsr (for E. faecalis regulator), is homologous in many respects to the Agr system of S. aureus. Enterococci lack a homologue of the staphylococcal AgrD protein but possess homologues of AgrABC. The lack of an AgrD homologue is consistent with the exquisite specificity seen with AIP QS systems for each species and subspecies. The Fsr quorum sensing system in E. faecalis activates gelE encoding gelatinase and sprE encoding a serine protease, which are two virulence factors shown to be important in both the invertebrate Caenorhabditis elegans and the mammalian mouse models of infection (60, 66, 67). Finally, E. faecalis also contains a luxS homologue (63, 64), although the role of this QS system in virulence has not been investigated for this species.
Yersinia species.
Two Yersinia species, Yersinia enterocolitica and Yersinia pseudotuberculosis, can cause diarrhea in humans. A pair of LuxR/LuxI homologues, called YenR and YenI, were first described in Y. enterocolitica (82). Production of two AIs, 3-oxo-C6-HSL and C6-HSL, was attributed to this locus, but a specific phenotype controlled by this locus could not be established. Subsequently, Y. pseudotuberculosis was shown to contain two pairs of LuxIR homologues that control motility and clumping (3). The LuxIR pairs in Y. pseudotuberculosis are YpsI/YpsR and YtbI/YtbR, and three AI molecules, C6-HSL, 3-oxo-C6-HSL, and C8-HSL, are produced from the two autoinducer synthases. YpsI is responsible for 3-oxo-C6-HSL and YtbI is responsible for C8-HSL, while both YpsI and YtbI can synthesize C6-HSL. Temperature appears to play a pivotal role in determining which autoinducer synthase is active, and various combinations of AI production were seen with ypsI or ytbI mutants at 22, 28, and 37°C. A mutation in ypsR results in overexpression of a major flagellin subunit and increased motility. The YpsIR and YtbIR systems appear to comprise a hierarchical QS cascade in which YpsR can help regulate YtbIR. Temperature plays an important role in the regulation of this cascade, which is particularly interesting in light of the temperature regulation of many Yersinia virulence genes.
C. perfringens.
C. perfringens causes gas gangrene and is also capable of causing food-borne illness. Ohtani et al. (51) demonstrated that the luxS-mediated AI-2 QS system enhances extracellular expression of alpha-, kappa-, and theta-toxins in a mechanism that appears to involve both transcriptional and posttranscriptional mechanisms. More than 20 years ago, C. perfringens was reported to produce an extracellular signaling molecule called substance A that activated theta-toxin expression, but this appears to be an additional, as-yet-uncharacterized autoinducer that is distinct from AI-2 (51).
S. flexneri.
Shigella species, the primary agents of bacillary dysentery, have a very low infectious dose and possess a TTS system that is essential for virulence. These two characteristics are shared with EHEC, and so Day and Maurelli (14) investigated QS in S. flexneri to determine if TTS in this species was regulated by the luxS/AI-2 system. They found that expression of the ipa, mxi, and spa loci of the TTS system that are responsible for invasion of host cells was enhanced by conditioned media derived from stationary-phase cultures, suggesting the presence of an AI molecule in the media. The AI-2 molecule was detected in culture supernatants, and mutation of the luxS locus resulted in decreased expression of VirB, a transcription factor that is essential for expression of these invasion loci. However, mutation of luxS did not affect invasion operon expression, and a luxS mutant was fully virulent in the tissue culture and guinea pig keratoconjunctivitis invasion assays tested. Day and Maurelli noted that in contrast to EPEC and EHEC, which persist in the intestinal lumen and are continuously exposed to high levels of AI-2 from normal flora, Shigella efficiently invades host cells and is therefore likely to be exposed to luminal AI-2 for only a short time.
C. jejuni.
Two studies have examined the role of luxS in C. jejuni, which has been the most frequently isolated bacterial agent of food-borne disease in several studies. A luxS mutant showed a comparable growth rate, a comparable resistance to oxidative stress, and a comparable ability to invade Caco-2 cell monolayers relative to the parent strain but showed decreased motility in semisolid media (20). The effect on motility was confirmed by another group of investigators, who also showed that mutation of luxS reduced transcription of flaA, the major flagellin gene in this species (30). These investigators also found reduced agglutination capability in a luxS mutant, suggesting that QS might be involved in formation of surface structures in C. jejuni.
Vibrio vulnificus.
V. vulnificus is the most frequent bacterial cause of death due to ingestion of seafood. Primary septicemia after ingestion of this species is particularly lethal for individuals with underlying hepatic diseases. Kim et al. (34) found that the 50% lethal dose of a V. vulnificus luxS mutant and the time required for death in a mouse model were significantly increased. Cytotoxicity for HeLa cells was also significantly decreased by the mutation. Mutation of luxS caused decreased protease activity and increased hemolysin activities, effects which were reversed by complementation with the wild-type luxS gene. These investigators found that V. vulnificus produced the AI-2 molecule but found no evidence for an AHL molecule similar to that produced by V. harveyi. However, a LuxR homologue was previously identified in this species by other investigators and named SmcR (65). Similar to a luxS mutant, an smcR mutant showed decreased protease and increased hemolytic activities, but notably, the virulence of the smcR mutant in mice was comparable to that of the wild type, indicating that SmcR is not required for virulence in this model. The similar effects on protease and hemolysin suggest an interaction or hierarchy involving these two QS systems in this species.
V. parahaemolyticus.
Although it is not as lethal as V. vulnificus, V. parahaemolyticus is a more frequently isolated bacterial agent of gastroenteritis due to ingestion of contaminated seafood. The genome sequence of V. parahaemolyticus revealed a TTS system, although the exact role of this system in disease is not yet known. Henke and Bassler (28) recently showed that this system, as well as a similar system in V. harveyi, is regulated by QS. V. parahaemolyticus possesses all of the Lux regulators present in V. harveyi that comprise system 1 (LuxM and LuxR) and system 2 (LuxS). Mutation of the LuxR homologue of V. parahaemolyticus, called OpaR, had a striking effect on TTS. In these studies the exact signaling molecule responsible for regulation of TTS remains to be established, given that the experiments were all performed using spent supernatants, which contained several signaling molecules. Notably, in contrast to the positive regulation by QS of the TTS system seen with EPEC and EHEC, QS acts to repress TTS in V. parahaemolyticus These results suggest a model in which V. parahaemolyticus secretes effector proteins at a low cell density and terminates secretion when the cell density is high. This scenario is similar to that seen with V. cholerae and may also play a role in preparing the transition of the vibrios from the intestine to the aquatic environment.
ADDITIONAL ASPECTS OF QUORUM SENSING IN THE INTESTINE
Prokaryotic-eukaryotic communication.
There are several examples of prokaryotic-eukaryotic communication in which bacterial signals can modulate expression of eukaryotic genes or vice versa in cross-kingdom communication. One of the AIs of P. aeruginosa (3-oxo-C12-HSL) has immunomodulatory activity, and it can downregulate tumor necrosis factor alpha and interleukin-12 production in leukocytes (81) and upregulate expression of the proinflammatory cytokine gamma interferon (70). This same molecule has also been shown to stimulate interleukin-8 production in human lung fibroblasts and epithelial cells through a mechanism involving the eukaryotic transcription factors NF-kappa B and activator protein 2 (69). Although not yet linked to modulating transcription of eukaryotic genes, the cytolysin of E. faecalis, which serves as an autoinducer of expression of the enterococcal cyl operon, also has toxic effects on erythrocytes, retinal tissues, intestinal epithelial cells, neutrophils, and macrophages (11). The converse situation (i.e., a eukaryotic factor affecting transcription of prokaryotic genes) is demonstrated by the effect of epinephrine and norepinephrine on transcription of genes encoding the TTS system and flagella in EHEC and EPEC, as described above. In E. faecalis infections, host cell membranes can selectively bind CylLL", thereby allowing the unbound CylLS" peptide to increase cytolysin expression via a QS mechanism (12). Additional examples of eukaryotic interference with bacterial QS in non-GI systems have been reported. Human airway epithelial cells produce a substance, as yet unidentified, that inactivates the 3-oxo-C12-HSL signaling molecule produced by P. aeruginosa (9). In yet another kingdom, the red alga Delisea pulchra produces halogenated furanones that inhibit QS mechanisms of the plant pathogen Erwinia carotovora (39). No doubt other examples of eukaryotic-prokaryotic communication involving QS systems will be discovered as additional hosts, body niches, and signaling molecules are examined
Bacterial QS can even play a role in the development of normal host tissue. A symbiotic relationship between the squid Euprymna scolopes and V. fischeri has been well characterized by Visick and colleagues, who showed that development of normal crypt epithelium of the squid light organ depends upon colonization with V. fischeri (84). Interestingly, isogenic luxI or luxR mutants were incapable of stimulating the normal crypt epithelium development seen with the QS-positive parent strain. Given the importance of commensal flora in the normal development of the mammalian intestine (5, 91), it would not be surprising if QS among commensal bacteria is also important in mammalian intestinal development.
QS in vivo.
The first evidence that QS signaling molecules are produced during human infection was found in the respiratory tract, where sputum samples from cystic fibrosis patients infected with P. aeruginosa produced the two principal AHL signaling molecules (19, 68). In the GI tract, Sperandio et al. (75) detected the presence of AI-2 in fecal specimens from 10 of 12 healthy individuals examined. In the same study the workers examined specimens from a continuous-flow anaerobic fecal culture system which was inoculated with a fecal specimen from a healthy subject. AI-3 signaling activity was detected in this simulated intestinal environment using the LEE1::lacZ fusion. As noted above, this continuous-flow culture system also contained signaling activity that activated expression of the major regulator of virulence gene expression in EAEC (61). There was also a report of multiple AHL AI molecules in the rumen contents of six of eight cattle examined, although interestingly, no pure cultures of bacteria isolated from the rumen contents had AHL activity and only the rumen contents directly obtained from the cattle had AHL activity (21).
In the study of AI-2 activity in human fecal specimens, the concentrations of this signaling activity varied among the 10 subjects by as much as 1 log (75). This result suggests that different levels of signaling molecules in the intestines of different individuals may lead to various levels of QS activity and transcription of QS-regulated genes in the intestine. One speculation that arises from these results is that the course of disease may vary among different individuals due to variable levels of QS activity in their intestines.
QS-based therapy.
The discovery of QS in human pathogens has led to considerable interest in developing new therapeutic interventions to interfere with these signaling molecules. Thus, instead of using an antibiotic to kill pathogenic bacteria, a compound that interferes with the QS mechanism would be used to repress expression of the virulence genes responsible for the disease. Such an approach is particularly promising in light of increasing resistance to conventional antimicrobial agents, and encouraging results in animal models have been obtained with P. aeruginosa (7, 29, 71, 86) and S. aureus (41). However, the development of anti-QS therapy has been primarily directed towards nonintestinal pathogens, which do not have to deal with the huge numbers of commensal organisms present in the GI tract. The presence of this complex microbial flora and the variety of signaling molecules that might be produced by the microbial flora or even the host itself (see above) greatly complicate the application of this approach to GI pathogens. The occurrence of C. difficile-associated colitis after broad-spectrum antibiotic use is an example of the negative consequences that can potentially result from disruption of the normal intestinal flora.
CONCLUDING REMARKS
A fundamental property of QS is that the greater the density of bacteria, the greater the density of signaling molecules and the greater the opportunity for cell-to-cell communication. There is no environment in the human body with a greater density of bacteria and a greater potential for cell-to-cell signaling than the GI tract. The first report of QS regulating virulence factors of GI pathogens appeared only 6 years ago, and numerous reports documenting QS mechanisms in a variety of enteric pathogens have appeared since that initial report. There is clearly much more to be learned about QS in the GI tract, but with our increased understanding of this phenomenon, new insights will emerge regarding the virulence mechanisms of enteric pathogens, as well as the development and maintenance of our commensal intestinal microbial flora.
Acknowledgments
Work in the authors' laboratories is supported by grants AI21657, AI19716, and DK58957 (J.B.K.) and AI 053067 and AI 054468 (V.S.) from the National Institutes of Health.
Editor: A. D. O'Brien
REFERENCES
- 1.Ahmer, B. M. 2004. Cell-to-cell signalling in Escherichia coli and Salmonella enterica. Mol. Microbiol. 52:933-945. [DOI] [PubMed] [Google Scholar]
- 2.Ahmer, B. M., J. van Reeuwijk, C. D. Timmers, P. J. Valentine, and F. Heffron. 1998. Salmonella typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J. Bacteriol. 180:1185-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkinson, S., J. P. Throup, G. S. Stewart, and P. Williams. 1999. A hierarchical quorum-sensing system in Yersinia pseudotuberculosis is involved in the regulation of motility and clumping. Mol. Microbiol. 33:1267-1277. [DOI] [PubMed] [Google Scholar]
- 4.Bassler, B. L. 2002. Small talk. Cell-to-cell communication in bacteria. Cell 109:421-424. [DOI] [PubMed] [Google Scholar]
- 5.Berg, R. D. 1996. The indigenous gastrointestinal microflora. Trends Microbiol. 4:430-435. [DOI] [PubMed] [Google Scholar]
- 6.Borrellio, S. P. 2002. The normal flora of the gastrointestinal tract, p. 1-12. In M. A. Kamm (ed.), Gut ecology. Martin Dunitz, Ltd., London, United Kingdom.
- 7.Camara, M., P. Williams, and A. Hardman. 2002. Controlling infection by tuning in and turning down the volume of bacterial small-talk. Lancet Infect. Dis. 2:667-676. [DOI] [PubMed] [Google Scholar]
- 8.Chen, X., S. Schauder, N. Potier, A. Van Dorssealaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 9.Chun, C. K., E. A. Ozer, M. J. Welsh, J. Zabner, and E. P. Greenberg. 2004. Inactivation of a Pseudomonas aeruginosa quorum-sensing signal by human airway epithelia. Proc. Natl. Acad. Sci. USA 101:3587-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke, M. B., and V. Sperandio. Submitted for publication.
- 11.Coburn, P. S., and M. S. Gilmore. 2003. The Enterococcus faecalis cytolysin: a novel toxin active against eukaryotic and prokaryotic cells. Cell. Microbiol. 5:661-669. [DOI] [PubMed] [Google Scholar]
- 12.Coburn, P. S., C. M. Pillar, B. D. Jett, W. Haas, and M. S. Gilmore. 2004. Enterococcus faecalis senses target cells and in response expresses cytolysin. Science 306:2270-2272. [DOI] [PubMed] [Google Scholar]
- 13.Day, A. M., J. H. Cove, and M. K. Phillips-Jones. 2003. Cytolysin gene expression in Enterococcus faecalis is regulated in response to aerobiosis conditions. Mol. Genet. Genomics 269:31-39. [DOI] [PubMed] [Google Scholar]
- 14.Day, W. A., Jr., and A. T. Maurelli. 2001. Shigella flexneri LuxS quorum-sensing system modulates virB expression but is not essential for virulence. Infect. Immun. 69:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Kievit, T. R., R. Gillis, S. Marx, C. Brown, and B. H. Iglewski. 2001. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl. Environ. Microbiol. 67:1865-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng, W., J. L. Puente, S. Gruenheid, Y. Li, B. A. Vallance, A. Vazquez, J. Barba, J. A. Ibarra, P. O'Donnell, P. Metalnikov, K. Ashman, S. Lee, D. Goode, T. Pawson, and B. B. Finlay. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. USA 101:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan, K., C. Dammel, J. Stein, H. Rabin, and M. G. Surette. 2003. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 50:1477-1491. [DOI] [PubMed] [Google Scholar]
- 20.Elvers, K. T., and S. F. Park. 2002. Quorum sensing in Campylobacter jejuni: detection of a luxS encoded signalling molecule. Microbiology 148:1475-1481. [DOI] [PubMed] [Google Scholar]
- 21.Erickson, D. L., V. L. Nsereko, D. P. Morgavi, L. B. Selinger, L. M. Rode, and K. A. Beauchemin. 2002. Evidence of quorum sensing in the rumen ecosystem: detection of N-acyl homoserine lactone autoinducers in ruminal contents. Can. J. Microbiol. 48:374-378. [DOI] [PubMed] [Google Scholar]
- 22.Falcao, J. P., F. Sharp, and V. Sperandio. 2004. Cell-to-cell signaling in intestinal pathogens. Curr. Issues Intest. Microbiol. 5:9-17. [PubMed] [Google Scholar]
- 23.Fuqua, C., and E. P. Greenberg. 2002. Listening in on bacteria: acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 3:685-695. [DOI] [PubMed] [Google Scholar]
- 24.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 25.Furness, J. B. 2000. Types of neurons in the enteric nervous system. J. Auton. Nerv. Syst. 81:87-96. [DOI] [PubMed] [Google Scholar]
- 26.Haas, W., B. D. Shepard, and M. S. Gilmore. 2002. Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature 415:84-87. [DOI] [PubMed] [Google Scholar]
- 27.Hammer, B. K., and B. L. Bassler. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101-104. [DOI] [PubMed] [Google Scholar]
- 28.Henke, J. M., and B. L. Bassler. 2004. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J. Bacteriol. 186:3794-3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hentzer, M., and M. Givskov. 2003. Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections. J. Clin. Investig. 112:1300-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeon, B., K. Itoh, N. Misawa, and S. Ryu. 2003. Effects of quorum sensing on flaA transcription and autoagglutination in Campylobacter jejuni. Microbiol. Immunol. 47:833-839. [DOI] [PubMed] [Google Scholar]
- 31.Jobling, M. G., and R. K. Holmes. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 26:1023-1034. [DOI] [PubMed] [Google Scholar]
- 32.Kanamaru, K., I. Tatsuno, T. Tobe, and C. Sasakawa. 2000. SdiA, an Escherichia coli homologue of quorum-sensing regulators, controls the expression of virulence factors in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 38:805-816. [DOI] [PubMed] [Google Scholar]
- 33.Kaper, J., B., anad A. D. O'Brien. 1998. Escherichia coli and other Shiga-toxin producing E. coli strains. ASM Press, Washington D.C.
- 34.Kim, S. Y., S. E. Lee, Y. R. Kim, C. M. Kim, P. Y. Ryu, H. E. Choy, S. S. Chung, and J. H. Rhee. 2003. Regulation of Vibrio vulnificus virulence by the LuxS quorum-sensing system. Mol. Microbiol. 48:1647-1664. [DOI] [PubMed] [Google Scholar]
- 35.Kovacikova, G., and K. Skorupski. 2002. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol. Microbiol. 46:1135-1147. [DOI] [PubMed] [Google Scholar]
- 36.Lenz, D. H., K. C. Mok, B. N. Lilley, R. V. Kulkarni, N. S. Wingreen, and B. L. Bassler. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69-82. [DOI] [PubMed] [Google Scholar]
- 37.Lyon, W. R., J. C. Madden, J. C. Levin, J. L. Stein, and M. G. Caparon. 2001. Mutation of luxS affects growth and virulence factor expression in Streptococcus pyogenes. Mol. Microbiol. 42:145-157. [DOI] [PubMed] [Google Scholar]
- 38.Macfarlane, G. T., and S. Macfarlane. 1997. Human colonic microbiota: ecology, physiology and metabolic potential of intestinal bacteria. Scand. J. Gastroenterol. Suppl. 222:3-9. [DOI] [PubMed] [Google Scholar]
- 39.Manefield, M., M. Welch, M. Givskov, G. P. Salmond, and S. Kjelleberg. 2001. Halogenated furanones from the red alga, Delisea pulchra, inhibit carbapenem antibiotic synthesis and exoenzyme virulence factor production in the phytopathogen Erwinia carotovora. FEMS Microbiol. Lett. 205:131-138. [DOI] [PubMed] [Google Scholar]
- 40.Manna, A. C., and A. L. Cheung. 2003. sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect. Immun. 71:343-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayville, P., G. Ji, R. Beavis, H. Yang, M. Goger, R. P. Novick, and T. W. Muir. 1999. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. USA 96:1218-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 44.Michael, B., J. N. Smith, S. Swift, F. Heffron, and B. M. Ahmer. 2001. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol. 183:5733-5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 46.Miller, M. B., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303-314. [DOI] [PubMed] [Google Scholar]
- 47.Miller, S. T., K. B. Xavier, S. R. Campagna, M. E. Taga, M. F. Semmelhack, B. L. Bassler, and F. M. Hughson. 2004. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell. 15:677-687. [DOI] [PubMed] [Google Scholar]
- 48.Mitsumori, M., L. Xu, H. Kajikawa, M. Kurihara, K. Tajima, J. Hai, and A. Takenaka. 2003. Possible quorum sensing in the rumen microbial community: detection of quorum-sensing signal molecules from rumen bacteria. FEMS Microbiol. Lett. 219:47-52. [DOI] [PubMed] [Google Scholar]
- 49.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 51.Ohtani, K., H. Hayashi, and T. Shimizu. 2002. The luxS gene is involved in cell-cell signalling for toxin production in Clostridium perfringens. Mol. Microbiol. 44:171-179. [DOI] [PubMed] [Google Scholar]
- 52.Ozaki, E., H. Kato, H. Kita, T. Karasawa, T. Maegawa, Y. Koino, K. Matsumoto, T. Takada, K. Nomoto, R. Tanaka, and S. Nakamura. 2004. Clostridium difficile colonization in healthy adults: transient colonization and correlation with enterococcal colonization. J. Med. Microbiol. 53:167-172. [DOI] [PubMed] [Google Scholar]
- 53.Parsek, M. R., and E. P. Greenberg. 2000. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. USA 97:8789-8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parsek, M. R., D. L. Val, B. L. Hanzelka, J. E. Cronan, Jr., and E. P. Greenberg. 1999. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. USA 96:4360-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prouty, A. M., W. H. Schwesinger, and J. S. Gunn. 2002. Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect. Immun. 70:2640-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pruss, B. M., J. W. Campbell, T. K. Van Dyk, C. Zhu, Y. Kogan, and P. Matsumura. 2003. FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J. Bacteriol. 185:534-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pruss, B. M., D. Markovic, and P. Matsumura. 1997. The Escherichia coli flagellar transcriptional activator flhD regulates cell division through induction of the acid response gene cadA. J. Bacteriol. 179:3818-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pruss, B. M., and P. Matsumura. 1996. A regulator of the flagellar regulon of Escherichia coli, flhD, also affects cell division. J. Bacteriol. 178:668-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Purves, D., D. Fitzpatrick, S. M. Williams, J. O. McNamara, G. J. Augustine, L. C. Katz, and A. LaMantia. 2001. Neuroscience, 2nd ed. Sinauer Associates, Inc., Sunderland, Mass.
- 60.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 68:2579-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruiz-Perez, F., J. Sheikh, S. Davis, E. C. Boedeker, and J. P. Nataro. 2004. Use of a continuous-flow anaerobic culture to characterize enteric virulence gene expression. Infect. Immun. 72:3793-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rumpel, S., A. Razeto, C. M. Pillar, V. Vijayan, A. Taylor, K. Giller, M. S. Gilmore, S. Becker, and M. Zweckstetter. 2004. Structure and DNA-binding properties of the cytolysin regulator CylR2 from Enterococcus faecalis. EMBO J. 23:3632-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schauder, S., and B. L. Bassler. 2001. The languages of bacteria. Genes Dev. 15:1468-1480. [DOI] [PubMed] [Google Scholar]
- 64.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 65.Shao, C. P., and L. I. Hor. 2001. Regulation of metalloprotease gene expression in Vibrio vulnificus by a Vibrio harveyi LuxR homologue. J. Bacteriol. 183:1369-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sifri, C. D., E. Mylonakis, K. V. Singh, X. Qin, D. A. Garsin, B. E. Murray, F. M. Ausubel, and S. B. Calderwood. 2002. Virulence effect of Enterococcus faecalis protease genes and the quorum-sensing locus fsr in Caenorhabditis elegans and mice. Infect. Immun. 70:5647-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh, K. V., X. Qin, G. M. Weinstock, and B. E. Murray. 1998. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J. Infect. Dis. 178:1416-1420. [DOI] [PubMed] [Google Scholar]
- 68.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 69.Smith, R. S., E. R. Fedyk, T. A. Springer, N. Mukaida, B. H. Iglewski, and R. P. Phipps. 2001. IL-8 production in human lung fibroblasts and epithelial cells activated by the Pseudomonas autoinducer N-3-oxododecanoyl homoserine lactone is transcriptionally regulated by NF-kappa B and activator protein-2. J. Immunol. 167:366-374. [DOI] [PubMed] [Google Scholar]
- 70.Smith, R. S., S. G. Harris, R. Phipps, and B. Iglewski. 2002. The Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)homoserine lactone contributes to virulence and induces inflammation in vivo. J. Bacteriol. 184:1132-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smith, R. S., and B. H. Iglewski. 2003. Pseudomonas aeruginosa quorum sensing as a potential antimicrobial target. J. Clin. Investig. 112:1460-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sperandio, V., C. C. Li, and J. B. Kaper. 2002. Quorum-sensing Escherichia coli regulator A (QseA): a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic Escherichia coli. Infect. Immun. 70:3085-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sperandio, V., A. G. Torres, B. Jarvis, J. P. Nataro, and J. B. Kaper. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. USA 100:8951-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sperandio, V., A. G. Torres, and J. B. Kaper. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43:809-821. [DOI] [PubMed] [Google Scholar]
- 77.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Taga, M. E., and B. L. Bassler. 2003. Chemical communication among bacteria. Proc. Natl. Acad. Sci. USA 100:14549-14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taga, M. E., S. T. Miller, and B. L. Bassler. 2003. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol. Microbiol. 50:1411-1427. [DOI] [PubMed] [Google Scholar]
- 80.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42:777-793. [DOI] [PubMed] [Google Scholar]
- 81.Telford, G., D. Wheeler, P. Williams, P. T. Tomkins, P. Appleby, H. Sewell, G. S. Stewart, B. W. Bycroft, and D. I. Pritchard. 1998. The Pseudomonas aeruginosa quorum-sensing signal molecule N-(3-oxododecanoyl)-l-homoserine lactone has immunomodulatory activity. Infect. Immun. 66:36-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Throup, J. P., M. Camara, G. S. Briggs, M. K. Winson, S. R. Chhabra, B. W. Bycroft, P. Williams, and G. S. Stewart. 1995. Characterisation of the yenI/yenR locus from Yersinia enterocolitica mediating the synthesis of two N-acylhomoserine lactone signal molecules. Mol. Microbiol. 17:345-356. [DOI] [PubMed] [Google Scholar]
- 83.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 84.Visick, K. L., J. Foster, J. Doino, M. McFall-Ngai, and E. G. Ruby. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 182:4578-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang, X. D., P. A. de Boer, and L. I. Rothfield. 1991. A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes of Escherichia coli. EMBO J. 10:3363-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Williams, P. 2002. Quorum sensing: an emerging target for antibacterial chemotherapy? Expert Opin. Ther. Targets 6:257-274. [DOI] [PubMed] [Google Scholar]
- 87.Winzer, K., K. R. Hardie, N. Burgess, N. Doherty, D. Kirke, M. T. Holden, R. Linforth, K. A. Cornell, A. J. Taylor, P. J. Hill, and P. Williams. 2002. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148:909-922. [DOI] [PubMed] [Google Scholar]
- 88.Winzer, K., K. R. Hardie, and P. Williams. 2002. Bacterial cell-to-cell communication: sorry, can't talk now—gone to lunch! Curr. Opin. Microbiol. 5:216-222. [DOI] [PubMed] [Google Scholar]
- 89.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6:191-197. [DOI] [PubMed] [Google Scholar]
- 90.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]
- 91.Xu, J., and J. I. Gordon. 2003. Inaugural article: honor thy symbionts. Proc. Natl. Acad. Sci. USA 100:10452-10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yarwood, J. M., and P. M. Schlievert. 2003. Quorum sensing in Staphylococcus infections. J. Clin. Investig. 112:1620-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhu, J., and J. J. Mekalanos. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5:647-656. [DOI] [PubMed] [Google Scholar]
- 94.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]