Abstract
We identified a gene (atlA) encoding autolytic activity from Streptococcus mutans Xc. The AtlA protein predicted to be encoded by atlA is composed of 979 amino acids with a molecular weight of 107,279 and has a conserved β-1,4-N-acetylmuramidase (lysozyme) domain in the C-terminal portion. Sodium dodecyl sulfate extracts of strain Xc showed two major bacteriolytic bands with molecular masses of 107 and 79 kDa, both of which were absent from a mutant with inactivated atlA. Western blot analysis revealed that the 79-kDa band was derived from the 107-kDa peptide by cleavage of its N-terminal portion. The inactivation of atlA resulted in a marked decrease in autolysis and the formation of very long chains of cells compared to the case for the parent strain. Although both the parent and mutant strains formed biofilms in the presence of sucrose, the biofilms formed by the mutant had a sponge-like architecture with large gaps and contained 30% less biomass than those formed by the parent strain. Furthermore, strain Xc formed glucose-dependent, loose biofilms in the absence of sucrose, but the mutant lost this ability. These results suggest that AtlA may play an important role in biofilm formation by S. mutans. The antibody produced against the C-terminal peptide containing the β-1,4-N-acetylmuramidase domain drastically inhibited the autolytic activity of strain Xc. This inhibition was specific among the oral streptococci to S. mutans. These results indicate that the catalytic domain of AtlA is located at the C terminus, suggesting that further characterization of this domain may provide a means to control cariogenic dental plaque formation.
Most bacteria possess several autolysins that are able to degrade their cell walls. These enzymes are classified according to the chemical bond that they break down in the peptidoglycan substrate and include β-1,4-N-acetylmuramidases (lysozymes), β-1,4-N-acetylglucosaminidases, N-acetylmuramyl-l-alanine amidases, endopeptidases, and transglycosylases (29, 33). They have been implicated in various biological functions, including cell separation, cell wall turnover, restructuring of cell walls, and bacterial autolysis (induced by antibiotics or adverse physiological conditions) (29, 37). Certain autolysins have also been reported to contribute to the pathogenicity of gram-positive bacteria. An intact autolytic function is required for the full virulence of Streptococcus pneumoniae (1). An autolysis-defective mutant of Staphylococcus aureus showed attenuated virulence in a rat model of endocarditis (18). The autolysin of Listeria monocytogenes contributes to its adhesion to eukaryotic cells and its colonization of the liver (20). Although autolysins are believed to play an important role in cell wall metabolism and in the pathogenicity of bacteria, only a limited number of autolysins have been extensively investigated.
Streptococcus mutans is a primary pathogen of human dental caries in the oral cavity (17). S. mutans is capable of forming a biofilm known as dental plaque on the tooth surface (34). Dental plaque formation is initiated by cell-to-surface adherence, followed by bacterial accumulation with the development of cell-to-cell interactions. Within dental plaque, S. mutans can produce large amounts of acids from fermentable sugars. Acid accumulation can eventually dissolve the hard, crystalline structure of the teeth, resulting in carious lesions (27). The ability to form biofilms is one of the important virulence properties of S. mutans. This bacterium is also a causative agent of infective endocarditis (8). It is primarily associated with subacute (chronic) infective endocarditis, which involves the colonization of heart tissue having preexisting endothelial damage (22). The abilities to adhere to and to colonize host tissues are very important virulence factors of this pathogen. We are interested in understanding the contribution of autolysins to the virulence of S. mutans. The autolytic system of S. mutans has not yet been characterized.
In this report, we present data on the isolation and characterization of the first described autolysin-encoding gene, atlA, from S. mutans.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used for this study are listed in Table 1. These strains were maintained and grown routinely as described previously (32). Antibiotics were used at the following concentrations: 200 μg of erythromycin per ml and 50 μg of ampicillin per ml for Escherichia coli and 10 μg of erythromycin per ml for S. mutans.
TABLE 1.
Bacterial strains used for this study
| Strain | Relevant properties | Source or referencea |
|---|---|---|
| E. coli DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 30 |
| S. mutans strains | ||
| Xc | Serotype c wild-type strain | 10 |
| Xc91 | Emr; Xc carrying Emr gene inserted into open reading frame 1 | This study |
| Xc92 | Emr; Xc carrying Emr gene inserted into atlA | This study |
| Xc93 | Emr; Xc carrying Emr gene inserted into open reading frame 3 | This study |
| Xc94 | Emr; Xc carrying Emr gene inserted into open reading frame 4 | This study |
| MT8148 | Serotype c strain | 25 |
| GS5 | Serotype c strain | 11 |
| UA159 | Serotype c strain | 21 |
| LM7 | Serotype e strain | 24 |
| MT6219 | Serotype f strain | 24 |
| S. gordonii strains | ||
| 10558 | Type strain | ATCC |
| Challis | Wild-type strain | 35 |
| S. oralis 10557 | Type strain | ATCC |
| S. salivarius strains | ||
| HHT | Wild-type strain | 36 |
| HT9R | Wild-type strain | 38 |
| S. sanguis 10556 | Type strain | ATCC |
| S. sobrinus strains | ||
| OMZ176 | Serotype d strain | 24 |
| 6715 | Serotype g strain | 2 |
ATCC, American Type Culture Collection.
DNA manipulation.
Standard DNA recombinant procedures such as DNA isolation, endonuclease restriction, ligation, and agarose gel electrophoresis were performed as described by Sambrook and Russell (30). The transformation of S. mutans and E. coli was carried out as described previously (42). Protein sequence similarity searches were performed with the BLAST program via the National Center for Biotechnology Information server.
DNA amplification.
To improve the fidelity of the PCR assay, we used KOD DNA polymerase (Toyobo Co., Ltd., Osaka, Japan). PCR was performed with 0.05 U of KOD DNA polymerase/ml in 120 mM Tris-HCl buffer (pH 8.2) containing appropriate amounts of the primers, a 0.2 mM concentration of each deoxyribonucleoside triphosphate, 6 mM ammonium sulfate, 10 mM KCl, 1 mM MgCl2, 0.1% Triton X-100, and 0.001% bovine serum albumin. The reactions were carried out for 25 cycles under the following conditions: denaturation at 94°C for 15 s, annealing at 58°C for 2 s, and extension at 74°C for 30 s.
Southern blot analysis.
Southern blot analysis was performed with digoxigenin (DIG)-labeled PCR probes using a nonradioactive DIG DNA labeling and detection kit (Roche Diagnostics, Mannheim, Germany) according to the supplier's instructions.
Random mutagenesis of S. mutans.
The random mutagenesis of S. mutans was carried out as described previously (41). Briefly, we constructed an S. mutans Xc genomic library by inserting a complete Sau3AI digest of the S. mutans Xc chromosome into the BamHI site of pResEmBBN. pResEmBBN can be used as an integration vector for gene inactivation by a single crossover with the streptococcal chromosome because it has no replicon in streptococcal species. S. mutans Xc was randomly mutated by transformation with the S. mutans genomic library. Transformants were spread on brain heart infusion (BHI; Difco, Detroit, Mich.) agar plates containing 10 μg of erythromycin per ml and heat-inactivated, proteinase K-treated S. mutans Xc cells (final optical density at 550 nm [OD550] of 1.0). Transformants causing an attenuated lytic zone around the colony were selected by visual screening.
Preparation of crude autolysin-containing samples.
Autolysin-containing samples were prepared from cultures grown to an OD550 of 0.7. Cell cultures (50 ml) were harvested by centrifugation, and the pellet was resuspended in 500 μl of 4% (wt/vol) sodium dodecyl sulfate (SDS). The suspension was incubated for 30 min at room temperature before being centrifuged. An equal volume of 50 mM Tris-HCl (pH 6.5) containing 10% glycerol was then added to the supernatant.
Zymogram analysis.
A zymogram analysis of autolysins was carried out by using an SDS-10% polyacrylamide gel (14) containing 1% (wet weight) S. mutans cells. The preparation of S. mutans cells for incorporation into polyacrylamide gels was performed as described previously (28, 44). Briefly, cell cultures (800 ml) of S. mutans Xc were harvested by centrifugation, and the pellet was washed three times with distilled water and then resuspended in 60 ml of 4% SDS. After being boiled for 30 min, the bacterial cells were washed five times with distilled water and then centrifuged. The cells were resuspended in 8 ml of 46% hydrofluoric acid and incubated at 4°C for 12 h. The suspension was then centrifuged, and the cells were washed five times with distilled water.
The prepared bacterial cells were incorporated into polyacrylamide gels, autolysin-containing samples were applied to the gels, and the samples were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, the gels were washed twice with distilled water for 15 min and then incubated in 200 ml of 0.2 M sodium phosphate buffer (pH 7.0) for 12 h at room temperature.
Autolysis assays.
S. mutans cells in the exponential phase of growth (OD550 of 0.9) were harvested by centrifugation and washed twice with phosphate-buffered saline. The cells were resuspended in 20 mM potassium phosphate buffer (pH 6.5) containing 1 M KCl, 10 mM CaCl2, 1 mM MgCl2, and 0.04% sodium azide to an OD550 of 0.9. The cell suspension was incubated at several different temperatures (37°C to 50°C), and autolysis was monitored by measuring the OD550 of the cell suspension. For inhibition assays, a solution of purified immunoglobulin G antibodies (final concentration of 0.25 mg/ml) was added to the cell suspension.
Production of polyclonal antibodies against partial peptides of the autolytic enzyme.
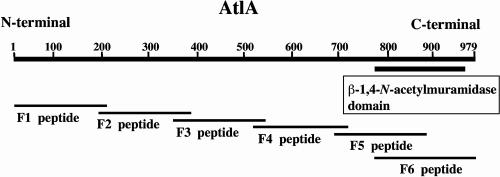
Partial DNA fragments (F1 to F6) of the atlA gene were amplified by PCRs using the primer pairs listed in Table 2. The amplified fragments, except for F6, were digested with BamHI and HindIII; the F6 fragment was digested with BamHI alone. Each fragment was then cloned in frame with a six-His tag into the pQE-80L vector (QIAGEN, Hilden, Germany). E. coli DH5α cells were transformed with the resulting plasmids. Each transformant was grown at 37°C until an OD550 of 0.6 was attained. Expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside. The cells were harvested 4 h after induction, and the partial proteins were purified by use of a Ni-nitrilotriacetic acid spin kit (QIAGEN) according to the supplier's instructions. The purities of the proteins were analyzed by SDS-PAGE. Antibodies against the F1, F2, F3, and F6 peptides (see Fig. 3) were raised in rabbits. Antibodies against the F4 and F5 peptides were raised in chickens because attempts to raise them in rabbits failed. The rabbit antibodies were purified by affinity chromatography on a protein G column (Amersham Biosciences, Piscataway, N.J.) and the chicken antibodies were purified with an Affi-Gel Hz immunoaffinity kit (Bio-Rad Laboratories, Hercules, Calif.). Each preimmune serum was purified in the same manner.
TABLE 2.
Primers used for this study
| Fragment | Primer | Sequence (5′ to 3′)a | Locationb |
|---|---|---|---|
| F1 | F1-forward | CGCGGATCCAAAAGCAAAACTTATTTG | 889-906 |
| F1-reverse | CGCAAGCTTTTAACCGTTATACTGTAG | 1485-1471 | |
| F2 | F2-forward | CGCGGATCCATGACCCCAGCAAGCAGC | 1366-1383 |
| F2-reverse | CGCAAGCTTTTAATTAGCAACAGATAT | 2025-2011 | |
| F3 | F3-forward | CGCGGATCCCATTATGAAGCTCATATT | 1906-1923 |
| F3-reverse | CGCAAGCTTTTAATAAAGCCCATAACC | 2505-2491 | |
| F4 | F4-forward | CGCGGATCCAATGGTCAAGATGATCTC | 2416-2433 |
| F4-reverse | CGCAAGCTTTTAAAGGTTAGATTGATC | 3015-3001 | |
| F5 | F5-forward | CGCGGATCCATAGATATGATTGTTAAA | 2926-2943 |
| F5-reverse | CGCAAGCTTTTAATTAATATTGTTCAC | 3525-3511 | |
| F6 | F6-forward | CGCGGATCCAGCTTGAATGCTGATAAA | 3166-3183 |
| F6-reverse | TTACTGTTGAGTAAATCGACC | 3825-3805 |
Nucleotides that are underlined in each primer sequence show the position of the restriction endonuclease site incorporated to facilitate cloning.
Numbers give positions on a HindIII-BglII fragment that was sequenced for this study (DDBJ data bank accession no. AB194064).
FIG. 3.
Schematic representation of AtlA and deletion mutants of AtlA. AtlA was divided into six parts, and antibodies were raised against each part in order to characterize the catalytic domain. Numbers indicate the positions of amino acids.
Biofilm formation and quantification.
To facilitate quantification and microscopy, we used 96-well polystyrene microtiter plates for the growth of biofilms. Growth was initiated by inoculating individual wells of a 96-well microtiter plate with 5 μl of cell suspension in 200 μl of BHI broth supplemented with 1% (wt/vol) sucrose or glucose. The microtiter plates were incubated at 37°C in 7.3% CO2 for 16 h without agitation. After the 16-h incubation, the liquid medium was removed and the wells were rinsed once with sterile distilled water. The plates were air dried, stained with 0.1% safranine for 10 min, rinsed with distilled water to remove excess dye, and then air dried for 3 h. The biofilms were quantified by measuring the absorbance of stained biofilms at 490 nm with an enzyme-linked immunosorbent assay microplate reader (Bio-Rad Laboratories). Each assay was performed in triplicate, and wells without biofilms were used as blank controls after safranine staining.
Microscopy.
The spatial distribution and architecture of biofilms of S. mutans strains were examined by scanning electron microscopy. Individual wells of a six-well microtiter plate were filled with 8 ml of BHI broth supplemented with 1% (wt/vol) sucrose. A sterile, cut-glass slide was added to each well, and each well was then inoculated with 100 μl of overnight culture. The plates were incubated at 37°C in 7.3% CO2 for 16 h without agitation. The glass slides were then removed from wells, rinsed briefly with 10 mM phosphate buffer, and fixed with 2% paraformaldehyde in 10 mM phosphate buffer for 3 h. Following dehydration in a series of increasing ethanol concentrations, the samples were freeze-dried with t-butyl alcohol (9) and sputter coated with platinum to a thickness of approximately 2 nm. The samples were examined with a Hitachi S-4300 FE scanning electron microscope.
Nucleotide sequence accession number.
The 7,166-bp nucleotide sequences determined in this study were deposited in the DDBJ data bank (http://www.ddbj.nig.ac.jp) under accession number AB194064.
RESULTS
Isolation of an autolysin-deficient mutant of S. mutans.
S. mutans Xc was randomly mutated by transformation with the S. mutans genomic library. Sixteen transformants showing an attenuated lytic zone around the colony were selected from 10,000 transformants by visual screening. The 16 transformants were spread on the same plates again, and one, designated Xc-AT, was confirmed to be completely defective in autolytic activity. Southern blotting with a DIG-labeled PCR probe specific for the erythromycin resistance (Emr) gene revealed that the probe hybridized with a 5.5-kb HindIII fragment, a 2.4-kb EcoRI fragment, and a 6.3-kb BglII fragment of Xc-AT but did not hybridize with any fragments of the wild-type strain Xc.
Cloning and sequencing of the region flanking the plasmid insertion site in strain Xc-AT.
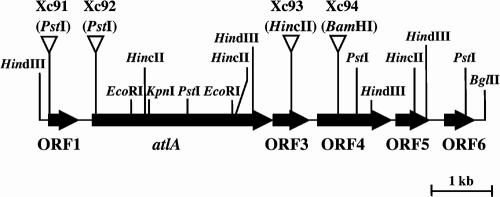
To recover the region flanking the plasmid insertion point in strain Xc-AT, we digested the Xc-AT chromosome with EcoRI and allowed it to self-ligate. E. coli DH5α was then transformed with this DNA. Emr transformants were isolated on BHI agar plates containing erythromycin. Almost all of the plasmids isolated from transformants were 2.4 kb long. One of these was designated pAT-E. In a similar manner, we constructed pAT-H (HindIII digestion, self-ligation, 5.5 kb) and pAT-B (BglII digestion, self-ligation, 6.3 kb). Nucleotide sequence analyses of these three plasmids revealed that six open reading frames (ORFs) were present in the region flanking the plasmid insertion point in strain Xc-AT (Fig. 1) and that the insert fragment of the integration plasmid was located within ORF2. ORF2 was designated atlA (Fig. 1). The predicted translational product of the atlA gene is a protein of 979 amino acids with a molecular weight of 107,279. The amino acid sequence deduced from the C-terminal portion (196 amino acid residues) of atlA has a high degree of similarity to the active domain of β-1,4-N-acetylmuramidase. A consensus promoter-like sequence, TTGAGA-N17-TATAAT, exists in the region upstream from atlA. The ORF4 gene product was homologous to peptidase T of S. pneumoniae. No proteins were found to be homologous to the gene products predicted by the other ORFs.
FIG. 1.
Restriction map of the atlA gene and its flanking region in S. mutans Xc. The arrows indicate the locations of the six ORFs. The Emr gene insertion sites for the insertional inactivation of the atlA gene and the flanking ORFs are indicated by inverted open triangles.
Insertional inactivation of atlA and flanking genes.
To confirm the role of atlA in autolytic activity, we insertionally inactivated atlA and the flanking genes (ORF1, ORF3, and ORF4) by interruption with the Emr gene at the restriction sites indicated in Fig. 1. The resultant mutants were designated Xc91, Xc92, Xc93, and Xc94 (Fig. 1).
Zymogram analysis.
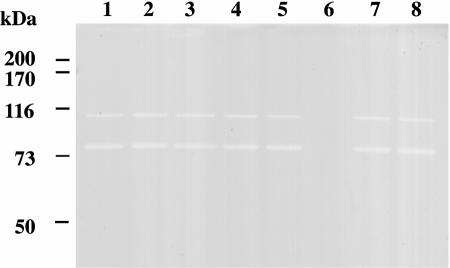
To determine whether the atlA mutation might affect the activity of autolysin, we examined the autolysin profiles of S. mutans Xc and Xc92 by zymogram analysis (Fig. 2). Two major autolysin bands were present in Xc whole-cell extracts (Fig. 2, lane 1). The lower band had a more pronounced clear zone of autolytic activity than the upper band. The proteins migrated with apparent masses of 107 and 79 kDa, and the mass of the larger protein, 107 kDa, corresponded to the size of the AtlA protein. Neither band was detected in the Xc92 whole-cell extract (Fig. 2, lane 6). The autolysin profiles of the other mutants (Xc91, Xc93, and Xc94) were the same as that of Xc (Fig. 2, lanes 5, 7, and 8). These results indicate that atlA encodes the major autolysin of S. mutans. Furthermore, other S. mutans serotype c strains had the same autolysin profiles as that of Xc (Fig. 2, lanes 2 to 4).
FIG. 2.
Autolysin profiles of S. mutans strains, determined by renaturing SDS-PAGE using a 10% polyacrylamide gel containing 1% (wet weight) S. mutans cells. For zymography, a 4% SDS extract of each strain was used. Lanes: 1, S. mutans Xc; 2, S. mutans MT8148; 3, S. mutans GS5; 4, S. mutans UA159; 5, S. mutans Xc91 (ΔORF1); 6, S. mutans Xc92 (ΔatlA); 7, S. mutans Xc93 (ΔORF3); 8, S. mutans Xc94 (ΔORF4). Prestained high-molecular-mass markers are indicated in kilodaltons on the left.
To determine whether the 79-kDa band was a proteolytic product of AtlA, we performed Western blotting by using antibodies raised against the F1 peptide (N-terminal portion) and the F6 peptide (C-terminal portion) of AtlA (Fig. 3). The antibody against the F6 peptide reacted with both the 107- and 79-kDa bands, while the antibody against the F1 peptide reacted with only the 107-kDa band (data not shown). These results indicate that the 79-kDa band is a proteolytic product of AtlA, created by the degradation of the N-terminal portion of the 107-kDa peptide.
Properties of the atlA mutant.
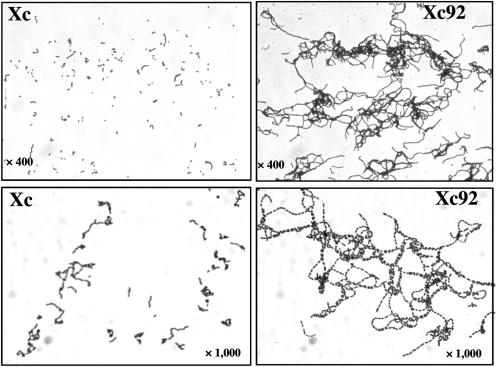
The growth rate of the cells was not affected by the atlA mutation; however, most Xc92 cells sedimented when grown overnight, while a broth culture of wild-type Xc was turbid, with just a few sedimented cells (data not shown). Light microscopic observations revealed that Xc92 formed very long chains of cells compared with those formed by the parental strain Xc (Fig. 4). These results clearly demonstrated that AtlA is involved in cell separation.
FIG. 4.
Light microscopic views of gram-stained Xc and Xc92 (ΔatlA) cells.
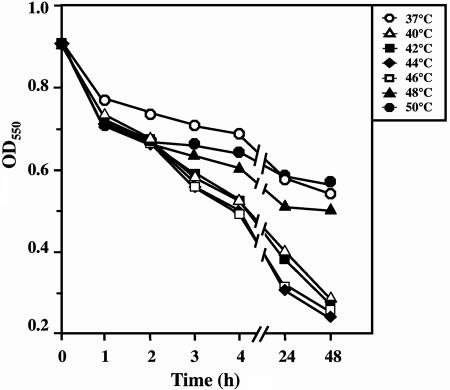
The autolytic activity of AtlA was examined at several different temperatures (37°C to 50°C) (Fig. 5). The optimum temperature for the autolytic activity of AtlA was 44°C. There was a distinct difference in the autolytic activities between 44°C and 37°C. However, the biological meaning of the fact that the optimum temperature is considerably higher than 37°C is not clear. The autolytic activity of the atlA mutant was compared with that of the wild-type Xc strain. As shown in Fig. 6, Xc92 exhibited a lower autolysis rate and lower extent of autolysis than did Xc. Thus, the inactivation of atlA resulted in a remarkable decrease in autolytic activity.
FIG. 5.
Effect of temperature on autolytic activity of AtlA. An autolysis assay of bacterial cells was carried out as described in Materials and Methods. The cell suspension was incubated at several different temperatures (37°C to 50°C), and autolysis was monitored by measuring the OD550 of the cell suspension.
FIG. 6.
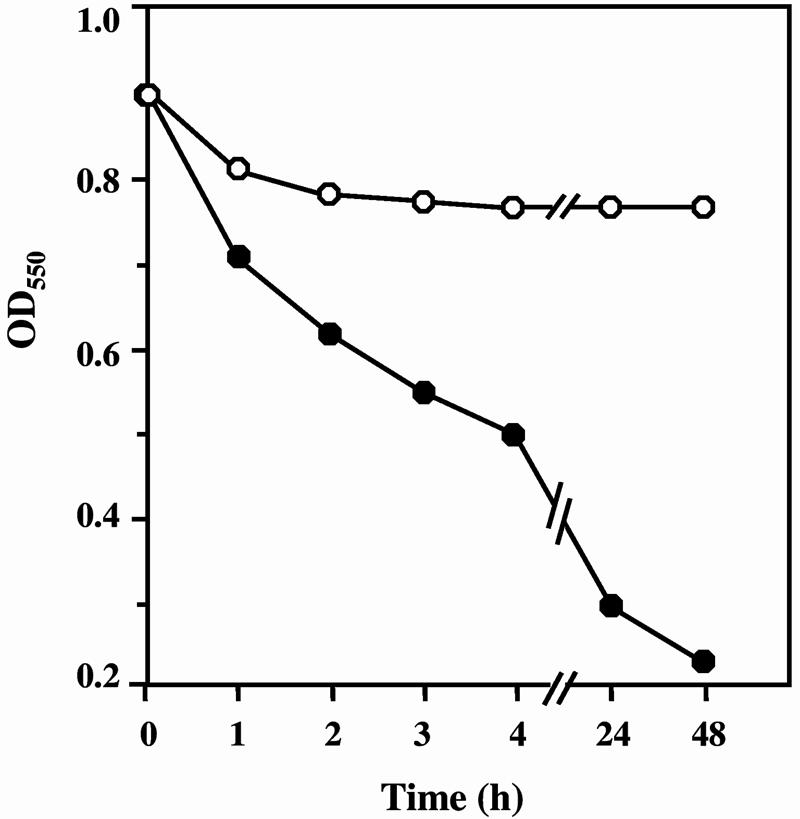
Comparison of autolytic activities of Xc92 (ΔatlA) and the wild-type strain Xc. An autolysis assay of bacterial cells was carried out as described in Materials and Methods. The cell suspension was incubated at the optimum temperature for the autolytic activity of AtlA (44°C). Open circles represent Xc92 (ΔatlA); closed circles represent Xc.
Role of the atlA gene in biofilm formation.
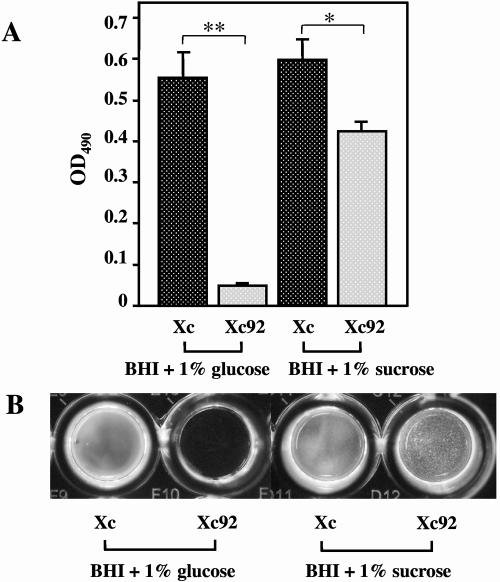
We next determined whether there were any alterations in the biofilm structure as a result of the loss of AtlA production. The capacity of the Xc and Xc92 strains to form biofilms in the wells of 96-well microtiter plates was evaluated (Fig. 7). Strains Xc and Xc92 both formed tight biofilms in the presence of sucrose, but the biofilm formed by Xc92 had a nearly 30% less biomass than that of Xc. Upon visual inspection of the microtiter wells, there was a noticeable difference in the biofilm structures of Xc92 and Xc. Macroscopically, the Xc biofilm generally had a very confluent appearance with no major discernible features. In contrast, the Xc92 biofilm had a very rough texture. In addition, the Xc strain formed very loose but obvious biofilms in the presence of glucose alone (no sucrose), while Xc92 had negligible biofilm formation under the same conditions.
FIG. 7.
Biofilm formation and quantification of wild-type Xc and Xc92 (ΔatlA) on polystyrene surfaces. Biofilms were cultivated in BHI broth with 1% glucose or 1% sucrose. Biofilms were stained and the OD490 was measured. (A) Mean OD490 values ± standard deviations. *, P < 0.01; **, P < 0.001 (Student's t test). (B) Photograph of typical biofilms grown on polystyrene microtiter plates.
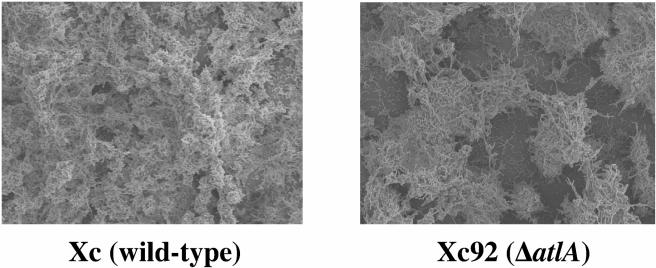
A closer examination of the biofilms by scanning electron microscopy revealed that the sucrose-dependent biofilms of the Xc and mutant strains had very different appearances. The mutant biofilm appeared to have a sponge-like architecture with what appeared to be large gaps (Fig. 8).
FIG. 8.
Scanning electron micrographs showing the spatial distribution and architecture of sucrose-dependent biofilms formed by S. mutans Xc (left) and Xc92 (ΔatlA) (right). Magnification, ×1,000.
Inhibition assay.
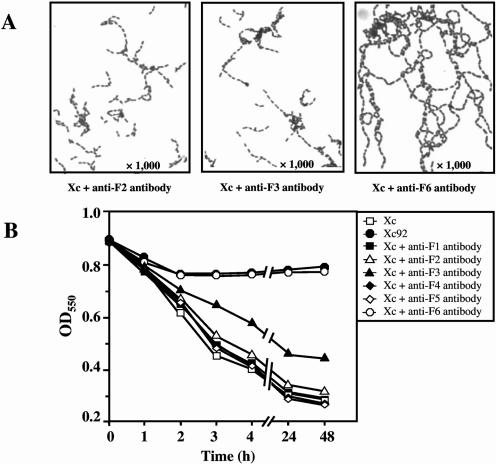
To determine the active center of AtlA, we performed an inhibition assay, using the six polyclonal antibodies raised against partial peptides within the AtlA protein and the respective preimmune sera (Fig. 3). As shown in Fig. 9A, when Xc was grown with the antibody produced against the F2, F3, or F6 peptide, the cells formed longer chains than when they were grown with the respective preimmune sera. The presence of the antibody against the F6 peptide, which corresponds to the C-terminal portion of the protein and contains a β-1,4-N-acetylmuramidase domain, led to the formation of very long chains. Furthermore, when the anti-F6 antibody was added to the Xc cell suspension, Xc showed a drastically reduced autolytic activity (Fig. 9B), with the same autolytic pattern as that of Xc92. The antibody against the F3 peptide also affected the autolytic activity of Xc, but the extent of this inhibition was about 30% that shown by the anti-F6 antibody. Based on these results, we concluded that the C-terminal portion of AtlA, which contains the β-1,4-N-acetylmuramidase domain, behaves as a true catalytic domain. In addition, the specificity of inhibition by the anti-F6 antibody was examined with several oral bacteria. The bacterial strains used for this experiment are listed in Table 1. No oral streptococci except for S. mutans formed longer chains when grown with the antibody (data not shown). Furthermore, the antibody did not affect the autolytic activities of oral streptococci other than S. mutans (data not shown). These results indicate that the inhibition by the anti-F6 antibody was specific to S. mutans.
FIG. 9.
Effect of antibodies raised against partial peptides within the AtlA protein on chain length and autolytic activity. (A) Light microscopic views. Xc was grown in BHI broth including each purified antibody (final concentration of 0.25 mg/ml). (B) Autolysis of Xc cells incubated with each antibody. Xc cells were incubated in autolysis buffer including each purified antibody (final concentration of 0.25 mg/ml). No preimmune sera affected both the chain length and the autolytic activity of the wild-type strain Xc.
DISCUSSION
In a standardized assay to detect autolytic activity in a denaturing polyacrylamide gel, two major lytic bands were found in an SDS extract of S. mutans cells by using S. mutans cell walls as a substrate. The same autolysin profile was detected for all S. mutans strains used. In this report, we present the first cloning of a gene (atlA) encoding an autolysin in S. mutans. A comparison of the wild-type strain with an atlA deletion mutant revealed that the two major autolytic protein bands originated from atlA, as both bands disappeared with the atlA deletion mutant. Western blot analysis with an antibody against the N-terminal region of AtlA confirmed that the deletion occurred in this region. The degradation of cell wall hydrolases without a loss of enzymatic activity has been observed previously for Bacillus licheniformis (23), Bacillus subtilis (13, 26), and Lactococcus lactis (3). These results suggest that the atlA gene cloned for this study encodes the major autolysin of S. mutans.
Autolysins have been extensively investigated from S. pneumoniae, one of the gram-positive bacteria (Table 3). Three kinds of autolysins (LytA, LytB, and LytC) from this organism have been identified and characterized (5, 6, 7). LytA, the main autolytic enzyme of S. pneumoniae, is an N-acetylmuramoyl-l-alanine amidase and is responsible for most autolysis in S. pneumoniae. LytB and LytC have been identified as an endo-β-N-acetylglucosaminidase and a β-1,4-N-acetylmuramidase, respectively. Inactivation of the lytB gene does not affect cell wall hydrolytic activity, and LytC is responsible for autolytic activity in a lytA mutant. Thus, each autolysin plays a different role in the autolytic system of S. pneumoniae. The results of homology studies showed that a sequence of 196 amino acid residues in the C-terminal region of AtlA had a high degree of similarity to the consensus sequence of the active domain of β-1,4-N-acetylmuramidase, which was also detected in LytC. The peptidoglycan of S. mutans consists of a repeating disaccharide, N-acetylmuramic acid-(β-1,4)-N-acetylglucosamine, that contains the site of hydrolysis for β-1,4-N-acetylmuramidase. Furthermore, most bacterial peptidoglycan hydrolases, including LytA, LytB, and LytC, have a substrate-binding domain in addition to a catalytic domain. However, there is no region in AtlA that is homologous to any substrate-binding domain.
TABLE 3.
Comparison of biochemical and biological properties of the AtlA enzyme and the LytA, LytB, and LytC enzymes
| Enzyme | Catalytic activity | Molecular massa (kDa) | Optimum temperature (°C) | Autolysis | Mutant phenotype | Biofilm effectb |
|---|---|---|---|---|---|---|
| AtlA | Muramidase homologue | 107 | 44 | Yes | Long chain | Important |
| LytA | Amidase | 36 | 37 | Yes | Small chain | NR |
| LytB | Glucosaminidase | 82 | 37 | No | Long chain | NR |
| LytC | Muramidase | 59 | 30 | Yes | NAc | NR |
Molecular masses were calculated from the deduced amino acid sequences.
NR, not reported.
NA, not affected.
In order to further characterize the AtlA protein, we compared AtlA with LytC, which is known to be a β-1,4-N-acetylmuramidase (Table 3). AtlA consists of 979 amino acids with a molecular weight of 107,279; LytC has 501 amino acids and a molecular weight of 58,682. AtlA and LytC each possess a conserved β-1,4-N-acetylmuramidase domain at the C-terminal end. The identity between these two domains is 24.5%. LytC has a choline-binding domain in the N-terminal portion, in addition to the catalytic domain, but there is no substrate-binding domain in the AtlA protein. As for biological function, LytC is responsible for the remaining autolytic activity in a lytA mutant, and inactivation of the atlA gene resulted in a marked decrease in autolysis. The optimum temperatures for the autolytic activities of AtlA and LytC were 44°C and 30°C, respectively. In S. pneumoniae, the inactivation of the lytB gene leads to the formation of long chains of cells (4), the inactivation of the lytA gene results in the formation of small chains (6 to 10 cells in length) (31), and the mutation of the lytC gene does not affect the chain length of S. pneumoniae cells (7). In contrast, the inactivation of atlA led to the formation of long chains in S. mutans. AtlA was involved in both cellular autolysis and cell separation. These results suggest that AtlA may have a biological function different from those of the previously identified autolysins. Considering that AtlA had the functions of both LytA and LytB, it may have an N-acetylmuramyl-l-alanine amidase and/or β-1,4-N-acetylglucosaminidase activity, although it does not contain these enzymatic domains. Unfortunately, we failed to purify the AtlA protein and therefore could not determine the site of hydrolysis of AtlA.
The ability to form biofilms is one of the virulence properties of S. mutans. This ability derives from two distinct processes, an initial sucrose-independent attachment and an enhancement of the attachment by a sucrose-dependent mechanism (12). It is well known that the sucrose-dependent biofilm plays an important role in the formation of dental caries (40). The mechanisms of sucrose-dependent biofilm formation, including the role of glucosyltransferase, have been well established. On the other hand, the mechanisms of sucrose-independent initial biofilm formation have not yet been elucidated. Recently, a genetic investigation of the initial sucrose-independent biofilm formation of S. mutans revealed the involvement of several genes associated with a quorum-sensing signaling system. com mutants formed sucrose-independent biofilms with an altered architecture or reduced biomass in the presence of glucose (16), whereas sucrose-dependent biofilms formed by com mutants were very similar to those formed by the wild-type strain (43). Deletion of the hk-11 or rr-11 gene, both of which are involved in a two-component regulatory system, also resulted in the formation of a biofilm with a reduced biomass and an abnormal structure in the absence of sucrose (15). Conversely, a luxS mutant formed altered biofilms with large gaps compared with the biofilms of the wild-type strain in the presence of sucrose (19), but there was no significant difference in the formation of sucrose-independent biofilms by the mutant and wild-type strains (43). Interestingly, for the atlA mutant, sucrose-dependent biofilm formation as well as sucrose-independent biofilm formation was altered, with sucrose-independent biofilm formation being extensively reduced. Furthermore, biofilms formed by the atlA mutant had a sponge-like architecture, while those of the parent strain had a relatively smooth and confluent structure. Recent studies have indicated that cells in mutant biofilms (the comC mutant, the brpA mutant, and the hr/rr11 mutants) with abnormal structures tend to form long chains, suggesting that the variation in biofilm structure might result from the formation of extremely long chains of cells (15, 16, 39). In the present study, the inactivation of the atlA gene led to the formation of long chains. The regulation of the chain length of Streptococcus cells may play an important role in biofilm formation.
The results of inhibition assays indicated that the active center of AtlA is located in the C-terminal portion of the protein, which contains the β-1,4-N-acetylmuramidase catalytic domain. The use of the antibody raised against the C-terminal peptide (F6 peptide) drastically attenuated the autolytic activity of S. mutans and led to the formation of long chains of S. mutans cells. These results were similar to those seen for the atlA mutant. However, the F6 peptide exhibited no lytic activity in a zymogram analysis when it was synthesized in E. coli. Furthermore, the inhibition by the anti-F6 antibody was extremely specific to S. mutans. The F6 peptide prepared for the production of the antibody was composed of 219 amino acids. Further characterization might identify the active center within the peptide and provide us with a novel target for antibacterial drugs. In addition to the anti-F6 antibody, the antibody raised against the F3 peptide had a small effect on the autolytic activity and chain length. A catalytic domain similar to those of other autolysins (amidases, glucosaminidases, etc.) may exist in the F3 region, which possesses a different structure from those identified previously. Much remains to be learned about this enzymatic activity, and further characterization of AtlA is required to elucidate the autolytic system of S. mutans.
Acknowledgments
This work was supported in part by a grant from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to promote a Multi-Disciplinary Research Project and the Promotion and Mutual Aid Corporation for Private Schools of Japan and by a grant-in-aid for developmental scientific research (12557186) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Editor: J. T. Barbieri
REFERENCES
- 1.Berry, A. M., R. A. Lock, D. Hansman, and J. C. Paton. 1989. Contribution of autolysin to virulence of Streptococcus pneumoniae. Infect. Immun. 57:2324-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowen, W. H., K. M. Madison, and S. K. Pearson. 1988. Influence of desalivation in rats on incidence of caries in intact cagemates. J. Dent. Res. 67:1316-1318. [DOI] [PubMed] [Google Scholar]
- 3.Buist, G., J. Kok, K. J. Leenhouts, M. Dabrowska, G. Venema, and A. J. Haandrikman. 1995. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J. Bacteriol. 177:1554-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Las Rivas, B., J. L. Garcia, R. Lopez, and P. Garcia. 2002. Purification and polar localization of pneumococcal LytB, a putative endo-beta-N-acetylglucosaminidase: the chain-dispersing murein hydrolase. J. Bacteriol. 184:4988-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia, E., J. L. Garcia, C. Ronda, P. Garcia, and R. Lopez. 1985. Cloning and expression of the pneumococcal autolysin gene in Escherichia coli. Mol. Gen. Genet. 201:225-230. [DOI] [PubMed] [Google Scholar]
- 6.Garcia, P., M. P. Gonzalez, E. Garcia, R. Lopez, and J. L. Garcia. 1999. LytB, a novel pneumococcal murein hydrolase essential for cell separation. Mol. Microbiol. 31:1275-1277. [DOI] [PubMed] [Google Scholar]
- 7.Garcia, P., M. Paz Gonzalez, E. Garcia, J. L. Garcia, and R. Lopez. 1999. The molecular characterization of the first autolytic lysozyme of Streptococcus pneumoniae reveals evolutionarily mobile domains. Mol. Microbiol. 33:128-138. [DOI] [PubMed] [Google Scholar]
- 8.Horaud, T., and F. Delbos. 1984. Viridans streptococci in infective endocarditis: species distribution and susceptibility to antibiotics. Eur. Heart J. 5(Suppl. C):39-44. [DOI] [PubMed] [Google Scholar]
- 9.Inoue, T., and H. Osatake. 1988. A new drying method of biological specimens for scanning electron microscopy: the t-butyl alcohol freeze-drying method. Arch. Histol. Cytol. 51:53-59. [DOI] [PubMed] [Google Scholar]
- 10.Koga, T., H. Asakawa, N. Okahashi, and I. Takahashi. 1989. Effect of subculturing on expression of a cell-surface protein antigen by Streptococcus mutans. J. Gen. Microbiol. 135:3199-3207. [DOI] [PubMed] [Google Scholar]
- 11.Kuramitsu, H. K. 1975. Characterization of extracellular glucosyltransferase activity of Streptococcus mutans. Infect. Immun. 12:738-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuramitsu, H. K. 2000. Streptococcus mutans: molecular genetic analysis, p. 280-286. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 13.Kuroda, A., and J. Sekiguchi. 1991. Molecular cloning and sequencing of a major Bacillus subtilis autolysin gene. J. Bacteriol. 173:7304-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Li, Y. H., P. C. Lau, N. Tang, G. Svensater, R. P. Ellen, and D. G. Cvitkovitch. 2002. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J. Bacteriol. 184:6333-6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, Y. H., N. Tang, M. B. Aspiras, P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mani, N., L. M. Baddour, D. Q. Offutt, U. Vijaranakul, M. J. Nadakavukaren, and R. K. Jayaswal. 1994. Autolysis-defective mutant of Staphylococcus aureus: pathological considerations, genetic mapping, and electron microscopic studies. Infect. Immun. 62:1406-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milohanic, E., R. Jonquieres, P. Cossart, P. Berche, and J. L. Gaillard. 2001. The autolysin Ami contributes to the adhesion of Listeria monocytogenes to eukaryotic cells via its cell wall anchor. Mol. Microbiol. 39:1212-1224. [DOI] [PubMed] [Google Scholar]
- 21.Murchison, H. H., J. F. Barrett, G. A. Cardineau, and R. Curtiss III. 1986. Transformation of Streptococcus mutans with chromosomal and shuttle plasmid (pYA629) DNAs. Infect. Immun. 54:273-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mylonakis, E., and S. B. Calderwood. 2001. Infective endocarditis in adults. N. Engl. J. Med. 345:1318-1330. [DOI] [PubMed] [Google Scholar]
- 23.Oda, Y., R. Nakayama, A. Kuroda, and J. Sekiguchi. 1993. Molecular cloning, sequence analysis, and characterization of a new cell wall hydrolase, CwlL, of Bacillus licheniformis. Mol. Gen. Genet. 241:380-388. [DOI] [PubMed] [Google Scholar]
- 24.Oho, T., Y. Yamashita, Y. Shimazaki, M. Kushiyama, and T. Koga. 2000. Simple and rapid detection of Streptococcus mutans and Streptococcus sobrinus in human saliva by polymerase chain reaction. Oral Microbiol. Immunol. 15:258-262. [DOI] [PubMed] [Google Scholar]
- 25.Ohta, H., H. Kato, N. Okahashi, I. Takahashi, S. Hamada, and T. Koga. 1989. Characterization of a cell-surface protein antigen of hydrophilic Streptococcus mutans strain GS-5. J. Gen. Microbiol. 135:981-988. [DOI] [PubMed] [Google Scholar]
- 26.Potvin, C., D. Leclerc, G. Tremblay, A. Asselin, and G. Bellemare. 1988. Cloning, sequencing and expression of a Bacillus bacteriolytic enzyme in Escherichia coli. Mol. Gen. Genet. 214:241-248. [DOI] [PubMed] [Google Scholar]
- 27.Quivey, R. G., W. L. Kuhnert, and K. Hahn. 2001. Genetics of acid adaptation in oral streptococci. Crit. Rev. Oral Biol. Med. 12:301-314. [DOI] [PubMed] [Google Scholar]
- 28.Ries, W., C. Hotzy, I. Schocher, U. B. Sleytr, and M. Sara. 1997. Evidence that the N-terminal part of the S-layer protein from Bacillus stearothermophilus PV72/p2 recognizes a secondary cell wall polymer. J. Bacteriol. 179:3892-3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogers, H. J., H. R. Perkins, and J. B. Ward. 1980. The bacterial autolysins, p. 437-460. In H. J. Rogers, H. R. Perkins, and J. B. Ward (ed.), Microbial cell walls and membranes. Chapman & Hall Ltd., London, United Kingdom.
- 30.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Sanchez-Puelles, J. M., C. Ronda, J. L. Garcia, P. Garcia, R. Lopez, and E. Garcia. 1986. Searching for autolysin functions. Characterization of a pneumococcal mutant deleted in the lytA gene. Eur. J. Biochem. 158:289-293. [DOI] [PubMed] [Google Scholar]
- 32.Shibata, Y., K. Ozaki, M. Seki, T. Kawato, H. Tanaka, Y. Nakano, and Y. Yamashita. 2003. Analysis of loci required for determination of serotype antigenicity in Streptococcus mutans and its clinical utilization. J. Clin. Microbiol. 41:4107-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shockman, G. D., and J.-V. Holtje. 1994. Microbial peptidoglycan (murein) hydrolases, p. 131-166. In J.-M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall. Elsevier, Amsterdam, The Netherlands.
- 34.Tanzer, J. M., J. Livingston, and A. M. Thompson. 2001. The microbiology of primary dental caries in humans. J. Dent. Educ. 65:1028-1037. [PubMed] [Google Scholar]
- 35.Tardif, G., M. C. Sulavik, G. W. Jones, and D. B. Clewell. 1989. Spontaneous switching of the sucrose-promoted colony phenotype in Streptococcus sanguis. Infect. Immun. 57:3945-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terleckyj, B., and G. D. Shockman. 1975. Amino acid requirements of Streptococcus mutans and other oral streptococci. Infect. Immun. 11:656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomasz, A. 1984. Building and breaking of bonds in the cell wall of bacteria—the role for autolysins, p. 3-12. In C. Nombela (ed.), Microbial cell wall synthesis and autolysis. Elsevier, Amsterdam, The Netherlands.
- 38.Tsukioka, Y., Y. Yamashita, T. Oho, Y. Nakano, and T. Koga. 1997. Biological function of the dTDP-rhamnose synthesis pathway in Streptococcus mutans. J. Bacteriol. 179:1126-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wen, Z. T., and R. A. Burne. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamashita, Y., W. H. Bowen, R. A. Burne, and H. K. Kuramitsu. 1993. Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infect. Immun. 61:3811-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamashita, Y., Y. Shibata, Y. Nakano, H. Tsuda, N. Kido, M. Ohta, and T. Koga. 1999. A novel gene required for rhamnose-glucose polysaccharide synthesis in Streptococcus mutans. J. Bacteriol. 181:6556-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamashita, Y., T. Takehara, and H. K. Kuramitsu. 1993. Molecular characterization of a Streptococcus mutans mutant altered in environmental stress responses. J. Bacteriol. 175:6220-6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshida, A., and H. K. Kuramitsu. 2002. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 68:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshimura, G., H. Komatsuzawa, J. Kajimura, T. Fujiwara, M. Ohara, K. Kozai, and M. Sugai. 2004. Zymographic characterization of bacteriolytic enzymes produced by oral streptococci. Microbiol. Immunol. 48:465-469. [DOI] [PubMed] [Google Scholar]