Abstract
The American lobster, Homarus americanus, is not only of considerable economic importance but has also emerged as a premier model organism in neuroscience research. Neuropeptides, an important class of cell-to-cell signaling molecules, play crucial roles in a wide array of physiological and psychological processes. Leveraging the recently sequenced high-quality draft genome of the American lobster, our study sought to profile the neuropeptidome in this model organism. Employing advanced mass spectrometry techniques, we identified 24 neuropeptide precursors and 101 unique mature neuropeptides in Homarus americanus. Intriguingly, 67 of these neuropeptides were discovered for the first time. Our findings provide a comprehensive overview of the peptidomic attributes of the lobster’s nervous system and highlight the tissue-specific distribution of these neuropeptides. Collectively, this research not only enriches our understanding of the neuronal complexities of the American lobster but also lays a foundation for future investigations into the functional roles that these peptides play in crustacean species. The mass spectrometry data have been deposited in the PRIDE repository with the identifier PXD047230.
Keywords: peptidomics, neuropeptide, American lobster, Homarus americaus, mass spectrometry, peptide identification
Graphical Abstract

Introduction
The American lobster, Homarus americanus, is globally acknowledged as a prominent crustacean species, primarily attributed to its culinary appeal. While it is one of the largest and most valuable fisheries along the east coast of North America1, 2, this decapod is also an important model organism in neuroscience research. Due to its relatively simple and accessible nervous system, scientists have been using this valuable model organism to study basic neural mechanisms and the relationship between neuronal circuits and behavior.3 One of the pioneering and prolific neuroscience research area involves the use of the crustacean stomatogastric nervous system (STNS), a simple neural network controlling the motion of gut and foregut, to study neural circuit function and dynamic regulation, as well as neuroplasticity and neuromodulation.3–5 Meanwhile, the STNS in crustaceans is significantly influenced by different modulators, with neuropeptides comprising the largest and most diverse class of these signaling molecules.6
Neuropeptides are small chains of amino acids and play an important role in various physiological and psychological processes, regulating energy homeostasis, pain, mood, memory and circadian rhythm.7, 8 These signaling peptides are derived from prohormones, which are large protein precursors, via post-translational enzymatic processing at cleavage sites that contain mono/dibasic amino acids such as lysine and arginine.9 Additionally, some neuropeptides may undergo post-translational modifications (PTMs), such as N-terminal pyro-glutamination formation and C-terminal amidation, to further fine-tune their biological functions, resulting in chemically distinct peptides with the same amino acid sequences, adding to their complexity.10–12
Mass spectrometry (MS)-based neuropeptidomics provides a powerful tool for profiling the neuropeptides at a global scale with high sensitivity and unparalleled molecular specificity.13–17 Compared to traditional study methods such as radioimmunoassay, immunohistochemistry, and Edman degradation, the MS-based strategy can simultaneously detect a large number of neuropeptides in a high-throughput manner, also with successful identification of the neuropeptides carrying PTM. This method has been effectively applied across a broad spectrum of species within the animal kingdom, demonstrating its versatility and effectiveness.15, 18–22 Confined to the crustacean, our group, along with others, has leveraged mass spectrometry for the discovery and profiling of various neuropeptides.23–27 However, the potential of this technique in crustacean research is somewhat hampered by the current limitations of the crustacean proteome databases. These databases are crucial as the identification of neuropeptides typically involves comparing the mass spectra of unknown peptides with the predicted spectra of known peptides, which are derived from these databases. Due to the historical lack of comprehensive, high-quality genome and proteome databases for crustaceans, the scope of identified neuropeptides and their families, including those in species like the Homarus americanus, remains significantly constrained.
In 2021, a landmark achievement was made with the publication of a high-quality draft assembly of the Homarus americanus genome, which included 25,284 predicted gene models.28 This significant advancement was further complemented by the annotation of the genome-encoded, yet previously uncharacterized, protein sequences using Google’s ProtNLM (Protein Natural Language Model).29 The high-quality genome and the detailed proteome database together are a great asset for studying the neural systems of the American lobster. Leveraging these resources, our study employed mass spectrometry combined with ion mobility spectrometry techniques to conduct an extensive profiling of neuropeptides, peptide hormones, and endogenous peptides within the lobster’s neural system.
The goal of our current study is to unravel the complexities of the peptidergic signaling system in the American lobster, focusing on the diversity, distribution, and functional implications of various neuropeptides. Furthermore, this research aims to discover novel neuropeptides, potentially expanding the pool of neuropeptide candidates for physiological functional investigation.
Materials and Methods
Chemicals
The solvents and reagents utilized, unless otherwise noted, were acquired from Thermo Fisher Scientific (Pittsburgh, PA) or MilliporeSigma (St. Louis, MO).
Animals and Tissue collection
The UW-Madison Animal Care and Use Committee granted approval for the protocols pertaining to animal care and study. American lobsters, Homarus americanus, were purchased from the local Global Market and Food Hall (Madison, WI) and maintained in artificial seawater tanks (made with Instant Ocean Sea Salt, 10°C, alternating 12-hour light/dark cycle). The lobsters were allowed to acclimate to the tanks for at least two weeks before conducting any experiments. Lobsters were cold anesthetized on ice for 30 min prior to the dissection conducted in chilled physiological saline solution (479 mM NaCl; 12.74 mM KCl; 20 mM MgSO4, 3.91 mM Na2SO4, 13.67 mM CaCl2, 5 mM HEPES, pH 7.45). Tissues of interest (a pair of sinus glands, the brain, the oesophageal ganglion, the stomatogastric ganglion, and the thoracic ganglion) were removed, placed in 200 μL ice-cold acidified methanol (90% methanol, 9% acetic acid, and 1% water), and stored at −80 °C until needed. Three lobsters were used for biological replicates.
Neuropeptide Extraction
Tissues in 200 μL ice-cold acidified methanol were homogenized in an ice water bath using a Fisher Scientific sonic dismembrator (Pittsburgh, PA) with an 8 sec on, 15 sec off cycle, 50% AMP until no tissue chunk remained. The homogenized samples were centrifuged at 20,000 xg at 4°C for 40 min, and the supernatant was collected and evaporated in a SpeedVac concentrator. Peptide samples were further dissolved in 100 μL 0.1% formic acid water and desalted by C18 ZipTip pipette tips following the manufacturer’s instructions. The desalted samples were then resuspended in 40 μL 0.1% formic acid in water. The peptide concentration of each reconstituted sample was measured with Thermo Scientific NanoDrop One microvolume spectrophotometer (Figure S1). The samples were then transferred into sample vials and 2 μL were injected into a mass spectrometer for analysis.
Mass Spectrometry Analysis
A Bruker timsTOF fleX mass spectrometer coupled with a Waters Acquity UPLC M-Class nano-LC system was used for LC-MS analysis. Samples were loaded on a homemade microcapillary column with an integrated emitter tip, packed with 15 cm of Bridged Ethylene Hybrid C18 particles (1.7 μm, 130 Å, Waters). The chromatographic separation was carried out via mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile). The flow rate was kept at 0.3 μL/min. The LC separation occurred during a 75-minute gradient elution with 2–13% mobile phase B for 42 min, 13–20% mobile phase B for 23 min, 20–30% mobile phase B for 5 min, 30–85% mobile phase B for 5 min. MS data were collected in a m/z range of 100–1700. Parallel accumulation serial fragmentation (PASEF) mode data collection was performed based on the following parameters: Ion mobility coefficient (1/K0) value was set from 0.6 to 1.6 cm2 V−1 s−1; data dependent mode was set as 10 MS2 PASEF scans. The collision cross section was calibrated with Agilent ESI-L Tuning Mix (m/z 622, 922, 1222).30
Database search and data analysis
The mass spectrometry raw data were searched by PEAKS Xpro studio (Bioinformatics Solutions Inc). The Homarus americanus database, which contains 24677 protein entries, was downloaded from the UniProt website on Oct 13th, 2022. The parameters for database searching include 10 ppm parent mass error tolerance, 0.02 Da fragment mass error tolerance, and an unspecific digest mode. The variable modifications include oxidation on the methionine, pyro-glu on the N-term glutamic acid and glutamine, and amidation on the C-termini. The identified peptides were filtered by these criteria: a false discovery rate (FDR) of less than 1%. Peptides were considered positively identified if they were detected in at least two of the three biological replicate experiments. Specifically, 10 peptides with C-terminal amidation followed by a non-glycine residue were also excluded from the identification list31, because amidation at the C-terminus of a neuropeptide typically requires the presence of a glycine residue. Signal peptides in the prohormone precursors were predicted with SignalP 6.0 tool.32 The cleavage sites in neuropeptide precursors were predicted through NeuroPred.33 The peptide intensity values were obtained from the PEAKS X Pro studio database search results and reported as the values in the ‘Area’ column of Supplementary Table 1 (Table S1), which is a good indicator of peptide abundance. To determine the cumulative abundance of peptides across different neural tissue regions, we averaged the peak area values for each peptide and summed these averages for each region. The data were analyzed using Microsoft Excel and in-house-built Python scripts.
Results
Mass spectrometry-based peptidome features
To globally profile the peptidergic signaling system in the American lobster, we dissected and collected five peptide-containing neural tissues (Figure 1A) including supraesophageal ganglion (brain), sinus gland (SG), thoracic ganglion (TG), oesophageal ganglion (OG) and stomatogastric ganglion (STG). The central nervous system (CNS) of the American lobster is similar to that of all other arthropods, starting with the brain and further connected by a longitudinal ventral nerve cord. In decapods, numerous behavioral responses are orchestrated through the neural pathways situated within the ganglia of the CNS.2 For example, the thoracic ganglion, a central component of the ventral nerve cord, is crucial in mediating motor functions and reflexes.4 Furthermore, the esophageal ganglion (OG) and the stomatogastric ganglion (STG) reside within the stomatogastric nervous system (STNS), constituting a specialized subsystem derived from the CNS.3 In addition to the regions of direct synaptic communication, the crustacean nervous system also contains neuroendocrine organs, such as the sinus glands located in the eyestalk, which can directly release the hormone into the hemolymph, a fluid in the circulatory system.34 The neuropeptides and peptide hormones released from the neuropil of ganglia or endocrine sites can act as strong neuromodulators of different physiological patterns and behavior.
Figure 1.
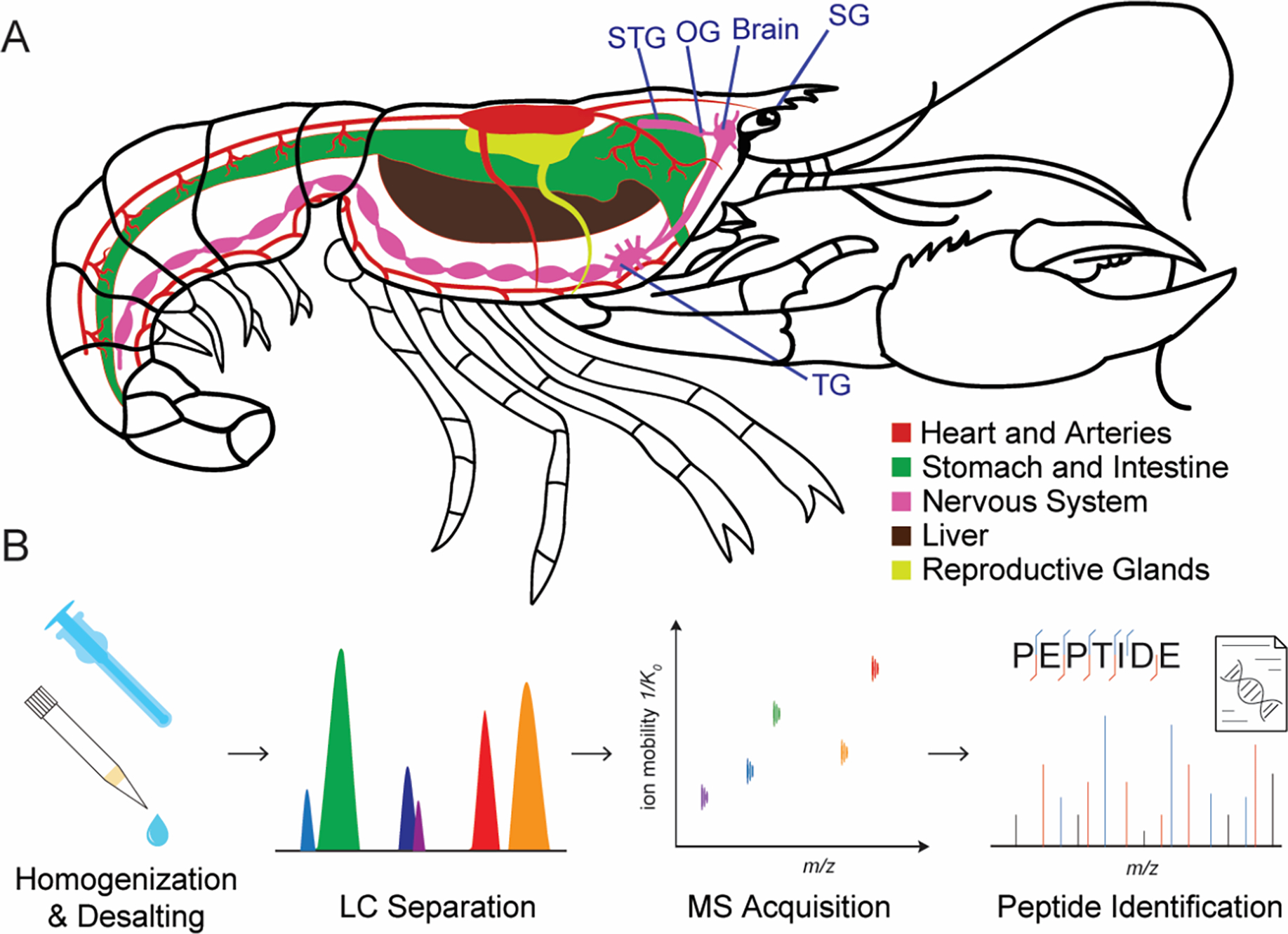
(A) The anatomy of an American lobster showing the location of the nervous system including sinus gland (SG), brain, oesophageal ganglion (OG) and stomatogastric ganglion (STG), and thoracic ganglion (TG). (B) Overall workflow for in-depth mass spectrometry analysis of lobster peptidomes.
The analysis of the endogenous peptidome inherently presents difficulties due to the low in vivo concentrations and extensive dynamic range of peptide abundance. To deal with these challenges, we devised a workflow (Figure 1B) specifically tailored to address these issues. Upon dissection, the tissues were immediately immersed in acidified methanol to deactivate the proteases and peptidases to minimize the peptide degradation. After homogenization, the desalted samples were injected into the timsTOF fleX mass spectrometer (Bruker, MA) with parallel accumulation serial fragmentation (PASEF)35 with data dependent acquisition (DDA) to improve the detection sensitivity of peptides while obtaining the ion mobility information at the same time. A summary of all the peptides including neuropeptides, peptide hormones and other endogenous peptides identified in different tissues is provided in Table S1.
Our workflow was benchmarked across different nervous tissue regions. This involved searching the mass spectrometry raw data against a lobster proteome database, which contains 24,677 protein entries. This comprehensive search resulted in the identification of 4815 endogenous modification-specific peptide sequences in total, with 3594, 1750, 484, 378, 175 peptides identified in TG, brain, SG, STG, and OG, respectively (Figure 2A). The number of peptides identified in each tissue reflects the quantity of endogenous peptides present, which corresponds to the varied peptide concentrations measured from extracted and reconstituted peptides from different tissues following an identical sample preparation procedure (Figure S1). The TG, being the largest of these tissues, not surprisingly had both the largest peptide quantity and the highest number of identified peptides, with the range of peptide MS intensity spanning four orders of magnitude. Meanwhile, OG and STG are small tissues and only contain 14 and 30 neurons, respectively. Even for those tiny tissue sizes and therefore smaller sample amount, we can still identify hundreds of peptides. Our results also show that, even for the same peptide, differential expression dynamics can be observed in different tissues. For example, Val1-SIFamide, a well-established crustacean neuropeptide with cardiac and pyloric rhythm modulation functions36, 37, was detected with the highest MS intensity among endogenous peptides in the brain region. However, this peptide only exhibited medium intensity in the SG tissue. In addition, we plotted the cumulative abundances of peptides in five regions (Figure 2B, Figure S2). For example, there were 5 peptides that contributed to the first 25% of the total accumulative intensity in the brain regions. Notably, 3 out of those 5 neuropeptides have been well studied in the crustacean nervous system with the name of Val1-SIFamdie, His13-orcokinin and CrabTRP1a. This observation underscores the difficulties encountered in the analysis of signaling peptides in crustacean, attributed to the wide dynamic range of peptide abundances.
Figure 2.

(A) Dynamic range of lobster endogenous peptide intensities across five tissue regions. The identified bioactive neuropeptides were annotated with different symbols. (B) Cumulative abundance of peptides in the lobster brain from the highest to the lowest abundance. (C) Logo plot displaying the motifs present in the N-termini and C-termini of the top 75% of endogenous peptides (by accumulative intensity).
Regarding the generation process of endogenous peptides, the protease cleavage preferences are an important indicator. Through the linear sequence analysis of the top 75% intensity peptides (Figure 2C), we can find the most prevalent amino acid located near the N-term −1 position is the basic residue arginine (R). We can also observe the enrichment of dibasic residue lysine and arginine or monobasic residue lysine at the N terminus. The C-term +1 glycine followed by dibasic residue is also evident. The full bioactivity of the signaling peptide also requires the C-term amidation as the mature process with the help of the peptidyl-glycine α-amidating monooxygenase (PAM) to convert the C-terminal glycine to the mature des-glycine C-term amidated peptide. Those motif observations are aligned with the processing mechanism of the neuropeptide or peptide hormone generation and previous reports.9, 38 We also investigated the peptide lengths of the peptides across the 5 regions. Most of the peptides consist of 11 to 19 amino acid residues (Figure S3).
In our further investigation of the tissue distribution of these peptides, as depicted in Figure 3A, we observe that 74 peptides are ubiquitously expressed across all five examined tissue regions. Notably, a significant proportion, representing 78.0% of the total peptide count, exhibits exclusive expression in one specific tissue region (Figure S4). Additionally, there is a noticeable correlation between the number of uniquely expressed peptides and the total peptide count identified in each respective region. This pronounced prevalence of tissue-specific expression patterns underscores the importance of performing analyses tailored to individual tissue types.
Figure 3.

(A) Venn diagram overlaying the peptides identified in the five different lobster tissues. (B-F) The pie chart in the lower left illustrating the cumulative abundance of prohormone-derived and non-prohormone-derived peptides across the tissues, whereas the pie chart on the right delineating the cumulative abundance of peptides among different neuropeptide families within the prohormone category.
In our study, we identified a total of 4,815 endogenous peptides, originating from 705 protein precursors. Regarding the American lobster, prior research predicted 52 genes encoding secretory signaling peptides,28, 39 based on their homology to established prohormones within arthropod species. To ensure comprehensive coverage and avoid overlooking potential neuropeptide precursors not included in the initial list of 52 genes, we conducted a meticulous manual inspection of all 705 identified protein precursors. This inspection was guided by specific criteria: the names predicted by ProtNLM for these precursors should exhibit similarity to known neuropeptide names; the precursors should exhibit hallmark prohormone features, including the presence of signal peptides, conserved repetitive segments, and paired basic residue cleavage sites. Through this rigorous examination, we were able to validate 24 neuropeptide precursors out of the 705 protein precursors identified. In this study, we adopt a broad definition of a neuropeptide family, categorizing neuropeptides cleaved from the same protein precursor as members of the same family. The 24 neuropeptide families include Allatostatin-A (AST-A), Allatostatin-B (AST-B), Crustacean Hyperglycemic Hormone (CHH), Corazonin, Crustacean Cardioactive Peptide (CCAP), Diuretic Hormone (DH), Calcitonin-like, FLRFamide, GSEFLamide, Myosuppressin, Orcokinin, Pigment Dispersing Hormone (PDH), Pyrokinin, SIFamide, Tachykinin, Natalisin, RYamide, Gonadoliberin, Leucokinin, Prohormone-3, FMRFamide, Agatoxin-like peptide, Allatostatin-C (AST-C), Neuropeptide F, and Sulfakinin.
In addition to these well-known neuropeptide families, we have also identified proteins that play crucial roles in neuropeptide processing and synaptic communication, such as Neuroendocrine Protein 7B2, Peptidylglycine Monooxygenase, Cathepsin L, Carboxypeptidase B, Noggin, Synapsin, and Synaptobrevin. The identification of peptides originating from peptidases/proteases is attributed to the natural degradation process within neural tissue. Proteases and peptidases can be cleaved into smaller fragments by other proteases/peptidases. Due to the high abundance of these enzymes in neural tissue, their degradation products are detectable by the highly sensitive mass spectrometry techniques utilized in our study. This comprehensive identification of neuropeptide families and related processing proteins expands our understanding of neurochemical complexity and diversity in crustaceans.
We further analyzed neuropeptide families by accumulating MS-based peptide intensities, distinguishing between peptides cleaved from prohormone precursors and others (Figure 3B–F). Accumulating peptide intensity data offers a more accurate assessment of peptide abundance than solely counting peptide identification numbers, despite variances in ionization efficiency among different peptide sequences. Given that the SG is the center for the storage and release of neurosecretory hormones in crustaceans,40 we found that the SG region has the most abundant peptide expression, with 95.6% signal intensity derived from the prohormone precursors. The ratio of prohormone-derived peptide intensity decreases in the order of Brain, STG, OG, and TG. Specifically for the prohormone portion, we observed that the orcokinin family is the most abundant neuropeptide family based on MS signal responses across all tissues. For CHH, it is highly abundant in the SG region, while it remains low in abundance in other tissues. Other high-abundance peptide families include SIFamide, Tachykinin, Allatostatin B, FLRFamide, and RYamide.
Leveraging the advancement of the ion mobility feature in trapped ion mobility instrumentation41, we also collected the ion mobility value of the prohormone-related peptides and plotted collision cross section (CCS) values as a function of m/z under different charge states as shown in Figure 4. We observed that peptides with a charge state of 2 show a tight correlation between CCS and m/z values. However, as the charge state increases, the relationship between CCS and m/z becomes more scattered. This dispersion could be attributed to peptides with higher charge states having greater mass. Consequently, these highly charged peptides exhibit a wider variety of amino acid sequences and more complex structures. CCS is a robust and reproducible physicochemical property of ions in the gas phase, demonstrating high consistency across different ion mobility platforms.30, 41 The measured CCS value of neuropeptides is beneficial for future neuropeptide library matching with ion mobility as an additional dimension. To the best of our knowledge, this is also the first time that ion mobility spectrometry has been utilized for the discovery and identification of crustacean neuropeptides. The trapped ion mobility spectrometry-based instrumentation not only enhances detection sensitivity but also provides another dimension for separation and the CCS value measurement.41
Figure 4.

The different charge states of the prohormone-derived peptides are plotted in the collision cross-section versus m/z space. The Pearson correlation coefficients (r) are denoted.
Mature neuropeptide distribution and the validation of predicted neuropeptides
Previously, many of the American lobster neuropeptides were predicted through the genome and transcriptome in silico mapping. However, the predicted results cannot replace experimental validation. In this study, based on mass spectrometry analysis, we not only validated the existence of the neuropeptides but also revealed their MS intensity-based abundance and tissue distribution. Based on the neuropeptide precursor identified and the mature neuropeptide cleavage site prediction tool, we have identified a total of 101 mature neuropeptides (Tabel 1, Figure S5) and revealed their tissue distribution. Among these, 67 neuropeptides have been detected for the first time. Notably, of these 67 novel peptides, 34 were both newly predicted and experimentally validated through mass spectrometry in this study, marking their first-ever observation and confirmation.
Tabel 1.
Neuropeptides detected in the peptidergic signaling system of the American lobster Homarus americanus.
| Family | Peptide Sequence | Mass | TG | Brain | SG | STG | OG |
|---|---|---|---|---|---|---|---|
|
| |||||||
| AST-A | GKR.HSNYGFGL(-.98).GKR | 892.42 | X | X | |||
| GKR.SVGDLPEVSKVEDGASPRT.KRD | 1941.96 | X | X | X | X | ||
| NKR.SKLYGFGL(-.98).GKR | 882.50 | X | X | ||||
| EKR.PRNYAFGL(-.98).GKR | 935.50 | X | |||||
| DKR.PRDYAFGL(-.98).GKR | 936.48 | X | |||||
| GKR.ASSDEDDEERYYAYEQ(-.98).GKR | 1967.77 | X | X | ||||
| GKR.AGRYAFGL(-.98).GKR | 852.46 | X | X | ||||
| GKK.AGHYAFGL(-.98).GKR | 833.42 | X | |||||
| GKR.SDSDSDQYTL(-.98).GRR | 1128.46 | X | |||||
|
| |||||||
| AST-B | VSS.SSSSPQQDDPASSPSHIEE.KRV | 1983.83 | X | X | X | X | X |
| EKR.VGWSSMHGTW(-.98).GKR | 1145.51 | X | X | X | X | X | |
| EKR.VGWSSM(+15.99)HGTW(-.98).GKR | 1161.50 | X | X | ||||
| GKR.PHLEDAQLDAAEV.KRT | 1406.67 | X | X | X | X | X | |
| VKR.TNWNKFQGSW(-.98).GKR | 1265.59 | X | X | X | X | X | |
| GKR.NNWRSLQGSW(-.98).GKR | 1245.60 | X | X | X | X | X | |
| GKR.AWNKLQGAW(-.98).GKR | 1071.56 | X | X | X | X | X | |
| SPR.STNWSSLRGTW(-.98).GKR | 1292.63 | X | X | X | |||
| GKR.SADWNKLRGAW(-.98).GKR | 1301.66 | X | X | X | X | X | |
| GKR.ASDWGQFRGSW(-.98).GKR | 1294.58 | X | X | X | |||
| GKR.APDMMSVAAPNQA | 1301.57 | X | X | X | X | ||
| GKR.APDM(+15.99)MSVAAPNQA | 1317.57 | X | |||||
|
| |||||||
| CHH-A | VGG.RSVEGASRMEKLLSSSNSPSSTPLGFLSQDHSVN.KRQ | 3603.76 | X | ||||
| VGG.RSVEGASRM(+15.99)EKLLSSSNSPSSTPLGFLSQDHSVN.KRQ | 3619.75 | X | X | ||||
|
| |||||||
| CHH-B | VGG.RSVEGVSRMEKLLSSISPSSTPLGFLSQDHSVN.KRQ | 3543.80 | X | X | |||
| VGG.RSVEGVSRM(+15.99)EKLLSSISPSSTPLGFLSQDHSVN.KRQ | 3559.79 | X | |||||
|
| |||||||
| Corazonin | AAA.Q(-17.03)TFQYSRGWTN(-.98).GRK | 1368.62 | X | ||||
|
| |||||||
| CCAP | AKR.DIGDLLEGKD.KRP | 1073.52 | X | X | X | X | |
| RKR.STPHTQPRQHLTSTPQQKVETEKQ | 2785.41 | X | X | ||||
|
| |||||||
| Diuretic Hormone | EAR.AVVQIEDPDYVLELLTRLGHSIIRANELEKFVRSSGSA.KRG | 4224.25 | X | ||||
| AKR.GLDLGLGRGFSGSQAAKHLMGLAAANFAGGP(-.98).GRR | 2939.52 | X | X | X | |||
| RRR.SSDDGLDLHHDDNLYAQDQAADLAESS.R | 2901.22 | X | X | X | |||
|
| |||||||
| FLRFamide | EKR.LLKYFLPASQAWGGDAYPIGQEGT.KRG | 2581.29 | X | X | |||
| TKR.GYSDRNYLRF(-.98).GRS | 1288.63 | X | X | X | X | ||
| FGR.SDTNDYEGEEMPESPE.KRN | 1827.66 | X | X | X | |||
| EKR.NRNFLRF(-.98).GRD | 964.54 | X | X | X | X | ||
| FGR.DQNRNFLRF(-.98).GRS | 1207.62 | X | X | X | X | ||
| FGR.SGSPMEFATDLQEDVELPVEE.KRG | 2321.03 | X | |||||
| EKR.GAHKNYLRF(-.98).GRG | 1103.60 | X | X | X | X | ||
| FGR.GNRNFLRF(-.98).GRG | 1021.56 | X | X | ||||
| FGR.GDRNFLRF(-.98).GRS | 1022.54 | X | X | X | X | ||
| AKR.FSHDRNFLRF(-.98).GKR | 1336.68 | X | X | X | X | ||
| GKR.DGSDDYPSSSSSAESPAPVVVVRPVEYPRYV.RAP | 3310.56 | X | |||||
| YVR.APSKNFLRF(-.98).G | 1077.61 | X | X | X | X | ||
|
| |||||||
| GSEFLamide | SAA.LPTHLPDELDDPVV.KRL | 1558.79 | X | X | |||
| VKR.LAGTPHESMIRYFLMAMSNPAGRYKSPQLLNRGV.RRI | 3804.94 | X | |||||
| GKR.QYEPEFAHTLDYDT.KRA | 1727.73 | X | X | ||||
|
| |||||||
| Myosuppressin | VKR.QDLDHVFLRF(-.98).GRS | 1287.67 | X | X | X | ||
| VKR.Q(-17.03)DLDHVFLRF(-.98).GRS | 1270.65 | X | X | X | X | X | |
|
| |||||||
| Orcokinin | AAA.GPIKVRFLSAIFIPIAAPARSSPQQDAAAGYTDGAPV.KRF | 3752.00 | X | ||||
| AAA.GPIKAAPARSSPQQDAAAGYTDGAPV.KRF | 2495.24 | X | X | X | X | X | |
| VKR.FDAFTTGFGHN.KRS | 1212.52 | X | X | X | X | X | |
| NKR.SSEDMDRLGFGFN.KRN | 1473.62 | X | X | X | X | X | |
| NKR.SSEDM(+15.99)DRLGFGFN.KRN | 1489.61 | X | X | X | X | ||
| NKR.NFDEIDRSGFGFH.KRN | 1539.67 | X | X | X | X | X | |
| HKR.NFDEIDRSGFGFN.KRN | 1516.66 | X | X | X | X | X | |
| HKR.GDYDVYPE.KRN | 956.38 | X | X | ||||
| EKR.NFDEIDRSGFGFV.KRV | 1501.68 | X | X | X | X | X | |
| VKR.VYGPRDIANLY.KRN | 1279.66 | X | X | X | X | X | |
|
| |||||||
| PDH | IQA.Q(-17.03)ELKYPEREVVADMAAQILRVALGPWGSVAAVPR.KRN | 3701.97 | X | ||||
| RKR.NSELINSLLGIPKVMNDA(-.98).GRR | 1926.02 | X | X | ||||
| RKR.NSELINSLLGIPKVM(+15.99)NDA(-.98).GRR | 1942.02 | X | X | ||||
| HKR.NSELINSILGLPKVMNDA(-.98).GRD | 1926.02 | X | X | ||||
| HKR.NSELINSILGLPKVM(+15.99)NDA(-.98).GRD | 1942.02 | X | X | ||||
| HKR.NSEILNTLLGSQDLSNMRSA(-.98).GRR | 2161.08 | X | |||||
|
| |||||||
| Pyrokinin | GKR.GDGFAFSPRL(-.98).GKR | 1064.54 | X | X | |||
| GKR.GADFAFSPRL(-.98).GRR | 1078.56 | X | |||||
| GRR.SEFVFSSRP(-.98).GKK | 1053.52 | X | |||||
| GKK.SDFAFSPRL(-.98).GKK | 1037.53 | X | X | ||||
|
| |||||||
| SIFamide | VSA.VYRKPPFNGSIF(-.98).GKR | 1422.78 | X | X | X | X | |
| AVY.RKPPFNGSIF(-.98).GKR | 1160.65 | X | X | ||||
|
| |||||||
| Tachykinin | ERR.APSGFLGMR(-.98).GKK | 933.49 | X | X | X | X | X |
| ERR.APSGFLGM(+15.99)R(-.98).GKK | 949.48 | X | X | X | X | ||
| GKK.SDEEVFSDATADNDLEILL.KRA | 2094.95 | X | |||||
| GKK.YYDDDSDMDAYIQALTAVVDGQQQQ.KRA | 2851.21 | X | |||||
| GKK.AYYSENPDEEISM(+15.99)TGVD.KRT | 1934.77 | X | X | X | |||
| GKK.AYYSENPDEEISMTGVD.KRT | 1918.78 | X | X | X | X | ||
| DKR.TPSGFLGMR(-.98).G | 963.50 | X | X | X | X | ||
| DKR.TPSGFLGM(+15.99)R(-.98).G | 979.49 | X | X | ||||
|
| |||||||
| Natalisin | GKR.PSSELLHQHHQ.KRS | 1311.63 | X | X | |||
| GKR.DGGGPFWIAR(-.98).GKR | 1073.54 | X | X | ||||
| GKR.Q(-17.03)ETEGNGGPFWIAR(-.98).GKK | 1542.72 | X | |||||
| GKR.QETEGNGGPFWIAR(-.98).GKK | 1559.75 | X | |||||
| HKR.Q(-17.03)DGGPFWISR(-.98).GKK | 1143.55 | X | |||||
| HKR.QDGGPFWISR(-.98).GKK | 1160.57 | X | |||||
| GRK.SEDERTFWVAR(-.98).GKK | 1393.67 | X | |||||
| GKK.ENKGNESELFWISR(-.98).GKR | 1706.84 | X | |||||
| GKR.EGEAPPFWVSR(-.98).GKK | 1272.63 | X | X | ||||
| GKK.EGEETHPFWVSR(-.98).GKK | 1471.68 | X | X | X | |||
| GKK.E(-18.01)GEETHPFWVSR(-.98).GKK | 1453.67 | X | |||||
| GKK.DTTYGPIDDPFVKGFLALR(-.98).G | 2123.11 | X | |||||
|
| |||||||
| RYamide | TAA.Q(-17.03)GFYTQRY(-.98).GKR | 1043.48 | X | X | X | X | |
| TVR.SGFYANRN(-.98).GRS | 926.44 | X | |||||
| NGR.SSPSQGLPEIKIRSSRFIGGSRY(-.98).GKR | 2520.36 | X | |||||
| KIR.SSRFIGGSRY(-.98).GKR | 1127.58 | X | X | ||||
|
| |||||||
| Gonadoliberin | ATS.Q(-17.03)IHWNRGWGAGGSM(+15.99)(-.98).GKR | 1553.69 | X | X | |||
| ATS.Q(-17.03)IHWNRGWGAGGSM(-.98).GKR | 1537.70 | X | X | ||||
|
| |||||||
| Leucokinin | GKR.SDPLLPASQHEPNT.KRN | 1504.72 | X | X | |||
| GKR.SPSMDLSGNQD.KRT | 1149.46 | X | |||||
| GKR.SSGDELDDHFLD.KKT | 1348.54 | X | X | ||||
| GKR.AEGTLSRLSESTLKAALDENSPEDNVDNH.KRP | 3111.46 | X | |||||
| GKR.ASPISEDSQLSDLYTSQL | 1952.92 | X | X | ||||
TG, thoracic ganglion; SG, sinus gland; STG, stomatogastric ganglion; OG, oesophageal ganglion; AST, allatostatin; CHH, crustacean hyperglycemic hormone; CCAP, crustacean cardioactive peptide; PDH, pigment dispersing hormone.
(−.98) indicates amidated C-terminal; (−17.03) and (−18.01) indicate the N-terminal pyroglutamate formation; (+15.99) indicates the oxidation on methionine.
Previously known H. americanus peptides are shown in normal font; peptides previously predicted in literature39 but detected by MS for the first time are shown in bold; peptides that have not been predicted before but detected for the first time are shown in bold and italics.
For example, Natalisin was initially recognized as a factor altering mating behavior in fruit flies, Drosophila melanogaster.42 Later, the transcriptome of the crustaceans, including the crayfish Procambarus clarkii and spiny lobster Panulirus argus, was also used to predict the existence of Natalisin.43, 44 However, the actual existence of the mature neuropeptide resulting from proteolytic cleavage from Natalisin is still unknown. By referring to the American lobster genome entries, we can obtain the full sequence of the putative Natalisin precursor. The signaling peptide and the double basic residue for the peptidase cleavage can further be recognized, as shown in Figure 5A. In total, 12 peptides with the classical cleavage pattern have been identified as belonging to the Natalisin precursor. It can be observed that the majority of peptide sequences exhibit conservation of the FWxxRamide motif at their C-termini (Figure 5B) with the exception of the two specific peptides PSSELLHQHHQ and DTTYGPIDDPFVKGFLALRamide. Among those Natalisin neuropeptides, the peptide EGEETHPFWVSRamide has the highest MS intensity, and the fragment ions of this peptide match the amino acid sequence, which also demonstrates the accuracy of the mass spectrometry identification (Figure 5C). Notably, Natalisin peptides were predominantly detected in brain samples, with only a few being present in the TG and SG regions. This distribution pattern underscores the need for further research to understand the biological functions of Natalisin neuropeptides in the crustacean nervous system. Additionally, the study opens avenues for the functional characterization of any neuropeptides newly discovered in this work.
Figure 5.

(A) The putative H. americanus Natalisin sequence, marked with the identified peptides (red single-letter code amino acid sequence and blue bar underneath the sequence). Dibasic motifs are marked by bold. The putative signal peptide at the N terminus is marked in italics and underlined. (B) Sequence alignment showcasing the consensus sequence among seven Natalisin-derived peptides, with identical amino acids within aligned sequences highlighted on a black background. (C) The MS/MS spectrum of a 2+ charged peptide ions identified as EGEETHPFWVSRamdie from the neuropeptide precursor Natalisin.
To elucidate the potential biological functions of uncharacterized peptides, we refined the data by considering the double basic residue cleavage motif and C-terminal amidation, a well-known post-translational modification in bioactive neuropeptides.45 A list of potential biologically active peptides, in addition to the previously listed neuropeptides, was created (Supplementary Table S2), using criteria that included having K/R at position −1 before the N-terminus and K/R at positions +1 and +2 following the C-terminus, or C-terminal amidation preceded by G at position +1. Intriguingly, this list includes some notable protein precursors, such as histone and peptidylglycine monooxygenase. For instance, certain antimicrobial peptides in crustaceans are known to be derived from histone.46–48 Peptidylglycine monooxygenase is also implicated in the processing of bioactive peptides. This curated list could serve as a valuable resource for future bioactive peptide discovery.
Discussion
Even though high-throughput nucleotide sequencing has provided valuable genetic insights into lobster neuropeptidergic signaling in previous studies39, 44, 49, mass spectrometry remains essential for validating these predictions, revealing novel enzymatic processing, identifying post-translational modifications, and discerning the peptides’ relative abundances in various neural tissues. Earlier research demonstrated the potential of using mass spectrometry for profiling Homarus americanus neuropeptides. However, these studies typically depended on either an in-house-constructed database with a limited range of peptide/protein sequences for database searches23–27 or focused exclusively on the targeted characterization of specific peptide families through cDNA cloning50–52. Consequently, the prior identification of neuropeptides in lobsters was notably constrained and limited in both numbers and families. The American lobster genome database used in this research stands out as the highest-quality and best-annotated database for crustacean species available. Thanks to this advanced genomic resource, we can now match peptide sequences with a level of confidence that was unattainable with earlier de novo sequencing efforts or when matching with less developed in-house databases. In this research, we showcased the most extensive neuropeptidome database within the crustacean category. We have identified 24 neuropeptide precursors and 101 unique mature neuropeptides, with 67 being newly discovered, underscoring the extensive nature of this neuropeptidome database. In addition, we have explored the neuropeptidome’s characteristics and examined the variations in neuropeptide distribution across different neural tissues, revealing tissue-specific expression patterns for many of these neuropeptide families.
However, certain limitations were encountered. During the study, some neuropeptide families could be identified, but their mature neuropeptides in this family were not detected. For example, for the crustacean hyperglycemic hormone precursor, we have successfully identified the crustacean hyperglycemic hormone precursor-related peptides; however, the mature form of the CHH (CHH-A and CHH-B), which are larger hormones with molecular weights over 8 kDa, could not be detected. This is due to the large size of the hormone and their intramolecular disulfide bonds which hinder collision-based dissociation. Discovery failures attributed to large size and the presence of intramolecular disulfide bonds may also extend to other neuropeptide families such as the agatoxin-like peptide, Bursicon, Neuroparsin, and Trissin. To address this problem, future experiments including the reduction and alkylation of the disulfide bonds and the enzymatic digestion of large neuropeptides into smaller peptide fragments will be explored for mass spectrometry analysis. An alternative way is to use the top-down method to directly analyze the large neuropeptides.53
Some neuropeptides, such as sulfakinin, may contain unique post-translational modifications such as sulfation on the tyrosine residue. In this family, we can detect the EFDEYGHMRFamide, but in predictions, the tyrosine residue of this peptide should carry the sulfation group, thus, we did not list this peptide in the identified peptide list. The labile sulfation group on the tyrosine is unstable during collision-induced dissociation. To overcome this problem, an alternative electron-based fragmentation strategy would facilitate the identification of this neuropeptide and localization of the sulfation site successfully.
Recently, glycosylated neuropeptides have garnered increasing attention and several studies have reported the glycosylation of crustacean neuropeptides.54–56 However, these glycosylated neuropeptides are often found in lower abundance compared to their non-glycosylated counterparts, making their detection and analysis challenging. Enrichment strategies are crucial for the effective discovery of glycosylated neuropeptides. In our research, we did not employ enrichment techniques for glycosylated neuropeptides, nor did we utilize customized MS methods tailored for their discovery. The glycosylated neuropeptides in lobster, therefore, warrant further investigation to elucidate their presence and functional roles. Overall, the data reported here provides a valuable resource for the study of the neuropeptidergic signaling system in the genus Homarus and crustaceans in general. The combination of a sequenced genome and the use of more advanced trapped ion mobility TOF MS/MS based platform substantially expanded the lobster neuropeptidome. The findings reported in this work will help advance our understanding of the American lobster’s neurochemistry and establish a foundation for future functional study of these neuropeptides in this important model organism.
Supplementary Material
Figure S1. The concentration of endogenous peptides extracted and reconstituted from lobster neural tissues.
Figure S2. The cumulative abundance of peptides in lobster SG, OG, STG and TG.
Figure S3. Analysis of identified peptide length distribution across five tissue regions.
Figure S4. The histogram quantifies the number of peptide segments occurring in various regions.
Figure S5. MS/MS spectra of neuropeptide sequences detected in the peptidergic signaling system of the American lobster Homarus americanus generated by PEAKS software.
Table S1. The endogenous peptide identification results across five lobster tissue regions.
Table S2. List of potential new bioactive peptides.
Acknowledgments
The study was supported in part by a National Science Foundation grant CHE-2108223 and National Institutes of Health grants R01DK071801 and R01NS029436. Some of the mass spectrometers were acquired using NIH shared instrument grants S10 OD028473, S10 RR029531, and S10 OD025084. L.L. wishes to acknowledge additional funding support from NIH grants R01AG078794 and R01AG052324, and the Vilas Distinguished Achievement Professorship, the Charles Melbourne Johnson Professorship, the Wisconsin Alumni Research Foundation, and the University of Wisconsin-Madison School of Pharmacy.
Footnotes
The authors declare no competing financial interest.
Data Availability
The mass spectrometry peptidomics data have been deposited to the ProteomeXchange Consortium via the PRIDE57 partner repository with the dataset identifier PXD047230.
References
- 1.Le Bris A; Mills KE; Wahle RA; Chen Y; Alexander MA; Allyn AJ; Schuetz JG; Scott JD; Pershing AJ, Climate vulnerability and resilience in the most valuable North American fishery. Proceedings of the National Academy of Sciences 2018, 115 (8), 1831–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Factor JR, Biology of the Lobster: Homarus americanus. Academic Press: 1995. [Google Scholar]
- 3.Marder E; Bucher D, Understanding circuit dynamics using the stomatogastric nervous system of lobsters and crabs. Annu Rev Physiol 2007, 69, 291–316. [DOI] [PubMed] [Google Scholar]
- 4.Hooper SL; DiCaprio RA, Crustacean motor pattern generator networks. Neurosignals 2004, 13 (1–2), 50–69. [DOI] [PubMed] [Google Scholar]
- 5.Blitz DM; Nusbaum MP, Neural circuit flexibility in a small sensorimotor system. Curr Opin Neurobiol 2011, 21 (4), 544–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hallberg M; Nyberg F, Neuropeptide conversion to bioactive fragments-an important pathway in neuromodulation. Current Protein and Peptide Science 2003, 4 (1), 31–44. [DOI] [PubMed] [Google Scholar]
- 7.Van Den Pol AN, Neuropeptide transmission in brain circuits. Neuron 2012, 76 (1), 98–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hökfelt T; Broberger C; Xu Z-QD; Sergeyev V; Ubink R; Diez M, Neuropeptides—an overview. Neuropharmacology 2000, 39 (8), 1337–1356. [DOI] [PubMed] [Google Scholar]
- 9.Hook V; Funkelstein L; Lu D; Bark S; Wegrzyn J; Hwang S-R, Proteases for processing proneuropeptides into peptide neurotransmitters and hormones. Annu. Rev. Pharmacol. Toxicol. 2008, 48, 393–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eggelkraut-Gottanka R. v.; Beck-Sickinger AG, Biosynthesis of peptide hormones derived from precursor sequences. Current medicinal chemistry 2004, 11 (20), 2651–2665. [DOI] [PubMed] [Google Scholar]
- 11.Secher A; Kelstrup CD; Conde-Frieboes KW; Pyke C; Raun K; Wulff BS; Olsen JV, Analytic framework for peptidomics applied to large-scale neuropeptide identification. Nat Commun 2016, 7, 11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Checco JW, Identifying and Measuring Endogenous Peptides through Peptidomics. ACS Chem Neurosci 2023, 14 (20), 3728–3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phetsanthad A; Vu NQ; Yu Q; Buchberger AR; Chen Z; Keller C; Li L, Recent advances in mass spectrometry analysis of neuropeptides. Mass Spectrometry Reviews 2023, 42 (2), 706–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vu NQ; DeLaney K; Li L, Neuropeptidomics: improvements in mass spectrometry imaging analysis and recent advancements. Current Protein and Peptide Science 2021, 22 (2), 158–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hellinger R; Sigurdsson A; Wu W; Romanova EV; Li L; Sweedler JV; Süssmuth RD; Gruber CW, Peptidomics. Nature Reviews Methods Primers 2023, 3 (1), 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falth M; Skold K; Svensson M; Nilsson A; Fenyo D; Andren PE, Neuropeptidomics strategies for specific and sensitive identification of endogenous peptides. Molecular & Cellular Proteomics 2007, 6 (7), 1188–1197. [DOI] [PubMed] [Google Scholar]
- 17.Hook V; Bandeira N, Neuropeptidomics mass spectrometry reveals signaling networks generated by distinct protease pathways in human systems. Journal of the American Society for Mass Spectrometry 2015, 26 (12), 1970–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svensson M; Sköld K; Svenningsson P; Andren PE, Peptidomics-based discovery of novel neuropeptides. Journal of proteome research 2003, 2 (2), 213–219. [DOI] [PubMed] [Google Scholar]
- 19.Petruzziello F; Fouillen L; Wadensten H; Kretz R; Andren PE; Rainer G; Zhang X, Extensive characterization of Tupaia belangeri neuropeptidome using an integrated mass spectrometric approach. Journal of Proteome Research 2012, 11 (2), 886–896. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X; Rauch A; Xiao H; Rainer G; Logothetis NK, Mass spectrometry-based neurochemical analysis: perspectives for primate research. Expert review of proteomics 2008, 5 (5), 641–652. [DOI] [PubMed] [Google Scholar]
- 21.Ye H; Wang J; Tian Z; Ma F; Dowell J; Bremer Q; Lu G; Baldo B; Li L, Quantitative mass spectrometry reveals food intake-induced neuropeptide level changes in rat brain: functional assessment of selected neuropeptides as feeding regulators. Molecular & Cellular Proteomics 2017, 16 (11), 1922–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu W; Ma M; Ibarra AE; Lu G; Bakshi VP; Li L, Global Neuropeptidome Profiling in Response to Predator Stress in Rat: Implications for Post-Traumatic Stress Disorder. Journal of the American Society for Mass Spectrometry 2023, 34 (8), 1549–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen R; Jiang X; Prieto Conaway MC; Mohtashemi I; Hui L; Viner R; Li L, Mass spectral analysis of neuropeptide expression and distribution in the nervous system of the lobster Homarus americanus. Journal of proteome research 2010, 9 (2), 818–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cape SS; Rehm KJ; Ma M; Marder E; Li L, Mass spectral comparison of the neuropeptide complement of the stomatogastric ganglion and brain in the adult and embryonic lobster, Homarus americanus. Journal of neurochemistry 2008, 105 (3), 690–702. [DOI] [PubMed] [Google Scholar]
- 25.Ma M; Chen R; Sousa GL; Bors EK; Kwiatkowski MA; Goiney CC; Goy MF; Christie AE; Li L, Mass spectral characterization of peptide transmitters/hormones in the nervous system and neuroendocrine organs of the American lobster Homarus americanus. General and comparative endocrinology 2008, 156 (2), 395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang X; Chen R; Wang J; Metzler A; Tlusty M; Li L, Mass spectral charting of neuropeptidomic expression in the stomatogastric ganglion at multiple developmental stages of the lobster Homarus americanus. ACS Chemical Neuroscience 2012, 3 (6), 439–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Q; Goy MF; Li L, Identification of neuropeptides from the decapod crustacean sinus glands using nanoscale liquid chromatography tandem mass spectrometry. Biochemical and biophysical research communications 2005, 337 (3), 765–778. [DOI] [PubMed] [Google Scholar]
- 28.Polinski JM; Zimin AV; Clark KF; Kohn AB; Sadowski N; Timp W; Ptitsyn A; Khanna P; Romanova DY; Williams P, The American lobster genome reveals insights on longevity, neural, and immune adaptations. Science Advances 2021, 7 (26), eabe8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gane A; Bileschi ML; Dohan D; Speretta E; Héliou A; Meng-Papaxanthos L; Zellner H; Brevdo E; Parikh A; Martin MJ, ProtNLM: model-based natural language protein annotation. Preprint at https://storage.googleapis.com/brain-genomics-public/research/proteins/protnlm/uniprot_2022_04/protnlm_preprint_draft.Pdf 2022.
- 30.Stow SM; Causon TJ; Zheng X; Kurulugama RT; Mairinger T; May JC; Rennie EE; Baker ES; Smith RD; McLean JA, An interlaboratory evaluation of drift tube ion mobility–mass spectrometry collision cross section measurements. Analytical chemistry 2017, 89 (17), 9048–9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anapindi KDB; Romanova EV; Southey BR; Sweedler JV, Peptide identifications and false discovery rates using different mass spectrometry platforms. Talanta 2018, 182, 456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teufel F; Almagro Armenteros JJ; Johansen AR; Gíslason MH; Pihl SI; Tsirigos KD; Winther O; Brunak S; von Heijne G; Nielsen H, SignalP 6.0 predicts all five types of signal peptides using protein language models. Nature biotechnology 2022, 40 (7), 1023–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Southey BR; Amare A; Zimmerman TA; Rodriguez-Zas SL; Sweedler JV, NeuroPred: a tool to predict cleavage sites in neuropeptide precursors and provide the masses of the resulting peptides. Nucleic acids research 2006, 34 (suppl_2), W267–W272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knowles SFG; Carlisle DB, Endocrine control in the Crustacea. Biological Reviews 1956, 31 (4), 396–467. [Google Scholar]
- 35.Meier F; Beck S; Grassl N; Lubeck M; Park MA; Raether O; Mann M, Parallel accumulation–serial fragmentation (PASEF): multiplying sequencing speed and sensitivity by synchronized scans in a trapped ion mobility device. Journal of proteome research 2015, 14 (12), 5378–5387. [DOI] [PubMed] [Google Scholar]
- 36.Dickinson PS; Samuel HM; Stemmler EA; Christie AE, SIFamide peptides modulate cardiac activity differently in two species of Cancer crab. Gen Comp Endocrinol 2019, 282, 113204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christie AE; Stemmler EA; Peguero B; Messinger DI; Provencher HL; Scheerlinck P; Hsu YW; Guiney ME; de la Iglesia HO; Dickinson PS, Identification, physiological actions, and distribution of VYRKPPFNGSIFamide (Val1)-SIFamide) in the stomatogastric nervous system of the American lobster Homarus americanus. The Journal of comparative neurology 2006, 496 (3), 406–21. [DOI] [PubMed] [Google Scholar]
- 38.Prigge ST; Mains RE; Eipper BA; Amzel*** LM, New insights into copper monooxygenases and peptide amidation: structure, mechanism and function. Cellular and Molecular Life Sciences CMLS 2000, 57, 1236–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christie AE; Chi M; Lameyer TJ; Pascual MG; Shea DN; Stanhope ME; Schulz DJ; Dickinson PS, Neuropeptidergic Signaling in the American Lobster Homarus americanus: New Insights from High-Throughput Nucleotide Sequencing. PLoS One 2015, 10 (12), e0145964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown FA Jr, The crustacean sinus gland and chromatophore activation. Physiological Zoology 1940, 13 (3), 343–355. [Google Scholar]
- 41.Delafield DG; Lu G; Kaminsky CJ; Li L, High-end ion mobility mass spectrometry: A current review of analytical capacity in omics applications and structural investigations. TrAC Trends in Analytical Chemistry 2022, 116761. [Google Scholar]
- 42.Jiang H; Lkhagva A; Daubnerova I; Chae HS; Simo L; Jung SH; Yoon YK; Lee NR; Seong JY; Zitnan D; Park Y; Kim YJ, Natalisin, a tachykinin-like signaling system, regulates sexual activity and fecundity in insects. Proc Natl Acad Sci U S A 2013, 110 (37), E3526–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veenstra JA, The power of next-generation sequencing as illustrated by the neuropeptidome of the crayfish Procambarus clarkii. Gen Comp Endocrinol 2015, 224, 84–95. [DOI] [PubMed] [Google Scholar]
- 44.Christie AE, Identification of putative neuropeptidergic signaling systems in the spiny lobster, Panulirus argus. Invert Neurosci 2020, 20 (1), 2. [DOI] [PubMed] [Google Scholar]
- 45.Tatemoto K; Carlquist M; Mutt V, Neuropeptide Y--a novel brain peptide with structural similarities to peptide YY and pancreatic polypeptide. Nature 1982, 296 (5858), 659–60. [DOI] [PubMed] [Google Scholar]
- 46.Sruthy KS; Nair A; Antony SP; Puthumana J; Singh ISB; Philip R, A histone H2A derived antimicrobial peptide, Fi-Histin from the Indian White shrimp, Fenneropenaeus indicus: Molecular and functional characterization. Fish & shellfish immunology 2019, 92, 667–679. [DOI] [PubMed] [Google Scholar]
- 47.Patat SA; Carnegie RB; Kingsbury C; Gross PS; Chapman R; Schey KL, Antimicrobial activity of histones from hemocytes of the Pacific white shrimp. European journal of biochemistry 2004, 271 (23–24), 4825–33. [DOI] [PubMed] [Google Scholar]
- 48.Chen B; Fan DQ; Zhu KX; Shan ZG; Chen FY; Hou L; Cai L; Wang KJ, Mechanism study on a new antimicrobial peptide Sphistin derived from the N-terminus of crab histone H2A identified in haemolymphs of Scylla paramamosain. Fish & shellfish immunology 2015, 47 (2), 833–46. [DOI] [PubMed] [Google Scholar]
- 49.Christie AE; Roncalli V; Cieslak MC; Pascual MG; Yu A; Lameyer TJ; Stanhope ME; Dickinson PS, Prediction of a neuropeptidome for the eyestalk ganglia of the lobster Homarus americanus using a tissue-specific de novo assembled transcriptome. General and comparative endocrinology 2017, 243, 96–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dickinson PS; Stemmler EA; Barton EE; Cashman CR; Gardner NP; Rus S; Brennan HR; McClintock TS; Christie AE, Molecular, mass spectral, and physiological analyses of orcokinins and orcokinin precursor-related peptides in the lobster Homarus americanus and the crayfish Procambarus clarkii. Peptides 2009, 30 (2), 297–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stevens JS; Cashman CR; Smith CM; Beale KM; Towle DW; Christie AE; Dickinson PS, The peptide hormone pQDLDHVFLRFamide (crustacean myosuppressin) modulates the Homarus americanus cardiac neuromuscular system at multiple sites. Journal of Experimental Biology 2009, 212 (24), 3961–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Christie AE; Cashman CR; Stevens JS; Smith CM; Beale KM; Stemmler EA; Greenwood SJ; Towle DW; Dickinson PS, Identification and cardiotropic actions of brain/gut-derived tachykinin-related peptides (TRPs) from the American lobster Homarus americanus. Peptides 2008, 29 (11), 1909–1918. [DOI] [PubMed] [Google Scholar]
- 53.Jia C; Hui L; Cao W; Lietz CB; Jiang X; Chen R; Catherman AD; Thomas PM; Ge Y; Kelleher NL; Li L, High-definition de novo sequencing of crustacean hyperglycemic hormone (CHH)-family neuropeptides. Mol Cell Proteomics 2012, 11 (12), 1951–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Polt R; Dhanasekaran M; Keyari CM, Glycosylated neuropeptides: a new vista for neuropsychopharmacology? Medicinal research reviews 2005, 25 (5), 557–585. [DOI] [PubMed] [Google Scholar]
- 55.Phetsanthad A; Roycroft C; Li L, Enrichment and fragmentation approaches for enhanced detection and characterization of endogenous glycosylated neuropeptides. Proteomics 2023, 23 (3–4), 2100375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao Q; Yu Q; Liu Y; Chen Z; Li L, Signature-ion-triggered mass spectrometry approach enabled discovery of N-and O-linked glycosylated neuropeptides in the crustacean nervous system. Journal of proteome research 2019, 19 (2), 634–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perez-Riverol Y; Bai J; Bandla C; García-Seisdedos D; Hewapathirana S; Kamatchinathan S; Kundu DJ; Prakash A; Frericks-Zipper A; Eisenacher M; Walzer M; Wang S; Brazma A; Vizcaíno JA, The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic acids research 2022, 50 (D1), D543–d552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The concentration of endogenous peptides extracted and reconstituted from lobster neural tissues.
Figure S2. The cumulative abundance of peptides in lobster SG, OG, STG and TG.
Figure S3. Analysis of identified peptide length distribution across five tissue regions.
Figure S4. The histogram quantifies the number of peptide segments occurring in various regions.
Figure S5. MS/MS spectra of neuropeptide sequences detected in the peptidergic signaling system of the American lobster Homarus americanus generated by PEAKS software.
Table S1. The endogenous peptide identification results across five lobster tissue regions.
Table S2. List of potential new bioactive peptides.
Data Availability Statement
The mass spectrometry peptidomics data have been deposited to the ProteomeXchange Consortium via the PRIDE57 partner repository with the dataset identifier PXD047230.


