Abstract
We describe here a role for quorum sensing in the detachment, or sloughing, of Serratia marcescens filamentous biofilms, and we show that nutrient conditions affect the biofilm morphotype. Under reduced carbon or nitrogen conditions, S. marcescens formed a classical biofilm consisting of microcolonies. The filamentous biofilm could be converted to a microcolony-type biofilm by switching the medium after establishment of the biofilm. Similarly, when initially grown as a microcolony biofilm, S. marcescens could be converted back to a filamentous biofilm by increasing the nutrient composition. Under high-nutrient conditions, an N-acyl homoserine lactone quorum-sensing mutant formed biofilms that were indistinguishable from the wild-type biofilms. Similarly, other quorum-sensing-dependent behaviors, such as swarming motility, could be rendered quorum sensing independent by manipulating the growth medium. Quorum sensing was also found to be involved in the sloughing of the filamentous biofilm. The biofilm formed by the bacterium consistently sloughed from the substratum after approximately 75 to 80 h of development. The quorum-sensing mutant, when supplemented with exogenous signal, formed a wild-type filamentous biofilm and sloughed at the same time as the wild type, and this was independent of surfactant production. When we removed the signal from the quorum-sensing mutant prior to the time of sloughing, the biofilm did not undergo significant detachment. Together, the data suggest that biofilm formation by S. marcescens is a dynamic process that is controlled by both nutrient cues and the quorum-sensing system.
Biofilms are increasingly recognized as the predominant form of growth in the environmental life cycle of bacteria. They are high-density, matrix-encased populations attached to surfaces. Biofilms are more resistant to environmental stresses (e.g., nutritional and oxidative stress) and to host-mediated responses (e.g., complement proteins and phagocytes) (17, 23). Due to the increased resistance, biofilms can cause problems in industrial and medical settings. Therefore, it is increasingly important to understand how biofilms form and detach, including the internal and external factors that control these processes.
Two biofilm morphologies are typically reported, a flat, undifferentiated biofilm and the classic, differentiated biofilm described for Pseudomonas aeruginosa, consisting of towers and mushroom shapes with channels and voids which presumably serve to deliver nutrients to, and remove waste products from, the cells (10, 36). Factors such as alginate expression, rhamnolipid production, and quorum sensing (QS) have been shown to be important for control of the three-dimensional architecture and biofilm stress resistance (9, 21).
Quorum sensing has been shown to play a role in the control of biofilm architecture. Quorum-sensing mutants of P. aeruginosa form flat, undifferentiated biofilms that adhere less tightly to the substratum than the wild-type biofilm (9). Similar findings have been reported for the control of biofilm formation in Burkholderia cepacia and Aeromonas hydrophila (25, 30). It has recently become apparent that nutrient cues also play a role in biofilm formation. For example, Christensen et al. (5) demonstrated that the architecture in a two-species biofilm depended on the substrate. It has also been shown that the role of quorum sensing in the control of biofilm formation is nutrient dependent (7, 11, 22). Thus, it appears that control of biofilm architecture is a multifaceted process involving a combination of nutrient components and master regulators, such as quorum sensing.
Serratia marcescens (Serratia liquefaciens) is emerging as an important opportunistic pathogen, particularly for immunocompromised patients and in ocular infections. During preparation of this paper, we performed 16S rRNA gene sequencing of the S. liquefaciens MG1 strain and determined that it is in fact a nonpigmented S. marcescens strain (27), and this organism is referred to here as S. marcescens. This organism has been shown to utilize quorum sensing to control swarming motility through the production of the surfactant serrawettin (15, 29). S. marcescens forms biofilms through a series of defined stages that culminate in a highly porous, filamentous biofilm composed of cell chains, filaments, and cell clusters (26), which is dependent on the quorum-sensing system. A signal synthase mutant forms a flat biofilm (26) lacking the characteristic cell chains and clusters and also attaches poorly to abiotic surfaces (Labbate, unpublished data). Thus, it appears that several aspects of the biofilm life cycle in S. marcescens, including attachment, swarming, and formation of the three-dimensional architecture, are quorum sensing regulated.
Results presented here indicate that the biofilm morphology is highly dependent on nutrient cues. The filamentous architecture was favored by high-nutrient conditions, while a classic microcolony biofilm was observed under low-nutrient conditions. It was also demonstrated that the role of quorum sensing in the formation of a filamentous biofilm could be overridden depending on the medium used. Furthermore, sloughing of the filamentous biofilm was determined to be quorum sensing controlled, thus indicating that quorum sensing is an important regulator for several stages of the biofilm life cycle of this organism. Based on these data, a model is presented for the biofilm life cycle of S. marcescens, focusing on the role of quorum sensing and nutrients in the control and maintenance of biofilm architecture.
MATERIALS AND METHODS
Bacterial strains, media, and growth.
The S. marcescens strains used in this study are listed in Table 1. The bacterial strains were routinely grown in Luria-Bertani (LB) medium (2) supplemented with 1.5% (wt/vol) agar or in minimal broth Davis (MBD) without dextrose (Difco Laboratories, Detroit, Mich.) supplemented with 0.2% (wt/vol) glucose and 0.5% (wt/vol) Casamino Acids (CAA). Streptomycin (200 μg/ml) and kanamycin (100 μg/ml) were added as needed. The cultures were incubated at 30°C and, for liquid cultures, with constant agitation at 200 rpm overnight.
TABLE 1.
Bacterial strains used
| Strain | Genotype and phenotypea | Reference |
|---|---|---|
| MG1 | Ampr Tcr; wild type | 18 |
| MG3 | MG1 flhD gene disrupted with a streptomycin cassette; Smr; swarming motility mutant due to failure to synthesize flagella | 14 |
| MG44 | MG1 swrI gene disrupted with a streptomycin cassette; Smr; AHL synthase mutant | 16 |
| MG3635 | MG44, swrA::Tn5:luxAB:Kanr:xylE; AHL synthase mutant unable to synthesize biosurfactant, which is required for swarming motility | 29 |
| MG3646 | MG44, bsmA::Tn5:luxAB:Kanr:xylE; AHL synthase mutant and surface attachment mutant | 29 |
| MG3651 | MG44, bsmBTn5:luxAB:Kanr:xylE; AHL synthase mutant, surface attachment mutant, and biofilm formation mutant | 29 |
Ampr, ampicillin resistance; Tcr, tetracycline resistance; Smr, streptomycin resistance; Kanr, kanamycin resistance.
Swarming assays.
Swarming motility was examined on the different media supplemented with 0.7% (wt/vol) Bacto agar (Difco Laboratories, Detroit, Mich.). Overnight cultures of different S. marcescens strains were stabbed into the center of the agar in 90-mm petri dishes (Sarstedt Inc., United States). The plates were then incubated at 30°C for 24 h. Swarming motility was determined by measuring the diameter of the swarming colony produced by each strain.
Flow cell biofilms.
Biofilms were cultivated in glass flow cells, which were incorporated into a biofilm flow cell system as described by Christensen et al. (6), with some modifications. The flow cells were constructed using a glass base plate (9 by 5.5 cm) that was glued together with two glass slides (7.5 by 2.5 by 0.1 cm), which were separated by two needles (20 gauge by 1 in.), thereby creating a 1-mm gap. A glass coverslip (6 by 2.4 cm) was then glued onto the glass slides to form an enclosed channel using UV-curing resin (Three Bond Co. Ltd., Japan). All remaining gaps on the flow cell were sealed with silicone (Plastic Putty Selleys Pty Ltd., Australia). The flow cell was connected with silicon tubing (Silastic laboratory tubing) via connectors and was autoclaved at 121°C for 15 min.
The inoculum for the flow cell system was prepared by concentrating an overnight culture four times with fresh medium (MBD supplemented with 0.05% [wt/vol] glucose and 0.05% [wt/vol] CAA). Four hundred microliters of the bacterial concentrate was then injected using a syringe into the inlet of the flow cell while the pump was switched off for 1 to 1.5 h to allow attachment to the flow cell. The biofilm flow cell system was maintained at a flow rate of 0.15 ml/min at room temperature. Independent triplicate experiments were run for 3 days (≤72 h), unless indicated otherwise. The different nutrient conditions used for the investigation of how nutritional cues can influence biofilm differentiation in wild-type S. marcescens MG1 and QS mutant S. marcescens MG44 biofilms in the biofilm flow cell system are described in Table 2. Further studies were carried out to investigate the effects of specific nutrient limitations on the biofilm architecture of the wild-type MG1 biofilm. The standard nutrient conditions used for cultivation of S. marcescens biofilms, MBD supplemented with 0.05% (wt/vol) glucose and 0.05% (wt/vol) CAA, was modified for various nutrient-limited conditions as shown in Table 2.
TABLE 2.
Media used to determine the effects of nutrients on biofilm ultrastructure development
| Medium composition | Biofilm morphology of MG1 | Specific growth rate (h−1) | Maximum optical density at 610 nm |
|---|---|---|---|
| MBD + 0.05% (wt/vol) glucose + 0.05% (wt/vol) CAA | Filamentous | 0.203 | 0.83 |
| MBD + 0.005% glucose + 0.005% CAA | Microcolony | 0.046 | 0.28 |
| 0.1× LB medium | Filamentous | 0.126 | 0.57 |
| MBD + 0.1% (wt/vol) sodium citrate + 0.05% (wt/vol) CAA | Microcolony | 0.161 | 0.76 |
| MBD + 0.1% (wt/vol) L-glutamic acid + 0.05% (wt/vol) CAA | Microcolony | 0.118 | 0.85 |
| MBD + 0.05% (wt/vol) glucose + 0.1% (wt/vol) ammonium sulfate | Microcolony | 0.142 | 0.55 |
| KH2PO4 and K2HPO4 concn reduced from 0.02 and 0.07% to 0.002 and 0.007%, respectively | Microcolony | NDa | |
| (NH4)2SO4 concn reduced from 0.1% to 0.01% | Microcolony | ND | |
| Addition of 0.004% 2,2-dipyridyl (Aldrich) (iron limiting) | Microcolony | ND |
ND, not determined.
Biofilm sloughing experiments.
Biofilms were cultivated in glass flow cell chambers as described above, with the following modifications. Multiple bubble traps were used to ensure that no bubbles could escape into the flow cell chamber. Five hundred nanomolar N-butanoyl-l-homoserine lactone (BHL) was added when necessary to complement the disrupted swrI N-acyl homoserine lactone (AHL) synthase gene. For one set of flow cells, the BHL was added to the mutant and was present for the duration of the experiment. For another set of flow cells, BHL was added to the mutant for the first 70 h of biofilm development, and then the medium was changed to MBD without BHL. Independent triplicate experiments were run for 5 days, and all flow cell experiments were conducted at room temperature. The biofilm effluent samples were vortexed at high speed for 1 min, and the cell density was determined at 610 nm with a NovaSpec II spectrophotometer (Pharmacia Biotech, United States).
Fixing and staining of biofilms.
Biofilms were prepared for confocal scanning laser microscopy (CSLM) by fixation with 2% (vol/vol) glutaraldehyde (Sigma) in phosphate-buffered saline (PBS), pH 7.4 (137 mM NaCl, 3 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4), for 15 min. Excess fixative was removed by washing the biofilms with PBS for 15 min. The biofilms were stained with 0.01% (wt/vol) acridine orange stain (Sigma) in PBS for 15 min, which was followed by washing with PBS for 30 min to remove excess stain.
Cell death within a biofilm was determined by staining the biofilm with 1 ml of LIVE/DEAD BacLight viability probes (Molecular Probes, Inc., Eugene, OR), prepared according to the manufacturer's specifications, without the glutaraldehyde fixation process. After 15 min of incubation, viable cells (stained green) and nonviable cells (stained red) were differentiated using the dual-channel option of CLSM.
Confocal scanning laser microscopy.
The stained biofilms were visualized in situ by CLSM with an Olympus LSMGB200 CLSM (Olympus Optical Co. Ltd., Tokyo, Japan). The CLSM used an argon ion laser at 488 nm for excitation, and a 522- to 535-nm band-pass filter or a 605- to 632-nm band-pass filter was used for emission. Images were captured using Olympus LSMGB200 CLSM bundled programs and were further processed for display by using the Adobe Photoshop software (version 6.0.1; Adobe Systems Inc., United States).
RESULTS
Effects of medium composition on biofilm morphology.
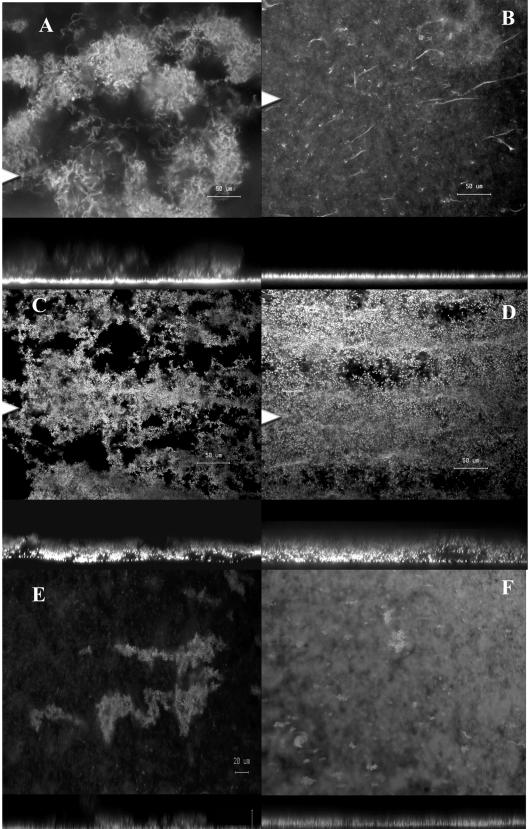
When grown under standard enrichment conditions for 3 days (MBD containing 0.05% [wt/vol] glucose and 0.05% [wt/vol] CAA), the wild-type strain S. marcescens MG1 formed a highly porous, filamentous biofilm composed of cell chains and clusters that were up to 80 μm deep (Fig. 1A). Under these conditions, the signal synthase mutant S. marcescens MG44 formed a flat, undifferentiated biofilm that appeared to stop development after the formation of a cell mat and the formation of polarly attached cells that extended up from the substratum as elongated filaments (Fig. 1B). In contrast, cultivation of both the wild-type strain and the QS mutant in 0.1× LB medium resulted in the formation of a biofilm that resembled the wild-type biofilm formed in MBD containing 0.05% glucose and 0.05% CAA (Fig. 1C and D). We have previously shown that two QS-controlled genes, bsmA and bsmB, are required for full development of a filamentous biofilm (26). Both of the mutants failed to progress past the stage of cell cluster formation; thus, the typical cell chains and filaments were not present. To determine if biofilm formation by these mutants was medium dependent, as observed for the QS mutant, they were allowed to form biofilms in 0.1× LB medium. Both mutants progressed to the stage of cell cluster formation, but, unlike the swrI mutant, they failed to develop further (Fig. 1E and F). To further test the suggestion that the medium may override the QS system, the S. marcescens wild-type strain, the signal synthase mutant MG44, and the nonswarming flhD mutant MG3 were tested for swarming ability on MBD containing glucose and on LB medium in the absence of exogenously added signal. As expected, the signal mutant MG44 did not swarm on MBD containing glucose, the wild-type strain swarmed on both media, and the MG3 mutant was deficient for swarming motility on both media (data not shown). However, MG44 did swarm on LB medium (data not shown), confirming that some QS phenotypes can be overridden depending on the type of medium used.
FIG. 1.
Effect of quorum sensing and media on filamentous biofilm formation in S. marcescens. (A and B) Confocal microscopic images of 3-day-old biofilms of S. marcescens wild-type strain MG1 and the quorum-sensing mutant MG44, respectively. (C to F) MG1 (C), MG44 (D), bsmA (E), and bsmB (F) mutant 3-day-old biofilms grown in 0.1× LB medium. In each panel the top image is the x-y plane, and the arrowhead indicates the position corresponding to the x-z cross section in the lower image. Magnification, ×377. (A to D) Bar, 50 μm. (E and F) Bar, 20 μm.
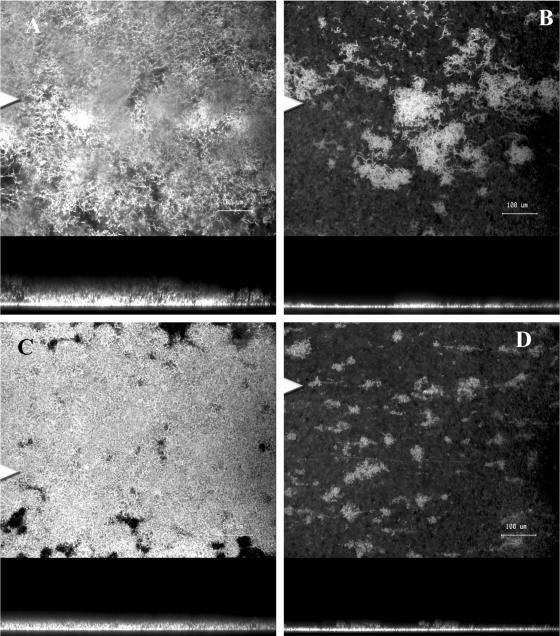
We also tested the effect of changing various components of standard MBD on biofilm formation and quorum sensing. When 0.1% citrate was substituted for glucose in the medium, the wild type formed a biofilm that consisted of a thin cell mat and microcolonies that were 20 to 25 μm in diameter. There was also a noticeable absence of cell filaments, clusters, and chains of cells (Fig. 2A). Under these conditions, it appeared that S. marcescens converted from the filamentous biofilm described above to a biofilm type more reminiscent of the microcolony biofilm described for P. aeruginosa and other organisms. Similarly, when glucose was substituted with 0.1% glutamate, the wild type again formed a biofilm that was thinner than the biofilm formed on MBD with 0.05% glucose and CAA, and the biofilm consisted of microcolonies (Fig. 2B). In the presence of glutamate, the wild type formed fewer microcolonies than when citrate was used. When the wild-type biofilm was formed in the presence of glucose and ammonium sulfate, microcolonies were again observed; however, their occurrence was more sporadic, with some areas consisting of only a monolayer of cells (data not shown). There was also significant variability in the size of the colonies; the colonies ranged from 20 to 400 μm in diameter under these conditions. In the presence of these modified MBD media, the QS mutant MG44 formed a flat, undifferentiated biofilm consisting of a monolayer of cells that did not develop the characteristic cell chains or clusters (data not shown).
FIG. 2.
Changes in medium composition lead to the formation of classic microcolony biofilms by S. marcescens: confocal microscopic images of 3-day-old wild-type biofilms of S. marcescens in MBD with 0.1% (wt/vol) sodium citrate plus 0.05% (wt/vol) CAA (A) and in MBD with 0.1% (wt/vol) l-glutamic acid plus 0.05% (wt/vol) CAA (B). In each panel the top image is the x-y plane, and the arrowhead indicates the position corresponding to the x-z cross section in the lower image. Magnification, ×400. Bar, 50 μm.
These data suggest that nutrient reduction or limitation may play a role in the switch between the filamentous type of biofilm and the microcolony type of biofilm. Therefore, we examined the effects of reducing the concentrations of specific medium components on biofilm formation. When the glucose and CAA concentrations in standard MBD were reduced 10-fold (final concentration of each, 0.005%), microcolonies were again observed (data not shown) and the size and frequency of microcolony formation were enhanced compared to the previously tested conditions (Fig. 2). Similarly, specific reductions in the concentrations of ammonia, phosphate, or iron resulted in the formation of microcolony biofilms (Table 2). However, in contrast to the 0.005% glucose-0.005% CAA conditions, these conditions led to the formation of fewer microcolonies and microcolonies that were more diffuse or less compact (data not shown).
Growth rate and biofilm morphology.
The planktonic growth rates of the wild type in the various media indicated that the specific growth rate varied depending on the medium composition. The standard medium, MBD containing 0.05% glucose and 0.05% CAA, produced the highest growth rate (0.203 h−1), and the lowest growth rate was observed when glucose and CAA were present at 10-fold-lower concentrations (0.046 h−1) (Table 2). It should be noted, however, that there was no obvious correlation between the growth rate or maximum final growth yield and the formation of a filamentous or microcolony type of biofilm, as the 0.1× LB medium biofilm (filamentous biofilm) had a growth rate of 0.126 h−1, which was slightly less than the growth rate in the presence of 0.05% glucose and 0.1% ammonium sulfate (0.142 h−1). Additionally, the growth yield in the diluted LB medium was lower than that obtained using any of the media with the exception of the medium containing 0.005% glucose and 0.005% CAA.
Biofilm morphology is reversible.
To determine if the biofilm is a static or a dynamic structure, the effect of altering medium conditions during biofilm development was investigated. Biofilms of the S. marcescens wild type were established for 24 h in MBD containing 0.05% glucose and 0.05% CAA, after which the medium was switched to MBD without glucose or CAA. At 24 h after the medium switch, the biofilm had the typical morphology of a filamentous biofilm (data not shown) and consisted of a thin layer of attached cells and polarly attached cell filaments extending up from the substratum into the medium. However, 48 h after the medium switch (at 72 h), the biofilm had formed large microcolony structures, similar to those shown in Fig. 2, that resembled typical microcolonies of other bacteria, such as P. aeruginosa (data not shown). Staining with the LIVE/DEAD BacLight stain indicated that the microcolonies also contained a mixture of live and dead cells, although, in contrast to what has been observed in P. aeruginosa (40), the dead cells did not appear to be localized to the center of the microcolony. Biofilm formation in the presence of MBD without glucose or CAA from time zero resulted in a flat, undifferentiated biofilm (data not shown), indicating that the microcolony formation in the medium switch experiment was likely to be the result of reorganization of the biofilm from the filamentous to the microcolony type. Similarly, an established microcolony biofilm was observed to switch to the filamentous morphology when the nutrient concentrations were increased from 0.005% glucose and 0.005% CAA to 0.05% glucose and 0.05% CAA (data not shown). Thus, it appears that the biofilm is a dynamic structure that can be altered in response to the prevailing conditions.
Quorum sensing control of biofilm sloughing.
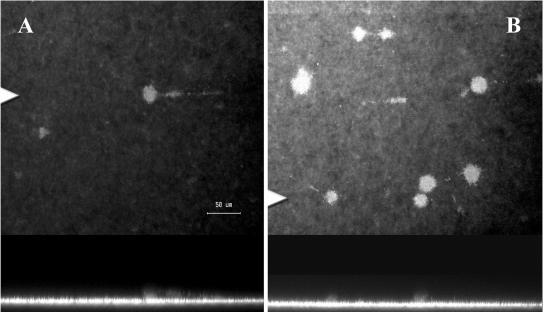
During these studies, it was observed that the mature filamentous biofilm detached or sloughed off from the surface after approximately 75 to 80 h (Fig. 3). Figures 3A and C, obtained after 70 h of development, show confocal images of the fully differentiated filamentous biofilms of the wild type and the swrI mutant (supplemented with 500 nM BHL for the duration of the experiment), respectively. Figures 3B and D show that there was a dramatic loss of biomass after sloughing; the biofilm consisted primarily of a single layer of cells, and only a few patches of differentiated filaments remained. After detachment, the biofilms of the wild type and the QS mutant supplemented with BHL resembled the undifferentiated biofilm formed by the QS mutant without BHL. The role of QS in sloughing was investigated by comparing sloughing of the wild type, the AHL synthase mutant MG44, MG44 plus exogenously added signal, and MG44 with exogenously added signal for the first 70 h of biofilm formation, at which time the signal was removed. Confocal microscopy and COMSTAT analysis of the wild type and MG44 complemented with BHL have demonstrated that the mutant can be restored to a wild-type morphology (26).
FIG. 3.
Detachment of S. marcescens biofilms: confocal microscope images of the wild type and the QS mutant grown with 500 nM BHL for the duration of the experiment prior to sloughing at 70 h (A and C) and after sloughing at 80 h (B and D). In each panel the top image is the x-y plane, and the arrowhead indicates the position corresponding to the x-z cross section in the lower image. Magnification, ×400. Bar, 50 μm.
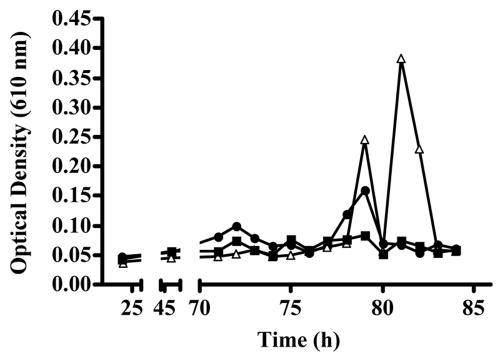
Sloughing was monitored by measuring the optical density at 610 nm of the effluent from the biofilm. Figure 4 shows typical sloughing profiles for the wild type, MG44 grown with BHL for the duration of the experiment, and MG44 grown with BHL for the first 70 h of biofilm development. BHL was added to restore the wild-type biofilm morphology and was removed for one set of biofilms at 70 h to test the role of quorum sensing in biofilm sloughing. Data for MG44 without BHL are not shown because it did not slough at all. For the first 70 h, there was no appreciable loss of biomass from any of the biofilms (Fig. 4), and the signal synthase mutant grown in the presence of BHL for the first 70 h showed little or no sloughing over the entire time course of the experiments. In contrast, the wild type and MG44 grown in the presence of BHL for the duration of the experiment showed high levels of sloughing (Fig. 3 and 4), which typically occurred at approximately 75 to 80 h. Interestingly, although the timing of sloughing was similar, the mutant grown in the presence of BHL for the duration of the experiment showed a higher degree of sloughing than the wild type.
FIG. 4.
Sloughing profiles of S. marcescens strains. Sloughing of the biofilm was measured by collecting effluent from the biofilm as described in Materials and Methods. Briefly, effluent from three flow cells per experiment was collected and vortexed to disperse clumps, and the optical density at 610 nm was determined. The sloughing profiles are the profiles for the wild type (circles), the swrI mutant MG44 grown with 500 nM BHL (triangles), and MG44 grown with 500 nM BHL for 70 h, after which the medium was switched to BHL-deficient MBD (squares). The data are data for triplicate flow cells from one experiment and are representative of three independent experiments.
It has been suggested that enzymes or surfactants, such as alginate lyase or rhamnolipids, may play a role in the detachment or dispersal of biofilms (3, 8). Therefore, we tested the effect of mutation of swrA, a quorum-sensing-controlled gene which is involved in the synthesis of serrawettin, a surfactant required for swarming (29), on sloughing in this system. Since this strain does not produce surfactant, it was expected that it might not detach as readily as the wild type. However, when we monitored sloughing of the swrA mutant, it was observed to slough at the same time and to the same degree as the wild type (data not shown), suggesting that the surfactant serrawettin did not play a significant role in the detachment of the filamentous biofilm.
DISCUSSION
In this study, we demonstrated that while S. marcescens displays quorum-sensing-dependent biofilm formation using MBD medium with 0.05% glucose and 0.05% Casamino Acids, this dependence can be circumvented by growing the quorum-sensing mutant in 0.1× LB medium. We also found that the quorum-sensing mutant failed to swarm on MBD plates but exhibited delayed swarming on LB medium. Quorum sensing has previously been shown to be involved in biofilm formation in P. aeruginosa, as well as in A. hydrophila (30) and B. cepacia (25). However, it has also been suggested that the dependence on quorum sensing is not absolute, and indeed, it has been reported that the nutrient conditions may affect whether quorum sensing is required for biofilm formation (22, 31). The effect of LB medium on the quorum-sensing system and biofilm formation of S. marcescens might be attributed to two possibilities. First, because LB medium is a complex medium, it may alter the central metabolic pathways of the cell and thus bypass the regulatory requirement for the quorum-sensing system. It has recently been shown that increasing nutrient levels can induce detachment of P. aeruginosa biofilms (34), and the degree of detachment was carbon source specific. The induction of detachment was associated with the induction of specific genes, such as flagellar, pilus, and denitrification genes (34). Thus, utilization of different metabolic pathways in S. marcescens may alter the expression of other regulatory factors that are also involved in the control of quorum-sensing phenotypes. A second possibility for the effect of LB medium on quorum-sensing phenotypes is that this complex medium contains either signals or other factors, such as surfactants, that are required for swarming and biofilm formation. It has been shown previously that quorum-sensing systems can be induced or repressed by cyclic dipeptides (24). While none of the cyclic dipeptides tested in the previous study induced swarming in S. marcescens, a wide range of cyclic dipeptides are found in complex media, and we have shown that extracts of LB medium contain numerous cyclic dipeptides (unpublished data).
The quorum-sensing-controlled genes bsmA and bsmB (26) were still required for filamentous biofilm formation when the cells were grown in 0.1× LB medium; biofilms of these mutants were unable to progress past cell cluster formation and did not develop the cell chains characteristic of the filamentous biofilm. This therefore suggests that, whatever factors that are present in LB medium that override the quorum-sensing system, they must act in such a way as to stimulate the quorum-sensing regulon to lead to biofilm development.
Medium composition was also shown to control biofilm morphology, resulting in either a filamentous or a “classic” microcolony biofilm. In particular, media with limiting or minimal nutrients, such as MBD with 0.005% glucose and 0.005% Casamino Acids, appeared to shift the biofilm toward a microcolony type of biofilm, and this effect appeared to be growth rate independent. Our observation that the biofilm type can shift from a filamentous biofilm to a microcolony biofilm and back to a filamentous biofilm demonstrates that the biofilm structure is dynamic and that it can be remodeled depending on the prevailing conditions.
There is growing interest in how bacteria escape from the biofilm and initiate colonization of new surfaces, which is important for the cells to find new sites for colonization. The process has been proposed to be either passive loss or removal of cells from the biofilm surface, as a consequence of high hydrodynamic forces on the biofilm, or an active process that controls the timing and extent of release of cells from the biofilm. Sauer et al. (34) have recently shown that increased nutrient levels can lead to biofilm detachment, and Thormann et al. (37) have shown that biofilm detachment is regulated by the oxygen concentration in Shewenella oneidensis. It has long been speculated that quorum sensing might be one such mechanism that controls release of cells (36), and it has been shown that diffusible factors are involved in the dispersal of cells from flocs formed by Rhodobacter sphaeroides (32) and Xanthomonas campestris (12). Furthermore, it was recently demonstrated that BHL controls detachment in P. aeruginosa and that this effect is mediated by the regulation of rhamnolipid production (35). We show here that BHL-mediated quorum sensing is involved in the sloughing of S. marcescens biofilms, suggesting that this is a regulated response to the prevailing biofilm conditions and that detachment is therefore an active process in the biofilm life cycle. Importantly, our data support the findings of Schooling et al. (35) and demonstrate that AHL-mediated quorum sensing may be more generally involved in dispersal phenotypes in bacteria. In contrast to the previous report (35), when we investigated the sloughing profile of a surfactant-deficient S. marcescens strain, MG3636, we observed no difference in sloughing. This suggests that quorum-sensing-mediated dispersal may involve a range of different gene products. Furthermore, our study demonstrates an extended role of quorum sensing in biofilm formation and development. In particular, we determined that quorum sensing plays key roles in at least four stages of surface colonization in S. marcescens, from attachment (Labbate, unpublished data) to swarming motility (15), biofilm development (26), and detachment.
The temporal separation of quorum-sensing control of attachment, development, and detachment may be the result of multiple regulators being required for the correct timing of expression of biofilm-related genes. For example, swarming motility requires the action of the quorum-sensing system and the independent FlhDC operon. The latter is known to be expressed in a growth-phase-dependent fashion, thus preventing swarming from occurring until the cells are physiologically prepared (19). This is supported by the data of Whitely et al. (41), who divided the quorum-sensing regulon of P. aeruginosa into early and late expressed genes. These authors also showed that gene expression was divided into expression of genes that were induced directly by N-3-oxo-dodecanoyl homoserine lactone and expression of genes that required both N-3-oxo-dodecanoyl homoserine lactone and N-butanoyl homoserine lactone. It has been reported that factors such as RpoS (28, 42), Vfr (1), GacA (4), polyphosphate kinase (33), cAMP (13), iron (20), HNS (39), and sigma 32 (38) are involved in regulating AHL-mediated phenotypes, further supporting the concept of cooperativity in the regulation of quorum-sensing-controlled genes. The inclusion of regulators such as cAMP, RpoS, and iron is particularly interesting as these regulators may also reflect our observations that the quorum-sensing system can be overridden depending on the medium composition, and the type of biofilm formed may also change depending on the prevailing nutrient conditions. For example, under low-iron conditions, we observed that S. marcescens shifted from a filamentous biofilm to a microcolony-type biofilm.
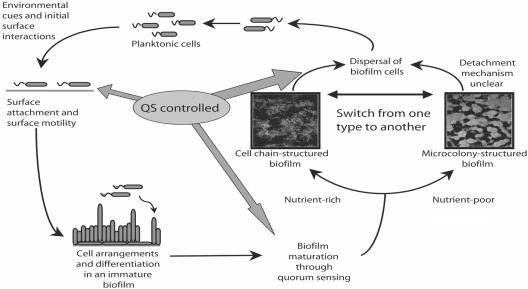
Based on the concept of coregulation and temporal separation of quorum-sensing-controlled genes involved in surface colonization, we present the model shown in Fig. 5, in which swrI/swrR-directed quorum sensing is involved in several key stages of surface colonization in S. marcescens, including attachment to a surface, surface motility, biofilm development, and finally detachment from the surface. The specific genes that are involved in surface colonization, detachment, and the type of biofilm formed and their role in medically and ecologically relevant settings are exciting areas that we are currently exploring using this system.
FIG. 5.
Model for quorum-sensing control of surface colonization by S. marcescens.
Acknowledgments
We thank Diane McDougald for critical reading of the manuscript and thank Michael Givskov for strains MG1, MG44, MG3635, MG3646, and MG3651.
This work was supported by the Centre for Marine Biofouling and Bio-Innovation at The University of New South Wales.
REFERENCES
- 1.Albus, A. M., E. C. Pesci, L. J. Runyen-Janecky, S. E. H. West, and B. H. Iglewski. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertani, G. 1951. Studies on lysogenesis. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, A., and A. M. Chakrabarty. 1994. Role of alginate lyase in cell detachment of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 60:2355-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chancey, S. T., D. W. Wood, and L. S. Pierson. 1999. Two-component transcriptional regulation of N-acyl-homoserine lactone production in Pseudomonas aureofaciens. Appl. Environ. Microbiol. 65:2294-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen, B. B., J. A. J. Haagensen, A. Heydorn, and S. Molin. 2002. Metabolic commensalism and competition in a two-species microbial consortium. Appl. Environ. Microbiol. 68:2495-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen, B. B., C. Sternberg, J. B. Andersen, R. K. Palmer, Jr., A. T. Nielsen, M. Givskov, and S. Molin. 1999. Molecular tools for study of biofilm physiology. Methods Enzymol. 310:20-42. [DOI] [PubMed] [Google Scholar]
- 7.Conway, B. A. D., V. Venu, and D. P. Speert. 2002. Biofilm formation and acyl homoserine lactone production in the Burkholderia cepacia complex. J. Bacteriol. 184:5678-5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davey, M. E., N. C. Caiazza, and G. A. O'Toole. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:1027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 10.Debeer, D., P. Stoodley, and Z. Lewandowski. 1996. Liquid flow and mass transport in heterogeneous biofilms. Water Res. 30:2761-2765. [Google Scholar]
- 11.De Kievit, T. R., R. Gillis, S. Marx, C. Brown, and B. H. Iglewski. 2001. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl. Environ. Microbiol. 67:1865-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dow, J. M., L. Crossman, K. Findlay, Y. Q. He, J. X. Feng, and J. L. Tang. 2003. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc. Natl. Acad. Sci. USA 100:10995-11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunlap, P. V., and E. P. Greenberg. 1985. Control of Vibrio fischeri luminescence gene expression in Escherichia coli by cyclic AMP and cyclic AMP receptor protein. J. Bacteriol. 164:45-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eberl, L., G. Christiansen, S. Molin, and M. Givskov. 1996. Differentiation of Serratia liquefaciens into swarm cells is controlled by the expression of the flhD master operon. J. Bacteriol. 178:554-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eberl, L., S. Molin, and M. Givskov. 1999. Surface motility of Serratia liquefaciens MG1. J. Bacteriol. 181:1703-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eberl, L., M. K. Winson, C. Sternberg, G. S. A. B. Stewart, G. Christiansen, S. R. Chhabra, B. Bycroft, P. Williams, S. Molin, and M. Givskov. 1996. Involvement of N-acyl-l-homoserine lactone autoinducers in controlling the multicellular behavior of Serratia liquefaciens. Mol. Microbiol. 20:127-136. [DOI] [PubMed] [Google Scholar]
- 17.Elkins, J. G., D. J. Hassett, P. S. Stewart, H. P. Schweizer, and T. R. McDermott. 1999. Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl. Environ. Microbiol. 65:4594-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Givskov, M., L. Olsen, and S. Molin. 1988. Cloning and expression in Escherichia coli of the gene for extracellular phospholipase from Serratia liquefaciens. J. Bacteriol. 170:5855-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Givskov, M., J. Ostling, L. Eberl, P. W. Lindum, A. B. Christensen, G. Christiansen, S. Molin, and S. Kjelleberg. 1998. Two separate regulatory systems participate in control of swarming motility of Serratia liquefaciens MG1. J. Bacteriol. 180:742-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haygood, M. G., and K. H. Nealson. 1985. Mechanisms of iron regulation of luminescence in Vibrio fischeri. J. Bacteriol. 162:209-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hentzer, M., G. M. Teitzel, G. J. Balzer, A. Heydorn, S. Molin, M. Givskov, and M. R. Parsek. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183:5395-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heydorn, A., B. K. Ersbøll, J. Kato, M. Hentzer, M. R. Parsek, T. A. Nielsen, M. Givskov, and S. Molin. 2002. Statistical analysis of Pseudomonas aeruginosa biofilm development: impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl. Environ. Microbiol. 68:2008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoiby, N., H. K. Johansen, C. Moser, Z. J. Song, O. Ciofu, and A. Kharazmi. 2001. Pseudomonas aeruginosa and the in vitro and in vivo biofilm mode of growth. Microb. Infect. 3:23-35. [DOI] [PubMed] [Google Scholar]
- 24.Holden, M. T. G., S. R. Chahabra, R. deNys, P. Stead, N. J. Bainton, P. J. Hill, M. Manefield, N. Kumar, M. Labbate, D. England, S. A. Rice, M. Givskov, G. Salmond, G. S. A. B. Stewart, B. W. Bycroft, S. Kjelleberg, and P. Williams. 1999. Quorum sensing cross talk: isolation and chemical characterisation of cyclic dipeptides from Pseudomonas aeruginosa and other Gram-negative bacteria. Mol. Microbiol. 33:1254-1266. [DOI] [PubMed] [Google Scholar]
- 25.Huber, B., K. Riedel, M. Hentzer, A. Heydorn, A. Gotschlich, M. Givskov, S. Molin, and L. Eberl. 2001. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147:2517-2528. [DOI] [PubMed] [Google Scholar]
- 26.Labbate, M., S. Y. Queck, K. S. Koh, S. A. Rice, M. Givskov, and S. Kjelleberg. 2004. Quorum sensing-controlled biofilm development in Serratia liquefaciens MG1. J. Bacteriol. 186:692-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labbate, M., H. Zhu, R. Bandara, J. S. Webb, M. D. P. Willcox, M. Givskov, S. A. Rice, and S. Kjelleberg. Submitted for publication.
- 28.Latifi, M. F., K. Tanaka, P. Williams, and A. Lazdunski. 1996. A hierarchial quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhlR (VsmR) to expression of the stationary phase sigma factor RpoS. Mol. Microbiol. 21:1137-1146. [DOI] [PubMed] [Google Scholar]
- 29.Lindum, P. W., U. Anthoni, C. Christopheersen, L. Eberl, S. Molin, and M. Givskov. 1998. N-Acyl-l-homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J. Bacteriol. 180:6384-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lynch, M. J., S. Swift, D. F. Kirke, C. W. Keevil, C. E. Dodd, and P. Williams. 2002. The regulation of biofilm development by quorum sensing in Aeromonas hydrophila. Environ. Microbiol. 4:18-28. [DOI] [PubMed] [Google Scholar]
- 31.Purevdorj, B., J. W. Costerton, and P. Stoodley. 2002. Influence of hydrodynamics and cell signaling on the structure and behavior of Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 68:4457-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puskas, A., E. P. Greenberg, S. Kaplan, and A. L. Schaefer. 1997. A quorum-sensing system in the free-living photosynthetic bacterium Rhodobacter sphaeroides. J. Bacteriol. 179:7530-7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rashid, M. H., K. Rumbaugh, L. Passador, D. G. Davies, A. N. Hamood, B. H. Iglewski, and A. Kornberg. 2000. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:9636-9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sauer, K., M. C. Cullen, A. H. Rickard, L. A. H. Zeef, D. G. Davies, and P. Gilbert. 2004. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 186:7312-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schooling, S. R., U. K. Charaf, D. G. Allison, and P. Gilbert. 2004. A role for rhamnolipid in biofilm dispersion. Biofilms 1:91-99. [Google Scholar]
- 36.Stoodley, P., K. Sauer, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 37.Thormann, K. M., R. M. Saville, S. Shukla, and A. M. Spormann. 2005. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J. Bacteriol. 187:1014-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ulitzer, S., and J. Kuhn. 1988. The transcription of bacterial luminescence is regulated by sigma 32. J. Biolumin. Chemilumin. 2:81-93. [DOI] [PubMed] [Google Scholar]
- 39.Ulitzur, S., A. Matin, C. Fraley, and E. Meighen. 1997. HNS protein represses transcription of the lux systems of Vibrio fischeri and other luminous bacteria cloned into Escherichia coli. Curr. Microbiol. 35:336-342. [DOI] [PubMed] [Google Scholar]
- 40.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:4585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whiteley, M., M. R. Parsek, and E. P. Greenberg. 2000. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J. Bacteriol. 182:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]