Abstract
Degradation of the mucus layer by Entamoeba histolytica is a prerequisite for invasion of the colonic mucosa. In this study, we demonstrate that amoeba-secreted products degrade 3H-labeled and native colonic mucin oligosaccharides independently of proteolytic activity. We conclude that E. histolytica degrades mucin oligosaccharides, which may facilitate parasite invasion of the colon.
Entamoeba histolytica is responsible for at least 50 million cases of diarrhea and an estimated 100,000 deaths per annum and ranks second only to malaria as a cause of mortality due to a protozoan parasite (12). Infection with the parasite leads to amebic colitis, colonic ulceration, and less frequently, dissemination to the liver, resulting in amebic liver abscess. The initial events leading to invasion of the colon by E. histolytica are poorly understood, and the mechanisms used by the parasite to overcome the innate host defenses of the gastrointestinal tract are currently under investigation. The parasite colonizes the colonic mucus layer by binding mucin oligosaccharides via a 170-kDa Gal/GalNAc lectin and must traverse this protective barrier in order to cause epithelial cell damage and colonic ulceration. Mucin oligosaccharides serve to protect the mucin core from proteases, preserving the integrity of the mucin polymer. Various O-linked glycan structures are attached to the apomucin via O-glycosidic linkage to serine and threonine residues, and these O-glycan branches contain N-acetylgalactosamine (GalNAc), N-acetylglucosamine (GlcNAc), fucose, galactose, and sialic acid. The oligosaccharide component of gastrointestinal mucin has been reported to account for up to 90% of its dry weight, and the densely packed oligosaccharides are responsible for many intrinsic physical properties of the mucus gel, including hydration, gel-forming capacity, protease resistance, and rigidity (3). Previous studies have identified numerous glycosidase activities in E. histolytica lysates and secretory products. More specifically, the parasite has been found to produce a sialidase, an α-glucosidase, and β-N-acetylhexosaminidase, enzymes which are released by the parasite and are hypothesized to be involved in amebic pathogenesis (7, 9, 14). These glycosidases may play a role in disrupting mucin by exposing the protein backbone to parasite proteases. Previously, we have shown that E. histolytica-secreted cysteine proteases degrade the poorly glycosylated regions of MUC2, and we hypothesize that the parasite may use the concerted actions of glycosidases and proteases to disassemble the mucin polymeric network (5).
In the present study, we determined whether E. histolytica-secreted glycosidases could degrade colonic mucin oligosaccharides. Parasite secretory products were collected from trophozoites incubated in Hanks' balanced salt solution for 2 h, and >95% of trophozoites were viable as determined by trypan blue exclusion assay (13). Secreted products were assayed for activity against a panel of glycosidase substrates as previously described with some modifications (2). Briefly, 20 μg of secreted components (representing ∼2 × 105 trophozoites) were assayed for glycosidase activities between pH 3.5 and pH 8.5 to determine optimal activity using various p-nitrophenyl glycoside substrates (2 mM) (EMD Biosciences Inc., San Diego, CA). One unit of enzyme activity was defined as the number of micromoles of substrate digested per minute per milligram of protein, and 1 U of activity was considered significant.
Highly purified 3H-labeled mucin, as well as native mucin, was collected from LS 174T colonic cells (American Type Culture Collection, Rockville, MD) grown to 80% confluence in minimal essential medium (Invitrogen Corporation, Burlington, Ontario, Canada) and purified by Sepharose 4B (S4B) gel filtration and/or cesium chloride density gradient centrifugation (CsCl mucin) as previously described (1, 5). Mucin oligosaccharide degradation was assessed with native mucin and was visualized by periodic acid-Schiff stain (PAS), in-gel staining of the mucin oligosaccharides using the GelCode glycoprotein staining kit according to the manufacturer's instructions (Pierce, Rockford, IL). Western blot analysis was performed using an antibody generated in New Zealand White rabbits against LS 174T cell mucin which was purified by gel filtration and density gradient centrifugation (1). The specificity of the antibody for mucin oligosaccharides was determined by oxidizing the mucin with 10 mM sodium metaperiodate (Sigma-Aldrich, Burlington, Ontario, Canada) in phosphate-buffered saline (PBS) (Invitrogen Corporation) in the dark for 1 h (11). In addition, degradation of 3H-labeled mucin glycoproteins was examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and fluorography, as well as S4B size exclusion chromatography, as previously described (5). Secreted products were pretreated with protease inhibitors or with the complete-mini EDTA-free protease inhibitor cocktail according to the manufacturer's instructions (Roche GmbH, Mannheim, Germany). Trypsin and papain were used as a control for proteolytic degradation in the absence of glycosidase activity (Roche GmbH).
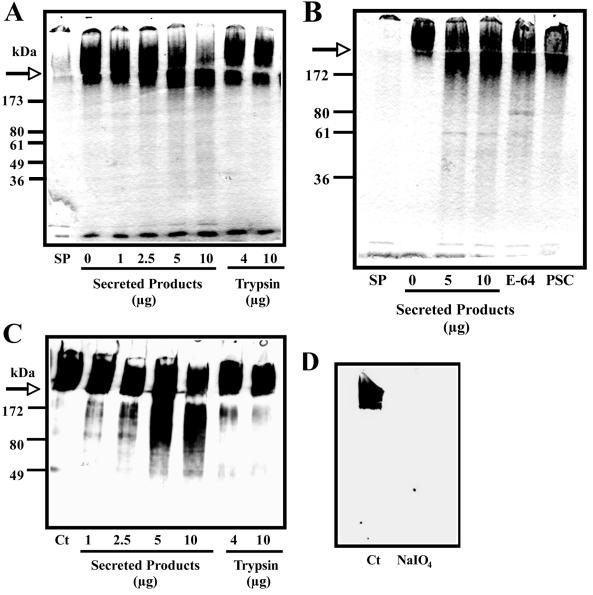
E. histolytica-secreted products were found to contain abundant amounts of activity against various glycoside substrates (Table 1). The highest level of activity detected was that of β-N-acetyl-d-glucosaminidase at pH 7.0. In addition, high levels of α-d-glucosidase activities were also detected, with maximal activity between pH 6.0 and 8.0, which is in agreement with a previous report (10). Modest levels of β-d-galactosidase, β-l-fucosidase, and α-N-acetyl-d-galactosaminidase were also detected. Based on the various structures of human intestinal mucin oligosaccharides, all of these enzymes would be required to break down mucin oligosaccharides by the parasite (6, 8). There has been no evidence to date that defines a role for these enzymes in mucin degradation, and previous methods used to examine oligosaccharide degradation by the parasite may have not been sensitive enough to detect minor changes in the structure of mucin due to its high molecular weight and polymeric nature. As shown in Fig. 1A, E. histolytica-secreted products degraded mucin and mucin oligosaccharides in a dose-dependent manner, as evidenced by the migration of PAS reactive material into an SDS-PAGE running gel. Incubation of the mucin with as little as 10 μg of secreted products resulted in an 87% decrease in high-molecular-weight stacking gel mucin, while trypsin did not alter the migration compared to control mucin. Since cysteine proteases are the major class of enzyme released by the parasite and have been shown to degrade the poorly glycosylated flanking regions of mucin (5), the involvement of these proteases in altering mucin oligosaccharide migration was assessed. Degradation of the mucin oligosaccharides by the parasite was not inhibited by the cysteine protease inhibitor E-64 or by the serine protease inhibitor Pefabloc-SC (Fig. 1B). These results are of particular interest, since E-64 has been shown to markedly inhibit the majority of proteolytic degradation of purified mucin by amoebae (5). Treatment of the secreted products with a protease inhibitor cocktail was also ineffective at inhibiting the liberation of mucin oligosaccharides into the running gel (data not shown). Western blot analysis of the digests with an antibody that recognizes purified colonic mucin oligosaccharides showed a 56% reduction in immunoreactive mucin remaining in the stacking gel (10 μg secreted products), while trypsin digestion of the mucin did not result in any significant loss of mucin carbohydrates from the stacking gel (Fig. 1C). The α-mucin antibody did not recognize mucin in which the sugars have been oxidized, indicating that the antibody specifically recognizes mucin oligosaccharides (Fig. 1D).
TABLE 1.
Glycosidase activity present in E. histolytica-secreted products
| Substrate | Activity (U) | Significancea |
|---|---|---|
| β-N-Acetyl-d-glucosamine | 62 | S |
| α-d-Glucose | 46 | S |
| β-d-Galactose | 3.6 | S |
| β-l-Fucose | 3.2 | S |
| α-N-Acetyl-d-galactosamine | 1.2 | S |
| α-l-Fucose | NS | |
| α-d-Mannose | NS | |
| α-d-Galactose | NS |
S, significant; NS, not significant.
FIG. 1.
PAS staining of native mucin treated with E. histolytica-secreted products. (A) Dose-dependent degradation of CsCl-purified mucin visualized by SDS-PAGE and PAS staining. (B) E. histolytica-secreted components (10 μg) were preincubated with E-64 and Pefabloc-SC prior to the digest. (C) Western blot analysis of mucin digests with a α-LS 174T cell mucin antibody. Ct, control. The arrow indicates border between stacking and running gel. (D) Western blot analysis showing specificity of the antibody for mucin oligosaccharides. Ct, control mucin; NaIO4, sodium metaperiodate treatment.
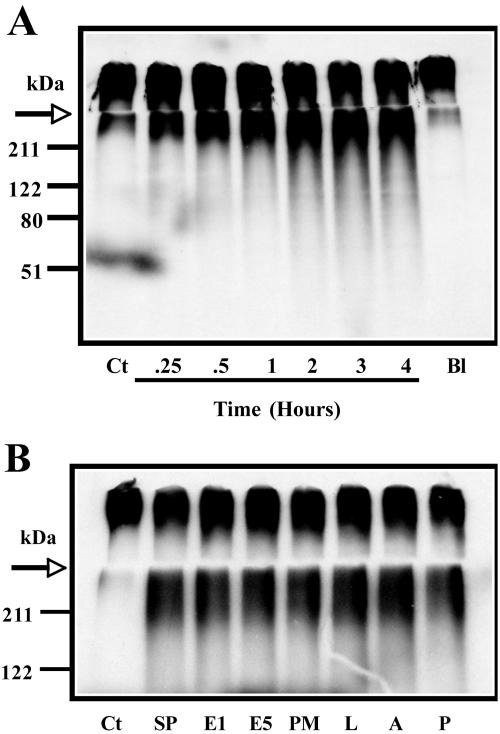
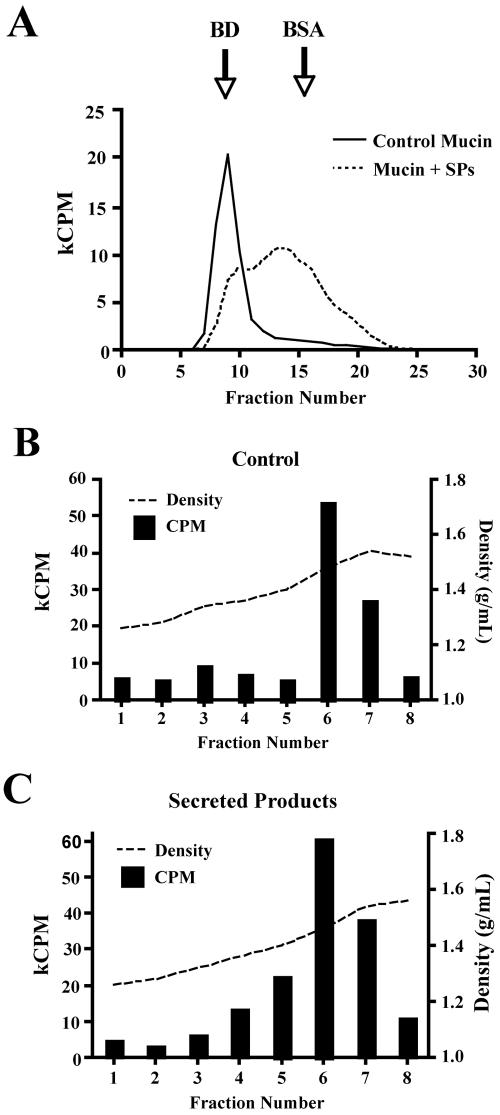
To address specific cleavage of mucin oligosaccharides, the mucin was metabolically labeled by incubating LS 174T cells with [3H]glucosamine. Under these conditions, the glucosamine is converted to various carbohydrates and is incorporated into mucin glycoproteins. This allows for the tracking of the sugars and is useful for detecting minor alterations in mucin structure. As shown in Fig. 2A, 3H-labeled S4B V0 (void volume) mucin was degraded by amoeba-secreted products in a dose-dependent fashion which was specifically inhibited by boiling. Amoebae glycosidase activity of 3H-labeled mucin was similar to the degradation pattern observed with native colonic mucin (Fig. 1A). More importantly, the addition of a variety of protease inhibitors had no effect on 3H-labeled mucin degradation (Fig. 2B), clearly demonstrating glycosidase activity. This method is more sensitive than PAS staining for detecting oligosaccharide degradation and confirmed that the mucin carbohydrates were being disrupted. As shown in Fig. 3A, 3H-labeled mucin purified by CsCl density gradient centrifugation elutes in the V0 of a S4B column. However, following incubation with amoeba-secreted products, there was a 51% decrease in void volume (fractions 6 to 11) mucin and an increase in 3H-labeled material in the included fractions (fractions 12 to 25). Although the size of the mucin molecule does change dramatically upon degradation with amoeba-secreted products, changes in the buoyant density of the molecule are not as dramatic but are evident (Fig. 3B and C). The degraded mucin does exhibit a wider range of densities and can be detected in fractions 5 to 7. Furthermore, mucin appeared to be solubilized by the secreted products, as evidenced by an increase in the total amount of recoverable 3H-labeled mucin in the presence of secreted products compared to controls (Fig. 3B).
FIG. 2.
SDS-PAGE and autoradiograph of 3H-labeled mucin degraded by amoeba-secreted products. (A) Time-dependent degradation of 3H-labeled mucin. Sepharose 4B-purified mucin was incubated with 50 μg of amoeba-secreted products for up to 4 h. Ct, control. Mucin was also incubated with secreted products that were inactivated by boiling (Bl). (B) Effect of protease inhibitors on the degradation of [3H]glucosamine-labeled mucins. Secreted products (SP) were treated with the following protease inhibitors prior to incubation with mucin: E-64 (E1 and E5), 100 and 500 μM; phenylmethylsulfonyl fluoride (PM), 10 mM; leupeptin (L), 10 mM; aprotinin (A), 10 mM; and pepstatin (P), 10 mM (Roche GmbH). The digests were performed for 6 h. The arrow indicates the border between stacking and running gel.
FIG. 3.
Sepharose 4B gel filtration of 3H-labeled CsCl mucin degraded by E. histolytica-secreted products. (A) Purified mucin (3 × 105 counts per minute) was incubated in PBS alone or with 200 μg of secreted products at 37°C for 18 h. The digests were separated by gel filtration, and aliquots of each fraction were analyzed by liquid scintillation counting. The column was calibrated with blue dextran (BD, 2,000 kDa; Pharmacia, Uppsala, Sweden) and bovine serum albumin (BSA, 68 kDa; Sigma-Aldrich). (B and C) CsCl density gradient centrifugation of 3H-labeled mucin incubated in PBS (B) or amoeba-secreted components (C). Data represent the results of one experiment repeated twice with similar results.
We have previously demonstrated that cysteine proteases secreted by E. histolytica are responsible for degrading the cysteine-rich regions of colonic mucin and the degraded mucin is less effective at inhibiting amebic adherence to target Chinese hamster ovary cells (5). Degradation of mucin by E. histolytica cysteine proteases is likely to result in destabilization of the mucin polymer, solation of mucus, and subsequent loss of the protective function of the mucus gel. Although mucin polymerization is an important and essential element required for its gel formation, mucin O-linked oligosaccharides are necessary for protecting the protein backbone, including the mucin domains, from degradation by proteases. Although some previous studies have determined that the parasite does not produce the proper range of glycosidases to degrade mucin, we have found that the parasite does produce a wide range of glycosidases and the products secreted by the parasite are capable of degrading colonic mucin. This indicates that the parasite uses both protease and glycosidase activity to disrupt the mucin polymeric network. The liver fluke Fasciola hepatica also secretes glycosidases into its environment, and the parasite's excretory-secretory products have been shown to degrade ovine mucin even in the absence of proteolytic activity (4). Our results indicate for the first time that E. histolytica releases abundant amounts of glycosidases into its environment, and this activity is sufficient to degrade colonic mucin oligosaccharides. In addition, the ability of these enzymes to degrade colonic mucin suggests that they play an important role in allowing the parasite to overcome the innate defense of the mucus barrier.
Acknowledgments
This study was supported by a grant from the Canadian Institutes of Health Research (CIHR). Research at the Institute of Parasitology is partially funded by the Fonds Pour la Formation de Chercheurs a l'Aide a la Recherche. D.M. is a recipient of a Ph.D. studentship from the CIHR.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Belley, A., K. Keller, J. Grove, and K. Chadee. 1996. Interaction of LS174T human colon cancer cell mucins with Entamoeba histolytica: an in vitro model for colonic disease. Gastroenterology 111:1484-1492. [DOI] [PubMed] [Google Scholar]
- 2.Connaris, S., and P. Greenwell. 1997. Glycosidases in mucin-dwelling protozoans. Glycoconj. J. 14:879-882. [DOI] [PubMed] [Google Scholar]
- 3.Herrmann, A., J. R. Davies, G. Lindell, S. Martensson, N. H. Packer, D. M. Swallow, and I. Carlstedt. 1999. Studies on the “insoluble” glycoprotein complex from human colon. Identification of reduction-insensitive MUC2 oligomers and C-terminal cleavage. J. Biol. Chem. 274:15828-15836. [DOI] [PubMed] [Google Scholar]
- 4.Irwin, J. A., P. E. Morrissey, J. P. Ryan, A. Walshe, S. M. O'Neill, S. D. Carrington, E. Matthews, E. Fitzpatrick, G. Mulcahy, A. P. Corfield, and J. P. Dalton. 2004. Glycosidase activity in the excretory-secretory products of the liver fluke, Fasciola hepatica. Parasitology 129:465-472. [DOI] [PubMed] [Google Scholar]
- 5.Moncada, D., K. Keller, and K. Chadee. 2003. Entamoeba histolytica cysteine proteinases disrupt the polymeric structure of colonic mucin and alter its protective function. Infect. Immun. 71:838-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moncada, D. M., and K. Chadee. 2002. Production, structure, and function of gastrointestinal mucins, p. 57-79. In M. J. Blaser, J. I. Ravdin, H. B. Greenberg, and R. L. Guerrant (ed.), Infections of the gastrointestinal tract, 2nd ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 7.Nok, A. J., and W. Rivera. 2003. Characterization of sialidase from Entamoaeba histolytica and possible pathogenic role in amebiasis. Parasitol. Res. 89:302-307. [DOI] [PubMed] [Google Scholar]
- 8.Podolsky, D. K. 1985. Oligosaccharide structures of human colonic mucin. J. Biol. Chem. 260:8262-8271. [PubMed] [Google Scholar]
- 9.Riekenberg, S., B. Flockenhaus, A. Vahrmann, M. C. Muller, M. Leippe, M. Kiess, and H. Scholze. 2004. The beta-N-acetylhexosaminidase of Entamoeba histolytica is composed of two homologous chains and has been localized to cytoplasmic granules. Mol. Biochem. Parasitol. 138:217-225. [DOI] [PubMed] [Google Scholar]
- 10.Spice, W. M., and J. P. Ackers. 1998. The effects of Entamoeba histolytica lysates on human colonic mucins. J. Eukaryot. Microbiol. 45:24S-27S. [DOI] [PubMed] [Google Scholar]
- 11.Tse, S. K., and K. Chadee. 1992. Biochemical characterization of rat colonic mucins secreted in response to Entamoeba histolytica. Infect. Immun. 60:1603-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization. 1997. Amoebiasis. Wkly. Epidemiol. Rec. 72:97-99.9100475 [Google Scholar]
- 13.Yu, Y., and K. Chadee. 1997. Entamoeba histolytica stimulates interleukin 8 from human colonic epithelial cells without parasite-enterocyte contact. Gastroenterology 112:1536-1547. [DOI] [PubMed] [Google Scholar]
- 14.Zamarripa-Morales, S., J. C. Villagomez-Castro, C. Calvo-Mendez, A. Flores-Carreon, and E. Lopez-Romero. 1999. Entamoeba histolytica: identification and properties of membrane-bound and soluble alpha-glucosidases. Exp. Parasitol. 93:109-115. [DOI] [PubMed] [Google Scholar]