Abstract
Salmonella enterica serotype Typhimurium causes human infections that can frequently be traced back through the food chain to healthy livestock whose intestine is colonized by the pathogen. Little is known about the genes important for intestinal carriage of S. enterica serotype Typhimurium in vertebrate animals. Here we characterized the role of 10 fimbrial operons, agf, fim, lpf, pef, bcf, stb, stc, std, stf, and sth, using competitive infection experiments performed in genetically susceptible (BALB/c) and resistant (CBA) mice. Deletion of agfAB, fimAICDHF, lpfABCDE, pefABCDI, bcfABCDEFG, stbABCD, stcABCD, stdAB, stfACDEFG, or sthABCDE did not reduce the ability of S. enterica serotype Typhimurium to colonize the spleen and cecum of BALB/c mice 5 days after infection. Similarly, deletion of agfAB, fimAICDHF, pefABCDI, and stfACDEFG did not result in reduced recovery of S. enterica serotype Typhimurium from fecal samples collected from infected CBA mice over a 30-day time period. However, S. enterica serotype Typhimurium strains carrying deletions in lpfABCDE, bcfABCDEFG, stbABCD, stcABCD, stdAB, or sthABCDE were recovered at significantly reduced numbers from the feces of CBA mice. There was a good correlation (R2 = 0.9626) between competitive indices in the cecum and fecal samples of CBA mice at 30 days after infection, suggesting that the recovery of S. enterica serotype Typhimurium from fecal samples closely reflected its ability to colonize the cecum. Collectively, these data show that six fimbrial operons (lpf, bcf, stb, stc, std, and sth) contribute to long-term intestinal carriage of S. enterica serotype Typhimurium in genetically resistant mice.
Nontyphoidal Salmonella serotypes are the leading cause of food-borne infections with a lethal outcome in the United States (30). S. enterica serotype Typhimurium is the serotype most frequently associated with this diarrheal disease (34). S. enterica serotype Typhimurium is commonly transmitted from animal to human through food products derived from livestock or domestic fowl (34). Intestinal carriage results in fecal contamination of equipment surfaces or workers hands with S. enterica serotype Typhimurium, thereby leading to contamination of carcasses and processed foods at processing plants. For this reason, the presence of S. enterica serotype Typhimurium in the intestine of healthy food animals arriving at slaughter is the main risk factor for introducing this pathogen into the human food supply. Although intestinal carriage of S. enterica serotype Typhimurium in livestock and domestic fowl is of considerable importance for food safety, the mechanisms that enable this pathogen to persist in the alimentary tract of vertebrate hosts remain poorly characterized.
Mice of genetically susceptible lineages (e.g., BALB/c) commonly used to study S. enterica serotype Typhimurium pathogenesis succumb to infection within 6 to 10 days after inoculation and are thus not well suited for studying mechanisms of long-term intestinal persistence. As a result, reports on genetic determinants that are required for persistent intestinal carriage of S. enterica serotype Typhimurium in vivo are sparse (21, 24). Fimbriae encoded by the lpf, fim, and agf (csg) operons mediate attachment of S. enterica serotype Typhimurium to epithelial cell lines in vitro (4, 12, 19, 37, 38, 41). These in vitro data suggest that S. enterica serotype Typhimurium fimbriae may be involved in intestinal colonization, which warrants further investigation of their role during host pathogen interaction in vivo.
Whole-genome sequencing has identified 13 operons containing fimbrial gene sequences in the S. enterica serotype Typhimurium genome, termed agf (csg), fim, lpf, pef, bcf, stb, stc, std, stf, sth, sti, saf, and stj (29). The S. enterica serotype Typhimurium fim operon directs the assembly of type 1 fimbriae on bacterial cells grown in standard broth culture (11, 27). Addition of type 1 fimbriated bacteria to a suspension of yeast or red blood cells results in an agglutination that can be inhibited by the addition of mannose (9). Type 1 fimbriae of S. enterica serotype Typhimurium bind the extracellular matrix glycoprotein laminin through its oligomannoside chains (26). The agf (csg) operon mediates expression of thin curled fimbriae (also known as thin aggregative fimbriae or curli) after growth of S. enterica serotype Typhimurium on agar plates (17, 36). Thin curled fimbriae mediate mannose-resistant agglutination of red blood cells (15, 16) and binding to the extracellular matrix protein fibronectin (8). The remaining 11 fimbrial operons of S. enterica serotype Typhimurium appear to be poorly expressed when bacteria are grown under standard laboratory conditions (18), and no information is available about the binding specificity of the encoded adhesins.
Expression of major fimbrial subunits encoded by nine fimbrial operons, including BcfA, FimA, LpfA, PefA, StbA, StcA, StdA, StfA, and StiA, is detectable upon recovery of S. enterica serotype Typhimurium 8 h after infection of bovine ligated ileal loops (18). Using genetically susceptible mouse lineages (e.g., BALB/c), the lpf, pef, and agf operons have been implicated in colonization of intestinal tissues (3, 5, 6, 40). However, it is currently unknown whether fimbrial operons identified by whole-genome sequencing contribute to long-term intestinal persistence of S. enterica serotype Typhimurium. Here we report on characterization of 10 S. enterica serotype Typhimurium strains, each with a deletion of a different fimbrial operon, in a mouse model of intestinal persistence.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
S. enterica serotype Typhimurium strain IR715 is a virulent, nalidixic acid-resistant derivative of American Type Culture Collection (ATCC) strain 14028 and has been described previously (35). AJB715 is a derivative of IR715 containing a kanamycin (Km) cassette inserted in the phoN open reading frame (21). Strain RAK48 is a derivative of IR715 with a deletion of the lpfABCDE operon (25). Strain AJB33 is a derivative of IR715 which carries a lpfABCDE::lacZYA transcriptional fusion generated by inserting a suicide plasmid (pMS1096) behind the lpfE stop codon by homologous recombination (33). Escherichia coli strain DH5α which was used for cloning has been described previously (13). Derivatives of Escherichia coli strain S17-1λpir carrying suicide plasmids for generating deletions in agfAB (pSF31), pefABCDI (pSF30), bcfABCDEFG (pEW1), stbABCD (pSF41), stcABCD (pSF42), stdAB (pEW5), stfACDEFG (pEW3), or sthABCDE (pSF37) have been described previously (18). Plasmids (pAH70, pAH77, pAH72, pAH69, pAH75, pAH76, pAH106, pAH108, and pAH60) containing cloned fragments of the bcfA, fimA, lpfA, pefA, stbA, sthA, stdA, stfA, and stcA open reading frames corresponding to the mature protein after cleavage of the signal peptide have been described previously (18).
Strains were routinely cultured aerobically at 37°C in Luria-Bertani (LB) broth (10g/liter Tryptone, 5g/liter yeast extract, 5g/liter NaCl) supplemented with antibiotics as appropriate at the following concentrations: chloramphenicol 30 mg/liter (LB+Cm), kanamycin 100 mg/liter (LB+Km), nalidixic acid 50 mg/liter (LB+Nal). LB agar plates contained 15 g/liter agar (Difco). For detection of alkaline phosphatase (PhoN) activity in S. enterica serotype Typhimurium, 30 mg/liter 5-bromo-4-chloro-3-indolyl phosphate (XP) was added to LB agar plates.
Animal experiments.
Female BALB/c mice (ByJ; Jackson Laboratories) (6 to 8 weeks old) and 8- to 12-week-old CBA mice (CBA/J; Jackson Laboratories) were used throughout this study. In all experiments the bacterial titer of the inoculum was determined by spreading serial 10-fold dilutions on LB agar plates containing XP and appropriate antibiotics. For competitive infection experiments groups of five mice were infected by oral gavage with an approximately 1:1 mixture of mutant and strain AJB715 at a dose of approximately 1 × 109 CFU/mouse for CBA/J and approximately 1 × 108 CFU/mouse for BALB/c mice. Fecal pellets were homogenized in 900 μl phosphate-buffered saline. Cecum and spleen were harvested aseptically and homogenized in 5 ml phosphate-buffered saline, pH 7.4. Dilutions of fecal pellets and homogenized organs were plated on LB agar plates containing XP and the appropriate antibiotics. For calculation of competitive indices, data were normalized by dividing the output ratio (CFU mutant/CFU wild type) by the input ratio (CFU mutant/CFU wild type). In case only one bacterial strain was recovered from fecal pellets, the limit of detection was determined for the missing strain and used to calculate the minimum mutant/wild type ratio. All data were converted logarithmically prior to the calculation of geometric means and statistical analysis. A Student's t test was used to determine whether the mutant/wild type ratio recovered from infected organs or fecal pellets was significantly different from that present in the inoculum.
Construction of mutants.
To generate a S. enterica serotype Typhimurium mutant with a deletion of the fimAICDHF genes, one DNA fragment originating upstream and one DNA fragment originating downstream of each fimbrial operon were PCR amplified using the primers 5′-GCACCAGAATACCGTCAATCGTG-3′, 5′-AACTGCAGCCAGCGACAAACATCAGACTCG-3′, 5′-AACTGCAGAACAACGGCAATGCTACC-3′, and 5′-ACGAATCAAATCTATCCAGG-3′. Each PCR product was cloned into vector pCR2.1 (TOPO TA cloning kit; Invitrogen). The two DNA fragments flanking the fim operon were then cloned into suicide vector pRDH10 where they were connected by a unique restriction site (PstI). A kanamycin resistance cassette (KSAC; Pharmacia) was cloned into this unique restriction site, and the resulting suicide plasmid was introduced into E. coli strain S1-7λpir and then conjugated into S. enterica serotype Typhimurium strain IR715. S. enterica serotype Typhimurium strains resulting from deletions of agfAB, pefABCDI, bcfABCDEFG, stbABCD, stcABCD, stdAB, stfACDEFG, or sthABCDE were constructed by conjugation between IR715 and E. coli strain S17-1λpir carrying suicide plasmids pSF31, pSF30, pEW1, pSF41, pSF42, pEW5, pEW3, or pSF37, respectively. Suicide plasmids pSF31, pSF30, pEW1, pSF41, pSF42, pEW5, pEW3, and pSF37 are derivatives of suicide vector pRDH10 in which a kanamycin cassette (KSAC; Pharmacia) is cloned between upstream and downstream DNA regions of each fimbrial operon (18). Exconjugants were selected on LB agar plates containing nalidixic acid (to select for the recipient IR715) and kanamycin (to select for the kanamycin cassette cloned on each suicide plasmid between upstream and downstream DNA regions of each fimbrial operon). Exconjugants, in which fimbrial operons had been replaced by homologous recombination with the kanamycin cassette, were identified by their sensitivity to chloramphenicol, the resistance marker on the vector pRDH10. The resulting strains were termed EHW2 (fimAICDHF), SF15 (agfAB), SF16 (pefABCDI), EHW1 (bcfABCDEFG), EHW12 (stbABCD), SF24 (stcABCD), EHW11 (stdAB), EHW3 (stfACDEFG), and SF22 (sthABCDE). To confirm deletion of fimbrial biosynthesis genes in each mutant, genomic DNA was prepared from S. enterica serotype Typhimurium strains by standard methods (2). Genomic DNA was digested with EcoRV, and samples were loaded on a 0.8% agarose gel. Subsequent to agarose gel electrophoresis, DNA was transferred to positively charged nylon membranes as previously described (2). To generate a DNA probe specific for agfA, part of the open reading frame was PCR amplified using primers 5′-ATAATCGGGCGAAAGTCG-3′ and 5′-CGGCAAGGAGCAATAAAG-3′ and cloned into pCR2.1 (Invitrogen) to give rise to plasmid pEW45. DNA probes specific for agfA, bcfA, fimA, lpfA, pefA, stbA, sthA, stdA, stfA, and stcA were generated from the inserts of plasmids pEW45, pAH70, pAH77, pAH72, pAH69, pAH75, pAH76, pAH106, pAH108, and pAH60, respectively, using a random prime labeling kit (Gene Images) according to protocols provided by the manufacturer. Hybridization was performed at 65°C in solutions without formamide. Hybrids were detected using a CDP-Star detection kit (Gene Images) according to protocols provided by the manufacturer.
A bacteriophage P22 HT105/1 int lysate propagated on strain AJB33 was used to transduce the chloramphenicol resistance marker of plasmid pMS1096 present in the AJB33 chromosome into strain RAK48 (lpfABCDE). Transductants were selected for by plating on LB+Cm containing 10 mM EGTA. Cotransduction of the wild-type lpf operon into strain RAK48 for complementation was tested for by replica plating transductants on LB+Cm and LB+Km. A phage P22-sensitive, chloramphenicol-resistant, kanamycin-sensitive transductant was designated EHW21 and purified from contaminating P22 phage by streaking to single colonies twice on Evans blue uranine plates. The presence of the lpf operon in EHW21 was confirmed by Southern hybridization.
A bacteriophage P22 HT105/1 int lysate propagated on strain SF24 (ΔstcABCD) or EHW1 (ΔbcfABCDEFG) was used to transduce kanamycin markers into the S. enterica serotype Typhimurium wild-type strain (IR715) as described above. Deletion of the fimbrial operon in the transductant was confirmed by Southern blotting.
RESULTS
Construction of fimbrial mutants.
To investigate the role of the agf, fim, lpf, pef, bcf, stb, stc, std, stf, or sth fimbrial operons in pathogenesis we used derivatives of the S. enterica serotype Typhimurium wild-type strain IR715 lacking portions of agfAB, fimAICDHF, lpfABCDE, pefABCDI, bcfABCDEFG, stbABCD, stcABCD, stdAB, stfACDEFG, or sthABCDE (Fig. 1). To generate a deletion of fimAICDHF, DNA regions upstream and downstream of the fim operon were PCR amplified and cloned into a suicide plasmid. A kanamycin resistance gene was cloned between both DNA fragments, and the resulting construct was introduced into IR715 by conjugation. Suicide vectors for constructing deletions in the S. enterica serotype Typhimurium fimbrial operons agfAB, pefABCDI, bcfABCDEFG, stbABCD, stcABCD, stdAB, stfACDEFG, or sthABCDE have been described previously (18) and were introduced into IR715 by conjugation. Mutants in which a fimbrial operon was replaced by a kanamycin resistance gene through homologous recombination were identified as exconjugants that were resistant to kanamycin but sensitive to the resistance marker on the suicide plasmid. Deletions of agfAB, fimAICDHF, lpfABCDE, pefABCDI, bcfABCDEFG, stbABCD, stcABCD, stdAB, stfACDEFG, or sthABCDE in individual mutants were confirmed by Southern blotting (Fig. 2) using genomic DNA of S. enterica serotype Typhimurium strains and DNA probes specific for genes encoding the major subunit of each operon. The name of each mutant and the precise location of each deletion are shown in Fig. 1.
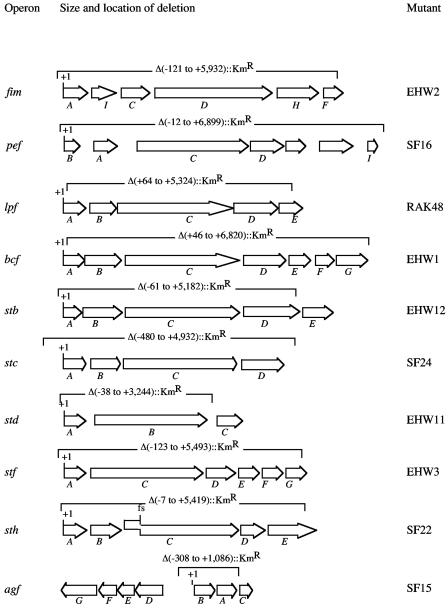
FIG. 1.
Size and location of fimbrial operon deletions present in S. enterica serotype Typhimurium 14028 derivatives used in this study. The name of fimbrial operons is given on the left. Arrows indicate the location of each gene in an operon. Brackets indicate the area deleted and replaced by a kanamycin resistance gene. The exact size of each deletion in base pairs is indicated relative to the first nucleotide (+1) of the first gene in each operon. The position of a frame shift (fs) mutation present in the sthC gene is indicated. The strain designation of S. enterica serotype Typhimurium mutants lacking a particular fimbrial operon is given on the right.
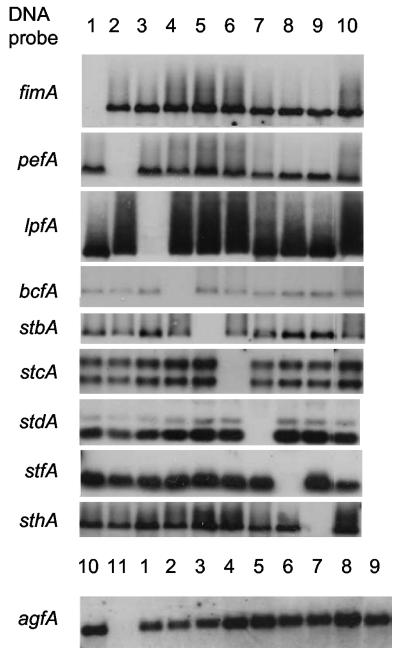
FIG. 2.
Southern blot of genomic DNA isolated from S. enterica serotype Typhimurium strains EHW2 (fimAICDHF, lane 1), SF16 (pefBACDI, lane 2), RAK48 (lpfABCDE, lane 3), EHW1 (bcfABCDEFG, lane 4), EHW12 (stbABCD, lane 5), SF24 (stcABCD, lane 6), EHW11 (stdAB, lane 7), EHW3 (stfACDEFG, lane 8), SF22 (sthABCDE, lane 9), IR715 (wild type, lane 10), and SF15 (agfAB, lane 11) detected with DNA probes specific for genes indicated on the left.
Role of fimbrial operons on colonization of spleen and cecum in BALB/c mice.
The ability of strains with deletions in individual fimbrial operons to colonize genetically susceptible mice (BALB/c) was assessed using competitive infection experiments. A mixture of equal amounts of each mutant and S. enterica serotype Typhimurium strain AJB715 was used to inoculate a group of five BALB/c mice intragastrically. Strain AJB715 carries a mutation in phoN, a gene encoding alkaline phosphatase. Inactivation of phoN abolishes the ability to cleave 5-bromo-4-chloro-3-indolyl phosphate (XP). Growth on Luria-Bertani (LB) agar plates supplemented with XP thus provided an easy means to distinguish between mutants with a deletion of a fimbrial operon (PhoN+ blue colonies) and AJB715 (PhoN− white colonies). Inactivation of phoN has been shown not to reduce the ability of S. enterica serotype Typhimurium to cause lethal infections in BALB/c mice (14). To verify that inactivation of phoN would not affect organ colonization, a group of five mice was infected with a mixture of equal amounts of the phoN mutant (AJB715) and its isogenic wild type (IR715). No significant differences between AJB715 and IR715 in bacterial numbers recovered from spleen and cecum 5 days after infection were observed in this control experiment (Fig. 3).
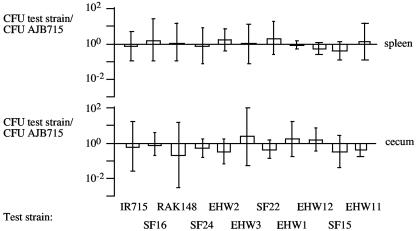
FIG. 3.
Competitive infection experiments performed with BALB/c mice. Mice were infected with a mixture of equal amounts of AJB715 (phoN) and one of the strains to be tested (IR715, EHW2, SF16, RAK48, EHW1, EHW12, SF24, EHW11, EHW3, SF22, or SF15). Competitive indices (i.e., ratios of test strain/AJB715 recovered from organs) for the spleen and cecum of mice at 5 days after infection are shown as geometric means (bars) ± standard deviations.
Competitive infection experiments with AJB715 and one of the 10 fimbrial mutants (EHW2, SF16, RAK48, EHW1, EHW12, SF24, EHW11, EHW3, SF22, or SF15) were performed with groups of five mice. No significant differences in bacterial numbers recovered were observed between AJB715 and any of the fimbrial mutants, suggesting that individual fimbrial operons were not required for S. enterica serotype Typhimurium colonization of spleen and cecum 5 days after infection of BALB/c mice (Fig. 3).
Role of fimbrial operons in intestinal persistence.
Susceptible mouse lineages (e.g., BALB/c) are not well suited to investigations of mechanisms of intestinal persistence, because these animals succumb to infection within 6 to 10 days. In contrast, genetically resistant mice (e.g., CBA) do not develop lethal morbidity but shed S. enterica serotype Typhimurium with their feces for a period of 2 to 3 months (21). To investigate the role of the agf, fim, lpf, pef, bcf, stb, stc, std, stf, or sth fimbrial operons in intestinal persistence, competitive infection experiments were performed with groups of five CBA mice. Mice were inoculated with a mixture of equal amounts of AJB715 (phoN) and one of the fimbrial deletion mutants (EHW2, SF16, RAK48, EHW1, EHW12, SF24, EHW11, EHW3, SF22, or SF15). Inactivation of phoN in AJB715 has been shown previously not to affect intestinal persistence in CBA mice (21). Fecal pellets were collected at 1, 3, 5, 7, 10, 15, 20, 25, and 30 days after infection, and numbers of AJB715 (PhoN− white colonies) and the fimbrial mutant (PhoN+ blue colonies) were determined by spreading serial 10-fold dilutions of fecal homogenates on LB agar plates supplemented with XP and appropriate antibiotics.
Over the 30-day time period of the experiments, strains SF15 (ΔagfAB), EHW2 (ΔfimAICDHF), SF16 (ΔpefABCDI), and EHW3 (ΔstfACDEFG) were recovered at numbers that were not significantly different from those of the competing strain (AJB715) (Fig. 4). The cecum was collected from three of the four groups (i.e., from mice infected with AJB715 and either SF15, SF16, or EHW3) at the end of the experiment. No significant differences in bacterial numbers of AJB715 (phoN) and SF15 (ΔagfAB), SF16 (ΔpefABCDI), or EHW3 (ΔstfACDEFG) were detected in the cecum at day 30 after infection (Fig. 4E). These data suggested that the agf, fim, pef, and stf fimbrial operons are not required for long-term intestinal persistence of S. enterica serotype Typhimurium in CBA mice.
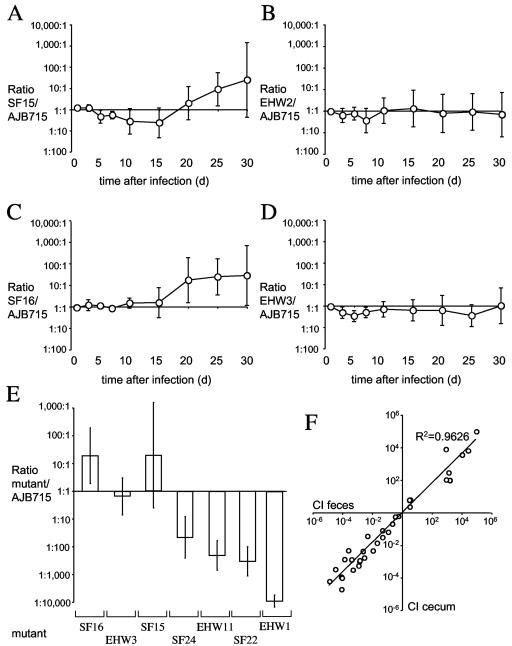
FIG. 4.
Competitive infection experiments performed with CBA mice. Mice were infected with a mixture of equal amounts of AJB715 (phoN) and one fimbrial deletion mutant. Ratios of SF15/AJB715 (A), EHW2/AJB715 (B), SF16/AJB715 (C), or EHW3/AJB715 (D) recovered from feces over a 30-day time period are plotted as geometric means (circles) ± standard errors. (E) Competitive indices (i.e., ratios of fimbrial deletion mutant/AJB715 recovered) for the cecum 30 days after infection are shown as geometric means (bar) ± standard errors. (F) Correlation of competitive indices (CI) for the cecum (y axis) and the feces (x axis) determined at 30 days after infection. Each circle represents data from an individual animal that was part of one of the following competitive infection experiments: SF16/AJB715, EHW3/AJB175, SF15/AJB175, SF24/AJB175, EHW11/AJB175, SF22/AJB175, or EHW1/AJB175. A trend line and the calculated R2 value are indicated.
One group of fimbrial mutants showed significantly reduced fecal colonization compared to strain AJB715 (phoN) starting at early times after infection. These included strains SF24 (ΔstcABCD), EHW11 (ΔstdAB), and SF22 (ΔsthABCDE), which were recovered in significantly lower numbers (P < 0.05) from fecal samples than strain AJB715 (phoN) at day 1 after infection and at all subsequent time points (Fig. 5). The differences were largest at day 30, when AJB715 was recovered on average in 60-fold-higher numbers than SF24 (ΔstcABCD) (Fig. 5C), in 410-fold-higher numbers than EHW11 (ΔstdAB) (Fig. 5B), and in 260-fold-higher numbers than SF22 (ΔsthABCDE) (Fig. 5A) from fecal pellets. Since the cecum has been shown to be the main reservoir for bacteria recovered from the feces of CBA mice (21), the cecum was collected at the end of the experiment and bacterial numbers in the organ were quantified. Significantly (P < 0.05) lower numbers of SF24 (ΔstcABCD), EHW11 (ΔstdAB), and SF22 (ΔsthABCDE) than of strain AJB715 (phoN) were recovered from the cecum collected at day 30 after infection (Fig. 4E).
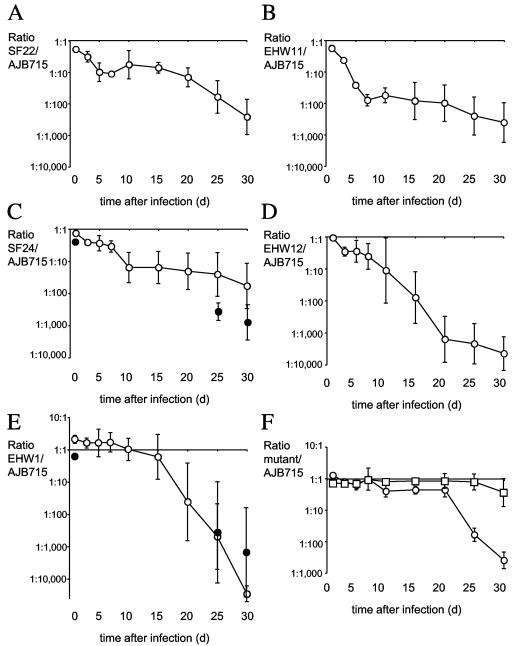
FIG. 5.
Competitive infection experiments performed with CBA mice. Mice were infected with a mixture of equal amounts of AJB715 (phoN) and one fimbrial deletion mutant. Ratios of SF22/AJB715 (A), EHW11/AJB715 (B), SF24/AJB715 (open circles) (C), EHW69/AJB715 (closed circles) (C), EHW12/AJB715 (D), EHW1/AJB715 (open circles) (E), or EHW65/AJB715 (closed circles) (E) recovered from feces over a 30-day time period are plotted as geometric means (circles) ± standard errors. (F) Ratios of RAK48/AJB715 (circles) or EHW21/AJB715 (squares) recovered from feces over a 30-day time period are plotted as geometric means (circles) ± standard errors.
Although strains SF24 (ΔstcABCD), EHW11 (ΔstdAB), and SF22 (ΔsthABCDE) were not defective for cecal colonization in BALB/c mice (Fig. 3), these three strains were recovered in significantly lower numbers than strain AJB715 (phoN) from fecal samples of CBA mice at day 5 after infection (Fig. 5). Specifically, AJB715 was on average recovered in approximately 27-fold-higher numbers from fecal pellets than EHW11 (ΔstdAB), which represented the largest difference detected at 5 days after infection of CBA mice (Fig. 5B). In contrast, AJB715 was on average recovered from the cecum 5 days after infection of BALB/c mice in approximately 2-fold-higher numbers than EHW11 (ΔstdAB), but this difference was not statistically significant (Fig. 3). To investigate whether deletion of stdAB would reduce cecal colonization at early times after infection of CBA mice, we infected a group of five animals with a mixture of equal amounts of AJB715 (phoN) and EHW11 (ΔstdAB) and determined bacterial numbers recovered from the cecum at 5 days after infection. At this time point, AJB715 was recovered in approximately ninefold-higher numbers (P < 0.05) from the cecum than strain EHW11 (ΔstdAB) (data not shown).
One group of fimbrial mutants, including EHW12 (ΔstbABCD), EHW1 (ΔbcfABCDEFG), and RAK48 (ΔlpfABCDE), showed significantly reduced fecal colonization compared to strain AJB715 (phoN) starting at later times after infection. Strain EHW12 (ΔstbABCD) was recovered in significantly lower numbers (P < 0.05) from fecal samples than strain AJB715 (phoN) at day 15 after infection and on all subsequent days (Fig. 5D). The differences were largest at day 30, when AJB715 (phoN) was recovered on average in 4,400-fold-higher numbers than EHW12 (ΔstbABCD). Strain EHW1 (ΔbcfABCDEFG) was recovered in significantly lower numbers (P < 0.05) from fecal samples than strain AJB715 (phoN) at days 25 and 30 after infection (Fig. 5E). The differences were largest at day 30, when AJB715 (phoN) was recovered on average in 29,000-fold-higher numbers than EHW1 (ΔbcfABCDEFG). Similarly, significantly (P < 0.05) lower numbers of EHW1 (ΔbcfABCDEFG) than of strain AJB715 (phoN) were recovered from the cecum collected at day 30 after infection (Fig. 4E). Finally, strain RAK48 (ΔlpfABCDE) was recovered in significantly lower numbers (P < 0.05) from fecal samples than strain AJB715 (phoN) at day 10 after infection, and the differences were largest at day 30 (Fig. 5F). However, some caution is warranted in distinguishing between mutants developing phenotypes early (at 5 days) or late (≥15 days) after infection of mice, because competitive indices of some mutants (e.g., EHW12 and SF24) differed in the size of the error bars rather than the magnitude of the colonization defect early after infection (Fig. 5C and 5D). In summary, our data suggested that the lpf, bcf, stb, stc, std, and sth fimbrial operons are required for long-term intestinal persistence of S. enterica serotype Typhimurium in CBA mice.
In this study, the composition of the bacterial population in the cecum 30 days after infection was analyzed in seven competitive infection experiments (i.e., competitive infections between AJB715 and SF14, SF22, SF16, SF15, EHW11, EHW3, or EHW1). Analysis of the data revealed a strong correlation (R2 = 0.9626) between the mutant/AJB715 ratios recovered from the feces of individual mice at 30 days after infection and the mutant/AJB715 ratios recovered from the cecum of individual animals at the same day (Fig. 4F). These data strongly supported the view that examination of the mutant/AJB715 ratio in fecal samples over time provided the opportunity to monitor cecal colonization by a noninvasive method.
To confirm that defects in intestinal persistence were due to deletion of a particular fimbrial operon, we transduced the insertions in strains EHW1 (ΔbcfABCDEFG) and SF24 (ΔstcABCD) into the S. enterica serotype Typhimurium wild type (IR715) using phage P22. The resulting transductants were designated EHW65 (ΔbcfABCDEFG) and EHW69 (ΔstcABCD). EHW65 and EHW69 were tested in competitive infection experiments with AJB715 using CBA mice, and bacteria were recovered from the feces at days 1, 25, and 30 after infection. This experiment revealed that at days 25 and 30 after infection strains EHW65 (ΔbcfABCDEFG) and EHW69 (ΔstcABCD) were shed with the feces in significantly lower numbers than strain AJB715, thus confirming the colonization defect observed with strains EHW1 (ΔbcfABCDEFG) and SF24 (ΔstcABCD) (Fig. 5C and 5E).
Complementation was performed to confirm that the defect in intestinal persistence of RAK48 (ΔlpfABCDE) was due to deletion of the lpf operon. We reasoned that introduction of a cloned copy of the lpf operon would present a problem, since it is difficult to ensure plasmid stability during growth in the mouse intestine for a 30-day period. We thus introduced a chromosomal copy of the lpf operon into strain RAK48 (ΔlpfABCDE) by homologous recombination. To this end we made a P22 HT105/1 int phage lysate of strain AJB33, which carries a lpfABCDE::lacZYA transcriptional fusion generated by inserting a suicide plasmid (pMS1096) behind the lpfE stop codon by homologous recombination (33). The lpfABCDE::lacZYA insertion was transduced into RAK48 (ΔlpfABCDE) with the AJB33 phage lysate by selecting for the chloramphenicol resistance of the suicide plasmid (pMS1096). A transductant into which the intact lpf operon had been cotransduced, thereby replacing the ΔlpfABCDE::Km deletion, was identified as a kanamycin-sensitive colony and termed EHW21.
A group of five CBA mice was infected with a mixture of equal amounts of strains AJB715 (phoN) and EHW21 (lpfABCDE::lacZYA). Both strains were recovered at approximately equal numbers from the feces of mice. At day 30, AJB715 was recovered on average in 2.7-fold-higher numbers from feces than EHW21, but this difference was not statistically significant. In contrast, in the mixed infection between AJB715 (phoN) and RAK48 (ΔlpfABCDE), strain AJB715 had been recovered on average in 390-fold-higher numbers from the feces than the lpf deletion mutant at day 30 after infection (Fig. 5F). These data showed that reintroduction of an intact lpf fimbrial operon into an lpfABCDE deletion mutant restored its ability to be persistently shed with the feces of mice at high numbers.
DISCUSSION
A previous study identified two genes, shdA and ratB, located on the CS54 island of S. enterica serotype Typhimurium that are involved in long-term intestinal persistence in genetically resistant mice (21). Here we provide evidence that six fimbrial operons, lpfABCDE, bcfABCDEFG, stbABCDE, stcABCD, stdABCD, and sthABCDE, contribute to the ability of S. enterica serotype Typhimurium to be shed with the feces and to colonize the cecum at 30 days after infection. Collectively these data suggest that intestinal persistence in mice is a complex trait, requiring the expression of at least 32 genes in S. enterica serotype Typhimurium.
Deletion of stdAB resulted in significantly reduced recovery of S. enterica serotype Typhimurium from the feces and cecum of CBA mice but not from the cecum of BALB/c mice at 5 days after infection. These data suggested either that the std operon is not required for cecal colonization of BALB/c mice or that the phenotype does not manifest at early time points (i.e., by 5 days after infection) in this mouse lineage. The reverse situation has been reported for the role of ShdA during colonization of CBA and BALB/c mice. In this case, a mutation in shdA significantly reduces cecal colonization observed 5 days after infection in BALB/c mice but not in CBA mice. However, a mutation in shdA significantly reduces cecal colonization in CBA mice at later time points (i.e., at day 21 after infection and on subsequent time points) (21). The contributions of type 1 fimbriae to intestinal colonization by S. enterica serotype Typhimurium also seem to differ between inbred mouse lineages. Type 1 fimbriae are required for long-term fecal shedding of S. enterica serotype Typhimurium in LAC Grey mice (10) and for colonization of the intestine of ICR mice (1). In contrast, type 1 fimbriae do not contribute to intestinal colonization of BALB/c mice (28) (Fig. 3) or persistent fecal shedding in CBA mice (Fig. 4).
It is not known why different S. enterica serotype Typhimurium adhesins contribute to colonization of different mouse lineages. In some cases the underlying mechanism may be that only a subset of mouse lineages expresses the cognate host receptor for a particular bacterial adhesin. Such differences in receptor expression are expected to be more common between different animal species than between inbred lineages belonging to the same animal species. It may thus not be unexpected that phenotypes reported for a fimbrial mutant in one animal model may differ from those described in a different animal species. For example, type 1 fimbriae did not contribute to intestinal carriage in BALB/c mice or CBA mice (Fig. 3 and 4) but are involved in colonizing intestinal tissues during S. enterica serotype Typhimurium infection of pigs and rats (1, 32). An S. enterica serotype Typhimurium mutant carrying a transposon insertion in the bcf operon exhibits a colonization defect for bovine but not for murine (BALB/c) intestinal tissue at 4 days after oral infection (39). Transposon insertions in agfD, sthB, or stbC result in an intestinal colonization defect after oral infection of chickens with S. enterica serotype Typhimurium, but none of these insertions reduce intestinal colonization in calves (31). Deletion of agfAB did not reduce the ability of S. enterica serotype Typhimurium to colonize the intestine of CBA or BALB/c mice (Figs. 3 and 4), thus exhibiting a phenotype similar to that of an agfD mutant in calves (31). In contrast, deletion of sthABCDE or stbABCD reduced cecal colonization in CBA mice (Fig. 4), thus resulting in a phenotype similar to that caused by transposon insertion in sthB or stbC during S. enterica serotype Typhimurium infection of chickens (31). Given the variability in the repertoires of adhesins required for colonization of different hosts, it could be speculated that the large number of genetic attachment factors present in the S. enterica serotype Typhimurium genome (e.g., 13 fimbrial operons) may represent an adaptation to colonizing hosts that differ with regard to the repertoire of receptors expressed on their mucosal surfaces.
Early studies show that during S. enterica serotype Enteritidis infection of mice the bulk of bacteria in the intestinal lumen reside in the cecum (7). More recently, mutations in shdA and ratB that reduce S. enterica serotype Typhimurium colonization of the cecum were shown to also reduce bacterial numbers recovered from feces. In contrast, a mutation in sivH reduces colonization of Peyer's patches but does not affect the ability of S. enterica serotype Typhimurium to colonize cecum or feces of mice (21). Based on these data it was hypothesized that the murine cecum may be the main reservoir for fecal shedding of Salmonella serotypes. In this study, colonization of Payer's patches was not investigated, because at 30 days after infection of CBA mice this organ is colonized at numbers that are often below the limit of detection. However, we found a good correlation (R2 = 0.9626) between the composition (i.e., the mutant/AJB715 ratio) of bacterial populations recovered from the cecum and the feces of mice 30 days after infection (Fig. 4F). This result further corroborated the notion that the cecum is an important intestinal reservoir that gives rise to bacteria shed with the feces.
In the case of the ShdA autotransporter protein, the mechanism for colonization of murine cecal tissue has been further investigated. The surface-localized passenger domain of ShdA binds cecal tissue at areas rich in extracellular matrix proteins (23). The ligands bound by ShdA in the extracellular matrix include fibronectin (23) and collagen I (22). S. enterica serotype Typhimurium encodes a second adhesin that binds fibronectin, termed thin curled fimbriae (also known as thin aggregative fimbriae or curli), which is encoded by the agf (csg) fimbrial gene cluster. Interestingly, deletion of agfAB did not reduce the ability of S. enterica serotype Typhimurium to persist in the murine cecum (Fig. 4A). Thin curled fimbriae do not mediate binding to collagen I and differ from ShdA in their binding affinity for fibronectin. That is, thin curled fimbriae bind fibronectin with low affinity (i.e., an estimated Kd of 74 μM) (8), while the affinity with which ShdA binds this extracellular matrix protein is approximately 600-fold higher (i.e., the estimated Kd is 0.12 μM) (20). It is currently not clear whether these differences in binding specificity and affinity may explain why mutations in shdA and agfAB have different effects on the ability of S. enterica serotype Typhimurium to colonize the cecum of mice.
Acknowledgments
We would like to thank R. A. Kingsley for helpful discussions and advice and R. Johnson for technical assistance.
Work in the laboratory of A.B. is supported by U.S. Department of Agriculture-National Research Initiative Competitive Grants Program grant 2002-35204-12247 and Public Health Service grants AI40124 and AI44170.
Editor: J. T. Barbieri
REFERENCES
- 1.Althouse, C., S. Patterson, P. Fedorka-Cray, and R. E. Isaacson. 2003. Type 1 fimbriae of Salmonella enterica serovar Typhimurium bind to enterocytes and contribute to colonization of swine in vivo. Infect. Immun. 71:6446-6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1994. Current protocols in molecular biology. J. Wiley & Sons, New York, N.Y.
- 3.Bäumler, A. J., R. M. Tsolis, F. Bowe, J. G. Kusters, S. Hoffmann, and F. Heffron. 1996. The pef fimbrial operon of Salmonella typhimurium mediates adhesion to murine small intestine and is necessary for fluid accumulation in the infant mouse. Infect. Immun. 64:61-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bäumler, A. J., R. M. Tsolis, and F. Heffron. 1996. Contribution of fimbrial operons to attachment to and invasion of epithelial cell lines by Salmonella typhimurium. Infect. Immun. 64:1862-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bäumler, A. J., R. M. Tsolis, and F. Heffron. 1996. The lpf fimbrial operon mediates adhesion of Salmonella typhimurium to murine Peyer's patches. Proc. Natl. Acad. Sci. USA 93:279-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bäumler, A. J., R. M. Tsolis, P. J. Valentine, T. A. Ficht, and F. Heffron. 1997. Synergistic effect of mutations in invA and lpfC on the ability of Salmonella typhimurium to cause murine typhoid. Infect. Immun. 65:2254-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter, P. B., and F. M. Collins. 1974. The route of enteric infection in normal mice. J. Exp. Med. 139:1189-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collinson, S. K., P. S. Doig, J. L. Doran, S. Clouthier, T. J. Trust, and W. W. Kay. 1993. Thin, aggregative fimbriae mediate binding of Salmonella enteritidis to fibronectin. J. Bacteriol. 175:12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duguid, J. P., E. S. Anderson, and I. Campbell. 1966. Fimbriae and adhesive properties in Salmonellae. J. Pathol. Bacteriol. 92:107-137. [DOI] [PubMed] [Google Scholar]
- 10.Duguid, J. P., M. R. Darekar, and D. W. F. Wheather. 1976. Fimbriae and infectivity in Salmonella typhimurium. J. Med. Microbiol. 9:459-473. [DOI] [PubMed] [Google Scholar]
- 11.Duguid, J. P., and R. R. Gillies. 1958. Fimbriae and haemagglutinating activity in Salmonella, Klebsiella, Proteus, and Chromobacterium. J. Path. Bacteriol. 75:519-520. [Google Scholar]
- 12.Ernst, R. K., D. M. Dombroski, and J. M. Merrick. 1990. Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of HEp-2 cells by Salmonella typhimurium. Infect. Immun. 58:2014-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant, S. G. N., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groisman, E. A., M. H. Saier, and H. Ochman. 1992. Horizontal transfer of a phosphatase gene as evidence for mosaic structure of the Salmonella genome. EMBO J. 11:1309-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grund, S., R. Helmuth, R. Stephan, and R. Meyer. 1988. [Fimbrial formation, antibiogram, plasmid content, lysotype and biotype of Salmonella]. Zentlbl. Vetmed. Reihe B. 35:138-151. [PubMed] [Google Scholar]
- 16.Grund, S., and A. Seiler. 1993. [Electron microscopic studies of fimbriae and lectin phagocytosis of Salmonella typhimurium variety copenhagen (STMVC)]. Zentlbl. Vetmed. Reihe B. 40:105-112. [PubMed] [Google Scholar]
- 17.Grund, S., and A. Weber. 1988. A new type of fimbriae on Salmonella typhimurium. Zentlbl. Vetmed. Reihe B. 35:779-782. [DOI] [PubMed] [Google Scholar]
- 18.Humphries, A. D., M. Raffatellu, S. Winter, E. H. Weening, R. A. Kingsley, R. Droleskey, S. Zhang, J. Figueiredo, S. Khare, J. Nunes, L. G. Adams, R. M. Tsolis, and A. J. Bäumler. 2003. The use of flow cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype Typhimurium fimbrial operons. Mol. Microbiol. 48:1357-1376. [DOI] [PubMed] [Google Scholar]
- 19.Jones, G. W., and L. A. Richardson. 1981. The attachment to, and invasion of HeLa cells by Salmonella typhimurium: the contribution of mannose-sensitive and mannose-resistant haemagglutinating activities. J. Gen. Microbiol. 127:361-370. [DOI] [PubMed] [Google Scholar]
- 20.Kingsley, R. A., D. Abi Ghanem, N. Puebla-Osorio, A. M. Keestra, L. Berghman, and A. J. Bäumler. 2004. Fibronectin binding to the Salmonella enterica serotype Typhimurium ShdA autotransporter protein is inhibited by a monoclonal antibody recognizing the A3 repeat. J. Bacteriol. 186:4931-4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kingsley, R. A., A. D. Humphries, E. H. Weening, M. R. De Zoete, S. Winter, A. Papaconstantinopoulou, G. Dougan, and A. J. Bäumler. 2003. Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype Typhimurium: identification of intestinal colonization and persistence determinants. Infect. Immun. 71:629-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kingsley, R. A., A. M. Keestra, M. R. de Zoete, and A. J. Bäumler. 2004. The ShdA adhesin binds to the cationic cradle of the fibronectin 13FnIII repeat module: evidence for molecular mimicry of heparin binding. Mol. Microbiol. 52:345-355. [DOI] [PubMed] [Google Scholar]
- 23.Kingsley, R. A., R. L. Santos, A. M. Keestra, L. G. Adams, and A. J. Bäumler. 2002. Salmonella enterica serotype Typhimurium ShdA is an outer membrane fibronectin-binding protein that is expressed in the intestine. Mol. Microbiol. 43:895-905. [DOI] [PubMed] [Google Scholar]
- 24.Kingsley, R. A., K. van Amsterdam, N. Kramer, and A. J. Bäumler. 2000. The shdA gene is restricted to serotypes of Salmonella enterica subspecies I and contributes to efficient and prolonged fecal shedding. Infect. Immun. 68:2720-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kingsley, R. A., E. H. Weening, A. M. Keestra, and A. J. Bäumler. 2002. Population heterogeneity of Salmonella enterica serotype Typhimurium resulting from phase variation of the lpf operon in vitro and in vivo. J. Bacteriol. 184:2352-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kukkonen, M., T. Raunio, R. Virkola, K. Lahteenmaki, P. H. Makela, P. Klemm, S. Clegg, and T. K. Korhonen. 1993. Basement membrane carbohydrate as a target for bacterial adhesion: binding of type I fimbriae of Salmonella enterica and Escherichia coli to laminin. Mol. Microbiol. 7:229-237. [DOI] [PubMed] [Google Scholar]
- 27.Lockman, H. A., and R. Curtiss III. 1992. Isolation and characterization of conditional adherent and non-type 1 fimbriated Salmonella typhimurium mutants. Mol. Microbiol. 6:933-945. [DOI] [PubMed] [Google Scholar]
- 28.Lockman, H. A., and R. Curtiss III. 1992. Virulence of non-type 1-fimbriated and nonfimbriated nonflagellated Salmonella typhimurium mutants in murine typhoid fever. Infect. Immun. 60:491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 30.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan, E., J. D. Campbell, S. C. Rowe, J. Bispham, M. P. Stevens, A. J. Bowen, P. A. Barrow, D. J. Maskell, and T. S. Wallis. 2004. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 54:994-1010. [DOI] [PubMed] [Google Scholar]
- 32.Naughton, P. J., G. Grant, S. Bardocz, E. Allen-Vercoe, M. J. Woodward, and A. Pusztai. 2001. Expression of type 1 fimbriae (SEF 21) of Salmonella enterica serotype enteritidis in the early colonisation of the rat intestine. J. Med. Microbiol. 50:191-197. [DOI] [PubMed] [Google Scholar]
- 33.Norris, T. L., R. A. Kingsley, and A. J. Bäumler. 1998. Expression and transcriptional control of the Salmonella typhimurium lpf fimbrial operon by phase variation. Mol. Microbiol. 29:311-320. [DOI] [PubMed] [Google Scholar]
- 34.Olsen, S. J., L. C. MacKinnon, J. S. Goulding, N. H. Bean, and L. Slutsker. 2000. Surveillance for foodborne-disease outbreaks—United States, 1993-1997. Morb. Mortal. Wkly. Rep. CDC Surveill. Summ. 49:1-62. [PubMed] [Google Scholar]
- 35.Stojiljkovic, I., A. J. Bäumler, and F. Heffron. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 177:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stolpe, H., S. Grund, and W. Schroder. 1994. Purification and partial characterization of type 3 fimbriae from Salmonella typhimurium var. copenhagen. Zentbl. Bakteriol. 281:8-15. [DOI] [PubMed] [Google Scholar]
- 37.Sukupolvi, S., R. G. Lorenz, J. I. Gordon, Z. Bian, J. D. Pfeifer, S. J. Normark, and M. Rhen. 1997. Expression of thin aggregative fimbriae promotes interaction of Salmonella typhimurium SR-11 with mouse small intestinal epithelial cells. Infect. Immun. 65:5320-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tavendale, A., C. K. H. Jardine, D. C. Old, and J. P. Duguid. 1983. Heamagglutination and adhesion of Salmonella typhimurium to HEp2 and HeLa cells. J. Med. Microbiol. 16:371-380. [DOI] [PubMed]
- 39.Tsolis, R. M., S. M. Townsend, E. A. Miao, S. I. Miller, T. A. Ficht, L. G. Adams, and A. J. Bäumler. 1999. Identification of a putative Salmonella enterica serotype Typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect. Immun. 67:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Velden, A. W. M., A. J. Bäumler, R. M. Tsolis, and F. Heffron. 1998. Multiple fimbrial adhesins are required for full virulence of Salmonella typhimurium in mice. Infect. Immun. 66:2803-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson, R. L., J. Elthon, S. Clegg, and B. D. Jones. 2000. Salmonella enterica serovars gallinarum and pullorum expressing Salmonella enterica serovar Typhimurium type 1 fimbriae exhibit increased invasiveness for mammalian cells. Infect. Immun. 68:4782-4785. [DOI] [PMC free article] [PubMed] [Google Scholar]