Abstract
Bacillus anthracis, the causative agent of anthrax, secretes numerous proteins into the extracellular environment during infection. A comparative proteomic approach was employed to elucidate the differences among the extracellular proteomes (secretomes) of three isogenic strains of B. anthracis that differed solely in their plasmid contents. The strains utilized were the wild-type virulent B. anthracis RA3 (pXO1+ pXO2+) and its two nonpathogenic derivative strains: the toxigenic, nonencapsulated RA3R (pXO1+ pXO2−) and the totally cured, nontoxigenic, nonencapsulated RA3:00 (pXO1− pXO2−). Comparative proteomics using two-dimensional gel electrophoresis followed by computer-assisted gel image analysis was performed to reveal unique, up-regulated, or down-regulated secretome proteins among the strains. In total, 57 protein spots, representing 26 different proteins encoded on the chromosome or pXO1, were identified by peptide mass fingerprinting. S-layer-derived proteins, such as Sap and EA1, were most frequently observed. Many sporulation-associated enzymes were found to be overexpressed in strains containing pXO1+. This study also provides evidence that pXO2 is necessary for the maximal expression of the pXO1-encoded toxins lethal factor (LF), edema factor (EF), and protective antigen (PA). Several newly identified putative virulence factors were observed; these include enolase, a high-affinity zinc uptake transporter, the peroxide stress-related alkyl hydroperoxide reductase, isocitrate lyase, and the cell surface protein A.
Bacillus anthracis, the etiologic agent of anthrax, is a gram-positive, rod-shaped, nonmotile facultative anaerobic and spore-forming bacterium (35). Anthrax is a disease of wildlife, livestock, and humans that begins when spores of B. anthracis enter a host, are engulfed by macrophages, germinate, and then cause septicemia. This pathogen's virulence, along with the spore's resistance to adverse environmental conditions and facility to be weaponized, has made it a significant agent of bioterrorism (1).
In addition to a 5.2-Mb circular chromosome, fully virulent strains of B. anthracis harbor two plasmids: pXO1 and pXO2 (35). The pXO1 plasmid (181.6 kb) encodes the toxins lethal factor (LF) and edema factor (EF) and the protective antigen (PA) responsible for the translocation of EF and LF into the host's cytosol (35). The cascade of events leading to toxin entry into host cells has been described (7). The genes encoding LF, EF, and PA are designated lef, cya, and pagA, respectively (8, 50, 63). Proteins required for a proteinous capsule biosynthesis (CapBCA) and depolymerization (Dep) are encoded on pXO2 (94.8 kb) (29, 60). Strains lacking either or both plasmids are attenuated in most animal hosts (22). The transcriptions of lef, cya, pagA, and capB have all been shown to be coordinately induced by the presence of bicarbonate-CO2, while temperature has been shown to be important for toxin, but not for CapB, production (55). High CO2 tension is believed to simulate conditions encountered within the mammalian host. When B. anthracis is cultured at 37°C in a bicarbonate-containing minimal medium, toxin production is enhanced, with peak levels occurring at the end of the exponential growth phase (55).
Bacterial pathogens must possess regulatory controls to ensure that their virulence factors are coordinately synthesized at the right time and place. This temporal and spatial regulation in B. anthracis genes occurs in response to environmental stimuli and appears to be controlled by a uniquely complex signal transduction cascade. The pXO1-encoded transcriptional activator AtxA has been reported to play a role in the CO2-dependent regulation of the exotoxin genes lef, cya, and pagA (12, 24, 59). AtxA has been shown to regulate S-layer genes (i.e., sap and eag) via the activity of PagR, a negative regulator of pagA, so that only eag is significantly expressed in the presence of CO2 (33).
A capsule gene activator (acpA) on pXO2 has also been identified (61). Studies have suggested that AcpA is a minor regulator with fewer gene targets than AtxA (15). A reexamination of the role of AcpA in B. anthracis gene regulation, however, led Bourgogne et al. (5) to propose that AtxA and AcpA act synergistically to regulate the expression of certain target genes in addition to the cap genes. Another newly discovered capsule gene regulator, AcpB, is the product of a gene located on pXO2 (13). Recent evidence suggests that AtxA is the major capsule synthesis regulator and that it indirectly controls capBCAD expression via positive regulation of acpA and acpB (13).
Acquiring a better understanding of the pathways controlling an organism's protein expression may be achieved via comparative proteomics. Such investigations often rely on the coupling of two-dimensional gel electrophoresis (2-DE) and matrix-assisted laser desorption ionization-mass spectrometry (MALDI-MS) to ensure accurate and sensitive analysis of proteins. In our laboratory, this approach was applied to a comprehensive study of the differences in protein expression between a fully virulent and a vaccinal strain of Brucella melitensis (14). Recently, focus has been on the analysis and characterization of the secretomes, i.e., the entire complement of secreted proteins, of pathogenic bacterial species. Secreted proteins are important in the course of infection, since they act as remote controls for direct contact with host cells, making them excellent as both biomarkers for diagnostic assays and targets for therapeutic agents. Recent secretome analyses of Bacillus subtilis (2, 18), Helicobacter pylori (9), and Bacillus cereus (16) have provided insights into the pathogenicity of these bacteria. Our laboratory recently conducted a comparative proteomics investigation of the secretomes of the virulent B. anthracis RA3 isolate grown under induced or noninduced conditions (41). LF, EF, and PA were among the 27 proteins identified (41).
In the present study, 2-DE and MALDI-MS were used to investigate the differences among the secretomes of the fully virulent strain RA3 (pXO1+ pXO2+), its partially cured derivative strain RA3R (pXO1+ pXO2−), and the plasmidless derivative strain RA3:00 (pXO1− pXO2−). Only protein spots that either had a difference in intensity across all three strains or were uniquely present in only one strain were targeted for mass spectroscopy analysis.
MATERIALS AND METHODS
Bacterial strains.
The virulent B. anthracis RA3 strain (pXO1+ pXO2+), which was isolated from a bovine spleen sample during an anthrax outbreak in France, was used as the reference strain in this study (39). Two attenuated B. anthracis RA3 derivative strains, RA3R (pXO1+ pXO2−) (39) and RA3:00 (pXO1− pXO2−), were also used. The plasmidless B. anthracis RA3:00 was derived from B. anthracis RA3R by repeated daily subculturing at 42.5°C in 20 ml brain heart infusion (BHI) broth (37). After 30 days, the culture was plated on BHI agar and incubated overnight at 37°C. RA3R isolates cured for pXO1, i.e., fully cured, were identified by testing crude vegetative cell lysates from isolated colonies in multiplex PCR assays. The multiplex PCR assay determines genomic content by amplifying specific genes on each plasmid (lef, cya, and pagA [pXO1] and capC [pXO2]) and the chromosome (marker Ba813) (46). Colonies tentatively deemed pXO1− pXO2− were cultured and centrifuged, and their total genomic DNA was extracted with the UltraClean Microbial Genomic DNA Isolation Kit (MoBio Laboratories, Solana Beach, CA) and used as the template for a high-stringency multiplex PCR. Variable-number tandem repeat analysis (39) and rpoB-specific fluorescent resonance energy transfer PCR assays (44) were performed on RA3R isolates deemed pXO1 cured to ensure that the chromosomal background was consistent with that of the RA3R mother cell. Glycerol stocks of this newly derived, double-cured B. anthracis strain, designated RA3:00, were stored at −80°C.
Medium preparation.
For all secretome isolation experiments, the bacteria were cultured in the protein-free minimal R medium, which was prepared as described by Ristroph and Ivins (49).
Inoculum preparation and culture conditions.
One loop (approximately 10 μl) of glycerol stock of B. anthracis RA3R (pXO1+ pXO2−) or RA3:00 (pXO1− pXO2−) was streaked for isolation on BHI agar and incubated overnight (16 h) at 37°C. Single colonies were transferred to 2 ml R medium. Approximately 1 ml (optical density at 600 nm = 0.5) of the resulting resuspension was immediately transferred to a flask containing 100 ml R medium supplemented with glucose (0.25% [wt/vol]). The flask was mixed gently and fitted with a BugStopper (Whatman Inc., Clifton, NJ) sterile venting closure. The culture inoculum was incubated at 37°C with shaking at 120 rpm for 5 to 6 h. Following this growth period, 5 ml of the culture was transferred to a 250-ml sterile, vented, canted Falcon tissue culture flask containing 70 ml R medium with glucose (0.25% [wt/vol]) and sodium bicarbonate (0.85% [wt/vol]). The cultures were incubated overnight at 37°C under 5% CO2 in a humid incubator, which corresponds in our study to an end point at 16 h and according to Ristroph and Ivins to a late exponential phase (49). Cells were collected at a state of minimum sporulation as confirmed by growth curve analyses (similar among all three strains) and spore counts (data not shown).
Isolation of secreted proteins.
All centrifugations were performed at 4,700 × g at 4°C for the specified time. The contents of the tissue culture flasks were centrifuged for 10 min, and the secretome-containing supernatant was passed through a 0.45-μm filter to remove any suspended vegetative cells. All bacterial cells were discarded. The method described by Rafie-Kolpin et al. (45) and adapted by Wagner et al. (62) was utilized to prepare the protein samples. 100% ice-cold trichloroacetic acid (TCA; Sigma Chemical Co., St. Louis, MO) was added to the supernatant for a final 10% TCA (vol/vol) concentration. Supernatant aliquots (∼45 ml) were chilled on ice for 45 min and centrifuged for 45 min. Each resultant pellet was washed with 3 ml acetone, incubated on ice for 15 min, and centrifuged (15 min) at 4°C. Following acetone evaporation, each pellet was resuspended in 100 μl sample buffer 1 (0.3% sodium dodecyl sulfate [SDS], 0.2 M dithiothreitol [DTT], 50 mM Tri-HCl, pH 8.0) obtained from Genomic Solutions (Ann Arbor, MI). Protein extracts were stored at −80°C.
Total protein determination.
The protein concentration of the culture filtrate extract was determined using the Bio-Rad Protein Assay kit (Bio-Rad Laboratories, Hercules, CA.) which is based upon the method of Bradford (6). Bovine serum albumin served as the standard for protein concentration determinations.
Sample preparation for isoelectric focusing (IEF).
A volume of secretome solution containing 100 μg total protein was transferred to a microcentrifuge tube, and dissolved proteins were precipitated upon addition of ice-cold TCA (10% final concentration) and a 5-min incubation on ice. The samples were centrifuged at 6,800 × g for 3 min. The resulting pellet was washed with 200 μl acetone, placed on ice for 5 min, and centrifuged again. The pellet was resuspended in 40 μl sample buffer 1 and 4 μl sample buffer 2 (50 mM MgCl2, 8 U DNase I, 3 U RNase A, 0.5 M Tris-HCl, pH 8.0), and incubated on ice for 10 min. Then, 160 μl loading buffer {8 M urea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 40 mM Tris-base, 65 mM DTT, 0.01% bromophenol blue} and 200 μl rehydration buffer (8 M urea, 2% CHAPS, 10 mM DTT, 2% carrier ampholytes, 0.01% bromophenol blue) were added to each sample. All buffers were purchased from Genomic Solutions.
IEF.
B. anthracis culture filtrate extract proteins (100 μg; 400-μl total volume) were separated on the basis of charge using 18-cm immobilized pH gradient (IPG) strips (Amersham Pharmacia Biotech, Uppsala, Sweden) with a 4 to 7 linear pH gradient. The IPG strips were allowed to rehydrate with the protein solution overnight at ambient temperature. A 2-ml layer of nonconducting oil (Genomic Solutions) was applied to prevent evaporation of the protein solution during the rehydration period. IEF was conducted at 20°C for 24 h (maximum voltage of 5,000 V, maximum current of 80 μA/gel, 80,000 Vh, with an end-of-run hold at 125 V).
SDS-PAGE.
The IPG strips were prepared for SDS-polyacrylamide gel electrophoresis (PAGE) analysis by a 15-min wash with 10 ml of Genomic Solutions Equilibration Buffer 1 (6 M urea, 133 mM DTT, 30% glycerol, 50 mM Tris-acetate, pH 7.0), followed by a 15-min wash with Genomic Solutions Equilibration Buffer 2 (6 M urea, 2.5% iodoacetamide, 30% glycerol, 50 mM Tris-acetate, pH 7.0). The IPG strips were then applied to precast 10% Duracryl gels (22 cm by 23 cm by 1 mm; Tris-Tricine-SDS; Genomic Solutions, Inc.). Electrophoresis took place at 4°C with a maximum voltage of 500 V and a maximum power of 1,600 μW/gel. Separation was achieved in 18 to 20 h. The gels were transferred to an automated staining apparatus and protected from light from this point forward (Genomic Solutions) and fixed for 30 min in a solution of 40% methanol and 10% acetic acid. After a 5-min wash with deionized water, the gels were incubated with SYPRO Ruby fluorescent stain (Molecular Probes, Eugene, OR) to stain each gel for a period of 12 h with gentle rocking. After being stained, the gels underwent a 5-min wash with deionized water and were destained for 30 min using a 10% methanol and 6% acetic acid solution. Following a final wash with deionized water, the gels were stored in 2% glycerol in the dark at 4°C. Gel images were captured using the Fujifilm LAS-1000Plus imager under 470-nm light with an exposure time of 5 min.
Gel analysis.
All strains were analyzed using gels run in triplicate. All gels used were from two or three different batches of cells grown independently. Each culture filtrate was processed and analyzed separately. The Investigator HT Analyzer program (Nonlinear Dynamics, New Castle upon Tyne, United Kingdom) determined the protein spots present on each gel image, along with their experimental isoelectric point (pI) and molecular mass. Spots present on each subgel of the same strain were matched, and an average gel was created; only those spots appearing in at least two of three subgels were included in the average gel. The average gels of the RA3R and RA3:00 strains were matched to the recently generated average gel of the fully virulent B. anthracis RA3 strain (pXO1+ pXO2+) (41). To compensate for differences in staining quality, the volume of each spot was normalized by selecting one spot (base spot) whose shape and intensity were consistent in all six subgels of the RA3R-RA3 and RA3:00-RA3 gel comparisons. Spot normalization was accomplished for all six subgels by dividing each spot's volume by that of the base spot and multiplying the result by 100. Protein spots were considered unique to a given strain if those spots were absent from all three subgels of the compared strain. Protein spots had to have a reproducible pattern of being over- or underexpressed in at least two subgels to be assigned as being differentially expressed in a strain.
Spot excision.
Protein spots were excised from the SYPRO Ruby-stained 2-D gels by a UV box-equipped ProPic robot (Genomic Solutions). The picked plugs were stored at −80°C prior to digestion with trypsin.
In-gel trypsin digestion with o-MU modification.
In-gel trypsin digestion was performed following the default long trypsin digestion protocol for the ProGest Digestion Station (Genomic Solutions) but was altered to include modification of the tryptic peptides with o-methylisourea (o-MU) (62). Tryptic peptides were modified with 1 M o-MU in 100 mM ammonium bicarbonate at pH 10.0 for 1 h at 37°C as described previously (17). Modified tryptic peptides were recovered in a mixture of 10% (vol/vol) formic acid and acetonitrile (ACN) (51).
Preparation of tryptic peptides for MALDI analysis.
o-MU-modified tryptic peptides were dried under vacuum and resuspended in 100 μl of 10% formic acid. Sample cleanup was accomplished using the ACN Elution Protocol provided with the ProMS Sample Preparation Station (Genomic Solutions). The peptide mixtures were desalted using ZipTips (C18 media) from Millipore (Bedford, MA) conditioned with a 1:1 mixture of ACN and water. The desalted peptides were concentrated as they were eluted with 3 μl of matrix (10 mg α-cyanohydroxycinnamic acid per 1 ml of 50% ACN, 0.1% trifluoroacetic acid in water). Finally, 1 μl of the peptide and matrix mixture was spotted onto a 384-well stainless steel target plate (Kratos Analytical Ltd., Manchester, United Kingdom).
Mass spectral analysis.
All spectra were obtained with the Kratos Axima-CFR running in reflectron mode, as described previously (62), using the Kompact software package (Kratos Analytical, Ltd.).
The trypsin autolysis peaks at 842.509 and 2254.12 Da (the 2212.11 peak, modified by o-MU) were used as internal calibrants. Monoisotopic peaks were manually selected, and the Mascot software package from Matrix Science (London, United Kingdom) was used to search the peak lists against a customized in-house database. The database contained all described open reading frame (ORF) sequences from the completed genomes of three virulent B. anthracis strains: Ames (47), Sterne (unpublished data), and A2012 (48). Sequences from B. cereus (ATCC 14579) (21) and Bacillus thuringiensis (ATCC 35646) were also added to the in-house database and searched to increase the chance of protein annotation (http://www.ncbi.nlm.nih.gov/). The search parameters were a maximum of one missed cleavage by trypsin, fixed modifications of oxidized methionine and carbamidomethylated cysteine, variable modification of guanidinated lysine, a charge state of +1, and a mass tolerance range of ±0.2 Da.
RESULTS
General observations.
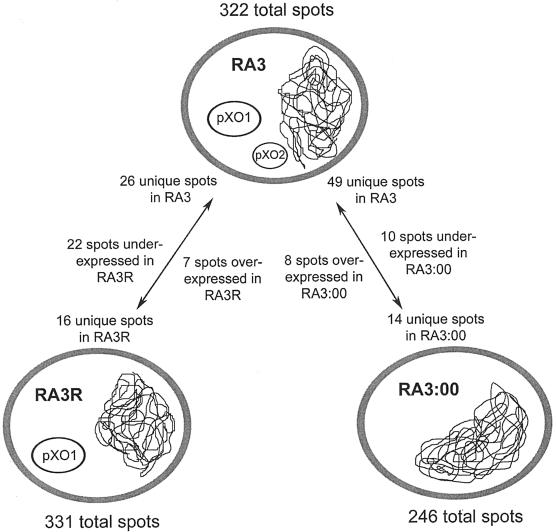
The secretomes of three B. anthracis RA3 derivative strains differing in plasmid content were analyzed in the pH range of 4 to 7. Gel images are presented in Fig. 1 and 2. The average gel of B. anthracis RA3R contained 331 protein spots having an experimental pI range of 4.08 to 6.85 and a mass range of 9.1 to 195.1 kDa. A total of 246 protein spots with pIs ranging from 4.09 to 6.84 and masses between 7.6 and 107.8 kDa were included in the average gel of strain RA3:00. Patra et al. (41) observed 322 spots, with masses between 9.0 and 178.3 kDa and pIs between 4.47 and 6.88, in the average gel of the virulent RA3 strain.
FIG. 1.
2-DE gel of the secreted proteins of B. anthracis RA3 (pXO1+ pXO2+) grown under inducing conditions (0.85% [wt/vol] bicarbonate, 5% CO2, 37°C) in minimal R medium. Samples containing 100 μg of TCA-precipitated extracellular proteins were focused on pH 4 to 7 IPG strips and separated on 10% Duracryl gels. The gels were stained with SYPRO Ruby fluorescent stain and imaged at 470 nm. Of the 42 identified spots, three (blue; **) are unique to RA3 compared to RA3R; six spots (blue; *) are exclusive to RA3 relative to RA3:00; two spots (*/**) are unique to RA3 compared to both RA3R and RA3:00. Molecular masses are indicated on the left.
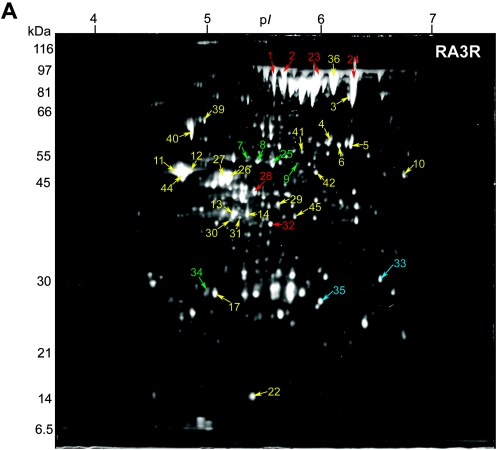
FIG. 2.
Identified secreted-protein spots in (A) B. anthracis RA3R (pXO1+ pXO2−) and (B) B. anthracis RA3:00 (pXO1− pXO2−), both grown under inducing conditions (0.85% [wt/vol] bicarbonate, 5% CO2, 37°C) in minimal R medium. Secretome samples containing 100 μg total protein were focused in the first dimension on pH 4 to 7 IPG strips and separated in the second dimension using 10% Duracryl gels. The gels were then stained with SYPRO Ruby fluorescent stain and imaged at 470 nm. Unique, up-regulated, and down-regulated protein spots in RA3R or RA3:00 compared to RA3 are labeled in blue, red, and green, respectively. Protein spots observed at similar expression levels (constitutive) in RA3R or RA3:00 relative to RA3 are labeled in yellow. Molecular masses are indicated on the left.
2-D gel comparison.
To investigate the variations in protein secretion between B. anthracis strains with variant plasmid contents, the average gels of the RA3R and RA3:00 strains grown under induced conditions were matched to the recently published B. anthracis RA3 (pXO1+ pXO2+) secretome average gel, also grown under CO2 induction (41). The use of SYPRO Ruby gel stain enabled quantification of protein staining intensities, allowing the accurate comparison of protein expression levels between the variant strains. Using the wild-type virulent RA3 strain as a reference, unique and differentially expressed proteins among the three strains in the pH range of 4 to 7 were observed (Fig. 3). These spots were further analyzed for protein identification by MALDI-time of flight.
FIG. 3.
Summary of the secretome differences between the virulent B. anthracis RA3 (pXO1+ pXO2+) strain and its RA3R (pXO1+ pXO2−) and RA3:00 (pXO1− pXO2−) derivative strains grown under CO2 induction conditions in defined medium. Only those spots exhibiting a >5.0-fold increase or decrease in expression level were regarded as differentially expressed. Comparisons were made with the aid of the HT Analyzer software.
A spot was considered to be differentially expressed if there was a fivefold or more change in spot volume. A total of seven spots with masses ranging from 39.3 to 102.6 kDa and pIs ranging from 5.36 to 6.43 were expressed at a significantly higher level (5.3- to 10.1-fold) in RA3R than in RA3. Their pIs ranged from 5.36 to 6.43. Twenty-two spots (mass = 22.4 to 82.4 kDa; pI = 4.97 to 6.58) were consistently underexpressed (5.1- to 56.6-fold) in RA3R; 10 of them exhibited a >20-fold decrease in expression compared to the virulent RA3 strain. Spots exhibiting decreases of 56.6- and 49.7-fold were previously identified as Sap proteins (41). Eight spots, with masses ranging from 9.2 to 83.4 kDa and pIs ranging from 5.01 to 6.42, exhibited increased expression (7.2- to 36.7-fold) in RA3:00 in comparison to RA3. One spot had a >20-fold increase, four spots had an increase between 10- and 20-fold, and three spots had an increase between 5- and 10-fold. There were 10 down-regulated spots (8.4- to 242.2-fold) in the doubly cured strain; they displayed masses from 25.1 to 54.3 kDa and pIs from 5.12 to 6.54. Two of these spots exhibited a >100-fold expression change.
Protein spots were considered unique if they were absent in all three subgels of the strain to which they were compared. Comparative image analysis between the RA3 and RA3R average gels allowed the detection of 16 spots (mass range, 26.0 to 96.3 kDa; pI range, 4.51 to 6.85) unique to RA3R, while 26 spots (mass range, 15.0 to 84.3 kDa; pI range, 4.55 to 6.67) were exclusive to RA3. Comparison of RA3:00/RA3 strains identified 14 spots unique to RA3:00 with masses between 22.1 and 107.0 kDa and pIs between 5.02 and 6.64. Forty-nine spots (mass range, 16.1 to 53.7 kDa; pI range, 4.63 to 6.79) were exclusive to RA3 relative to RA3:00. Included in these 49 unique spots were EF, LF, and Sap S-layer protein, all of which were identified previously by Patra et al. (41).
Protein identification.
Select protein spots that had quantitative differences from RA3 and its derivative strains were processed for MALDI-MS analysis. The protein identifications determined from the resultant tryptic fingerprint spectra are recorded in Tables 1, 2, and 3. Gel comparisons were performed with 100 μg total protein, although subsequent gels were loaded with 225 μg protein or more to enhance detection by MALDI-MS. Twenty-two, 16, and 20 protein spots were identified in the RA3, RA3R, and RA3:00 strains, respectively. However, identifications made in only one strain were correlated, when possible, with the remaining strains by positional overlap. Several RA3 identifications are in agreement with a recent investigation (41) (Table 1). The 57 identifications corresponded to 26 different proteins encoded on either the chromosome or pXO1. Of these protein identifications, 56 matched ORFs with predicted functions, whereas one matched an ORF encoding a hypothetical protein. No pXO2-encoded protein species were identified in this study.
TABLE 1.
B. anthracis RA3 (pXO1+ pXO2+)-secreted proteins identified by MALDI-MS
| Spot no.a | Protein name | Accession no.b | Signal peptidec | pId
|
Masse
|
Gene locationf | ||
|---|---|---|---|---|---|---|---|---|
| Exptl | Theoretical | Exptl | Theoretical | |||||
| Proteins identified directly by MS in RA3 | ||||||||
| 1 | EA1 | NP_654830 | + | 5.79 | 5.70 | 102.6 | 91.4 | Chr |
| 2 | EA1 | NP_654830 | + | 5.70 | 5.70 | 101.8 | 91.4 | Chr |
| 3 | Sap | NP_654829 | + | 6.42 | 6.89 | 83.4 | 86.7 | Chr |
| 4 | Oligopeptide ABC transporter | NP_843667 | + | 6.39 | 6.56 | 58.5 | 61.8 | Chr |
| 5 | Oligopeptide ABC transporter41 | NP_843667 | + | 6.25 | 6.34 | 57.6 | 61.7 | Chr |
| 6 | Oligopeptide ABC transporter | NP_843667 | + | 6.39 | 6.56 | 57.6 | 61.8 | Chr |
| 7 | Protective antigen precursor A41 | AAA22637 | + | 5.44 | 5.88 | 54.0 | 85.9 | pXO1 |
| 8 | Protective antigen precursor A | AAA22637 | + | 5.56 | 5.88 | 53.8 | 85.9 | pXO1 |
| 9 | Protective antigen precursor Ag | AAA22637 | + | 5.83 | 5.88 | 53.7 | 85.9 | pXO1 |
| 10 | Enolase | NP_653583 | − | 6.90 | 4.66 | 49.9 | 46.6 | Chr |
| 11 | Enolase | NP_653583 | − | 4.67 | 4.66 | 50.4 | 46.6 | Chr |
| 12 | Enolase41 | NP_653583 | − | 4.79 | 4.66 | 50.4 | 46.6 | Chr |
| 13 | S-layer homology domain | NP_657198 | + | 5.29 | 5.84 | 42.0 | 40.6 | Chr |
| 14 | S-layer homology domain | NP_657198 | + | 5.40 | 5.84 | 41.9 | 40.6 | Chr |
| 15 | Edema factor (EF)41g | AAA22374 | + | 5.36 | 6.95 | 36.6 | 92.5 | pXO1 |
| 16 | Edema factor (EF)gh | CAC93925 | + | 5.23 | 6.95 | 34.8 | 92.5 | pXO1 |
| 17 | Triosephosphate isomerase | NP_653585 | − | 5.15 | 5.00 | 28.4 | 26.7 | Chr |
| 18 | Lethal factor (LF)41gh | AAA22569 | + | 4.75 | 5.62 | 28.9 | 93.9 | pXO1 |
| 19 | Zinc uptake system protein, ZnuAg | NP_052826 | + | 5.75 | 5.69 | 27.4 | 27.9 | pXO1 |
| 20 | Hypothetical proteinh | NP_653819 | + | 4.75 | 5.06 | 27.7 | 23.2 | Chr |
| 21 | Zinc uptake system protein, ZnuAg | NP_052826 | + | 5.63 | 5.69 | 27.5 | 27.9 | pXO1 |
| 22 | Putative nucleoside diphosphate kinase | NP_655416 | − | 5.46 | 5.24 | 16.5 | 16.7 | Chr |
| Proteins identified by positional overlapi | ||||||||
| 23 | Sap | NP_654829 | + | 6.10 | 6.89 | 100.1 | 86.7 | Chr |
| 36 | Sap | NP_654829 | + | 6.24 | 6.89 | 95.8 | 86.7 | Chr |
| 24 | Sap | NP_654829 | + | 6.43 | 6.89 | 94.9 | 86.7 | Chr |
| 39 | Phosphoglycerate mutase | NP_834804 | − | 4.90 | 4.86 | 64.1 | 56.9 | Chr |
| 40 | Phosphoglycerate mutase | NP_981532 | − | 4.85 | 4.84 | 62.4 | 53.4 | Chr |
| 41 | EA1 | NP_654830 | + | 5.90 | 5.70 | 58.4 | 91.4 | Chr |
| 25 | MerA mercuric reductase | CAA70224 | − | 5.66 | 5.30 | 54.1 | 58.8 | Chr |
| 42 | Isocitrate lyase | NP_830914 | − | 6.02 | 5.27 | 52.0 | 47.1 | Chr |
| 11 | Enolase | NP_653583 | − | 4.67 | 4.66 | 50.8 | 46.6 | Chr |
| 44 | Enolase | NP_653583 | − | 4.72 | 4.66 | 50.3 | 46.6 | Chr |
| 26 | Phosphoglycerol transferase | NP_834895 | − | 5.25 | 5.37 | 49.3 | 44.9 | Chr |
| 27 | Cell surface protein A | NP_658575 | + | 5.19 | 5.56 | 48.6 | 99.9 | Chr |
| 28 | Alanine dehydrogenase | NP_658654 | − | 5.48 | 5.22 | 46.6 | 40.3 | Chr |
| 29 | Glyceraldehyde 3-phosphate dehydrogenase | NP_834805 | − | 5.70 | 5.04 | 44.0 | 35.5 | Chr |
| 45 | Fructose-1,6-bisphosphatase | NP_847733 | − | 5.85 | 5.53 | 41.8 | 34.2 | Chr |
| 31 | Malate dehydrogenase | NP_847040 | − | 5.38 | 5.12 | 42.2 | 33.6 | Chr |
| 32 | Cysteine Synthase | NP_842636 | − | 5.64 | 5.37 | 41.1 | 33.0 | Chr |
| 47 | EA1 | NP_654830 | + | 6.06 | 5.7 | 35.5 | 91.4 | Chr |
| 34 | Triosephosphate isomerase | NP_653585 | − | 5.07 | 5.13 | 29.3 | 26.7 | Chr |
| 49 | Alkyl hydroperoxide reductase | NP_654276 | − | 4.73 | 4.81 | 24.2 | 20.9 | Chr |
Spot numbers refer to Fig. 1.
Accession numbers available through National Center for Biotechnology Information.
Presence of signal peptide predicted by SignalP 3.0 (+, yes; −, no).
Isoelectric point. Exptl, experimental.
Molecular Mass (kDa). Exptl, experimental.
Chr, chromosome.
Unique to RA3 compared to RA3:00.
Unique to RA3 compared to RA3R.
Of proteins identified from RA3R and RA3:00 by MS.
TABLE 2.
B. anthracis RA3R (pXO1+ pXO2−)-secreted proteins identified by MALDI-MS
| Spot no.a | Protein name | Accession no.b | Signal peptidec | pId
|
Masse
|
Gene location | ||
|---|---|---|---|---|---|---|---|---|
| Exptl | Theoretical | Exptl | Theoretical | |||||
| Proteins identified directly by MS in RA3R | ||||||||
| 1 | EA1 | NP_654830 | + | 5.79 | 5.70 | 102.6 | 91.4 | Chrf |
| 2 | EA1 | NP_654830 | + | 5.70 | 5.70 | 101.8 | 91.4 | Chr |
| 23 | Sap | NP_654829 | + | 6.08 | 6.89 | 95.1 | 86.7 | Chr |
| 24 | Sap | NP_654829 | + | 6.43 | 6.89 | 96.0 | 86.7 | Chr |
| 25 | MerA mercuric reductase | CAA70224 | − | 5.64 | 5.30 | 52.9 | 58.8 | Chr |
| 26 | Phosphoglycerol transferase | NP_834895 | − | 5.28 | 5.37 | 47.8 | 44.9 | Chr |
| 27 | Cell surface protein A | NP_658575 | + | 5.19 | 5.56 | 44.5 | 99.9 | Chr |
| 28 | Alanine dehydrogenase | NP_658654 | − | 5.47 | 5.40 | 44.6 | 40.2 | Chr |
| 29 | Glyceraldehyde 3-phosphate dehydrogenase | NP_834805 | − | 5.71 | 5.04 | 44.0 | 35.5 | Chr |
| 13 | S-layer homology domain | NP_657198 | + | 5.28 | 5.84 | 40.3 | 40.6 | Chr |
| 30 | S-layer homology domain | NP_657198 | + | 5.28 | 5.84 | 39.7 | 40.6 | Chr |
| 31 | Malate dehydrogenase | NP_847040 | − | 5.33 | 5.12 | 40.4 | 33.6 | Chr |
| 32 | Cysteine Synthase | NP_842636 | − | 5.62 | 5.37 | 39.3 | 33.0 | Chr |
| 33 | Putative lipoprotein, VanW-related | NP_845858 | + | 6.66 | 6.85 | 31.2 | 32.6 | Chr |
| 34 | Triosephosphate isomerase | NP_653585 | − | 5.07 | 5.13 | 29.4 | 26.7 | Chr |
| 35 | S-layer protein A | NP_652888 | + | 6.12 | 9.16 | 25.9 | 76.3 | pXO1 |
| Proteins identified by positional overlapg | ||||||||
| 36 | Sap | NP_654829 | + | 6.26 | 6.89 | 87.3 | 86.7 | Chr |
| 24 | Sap | NP_654829 | + | 6.45 | 6.89 | 92.2 | 86.7 | Chr |
| 3 | Sap | NP_654829 | + | 6.40 | 6.89 | 76.1 | 86.7 | Chr |
| 39 | Phosphoglycerate mutase | NP_834804 | − | 5.00 | 4.86 | 64.7 | 56.9 | Chr |
| 40 | Phosphoglycerate mutase | NP_981532 | − | 4.89 | 4.84 | 62.4 | 53.4 | Chr |
| 4 | Oligopeptide ABC transporter | NP_843667 | + | 6.22 | 6.56 | 58.5 | 61.8 | Chr |
| 6 | Oligopeptide ABC transporter | NP_843667 | + | 6.30 | 6.56 | 56.8 | 61.8 | Chr |
| 5 | Oligopeptide ABC transporter | NP_843667 | + | 6.40 | 6.34 | 56.9 | 61.7 | Chr |
| 41 | EA1 | NP_654830 | + | 5.94 | 5.70 | 57.1 | 91.4 | Chr |
| 7 | Protective Antigen Precursor A | AAA22637 | + | 5.41 | 5.88 | 53.4 | 85.9 | pXO1 |
| 8 | Protective Antigen Precursor A | AAA22637 | + | 5.52 | 5.88 | 53.4 | 85.9 | pXO1 |
| 9 | Protective Antigen Precursor A | AAA22637 | + | 5.86 | 5.88 | 51.2 | 85.9 | pXO1 |
| 42 | Isocitrate Lyase | NP_830914 | − | 6.05 | 5.27 | 50.9 | 47.1 | Chr |
| 12 | Enolase | NP_653583 | − | 4.87 | 4.66 | 48.6 | 46.6 | Chr |
| 11 | Enolase | NP_653583 | − | 4.71 | 4.66 | 48.4 | 46.6 | Chr |
| 44 | Enolase | NP_653583 | − | 4.78 | 4.66 | 48.1 | 46.6 | Chr |
| 10 | Enolase | NP_653583 | − | 6.84 | 4.66 | 47.4 | 46.6 | Chr |
| 14 | S-layer homology domain | NP_657198 | + | 5.41 | 5.84 | 38.8 | 40.6 | Chr |
| 45 | Fructose-1,6-bisphosphatase | NP_847733 | − | 5.88 | 5.53 | 38.4 | 34.2 | Chr |
| 31 | Malate dehydrogenase | NP_658620 | + | 5.34 | 5.12 | 39.5 | 33.6 | Chr |
| 17 | Triosephosphate isomerase | NP_653585 | − | 5.12 | 5.00 | 27.6 | 26.7 | Chr |
| 22 | Putative nucleoside diphosphate kinase | NP_655416 | − | 5.49 | 5.24 | 15.7 | 16.7 | Chr |
Spot numbers refer to Fig. 2A.
Accession numbers available through National Center for Biotechnology Information.
Presence of signal peptide predicted by SignalP 3.0 (+, yes; −, no).
Isoelectric point. Exptl, experimental.
Molecular mass (kDa). Exptl, experimental.
Chr, chromosome.
Of proteins identified from RA3 and RA3:00 by MS.
TABLE 3.
B. anthracis RA3:00 (pXO1− pXO2−)-secreted proteins identified by MALDI-MS
| Spot no.a | Protein name | Accession no.b | Signal peptidec | pId
|
Masse
|
Gene location | ||
|---|---|---|---|---|---|---|---|---|
| Exptl | Theoretical | Exptl | Theoretical | |||||
| Proteins identified directly by MS in RA3:00 | ||||||||
| 36 | Sap | NP_654829 | + | 6.29 | 6.89 | 100.3 | 86.7 | Chrf |
| 24 | Sap | NP_654829 | + | 6.47 | 6.89 | 101.1 | 86.7 | Chr |
| 38 | Sap | NP_654829 | + | 6.71 | 6.89 | 85.6 | 86.7 | Chr |
| 3 | Sap | NP_654829 | + | 6.42 | 6.89 | 84.3 | 86.7 | Chr |
| 39 | Phosphoglycerate mutase | NP_834804 | − | 5.04 | 4.86 | 67.6 | 56.9 | Chr |
| 40 | Phosphoglycerate mutase | NP_981532 | − | 4.90 | 4.84 | 64.0 | 53.4 | Chr |
| 37 | Oligopeptide ABC transporter | NP_843667 | + | 6.00 | 6.34 | 60.4 | 61.7 | Chr |
| 5 | Oligopeptide ABC transporter | NP_843667 | + | 6.41 | 6.56 | 63.2 | 61.8 | Chr |
| 41 | EA1 | NP_654830 | + | 5.90 | 5.70 | 58.4 | 91.4 | Chr |
| 42 | Isocitrate Lyase | NP_830914 | − | 6.08 | 5.27 | 53.8 | 47.1 | Chr |
| 43 | Enolase | NP_653583 | − | 4.73 | 4.66 | 53.3 | 46.6 | Chr |
| 11 | Enolase | NP_653583 | − | 4.74 | 4.66 | 50.6 | 46.6 | Chr |
| 44 | Enolase | NP_653583 | − | 4.77 | 4.66 | 53.3 | 46.6 | Chr |
| 28 | Alanine dehydrogenase | NP_658654 | − | 5.37 | 5.22 | 42.6 | 40.3 | Chr |
| 45 | Fructose-1,6-bisphosphatase | NP_847733 | − | 5.79 | 5.53 | 39.7 | 34.2 | Chr |
| 31 | Malate dehydrogenase | NP_658620 | + | 5.39 | 5.12 | 42.0 | 33.6 | Chr |
| 46 | S-layer homology domain | NP_657198 | + | 5.40 | 5.84 | 41.9 | 40.6 | Chr |
| 47 | EA1 | NP_654830 | + | 6.01 | 5.70 | 32.9 | 91.4 | Chr |
| 48 | Bacillolysin | NP_654541 | + | 5.83 | 5.82 | 39.5 | 61.0 | Chr |
| 49 | Alkyl hydroperoxide reductase | NP_654276 | − | 4.68 | 4.81 | 23.6 | 20.9 | Chr |
| Proteins identified by positional overlapg | ||||||||
| 1 | Sap | NP_654829 | + | 5.73 | 6.89 | 103.9 | 86.7 | Chr |
| 2 | EA1 | NP_654830 | + | 5.81 | 5.70 | 102.5 | 91.4 | Chr |
| 23 | EA1 | NP_654830 | + | 6.10 | 5.70 | 101.6 | 91.4 | Chr |
| 24 | Sap | NP_654829 | + | 6.44 | 6.89 | 98.6 | 86.7 | Chr |
| 4 | Oligopeptide ABC transporter | NP_843667 | + | 6.20 | 6.56 | 62.9 | 61.8 | Chr |
| 6 | Oligopeptide ABC transporter | NP_843667 | + | 6.25 | 6.56 | 60.8 | 61.8 | Chr |
| 11 | Enolase | NP_653583 | − | 4.76 | 4.66 | 51.8 | 46.6 | Chr |
| 12 | Enolase | NP_653583 | − | 4.82 | 4.66 | 51.9 | 46.6 | Chr |
| 26 | Phosphoglycerol transferase | NP_834895 | − | 5.20 | 5.37 | 48.0 | 44.9 | Chr |
| 27 | Cell surface protein A | NP_658575 | + | 5.13 | 5.56 | 48.1 | 99.9 | Chr |
| 29 | Glyceraldehyde 3-phosphate dehydrogenase | NP_834805 | − | 5.73 | 5.04 | 44.1 | 35.5 | Chr |
| 13 | S-layer homology domain | NP_657198 | + | 5.31 | 5.84 | 41.8 | 40.6 | Chr |
| 17 | Triosephosphate isomerase | NP_653585 | − | 5.17 | 5.00 | 29.5 | 26.7 | Chr |
| 22 | Putative nucleoside diphosphate kinase | NP_655416 | − | 5.53 | 5.24 | 15.3 | 16.7 | Chr |
Spot numbers refer to Fig. 2B.
Accession numbers available through National Center for Biotechnology Information.
Presence of signal peptide predicted by SignalP 3.0 (+, yes; −, no).
Isoelectric point. Exptl, experimental.
Molecular mass (kDa). Exptl, experimental.
Chr, chromosome.
Of proteins identified from RA3 and RA3R by MS.
Proteins overexpressed in RA3R and RA3:00.
Six of the seven up-regulated protein spots (86%) in the pXO2-cured RA3R strain were identified unambiguously by peptide mass fingerprinting (Fig. 2A and Table 2). The identifications exhibiting overexpression in RA3R include the proteins Sap (spots 23 and 24; 5.3- and 17.1-fold), EA1 (spots 1 and 2; 6.2- and 6.9-fold), cysteine synthase A (spot 32; 6.1-fold), and alanine dehydrogenase (AlaDH; spot 28; 5.5-fold) (Fig. 4A). All four of these up-regulated proteins are chromosomally encoded. Sap (86.7 kDa) and EA1 (91.4 kDa) are the major protein components of the S layer in B. anthracis. Displaying a highly patterned ultrastructure, the B. anthracis S layer constitutes the outermost layer in nonencapsulated strains (35). Cysteine synthase A (33.0 kDa) is an enzyme responsible for the formation of cysteine from O-acetyl serine and sulfide with the concomitant release of acetic acid. AlaDH (40.2 kDa), which has been purified and characterized from different species of Bacillus, catalyzes the reversible conversion of alanine to pyruvate and ammonia (25).
FIG. 4.
Differentially expressed protein spots in (A) the pXO2-cured RA3R strain and (B) the plasmidless RA3:00 strain compared to the virulent RA3 (pXO1+ pXO2+) isolate. Bacteria were grown under inducing conditions (0.85% [wt/vol] bicarbonate, 5% CO2, 37°C). Expression corresponds to the ratio of the volumes of matched spots between RA3:00 or RA3R and RA3. Positive values correspond to protein overexpression; negative values refer to protein underexpression. Numbers in parentheses refer to spot numbers as in Fig. 2A or B. *, protective antigen (spot 7) was expressed at variable levels in RA3 but with consistent down-regulation in RA3R. The error bars indicate standard deviations.
Among the eight overexpressed proteins in RA3:00, four (50%) were identified by peptide mass fingerprinting (Fig. 2B and Table 3). These overexpressed proteins were the S-layer proteins Sap (36.7-fold) and EA1 (8.5-fold), isocitrate lyase (12.8-fold), and AlaDH (7.6-fold) (Fig. 4B). AlaDH was also overexpressed in the secretome of RA3:00. Isocitrate lyase catalyzes the reversible cleavage of isocitrate to glyoxylate and succinate. Recently, it was found that the presence of the isocitrate lyase gene is necessary for full virulence in the plant-pathogenic fungus Leptosphaeria maculans (20) and in Mycobacterium tuberculosis (31).
Proteins underexpressed in RA3R and RA3:00.
Of the 22 underexpressed spots in RA3R in comparison to RA3, five (28%) were identified (Fig. 2A and Table 2). These down-regulated proteins include the structural binding toxin PA (spots 7, 8, and 9; 20.8- to 27.2-fold), mercuric reductase (MerA) (spot 25; 12.7-fold), and triosephosphate isomerase (spot 34; 5.1-fold) (Fig. 4A). In RA3, PA was observed as a train of spots with various pI values (5.44 to 5.83). This is an indication of possible posttranslational modifications. The 58.8-kDa MerA, which is encoded on the chromosome, is an enzyme that reduces divalent mercury to the relatively nontoxic metallic mercury via an NADPH-dependent reaction (52). The chromosome-borne triosephosphate isomerase (26.7 kDa), a common glycolytic enzyme, is responsible for the interconversion of dihydroxyacetone phosphate and glyceraldehyde 3-phosphate (27). Of the 10 underexpressed spots in RA3:00 compared to RA3, only 1 (10%) was identified (spot 46; 11.4-fold) (Fig. 2B and 4B and Table 3).
Unique spots.
In the present investigation, unique proteins were present in small amounts and consequently were only weakly observed on the gels. This relative paucity of starting material made identification by MALDI-MS a challenge. The pH range 4 to 7 had one unique protein, oligopeptide binding protein A (OppA) (spot 5), that was identified in RA3:00 relative to the virulent RA3 strain (Fig. 2B and Table 3). A member of the oligopeptide transport system, OppA is tethered to the extracellular face of the cell membrane and facilitates peptide uptake using the energy generated from ATP hydrolysis (43). In RA3R relative to RA3, two unique proteins, a VanW-related lipoprotein (spot 33) and S-layer protein A (spot 35), were identified (Fig. 2A and Table 2). The VanW-related lipoprotein is a potential vancomycin B-type resistance protein. Spots identified as PA (spot 9), EF (spots 15 and 16), LF (spot 18), and the high-affinity zinc uptake protein A (ZnuA) (spots 19 and 21) were unique to RA3 compared to the doubly cured RA3:00 strain, while spots identified as EF (spot 16), LF (spot 18), and a hypothetical protein (spot 20) were absent from the pXO2-cured RA3R strain compared to RA3 (Fig. 1 and Table 1). ZnuA is a periplasmic binding protein that is one of the three members of the Znu Zn2+-specific uptake system (ZnuA, -B, and -C) (42).
Constitutively expressed spots.
Many identified spots were observed in all strains at expression levels similar to that in the virulent strain RA3, and the intensity of the staining or expression did not appear to be dependent on the plasmid content. Phosphoglycerate mutase is an enzyme common in gram-positive and spore-forming bacteria, such as Bacillus and Clostridium species (23). It catalyzes the transfer of phosphate groups between the carbon atoms of phosphoglycerates and is essential for glucose metabolism. Another commonly found secretome protein was the glycolytic enzyme enolase, which is also found on the cell surface in many bacteria (53). Other spots belonging in the category of constitutively expressed spots include those involved in the glycolytic pathway (glyceraldehyde 3-phosphate dehydrogenase and triosephosphate isomerase), the gluconeogenesis pathway (fructose-1,6-bisphosphatase), and NTP synthesis (nucleoside diphosphate kinase).
DISCUSSION
An organism's secretome is the proteome subset defined by active exportation from the cell. The secretome includes the native secreted proteins, as well as components of some protein secretion pathways (58). Since they come into direct contact with host compartments during the course of the infection, many secreted proteins mediate host-pathogen interactions, making them potential targets for immunodetection, immunoprotection, and innovative therapeutic approaches. B. anthracis, as a member of the genus Bacillus, secretes large quantities of proteins into the extracellular environment (58). A recent publication by our laboratory described the secretome of the virulent B. anthracis RA3 strain grown under both host-simulated (CO2 induction) and standard aerobic conditions, using 2-DE coupled with MALDI-MS (41). Numerous proteins, including the pXO1-encoded toxins and the chromosome-encoded Sap and EA1, were identified. Here, we report a follow-up comparative proteomic study whose objective was the characterization of the differences between the secretomes of three B. anthracis RA3 genotype strains differing in plasmid content. By using a comparative proteomic approach that relied on 2-DE in conjunction with MALDI-MS, it was possible to determine genotype-specific and differentially expressed proteins between the strains.
The present study presents a snapshot of the secretomes of three genotypes of B. anthracis grown under host-simulated conditions. Tjalsma et al. recently reported that 47% of the proteins contained in the B. subtilis secretome were not predicted by bioinformatics analysis to be secreted (58). Furthermore, 13 of the 26 identified protein species exhibited putative signal peptide sequences based on SignalP 3.0 prediction (3). Thus, absence of a signal peptide sequence does not necessarily indicate that a protein cannot be secreted; many nonclassically secreted proteins are secreted despite the lack of a currently described sequence motif. As revealed by the average 2-DE gel images (Fig. 1 and 2), there are significant differences between the secretomes of the virulent, wild-type RA3 (pXO1+ pXO2+) strain and its two derivative strains: the toxigenic but nonencapsulated RA3R (pXO1+ pXO2−) and the plasmidless nontoxigenic, nonencapsulated strain RA3:00 (pXO1− pXO2−). To ensure isogenicity among the strains, the RA3:00 strain was generated from the pXO2-cured RA3R strain by a 30-day subculturing process at the elevated temperature of 42.5°C (35). Thus, all observed 2-D gel differences are most likely the result of secretome variation due to plasmid content as opposed to altered chromosomal backgrounds, if any, during the plasmid-curing process (40).
The loss of pXO2 seems to be correlated with a down-regulation or nonappearance of numerous pXO1-derived gene products. Spots observed in RA3 and subsequently determined to be the pXO1-encoded toxins, such as EF and LF, were not observed in RA3R. Three RA3 spots identified as PA were expressed at significantly lower levels (20.8- to 27.2-fold decrease) in RA3R, perhaps in part due to the loss of the pXO2-borne gene regulator. Studies have sought to identify and characterize possible gene regulators encoded on pXO2 (13, 61). In light of the results presented here, future studies should seek to determine the effect of AcpB, a pXO2-encoded gene regulator, on pXO1 genes, especially the toxin genes.
Alternatively, relative decreased toxin expression levels in RA3R may be the result of altered efficiency of the secretion mechanism, leading to decreased toxin export in this nonencapsulated strain. There may also be a misregulation of protein folding. It is known that PA and the other toxins are secreted from B. anthracis via the Sec-dependent secretion pathway, which requires the proteins to be translocated across the cytoplasmic membrane in an unfolded conformation (64). One membrane-bound molecular chaperone, PrsA, was recently discovered to have a role in PA production (19, 64). Increasing the concentration of PrsA led to an increase in recombinant PA production in B. subtilis (64). It was found that several PrsA homologues present in the B. anthracis genome were able to complement the activity of B. subtilis PrsA with respect to recombinant PA secretion (64). Hence, the lower levels of PA in this study may be a consequence of reduced expression of PrsA in strain RA3R.
Another particularly striking observation is that, among the proteins whose levels increased in RA3R and RA3:00, several are potentially involved in processes leading to sporulation (32). AlaDH, whose expression was enhanced 6.1-fold in RA3R and 7.6-fold in RA3:00 relative to RA3, has been found to be important for normal sporulation in B. subtilis (54). This enzyme catalyzes the reversible conversion of alanine to pyruvate and ammonia, and it is postulated that pyruvate is then used to generate energy for sporulation activity via the TCA cycle (10). Since the amino acid alanine is missing in R medium, the presence of these enzymes can also be viewed as corresponding to the anabolism of this amino acid by B. anthracis. Furthermore, sporulation defects caused by null mutations in the alanine dehydrogenase gene (ald) were lessened by the addition of high concentrations of pyruvate (54). Meanwhile, the protein isocitrate lyase, which in strain RA3:00 displayed a 12.8-fold increase in expression relative to the virulent RA3 strain, may also be involved in the mechanism of sporulation. Isocitrate lyase is one of the two enzymes in the glyoxylate pathway that function in the metabolism of two-carbon compounds, such as acetate.
The exact reason for the presence of these sporulation-associated enzymes cannot be stated, but it has been shown that vegetative cells of the B. anthracis Sterne strain (lacking the pXO2 plasmid, like RA3R and RA3:00) loads itself with sporulation-associated enzymes, effectively preparing it for circumstances it has not yet encountered in vegetative growth (28). In the present study, therefore, the observance of numerous sporulation-associated enzymes can be attributed to the non-growth-phase-dependent synthesis of spore constituents in these vegetative cells. The observation that these spore-related enzymes were overexpressed in both RA3R and RA3:00 compared to the virulent RA3 strain is not unexpected, since differences in sporulation characteristics between cured and uncured cells exist (35). Cured strains are known to sporulate both earlier and at higher frequencies than uncured isolates (35). In our study, the strain RA3:00 sporulation rate is approximately nine times less than that of strain RA3R (data not shown).
The experimental mass and pI values of the majority of identified proteins do not significantly deviate from the corresponding theoretical values for the mature protein. The exceptions are the exotoxins (PA-EF/LF), which in RA3 (and RA3R by positional overlap) exhibited considerably lower experimental than theoretical masses (Table 1), a phenomenon that has been previously observed (41). PA, for example, has a predicted mass of 86 kDa; however, in this study, it was observed at ∼54 kDa by SDS-PAGE. This disparity is likely a result of protease digestion, since PA is known to be sensitive to proteolysis at both cell-associated and extracellular locations (57). Cleavage of the PA monomer in vivo by furin-like proteases has been shown to result in the release of a 20-kDa (PA20) and a 63-kDa (PA63) PA fragment (7). In fact, PA proteolysis resulting in the formation of PA20 and PA63 is critical in anthrax pathogenesis, since it is PA63 that heptamerizes and competitively binds LF and EF, allowing their cellular internalization in the host (7).
Furthermore, a PA proteolysis-induced form of the mature PA appeared as a series of isoelectric forms (in the range of 5.44 to 5.83 on 2-D gels) (Fig. 2A, spots 7, 8, and 9). The series of isoelectric forms suggests that PA may have undergone posttranslational modifications resulting in a series of charge alterations. Phosphorylations, methylations, and/or deamidations are the most likely PA modifications, since the various isoelectric points are not accompanied by a significant change in mass. Disagreement of EF and LF with their theoretical values (both mass and pI) may be due to proteolysis (Table 1), as described for PA. In both cases, trypsin-digested peptides were found to match sequences from the N-terminal portions (corresponding to the PA binding domain) of the intact, mature proteins.
Proteins containing S-layer homology domains were the most prevalent class of proteins identified. In total, Sap and EA1 accounted for 13 of the 57 protein identifications. As components of the S layer, the paracrystalline sheath completely overlying the peptidoglycan of vegetative cells, Sap and EA1 were found to represent more than 75% of the B. anthracis membrane fraction (11). It has been reported that both EA1 and Sap undergo posttranslational modifications, since both proteins were found to exist in a multiplicity of isoelectric forms and, for EA1, in numerous mass forms (11). EA1 was observed in this investigation to be present in at least four different isoelectric and three different mass forms. Sap also appeared as a series of at least four isoelectric forms and two different mass forms. It has been postulated that subpopulations of differently glycosylated Sap and EA1 may exist (11), which may account for the array of observed masses in this study. The reason for possible posttranslational modifications is not known. Expression of the pXO1 S-layer protein A is in addition to the seven B. anthracis S-layer proteins characterized in a recent proteomics approach (11). The contribution of this particular S-layer protein to the physiology of B. anthracis will have to be investigated, along with the regulation of its expression in vitro and in vivo, keeping in mind that the S-layer proteins are not secreted but rather cell wall displaced proteins. This phenomenon may be seen as a strategy used by B. anthracis to introduce surface-exposed S-layer variations throughout the phases of vegetative cell growth inside the host (15, 33).
The proteins observed in this study represent only a portion of the entire repertoire of B. anthracis secretome proteins. N-terminal signal sequence analysis of the proteome of B. anthracis A2012 revealed a total of 970 secretory-protein candidates on the chromosome, pXO1, and pXO2. The vast majority of these potentially secreted proteins are located on the chromosome, with pXO1 and pXO2 contributing only 25 and 20 protein candidates, respectively. As expected, in our present study involving the comparison of the fully virulent strain RA3 to the two cured strains RA3:00 and RA3R, both lacking pXO2, no identified pXO2-encoded proteins were observed. The secretome of a strain harboring only pXO2 will be investigated in future studies. Proteins lacking an obvious signal sequence have also been observed in the secretomes of several other bacterial species (2, 9). Some of the 970 putative secreted proteins are on the basic end of the spectrum with theoretical pIs between 7 and 10. Many of these proteins, however, may be posttranslationally modified to produce mature proteins with pIs within our experimental range. It is also worth noting that our system does not effectively resolve very high- or very low-mass proteins, since the 10% Duracryl gels used optimally resolve proteins between approximately 15 and 116 kDa. Moreover, our method of protein isolation, TCA precipitation coupled with an acetone wash, may cause the loss of low-abundance and/or low-mass and hydrophobic proteins.
This investigation has enhanced our understanding of the differences in sporulation and regulatory activities among strains of the potentially deadly bacterium B. anthracis with variant plasmid contents. The comparative proteomic analysis suggests that the presence of the pXO2 plasmid is necessary for maximal toxin (EF, LF, and PA) expression. However, it cannot be definitively stated whether this observation is a consequence of a toxin-controlling, pXO2-borne regulatory gene product; the misregulation of protein secretion pathways in strains lacking pXO2; or the disruption of folding pathways in nonencapsulated isolates. Moreover, the presence of a wide array of sporulation-associated enzymes in the secretomes of plasmid-cured strains suggests that these strains prepare themselves for adverse environmental conditions to a greater extent than genetically complete (virulent) B. anthracis strains. Dialogue among the chromosome, pXO1, and pXO2 takes place within the cell via pleiotropic regulators, such as AtxA in B. anthracis or PlcR in B. thuringiensis and B. cereus, but can also take place via a cell-to-cell signaling mechanism involving the Opp oligopeptide permease. These data hint at a mechanism of gene regulation via a reimported small peptide in B. anthracis (56).
Four proteins present in the B. anthracis secretome may be putative virulence factors: enolase may contribute to pathogenicity by binding the infected host's plasminogen, potentially allowing the bacteria to acquire surface-associated proteolytic activity (4, 38). Enolase is also part of the glycolytic pathways, and as such, may be categorized as a bifunctional or “moonlighting” protein (36). The pXO1-encoded protein corresponding to a high-affinity zinc uptake protein (ZnuA) may play an important role in the biological activity of the lethal factor (35) and other zinc metalloproteases (47). It is interesting that in Haemophilus influenzae the presence of ZnuA enhances virulence (26). Like zinc, iron is necessary to obtain full virulence of B. anthracis. The identification of the cell surface protein A (BA_5215) harboring an iron transport-associated domain may correspond to one of the strategies for iron uptake by B. anthracis (47). These two proteins may have an important role in zinc and iron scavenging when B. anthracis is present in a mammalian host. Also identified was an alkylhydroperoxide reductase (AhpC-TSA), a protein involved in oxidative stress response, which may offer resistance to peroxynitrite within the macrophages. As such, AhpC-TSA may be seen as a putative virulence factor in B. anthracis (30). Ultimately the identification of all proteins expressed and/or secreted by the plasmids pXO1 and pXO2 will provide new clues regarding the contributions of these plasmids to B. anthracis pathogenicity.
Acknowledgments
We thank Cesar V. Mujer for advice and helpful discussions, Frank Estock for expert preparation of the figure graphics, and Josée Vaissaire and Michèle Mock for providing Bacillus anthracis strains. We also express our gratitude to Christian Merz and Edward C. Evans for their invaluable technical assistance.
This study was supported by grant DE-FG-02-00ER62773 from the U.S. Department of Energy.
Editor: J. D. Clements
REFERENCES
- 1.Alexander, D. A. 2003. Bioterrorism: preparing for the unthinkable. J. R. Army Med. Corps 149:125-130. [DOI] [PubMed] [Google Scholar]
- 2.Antelmann, H., H. Tjalsma, B. Voigt, S. Ohlmeier, S. Bron, J. M. van Dijl, and M. Hecker. 2001. A proteomic view on genome-based signal peptide predictions. Genome Res. 11:1484-1502. [DOI] [PubMed] [Google Scholar]
- 3.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann, S., M. Rohde, G. S. Chhatwal, and S. Hammerschmidt. 2001. α-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40:1273-1287. [DOI] [PubMed] [Google Scholar]
- 5.Bourgogne, A., M. Drysdale, S. G. Hilsenbeck, S. N. Peterson, and T. M. Koehler. 2003. Global effects of virulence gene regulators in a Bacillus anthracis strain with both virulence plasmids. Infect. Immun. 71:2736-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Bradley, K. A., J. Mogridge, M. Mourez, R. J. Collier, and J. A. Young. 2001. Identification of the cellular receptor for anthrax toxin. Nature 414:225-229. [DOI] [PubMed] [Google Scholar]
- 8.Bragg, T. S., and D. L. Robertson. 1989. Nucleotide sequence and analysis of the lethal factor gene (lef) from Bacillus anthracis. Gene 81:45-54. [DOI] [PubMed] [Google Scholar]
- 9.Bumann, D., S. Aksu, M. Wendland, K. Janek, U. Zimny-Arndt, N. Sabarth, T. F. Meyer, and P. R. Jungblut. 2002. Proteome analysis of secreted proteins of the gastric pathogen Helicobacter pylori. Infect. Immun. 70:3396-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carls, R. A., and R. S. Hanson. 1971. Isolation and characterization of tricarboxylic acid cycle mutants of Bacillus subtilis. J. Bacteriol. 106:848-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chitlaru, T., N. Ariel, A. Zvi, M. Lion, B. Velan, A. Shafferman, and E. Elhanany. 2004. Identification of chromosomally encoded membranal polypeptides of Bacillus anthracis by a proteomic analysis: prevalence of proteins containing S-layer homology domains. Proteomics 4:677-691. [DOI] [PubMed] [Google Scholar]
- 12.Dai, Z., J. C. Sirard, M. Mock, and T. M. Koehler. 1995. The atxA gene product activates transcription of the anthrax toxin genes and is essential for virulence. Mol. Microbiol. 16:1171-1181. [DOI] [PubMed] [Google Scholar]
- 13.Drysdale, M., A. Bourgogne, S. G. Hilsenbeck, and T. M. Koehler. 2004. atxA controls Bacillus anthracis capsule synthesis via acpA and a newly discovered regulator, acpB. J. Bacteriol. 186:307-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eschenbrenner, M., M. A. Wagner, T. A. Horn, J. A. Kraycer, C. V. Mujer, S. Hagius, P. Elzer, and V. G. DelVecchio. 2002. Comparative proteome analysis of Brucella melitensis vaccine strain Rev 1 and a virulent strain, 16M. J. Bacteriol. 184:4962-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fouet, A., and M. Mock. 1996. Differential influence of the two Bacillus anthracis plasmids on regulation of virulence gene expression. Infect. Immun. 64:4928-4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gohar, M., O. A. Økstad, N. Gilois, V. Sanchis, A.-B. Kolstø, and D. Lereclus. 2002. Two-dimensional electrophoresis analysis of the extracellular proteome of Bacillus cereus reveals the importance of the PlcR regulon. Proteomics 2:784-791. [DOI] [PubMed] [Google Scholar]
- 17.Hale, J. E., J. P. Butler, M. D. Knierman, and G. W. Becker. 2000. Increased sensitivity of tryptic peptide detection by MALDI-TOF mass spectrometry is achieved by conversion of lysine to homoarginine. Anal. Biochem. 287:110-117. [DOI] [PubMed] [Google Scholar]
- 18.Hirose, I., K. Sano, I. Shioda, M. Kumano, K. Nakamura, and K. Yamane. 2000. Proteome analysis of Bacillus subtilis extracellular proteins: a two-dimensional protein electrophoretic study. Microbiology 146:65-75. [DOI] [PubMed] [Google Scholar]
- 19.Hyyryläinen, H.-L., M. Vitikainen, J. Thwaite, H. Wu, M. Sarvas, C. R. Harwood, V. P. Kontinen, and K. Stephenson. 2000. d-Alanine substitution of teichoic acids as a modulator of protein folding and stability at the cytoplasmic membrane/cell wall interface of Bacillus subtilis. J. Biol. Chem. 275:26696-26703. [DOI] [PubMed] [Google Scholar]
- 20.Idnurm, A., and B. J. Howlett. 2002. Isocitrate lyase is essential for pathogenicity of the fungus Leptosphaeria maculans to canola (Brassica napus). Eukaryot. Cell 1:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 22.Ivins, B. E., J. W. Ezzell, Jr., J. Jemski, K. W. Hedlund, J. D. Ristroph, and S. H. Leppla. 1986. Immunization studies with attenuated strains of Bacillus anthracis. Infect. Immun. 52:454-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jedrzejas, M. J. 2002. Three-dimensional structure and molecular mechanism of novel enzymes of spore-forming bacteria. Med. Sci. Monit. 8:RA183-RA190. [PubMed] [Google Scholar]
- 24.Koehler, T. M., Z. Dai, and M. Kaufman-Yarbray. 1994. Regulation of the Bacillus anthracis protective antigen gene: CO2 and trans-acting element activate transcription from one of two promoters. J. Bacteriol. 176:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuroda, S., K. Tanizawa, Y. Sakamoto, H. Tanaka, and K. Soda. 1990. Alanine dehydrogenase from two Bacillus species with distinct thermostabilities: molecular cloning, DNA and protein sequence determination, and structural comparison with other NAD(P)+-dependent dehydrogenases. Biochemistry 29:1009-1015. [DOI] [PubMed] [Google Scholar]
- 26.Lewis, D. A., J. Klesney-Tait, S. R. Lumbley, C. K. Ward, J. L. Latimer, C. A. Ison, and E. J. Hansen. 1999. Identification of the znuA-encoded periplasmic zinc transport of Haemophilus ducreyi. Infect. Immun. 67:5060-5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leyva-Vazquez, M. A., and P. Setlow. 1994. Cloning and nucleotide sequences of the genes encoding triose phosphate isomerase, phosphoglycerate mutase, and enolase from Bacillus subtilis. J. Bacteriol. 176:3903-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, H., N. H. Bergman, B. Thomason, S. Shallom, A. Hazen, J. Crossno, D. A. Rasko, J. Ravel, T. D. Read, S. N. Peterson, J. Yates III, and P. C. Hanna. 2004. Formation and composition of the Bacillus anthracis endospore. J. Bacteriol. 186:164-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makino, S., I. Uchida, N. Terakado, C. Sasakawa, and M. Yoshikawa. 1989. Molecular characterization of the cap region, which is essential for encapsulation in Bacillus anthracis. J. Bacteriol. 171:722-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Master, S. S., B. Springer, P. Sander, E. C. Boettger, V. Deretic, and G. S. Timmins. 2002. Oxidative stress response genes in Mycobacterium tuberculosis: role of ahpC in resistance to peroxynitrite and stage-specific survival in macrophages. Microbiology 148:3139-3144. [DOI] [PubMed] [Google Scholar]
- 31.McKinney, J. D., K. Höner zu Bentrup, E. J. Muñoz-Elias, A. Micak, B. Chen, W.-T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 32.Megraw, R. E., and R. J. Beers. 1964. Glyoxylate metabolism in growth and sporulation of Bacillus cereus. J. Bacteriol. 87:1087-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mignot, T., M. Mock, and A. Fouet. 2003. A plasmid-encoded regulator couples the synthesis of toxins and surface structures in Bacillus anthracis. Mol. Microbiol. 47:917-927. [DOI] [PubMed] [Google Scholar]
- 34.Mikesell, P., B. E. Ivins, J. D. Ristroph, and T. M. Dreier. 1983. Evidence for plasmid-mediated toxin production in Bacillus anthracis. Infect. Immun. 39:371-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55:647-671. [DOI] [PubMed] [Google Scholar]
- 36.Moore, B. 2004. Bifunctional and moonlighting enzymes: lighting the way to regulatory control. Trends Plant Sci. 9:221-228. [DOI] [PubMed] [Google Scholar]
- 37.Nakata, H. M., and H. O. Halvorson. 1960. Biochemical changes occurring during growth and sporulation of Bacillus cereus. J. Bacteriol. 80:801-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pancholi, V., and V. A. Fischetti. 1998. α-Enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273:14503-14515. [DOI] [PubMed] [Google Scholar]
- 39.Patra, G., J. Vaissaire, M. Weber-Levy, C. Le Doujet, and M. Mock. 1998. Molecular characterization of Bacillus strains involved in outbreaks of anthrax in France in 1997. J. Clin. Microbiol. 36:3412-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patra, G., A. Fouet, J. Vaissaire, J. L. Guesdon, and M. Mock. 2002. Variation in rRNA operon number as revealed by ribotyping of Bacillus anthracis strains. Res. Microbiol. 153:139-148. [DOI] [PubMed] [Google Scholar]
- 41.Patra, G., L. E. Williams, K. G. Dwyer, and V. G. Delvecchio. 2003. Bacillus anthracis secretome p. 53-66. In V. G. DelVecchio and V. Krcmery (ed.), Applications of genomics and proteomics for analysis of bacterial biological agents. NATO Science Series, vol. 352. IOS Press, Amsterdam, The Netherlands.
- 42.Patzer, S. I., and K. Hantke. 2000. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J. Biol. Chem. 275:24321-24332. [DOI] [PubMed] [Google Scholar]
- 43.Perego, M., C. F. Higgins, S. R. Pearce, M. P. Gallagher, and J. A. Hoch. 1991. The oligopeptide transport system of Bacillus subtilis plays a role in the initiation of sporulation. Mol. Microbiol. 5:173-185. [DOI] [PubMed] [Google Scholar]
- 44.Qi, Y., G. Patra, X. Liang, L. E. Williams, S. Rose, R. J. Redkar, and V. G. DelVecchio. 2001. Utilization of the rpoB gene as a specific chromosomal marker for real-time PCR detection of Bacillus anthracis. Appl. Environ. Microbiol. 67:3720-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rafie-Kolpin, M., R. C. Essenberg, and J. H. Wyckoff III. 1996. Identification and comparison of macrophage-induced proteins and proteins induced under various stress conditions in Brucella abortus. Infect. Immun. 64:5274-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramisse, V., G. Patra, H. Garrigue, J. Guesdon, and M. Mock. 1996. Identification and characterization of Bacillus anthracis by multiplex PCR analysis of sequences on plasmids pXO1 and pXO2 and chromosomal DNA. FEMS Microbiol. Lett. 145:9-16. [DOI] [PubMed] [Google Scholar]
- 47.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulson, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Elsen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. Deboy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weldman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. Kolstø, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 48.Read, T. D., S. L. Salzberg, M. Pop, M. Shumway, L. Umayam, L. Jiang, E. Holtzapple, J. D. Busch, K. L. Smith, J. M. Schupp, D. Solomon, P. Keim, and C. M. Fraser. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296:2028-2033. [DOI] [PubMed] [Google Scholar]
- 49.Ristroph, J., and B. Ivins. 1983. Elaboration of Bacillus anthracis antigens in a new, defined culture medium. Infect. Immun. 39:483-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robertson, D. L., M. T. Tippets, and S. H. Leppla. 1988. Nucleotide sequence of the Bacillus anthracis edema factor gene (cya): a calmodulin-dependent adenylate cyclase. Gene 73:363-371. [DOI] [PubMed] [Google Scholar]
- 51.Rosenfeld, J., J. Capdevielle, J. C. Guillemot, and P. Ferrara. 1992. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 203:173-179. [DOI] [PubMed] [Google Scholar]
- 52.Rugh, C. L., H. D. Wilde, N. M. Stack, D. M. Thompson, A. O. Summers, and R. B. Meagher. 1996. Mercuric ion reduction and resistance in transgenic Arabidopsis thaliana plants expressing a modified bacterial merA gene. Proc. Natl. Acad. Sci. USA 93:3182-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sha, J., C. L. Galindo, V. Pancholi, V. L. Popov, Y. Zhao, C. W. Houston, and A. K. Chopra. 2003. Differential expression of the enolase gene under in vivo versus in vitro growth conditions of Aeromonas hydrophila. Microb. Pathog. 34:195-204. [DOI] [PubMed] [Google Scholar]
- 54.Siranosian, K. J., K. Ireton, and A. D. Grossman. 1993. Alanine dehydrogenase (ald) is required for normal sporulation in Bacillus subtilis. J. Bacteriol. 175:6789-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sirard, J. C., M. Mock, and A. Fouet. 1994. The three Bacillus anthracis toxin genes are coordinately regulated by bicarbonate and temperature. J. Bacteriol. 176:5188-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slamti, L., and D. Lereclus. 2002. A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J. 21:4550-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thwaite, J. E., L. W. J. Baillie, N. M. Carter, K. Stephenson, M. Rees, C. R. Harwood, and P. T. Emmerson. 2002. Optimization of the cell wall microenvironment allows increased production of recombinant Bacillus anthracis protective antigen from B. subtilis. Appl. Environ. Microbiol. 68:227-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tjalsma, H., A. Bolhuis, J. D. H. Jongbloed, S. Bron, and J. M. van Dijl. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uchida, I., M. Hornung, C. B. Thorne, K. R. Klimpel, and S. H. Leppla. 1993. Cloning and characterization of a gene whose product is a trans-activator of anthracis toxin synthesis. J. Bacteriol. 175:5329-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uchida, I., S. Makino, C. Saskawa, M. Yodhikawa, C. Sugimoto, and N. Terakado. 1993. Identification of a novel gene, dep, associated with depolymerization of the capsular polymer in Bacillus anthracis. Mol. Microbiol. 9:487-496. [DOI] [PubMed] [Google Scholar]
- 61.Vietri, N. J., R. Marrero, T. A. Hoover, and S. L. Welkos. 1995. Identification of a trans-activator involved in the regulation of encapsulation by Bacillus anthracis. Gene 152:1-9. [DOI] [PubMed] [Google Scholar]
- 62.Wagner, M. A., M. Eschenbrenner, T. A. Horn, J. A. Kraycer, C. V. Mujer, S. Hagius, P. Elzer, and V. G. DelVecchio. 2002. Global analysis of the Brucella melitensis proteome: identification of proteins expressed in laboratory-grown culture. Proteomics 2:1047-1060. [DOI] [PubMed] [Google Scholar]
- 63.Welkos, S. L., J. R. Lowe, F. Eden-Mccutchan, M. Vodkin, S. H. Leppla, and J. J. Schmidt. 1988. Sequence and analysis of the DNA encoding protective antigen of Bacillus anthracis. Gene 69:287-300. [DOI] [PubMed] [Google Scholar]
- 64.Williams, R. C., M. L. Rees, M. F. Jacobs, Z. Pragai, J. E. Thwaite, L. W. J. Baillie, P. T. Emmerson, and C. R. Harwood. 2003. Production of Bacillus anthracis protective antigen is dependent on the extracellular chaperone, PrsA. J. Biol. Chem. 278:18056-18062. [DOI] [PubMed] [Google Scholar]