Abstract
In addition to causing fulminant disease, Streptococcus pyogenes may be asymptomatically carried between recurrent episodes of pharyngitis. To better understand streptococcal carriage, we characterized in vitro long-term stationary-phase survival (>4 weeks) of S. pyogenes. When grown in sugar-limited Todd-Hewitt broth, S. pyogenes cells remained culturable for more than 1 year. Both Todd-Hewitt supplemented with excess glucose and chemically defined medium allowed survival for less than 1 week. After 4 weeks of survival in sugar-limited Todd-Hewitt broth, at least 103 CFU per ml remained. When stained with fluorescent live-dead viability stain, there were a number of cells with intact membranes that were nonculturable. Under conditions that did not support persistence, these cells disappeared 2 weeks after loss of culturability. In persistent cultures, these may be cells that are dying during cell turnover. After more than 4 weeks in stationary phase, the culturable cells formed two alternative colony phenotypes: atypical large colonies and microcolonies. Protein expression in two independently isolated microcolony strains, from 14-week cultures, was examined by use of two-dimensional electrophoresis. The proteomes of these two strains exhibited extensive changes compared to the parental strain. While some of these changes were common to the two strains, many of the changes were unique to a single strain. Some of the common changes were in metabolic pathways, suggesting a possible alternate metabolism for the persisters. Overall, these data suggest that under certain in vitro conditions, S. pyogenes cells can persist for greater than 1 year as a dynamic population.
Streptococcus pyogenes (group A streptococci) is a gram-positive human pathogen which is the causative agent of a diverse range of human disease. It can cause both mild infections, such as impetigo and pharyngitis, and more severe disease, such as toxic shock syndrome and necrotizing fasciitis (12). Infections can also result in secondary sequelae, such as rheumatic fever and glomerulonephritis (12). Further, a small percentage of patients will experience recurrent tonsillitis caused by S. pyogenes, with intermittent asymptomatic periods ranging from weeks to months (4, 36, 37, 43). These chronic infections may be attributed to a bacterial subpopulation surviving beyond the primary episode. Molecular typing studies have confirmed the clonal nature of the bacteria associated with recurrent tonsillitis (4, 37, 43). These streptococci may persist extracellularly by embedding themselves in the matrix proteins of the human mucosa, where they may evade the human immune response (15); alternatively, these streptococci may persist intracellularly, where they may be protected from both antibiotic therapy and the human immune response via residence in an intracellular reservoir where they can fuel future acute infections (17, 33, 36, 37, 43).
Some studies suggest that intracellular bacteria could possibly constitute a reservoir for the streptococcal carrier state. Though S. pyogenes can induce eucaryotic cell lysis, it has also been demonstrated that S. pyogenes can stably infect eucaryotic cells for periods exceeding 1 month (26, 41, 47). After the establishment of stable intracellular residence, these streptococci began to lose their cell wall, including associated virulence factors (41). However, these phenotypically altered bacterial cells were able to revert to their original forms (41). In agreement with this, in vivo studies have shown that cell wall-defective streptococci were able to persist in a rat model of infection at least twice as long as their parental forms (29). The relevance of these observations to the mechanism of chronic infection seems to be borne out by a direct association between intracellular S. pyogenes and patients suffering from recurrent tonsillitis (36, 37). In vitro studies have confirmed that S. pyogenes cells undergo phenotypic switching (8, 28) and can invade eucaryotic cells (7, 11, 31, 32).
During persistence in the eucaryotic host environment, S. pyogenes cells may enter a quiescent state, reminiscent of the stationary phase of laboratory cultures, due to conditions which do not support the rapid bacterial multiplication seen in fulminant infections. For this reason, the persistence of S. pyogenes in an in vitro stationary phase environment has been of interest. Trainor et al. have described a chemically defined media system for the study of stationary-phase S. pyogenes (46). They found survival of S. pyogenes for approximately 3 weeks, at which time they terminated their studies (46).
In this study, we wished to determine whether S. pyogenes cells could survive in stationary phase for extended periods (>4 weeks). When grown in carbon-limited, complex medium, such as Todd-Hewitt broth, S. pyogenes cells were able to survive for periods exceeding 1 year. Cells persisting in stationary phase for more than 4 weeks regrew on agar plates as stable phenotypic variants termed here either atypical large colonies or microcolonies. Two microcolonies, isolated from 14-week-old, independent cultures, were examined using both two-dimensional proteomics and Northern blots. Though these two strains exhibited common colony morphologies and shared common proteomic alterations, their proteomes also exhibited many unique changes compared to the parental strain and to each other.
MATERIALS AND METHODS
Bacterial strains and incubation conditions.
Strains used for this study include S. pyogenes serotype M49 strain CS101 (M49-CS101), serotype M49 strain 591 (M49-591), serotype M3 strain AM3 (M3-AM3), and serotype M6 strain JRS4 (M6-JRS4). S. pyogenes M3-AM3 and M6-JRS4 were provided by J. R. Scott and C. P. Moran, Jr., M49-591 was provided by A. Podbielski, and M49-CS101 was provided by P. P. Cleary. Strains Alt. 1 and Alt. 2 were also examined in this study and were isolated from independent 14-week stationary-phase M49-CS101 cultures grown in Todd-Hewitt broth. All S. pyogenes cultures were grown and maintained static at 37°C under a 5% CO2 atmosphere.
Bacterial survival assays.
Liquid Todd-Hewitt (TH), Todd-Hewitt yeast extract (THY) (DIFCO Laboratories, Detroit, Michigan), and chemically defined medium (CDM) containing final glucose concentrations of either 0.2% (wt/vol) or 1.0% (wt/vol) were inoculated with colonies of S. pyogenes M49-CS101 grown on TH agar plates. The CDM recipe used in this study is a modified version of that described by van de Rijn and Kessler, which has been used in our previous studies on persistence (28, 48). The following CDM components were used in different final concentrations in this formulation: l-arginine, 121 mg liter−1; glycine, 200 mg liter−1; l-histidine, 135 mg liter−1; l-leucine, 200 mg liter−1; l-lysine, 138 mg liter−1; l-proline, 200 mg liter−1; l-hydroxyproline, 200 mg liter−1; l-asparagine, 100 mg liter−1; (NH4)2SO4, 600 mg liter−1; p-aminobenzoic acid, 0.4 mg liter−1; biotin, 0.5 mg liter−1; pyridoxamine HCl, 1.44 mg liter−1; thiamine HCl, 2.0 mg liter−1; nicotinamide, 10 mg liter−1; adenine sulfate, 38.28 mg liter−1; guanine HCl, 27.28 mg liter−1; uracil, 22 mg liter−1; and sodium citrate, 66.7 mg liter−1. The following CDM components were eliminated from this formulation: vitamin B12 and niacinamide. CDM was made with final glucose concentrations of either 0.2% (wt/vol) or 1.0% (wt/vol). TH broth consisted of 30 g liter−1 TH powder and contained a glucose concentration of 0.2% (wt/vol). For TH broth (1.0% glucose), the medium was supplemented with 8 g liter−1 glucose. THY broth contained 30 g liter−1 TH powder and 5 g liter−1 yeast extract powder. Survival was assayed by spotting 20-μl culture aliquots, at 24-h time intervals postinoculation, onto TH agar plates, resulting in a lower limit of detection of 50 CFU ml−1. The formation of any S. pyogenes colonies within the culture spot after 24 h of incubation was scored positive for survival. The terminal pH for each culture was recorded within 24 h after survival tubes were determined to be nonculturable. pH was assayed by filtration of spent medium through a 0.2-μm syringe filter and measurement of filtrate with a pH meter. For TH and THY cultures (0.2% glucose) which were still surviving after 6 months, the pH was determined 1 week after inoculation and was subsequently stable. The preinoculation pHs of the liquid CDM were approximately 7.0, and the preinoculation pHs of the TH and THY broths were approximately 7.8.
Bacterial survival kinetics.
S. pyogenes M49-591, M3-AM3, M6-JRS4, M49-CS101, Alt. 1, and Alt. 2 were inoculated into and grown in sterile TH broth. Over a 12-week period, stationary-phase culture aliquots were removed, serially diluted in sterile TH broth, and plated on TH agar plates for the determination of the number of CFU per milliliter.
pH shift assays.
S. pyogenes M49-CS101 was inoculated into TH broth, grown to stationary phase, and aged 1 week. Exogenous HCl or lactic acid was then added from 2 M stocks to final culture concentrations of 25.0, 12.5, or 6.3 mM. Survival was assayed by spotting 20 μl of culture aliquots on TH agar plates in 24-h intervals after addition of exogenous acid. These plates were incubated at 37°C under a 5% CO2 atmosphere, and formation of any CFU in these culture spots was scored as positive for survival.
Phenotypic variants.
The identities of phenotypic variants (microcolonies and atypical large colonies) as S. pyogenes were confirmed through 16S rRNA sequence analysis. Genomic DNA was isolated from these cultures and was used as a template for PCR using “universal” eubacterial 16S primers EubA (5′-AAGGAGGTGATCCANCCRCA) and EubB (5′-AGAGTTTGATCMTGGCTCAG) (10). For confirmation of species identity, this PCR product was sequenced using the EubB primer, and this sequence was compared against the NCBI sequence database using the BLAST program.
Fluorescence imaging.
Bacterial viability was assessed through use of the Live/Dead BacLight bacterial viability stain (Molecular Probes, Eugene, Oregon). Three microliters of a solution containing a 1:1 ratio of SYTO-9 to propidium iodide was mixed with 500 μl of sterile water with 500 μl of culture and incubated at room temperature for approximately 25 min. Images were captured with Image-Pro Plus on a Nikon Eclipse TE300 microscope (Media Cybernetics, Silver Spring, Maryland) using filters with 465 to 495 excitation and 515 to 555 emission (green fluorescence) or a 510 to 560 excitation and 590 emission (red fluorescence). These were subsequently scored using Adobe Photoshop v. 7.0 (Adobe, San Jose, California).
Alternate growth media.
S. pyogenes M49-CS101 was inoculated from colonies grown on TH agar plates into TH broth cultures, grown to stationary phase, and aged 2 weeks. Culture aliquots were then removed, serially diluted, and plated on various agar types: 0.5× TH (15 g liter−1), TH (30 g liter−1), 2× TH (60 g liter−1), TH yeast extract, TH supplemented with 5% (vol/vol) sheep's blood, TH supplemented with 0.25 M sucrose, TH supplemented with 0.50 M sucrose, TH supplemented with 0.50 M sorbitol, TH supplemented with 40 mg of pyruvate per 20-ml plate, TH supplemented with 80 mg of pyruvate per 20-ml plate, TH supplemented with 500 U of catalase per 20-ml plate, and TH supplemented with 1,000 U of catalase per 20-ml plate. Sterile pyruvate and catalase solutions were made fresh before each experiment, and the desired amount was spread on agar plates before plating of bacteria. Sterile TH broth was used as a diluent for all culture aliquots except for those plated on TH with 0.50 M sucrose and TH with 0.50 M sorbitol. For each of these conditions, a sterile 0.50 M sucrose or sorbitol solution based on the plate medium type was used. Plates were incubated for at least 48 h before being scored for colonies.
Isolation and purification of cytoplasmic proteins.
To isolate protein extracts from strains M49-CS101, Alt. 1, and Alt. 2, 40 ml of sterile TH broth was inoculated with early-stationary-phase liquid culture and regrown to an optical density at 600 nm (OD600) between 0.50 and 0.65, at which point the cells were pelleted. Culture supernatant was then removed, and the cell pellet was resuspended in 2 ml of solubilization-rehydration solution (8 M urea, 2 M thiourea, 4% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 75 mM dithiothreitol). The cell suspension was chilled on ice, and 1.0 g of 0.1-mm zirconia-silica beads (Biospec, Bartlesville, OK) was added. This mix was lysed by five 20-s pulses at maximum speed using a mini-bead beater (Biospec). Tubes containing lysate were allowed to sit at room temperature on a rocking table for at least 10 h, at which time they were centrifuged (18 × g) to pellet cell debris. The supernatant was transferred to fresh Eppendorf tubes, where cytoplasmic proteins were purified using the PlusOne 2-D Clean-Up kit (Amersham Biosciences, San Francisco, CA) according to the manufacturer's directions. Purified protein extract was then quantitated using the PlusOne 2-D Quant kit (Amersham) according to the manufacturer's specifications.
Two-dimensional electrophoresis.
Isoelectric focusing was done with an IPGphor isoelectric focusing system using immobiline dry strips (24 cm) with a linear pH range of 4 to 7, as described by the manufacturer (Amersham). IPG strips were hydrated with 150 to 170 μg of protein in 450 μl of sample buffer for 12 h at 20°C. Isoelectric focusing was done with 500 V for 500 V · h, 1,000 V for 1,000 V · h, and 8,000 V for 96,000 V · h at 20°C. The strips were incubated in sodium dodecyl sulfate (SDS) equilibration buffer (50 mM Tris-Cl [pH 8.8], 6 M urea, 30% [vol/vol] glycerol, 2% SDS, and bromophenol blue) for 10 min. SDS-polyacrylamide gel electrophoresis (SDS-PAGE) separation was done with a DALT II six electrophoresis apparatus (Amersham) and 10% acrylamide resolving gels (0.1 by 23.4 by 19.5 cm) containing 1% SDS. Rhinohide (Molecular Probes) was added to the acrylamide solution as a strengthening agent, as instructed by the manufacturer. The running buffer consisted of 0.25 M Tris, 1.92 M glycine, and 1% (wt/vol) SDS, and electrophoresis was done for approximately 18 h at 75 V. Proteins were stained with Sypro Ruby (Molecular Probes), and digital images were acquired with a Typhoon imager (Amersham). Analysis of the gels, including protein spot detection and quantitation, was done with PDQuest software (Bio-Rad, Hercules, Calif.). Protein spots were quantitated by summing the values of pixels comprising each protein spot. Gels were normalized based on the sum of all protein spots detected in each sample.
Protein identification.
Proteins of interest were excised from the SDS-PAGE gels with a robotic spot cutter (Bio-Rad). The gel plugs were washed three times for 15 min in 400 μl of 25 mM NH4HCO3-50% acetonitrile (ACN; Aldrich, Milwaukee, Wis.), incubated in 100% ACN for 5 min, and lyophilized in a SpeedVac for 30 min. The dried gel plugs were hydrated with 25 mM NH4HCO3 containing 10 μg ml−1 of sequencing-grade trypsin (Sigma Chemical Co., St. Louis, Mo.). Following incubation at 37°C for approximately 16 h, the trypsin solution was aspirated to a 96-well microtiter plate, and additional peptides were recovered from the gel plugs by extracting twice with 50% ACN-5% trifluoroacetic acid for 1 h each. The extracted peptides were lyophilized in a SpeedVac and suspended in 20 μl of 1% formic acid-2% ACN. A portion of the peptide mixture (5 to 10 μl) was loaded onto a C18 reverse-phase column (13 by 25 μm; LC Packings, Sunnyvale, Calif.). Peptides were eluted directly into a Micromass electrospray ionization quadrupole-orthogonal time of flight mass (Q-ToF Micro) hybrid spectrometer with a 3 to 40% gradient of ACN-0.1% formic acid over 40 min and a flow rate of approximately 20 nl per min. Spectra were obtained in positive ion mode, deconvoluted, and analyzed with MassLynx 4.0 software (Micromass). Protein Lynx Global Server v. 1.2 (Micromass) was used to search databases consisting of S. pyogenes genome sequences and the NCBI nonredundant genomic databases. Proteins were identified by matching tandem mass spectrometry spectra from at least two tryptic peptides or by de novo peptide sequence determination when only one tandem mass spectrometry match was identified.
mRNA isolation, detection, and comparison.
RNA was prepared by growing cells overnight in TH broth. The following day, the cultures were diluted 1:10 into fresh prewarmed TH broth. Incubation was continued until cultures reached an OD600 between 0.50 and 0.65. These cells represent mid-exponential-phase cells, and total RNA was isolated from them using the hot phenol extraction method of Shaw and Clewell (44). The isolated RNA was quantitated by spectrophotometric determination using the OD260 of each sample. Denaturing agarose gel electrophoresis and Northern blotting were done on dilutions of the RNA representing 10, 5, and 2.5 μg of total cellular RNA using the method of Podbielski et al. (38). Even loading of total RNA was confirmed by visualization of ethidium bromide-stained gels. Digoxigenin-dUTP-labeled probes for genes of interest were generated by PCR using the following primer pairs: Spy0145 (Spy0145 FWD, 5′-TCTATACACCGCAGGCCAATTACC; and Spy0145 REV, 5′-TCTGGCGACAGCTTCGACTTCAAT); RNA polymerase alpha subunit (rpoA FWD, 5′-CCTGTGGGTACATTGGCAGTAGAT; and rpoA REV, 5′-GTCCGAGACCTAAGTCAGCAAGTT); NADH oxidase (nox FWD, 5′-GATATGGTTATCCTCGCCGTTGGT; and nox REV, 5′-CGCCTTCTTGGATAGCAAGTGAGA); phosphoglycerate kinase (pgk FWD, 5′-ATTCCCAGGTGTTACTCGTGGTTC; and pgk REV, 5′-GTCAAGACCAAGGAAGCCTTCTGA); and l-lactate dehydrogenase (ldh FWD, 5′-TCAGGTTTCCCTAAAGAGCGTGTC; and ldh REV, 5′-TACGAGCTAGGGCAACTGCAATAC). Hybrid-ization and visualization using disodium-3-(4-methoxyspiro[1,2-dioxetane-3′2′-(5-chloro)tricyclo(3.3.1.33,7)decan]-4-yl)phenylphosphate (CSPD) (Roche, Indianapolis, IN) were achieved as previously described (38). The Northern blots represent the result of at least two independent experiments using freshly isol-ated RNA for each blot.
RESULTS
S. pyogenes can survive in stationary phase for prolonged periods.
To determine if S. pyogenes could survive for extended periods of time (>4 weeks) and to ascertain the effects of growth medium on this survival, both complex and defined medium conditions were assayed for their ability to support a stationary-phase population. S. pyogenes M49-CS101 survival was assayed in the following medium types: (i) CDM, 0.2% final glucose concentration (wt/vol); (ii) CDM, 1.0% final glucose concentration (wt/vol); (iii) TH broth, 0.2% final glucose concentration (wt/vol); (iv) TH broth, 1.0% final glucose concentration (wt/vol); and (v) THY, 0.2% final glucose concentration (wt/vol). The CDM formulation used in this study has been previously used in our studies on short-term persistence (28). Cultures grown in media containing 1.0% glucose were determined to have excess glucose remaining during early stationary phase, while cultures grown in 0.2% glucose had exhausted their sugar supply by early stationary phase, as determined by the glucose detection kit (Sigma) (data not shown). TH broth containing either 0.2 or 1.0% glucose yielded stationary phase OD600s between 1.1 and 1.3, CDM containing 1.0% glucose yielded stationary phase OD600s between 1.2 and 1.4, and CDM containing 0.2% glucose yielded a stationary phase OD600 between 0.3 and 0.4. After inoculation into these five medium types, bacterial survival was monitored by colony regrowth on TH agar plates.
CDM did not support long-term stationary-phase survival of S. pyogenes. S. pyogenes M49-CS101 grown in CDM containing excess glucose (1.0%) survived for 1.5 ± 0.7 days. CDM containing growth-limiting glucose (0.2%) supported S. pyogenes survival for 5.2 ± 0.4 days after inoculation. In contrast, both TH and THY broths, where glucose was exhausted in late exponential phase, supported survival in excess of 1 year. The addition of exogenous glucose to TH broth, at a final concentration of 1.0%, caused S. pyogenes to become nonculturable after 1.0 ± 0.0 day postinoculation.
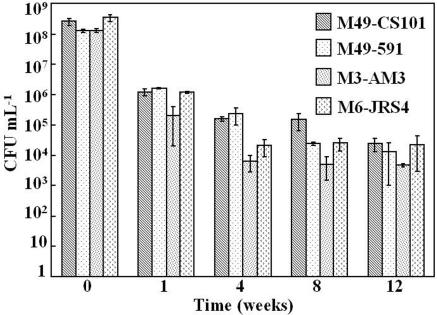
To determine whether long-term stationary-phase survival of S. pyogenes in TH broth was a strain-specific phenomenon, several strains were assayed for survival. In addition to S. pyogenes M49-CS101, strains M3-AM3, M6-JRS4, and M49-591 were assayed for the ability to survive in TH broth and for their survival kinetics. For this assay, survival kinetics were measured as CFU per milliliter over a 12-week period. At entry into stationary phase, all strains reached between 108 and 109 CFU ml−1 (Fig. 1). At 1 week in stationary phase, an average of between 105 and 106 CFU ml−1 were culturable, while at 12 weeks in stationary phase between 103 and 105 CFU ml−1 were culturable (Fig. 1). All strains maintained culturability for the duration of the assay (Fig. 1). These results demonstrate that long-term stationary-phase survival of S. pyogenes in TH broth is not limited to only M49-CS101.
FIG. 1.
The stationary-phase survival kinetics of four strains of S. pyogenes. Four different strains of S. pyogenes were inoculated into TH broth and monitored for exit from exponential phase (T0). Stationary-phase culture aliquots were then removed, serially diluted in sterile TH broth, and plated on TH agar plates to determine the number of CFU per milliliter. Bars in the graph represent the mean number of CFU per milliliter for each time point. Each data set is the mean of results from at least two independent experiments, quantitated in duplicate. Error bars represent the standard deviation between these experiments.
Culture pH is an important factor for persistence of S. pyogenes in stationary phase.
S. pyogenes is classified as a lactic acid bacterium, and cells produce large amounts of lactic acid in the presence of excess glucose. Since cultures grown in the presence of excess glucose survived for significantly shorter times than their cognate cultures, the terminal pH of each culture was assayed. The pH range for TH broth cultures containing excess glucose (1.0% glucose) ranged between 4.8 and 4.9, while the pH range for TH broth cultures containing limited glucose (0.2% [wt/vol] glucose) was between 5.6 and 6.2. Because TH broth cultures survive in excess of 1 year, the terminal pH was assayed after 1 week of stationary-phase survival and was subsequently stable. The pH range for CDM cultures containing excess glucose (1.0% glucose) was between 5.0 and 5.4, while the pH range for CDM cultures containing limited glucose (0.2% glucose) was between 6.5 and 6.9.
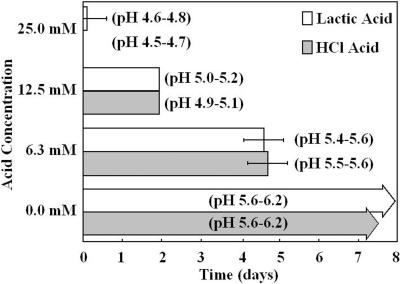
To test whether maintenance of pH is a precondition of survival, S. pyogenes cultures were grown in TH broth (0.2% glucose) to stationary phase and aged for 1 week. After 1 week of stationary phase survival, increasing concentrations of exogenous lactic acid and hydrochloric acid were added to each culture to artificially decrease the pH. Addition of either exogenous acid reduced culturability over time in a concentration-dependent manner (Fig. 2). Decreases in pH below 5.6, the minimum observed in TH broth cultures, caused culturability to be truncated from over 6 months to approximately 5 days (Fig. 2). Stationary-phase cultures with addition of no exogenous acid maintained the pH previously observed for TH broth cultures and were able to survive beyond the period of assay (Fig. 2). Taken together, these results indicate that pH maintenance is important for the long-term persistence of S. pyogenes in batch culture, though it is not the only factor affecting the outcome of stationary-phase survival.
FIG. 2.
The effects of pH decreases on S. pyogenes survival time. S. pyogenes M49-CS101 was inoculated into TH broth, grown to stationary phase, and aged 1 week. Exogenous HCl or lactic acid was then added from 2 M stocks to final concentrations of 25.0, 12.5, or 6.3 mM in these aged cultures. Survival was assayed by spotting culture aliquots on TH agar plates at 24-h time intervals after addition of exogenous acid. The formation of any S. pyogenes colonies within the culture spot was scored positive for survival. Since 20 μl of culture was plated to assay for survival, the lower limit of detection for this assay was 50 CFU ml−1. Bars in the graph represent the mean duration of survival for each culture condition. For this graph, T0 is at the point of addition of exogenous acid to 1-week-old cultures. The terminal pH range, after addition of exogenous acid, is noted next to each culture condition. Each data set is the mean of results from eight independent experiments. Error bars represent the standard deviation between these experiments.
S. pyogenes cells form alternate colony phenotypes after prolonged survival in stationary phase.
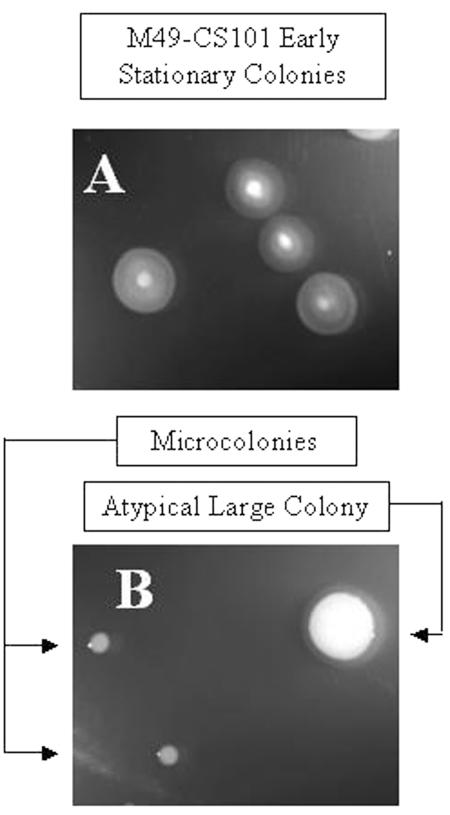
Previous studies have shown that phenotypic switching (i.e., changes in colony morphology) can occur in populations of S. pyogenes M49-CS101 if they are maintained in stationary phase in a complex medium, such as THY broth (28). To determine if phenotypic switching occurs in stationary-phase cultures grown and maintained in TH broth, colony morphology of stationary-phase persisters was monitored periodically over time in parallel cultures of both TH and THY broth. Cells aged for greater than 4 weeks in culture began to show phenotypic alterations when regrown on TH agar plates. These colony variants are termed here atypical large colonies and microcolonies (Fig. 3). The ratio of microcolonies to atypical large colonies was variable from culture to culture, with microcolonies making up anywhere from 10 to 100% of the bacterial colonies regrown on TH agar plates. Reversion to the original colony morphology was not observed after more than 10 passages (∼200 generations) on fresh TH agar. Both microcolonies and atypical large colonies regrew in TH broth without the addition of any osmotic support and stained gram positive, indicating that these colonies retained their cell wall (data not shown). Identical results were obtained with long-term survivors maintained in THY broth in stationary phase.
FIG. 3.
Phenotypic variants of surviving, stationary-phase S. pyogenes. S. pyogenes M49-CS101 was inoculated into TH broth, grown to stationary phase, and aged approximately 8 weeks. Culture aliquots were then removed, serially diluted, and plated on TH agar plates. S. pyogenes M49-CS101 microcolonies and alternative large colonies from the 8-week culture are pictured in panel B, while the early-stationary-phase (<12 h) precursor colonies are pictured in panel A. On plate medium, early-stationary-phase M49-CS101 cells grow with a dense center, yielding colonies with a “fried egg” appearance. These pictures are scaled relative to one another.
Nonculturable, membrane-intact S. pyogenes cells do not persist for more than 2 weeks after loss of culturability.
It has been suggested that conditions which are unfavorable for bacterial growth can lead to the formation of a viable but nonculturable (VBNC) state (30, 46, 49, 51). That is, cells retain membrane integrity and are capable of metabolism but are not culturable under standard laboratory conditions. During previous studies by Trainor et al. on surviving stationary-phase S. pyogenes, it was noted that only 0.01% of the cells remained culturable, while 33.0% of the cells had a functional membrane potential (46). For those studies, viability was assayed using rhodamine-123, a stain which measures membrane potential, and complete cell number was determined by counterstaining with 4′,6′-diamidino-2-phenylindole (DAPI) (46). These studies suggested a subpopulation of cells entered the VBNC state.
It has been demonstrated that cells which have entered a nonculturable state can be resuscitated if cultured under special growth conditions. Escherichia coli, Aeromonas hydrophila, and Vibrio vulnificus cells were resuscitated from an otherwise nonculturable state if they were grown in the presence of H2O2-degrading compounds, such as catalase or pyruvate, while regrowth of otherwise nonculturable V. vulnificus and Staphylococcus aureus required appropriate types and levels of nutrients (3, 14, 30, 49, 51). To address the possibility that a portion of stationary-phase S. pyogenes cells in our culture system may have entered the VBNC state, we aged M49-CS101 in TH broth in stationary phase for 2 weeks and then plated these cultures under a variety of culture conditions which have been known to resuscitate bacterial cells from the VBNC state (Table 1). No significant differences in culturability were observed (105 to 106 CFU ml−1) when 2-week-old stationary-phase cultures were regrown on media containing altered nutrient levels (diluted or concentrated TH, THY, or TH supplemented with sheep's blood) or antioxidants (catalase or pyruvate) (Table 1). Further, to eliminate the possibility that these streptococci were nonculturable as a result of the absence of osmotic support, we plated cells on TH supplemented with either sucrose or sorbitol. Neither condition increased the quantity of CFU above the levels observed if the cells were grown on TH agar alone (Table 1).
TABLE 1.
Regrowth of stationary-phase S. pyogenes
| Regrowth medium | CFU ml−1a | SD |
|---|---|---|
| Todd-Hewitt (1.0× concn)b | 4.4 × 105 | ±1.2 × 105 |
| Todd-Hewitt (0.5× concn)b | 4.3 × 105 | ±1.9 × 105 |
| Todd-Hewitt (2.0× concn)b | 1.6 × 105 | ±2.1 × 104 |
| Todd-Hewitt yeast extractb | 1.9 × 105 | ±6.0 × 104 |
| Todd-Hewitt with sheep's bloodb | 3.0 × 105 | ±1.2 × 105 |
| Todd-Hewitt-0.25 M sucroseb | 1.7 × 105 | ±2.1 × 104 |
| Todd-Hewitt-0.50 M sucrosec | 2.9 × 105 | ±8.4 × 104 |
| Todd-Hewitt-0.50 M sorbitold | 2.4 × 105 | ±7.0 × 104 |
| Todd-Hewitt-500 U catalaseb,e | 3.0 × 105 | ±5.5 × 104 |
| Todd-Hewitt-1,000 U catalaseb,e | 2.3 × 105 | ±1.0 × 104 |
| Todd-Hewitt-40 mg pyruvateb,f | 5.3 × 105 | ±1.4 × 105 |
| Todd-Hewitt-80 mg pyruvateb,f | 4.2 × 105 | ±1.3 × 105 |
The number of CFU mL−1 was determined by serial dilution and subsequent platings of S. pyogenes M49-CS101 cultures aged 2 weeks in stationary phase as described in Materials and Methods.
Serial diluent was TH broth.
Serial diluent was 0.50 M sucrose solution.
Serial diluent was 0.50 M sorbitol solution.
A freshly made catalase solution was spread onto TH agar before plating.
A freshly made pyruvate solution was spread onto TH agar before plating.
Previous studies demonstrated the presence of VBNC cells only as a subpopulation in the presence of culturable cells (46). Therefore, it is formally possible that these cells are not VBNC cells but damaged cells that are dying out during the slow turnover of the culture. To examine this, S. pyogenes M49-CS101 was inoculated into TH broth containing a final concentration of 1.0% glucose and grown into stationary phase. After 24 h in stationary phase, no culturable bacteria were recoverable. These cultures were also stained with the BacLight live-dead viability stain to determine the percentage of membrane-intact cells. The BacLight stain consisted of SYTO-9, which stains all cells regardless of membrane state, and propidium iodide, a membrane-impermeable fluorescent dye. Therefore, cell viability was based on membrane integrity; cells which stained with both fluorescent dyes were scored as nonviable, while cells which stained with SYTO-9 only were scored as viable. After 24 h in stationary phase, approximately 0.83% of the initial cell population was scored as viable using the BacLight stain even though no culturable cells were detected (data not shown). However, after 2 weeks in stationary phase, neither culturable cells nor membrane-intact cells were detected (data not shown).
Two microcolony strains have significantly altered proteomes.
Two microcolony strains (termed here Alt. 1 and Alt. 2) were isolated from independent 14-week-old cultures of M49-CS101, as described in Materials and Methods. To characterize these two microcolony strains, we employed a two-dimensional proteomic method to compare changes in the mid-exponential proteomes of M49-CS101, Alt. 1, and Alt. 2. Final protein levels can be affected by changes in transcription, mRNA stability, translation, proteolytic processing or degradation, and posttranslational modifications (5, 16). In addition, since our strains are long-term persisters, mutations could accumulate in the proteins that would change their pI or molecular weight. Though Alt. 1 and Alt. 2 exhibit similar colony morphologies, their mid-exponential proteomes showed changes in both common and unique pathways.
Sixty-seven independent protein spots were changed between M49-CS101 and Alt. 1 or Alt. 2. Thirty-three of these spots were positively identified as described in Materials and Methods (Table 2). These 33 spots represented 23 individual proteins, since some of the spots represented multiple isoforms of the same protein (Table 2). The remaining 34 spots were unidentified (data not shown). Functional categories encompassing proteins used for processes including division, metabolism, transcription, and translation were identified (Table 2). Additionally, proteins used in the heat shock response, proteolysis, and GTP binding and proteins of unknown function were identified (Table 2). There was significant variability in the absolute values of protein spot quantitation among experiments. In addition to standard experimental variation, additional variation in protein spot quantitation may have resulted from batch-to-batch variations in the preparation of rich media, which is likely to affect the abundance of metabolic enzymes. In addition, low levels of reversion from microcolony to parental phenotypes could occur and similarly influence the experimental variation in protein abundance.
TABLE 2.
Identified changes in the proteomes of S. pyogenes microcolonies
| Functional category and identity | Function | Quantitationa
|
||
|---|---|---|---|---|
| M49-CS101 | Alt. 1 | Alt. 2 | ||
| Division | ||||
| Spy1514 | Cell division initiation protein (DivIVAS) | 0 ± 0 | 3,181 ± 2,004 | (0; 0; 854) |
| Spy1514 | Cell division initiation protein (DivIVAS) | 0 ± 0 | (289; 283; 2,176)b | (0; 0; 1,868) |
| Spy1514 | Cell division initiation protein (DivIVAS) | 6,442 ± 907 | 1,239 ± 721 | (0; 0; 926) |
| Spy1520 | Cell division protein (FtsZ) | 12,835 ± 650 | 5,301 ± 1,173 | 4,380 ± 1,760 |
| Metabolic | ||||
| Spy0731 | Enolase | 0 ± 0 | 1,588 ± 205 | (0; 0; 1,155) |
| Spy0731 | Enolase | 0 ± 0 | 437 ± 198 | (0; 802; 168) |
| Spy0731 | Enolase | 20,778 ± 1,599 | 47,082 ± 2,448 | 28,886 ± 23,407 |
| Spy0927 | Adenine phosphoribosyltransferase | 730 ± 175 | (0; 737; 0) | 0 ± 0 |
| Spy1128 | Phosphotransacetylase | 6,337 ± 217 | 3,557 ± 570 | (618; 0; 2,135) |
| Spy1150 | NADH oxidase (NOX) | 0 ± 0 | 585 ± 256 | 352 ± 14 |
| Spy1151 | l-lactate dehydrogenase | 3,619 ± 1,195 | (0; 3,988; 1,918) | 0 ± 0 |
| Spy1541 | Carbamate kinase | 1,351 ± 1,059 | (0; 559; 0) | 0 ± 0 |
| Spy1541 | Carbamate kinase | 16,592 ± 3,443 | 2,906 ± 508 | 1,874 ± 1,189 |
| Spy1618 | Cysteine synthase/O-acetylserine lyase | 158 ± 38 | 759 ± 182 | (0; 0; 579) |
| Spy1745 | Biotin carboxylase | 45 ± 64 | 444 ± 157 | (0; 0; 325) |
| Spy1881 | Phosphoglycerate kinase | 789 ± 231 | 1,576 ± 832 | 0 ± 0 |
| Transcription | ||||
| Spy0080 | DNA-directed RNA polymerase alpha subunit | 615 ± 304 | (0; 668; 605) | 0 ± 0 |
| Spy0611 | Translation elongation factor EF-Tu | 4,914 ± 201 | 11,520 ± 2,873 | 3,915 ± 2,908 |
| Spy0611 | Translation elongation factor EF-Tu | 1,809 ± 582 | (802; 1,034; 0) | 0 ± 0 |
| Spy0611 | Translation elongation factor EF-Tu | 0 ± 0 | 1,424 ± 257 | 0 ± 0 |
| Spy2093 | Elongation factor Ef-Ts | 201 ± 138 | (1,059; 995; 0) | 0 ± 0 |
| Spy2093 | Elongation factor Ef-Ts | 4,448 ± 2,762 | 1,383 ± 441 | 0 ± 0 |
| Miscellaneous | ||||
| M18_1302 | Hypothetical phage protein | 4,252 ± 1,692 | 247 ± 233 | 0 ± 0 |
| Spy0022 | Fatty acid/phospolipid synthesis | 558 ± 115 | 1,300 ± 123 | 1,548 ± 844 |
| Spy0022 | Fatty acid/phospolipid synthesis | 42 ± 60 | 363 ± 111 | (0; 0; 159) |
| Spy0022 | Fatty acid/phospolipid synthesis | 1,959 ± 1,713 | (1,080; 2,064; 0) | 0 ± 0 |
| Spy0123 | Heat shock protein HSP33 | 9,700 ± 1,383 | 3,520 ± 726 | 4,628 ± 2,996 |
| Spy0145 | Conserved hypothetical protein | 2,188 ± 65 | 0 ± 0 | 0 ± 0 |
| Spy0416 | Cell envelope proteinase | 12,023 ± 3,354 | 78 ± 134 | 9,932 ± 8,623 |
| Spy1527 | GTP binding protein TypA/BipA | 2,523 ± 259 | 3,184 ± 179 | 1,300 ± 1,216 |
| Spy1760 | Heat shock protein 70 | 1,429 ± 1,164 | 32 ± 28 | 0 ± 0 |
| Spy1896 | Trigger factor (prolyl isomerase) | 2,318 ± 69 | 2,395 ± 1,558 | (0; 1,257; 0) |
| Spy2010 | C5a peptidase precursor | 379 ± 212 | 32 ± 55 | 0 ± 0 |
Quantitative values for protein spots are in pixels per spot area from two independent experiments for M49-CS101 and three independent experiments for Alt. 1 and Alt. 2. The values are given as a mean ± standard deviation for M49-CS101, Alt. 1, and Alt. 2 if the values obtained were consistent across all replicates. If the spot quantitation was inconsistent across the three replicates for either Alt. 1 or Alt. 2, individual values in pixels per spot area are given for the three independent replicates. These are considered lower-confidence values.
These data are given as three individual data points, rather than a mean ± standard deviation, for clarity. Since these Alt. 1 data points are consistently elevated above what is observed for M49-CS101, this spot is considered a high-confidence increase.
When compared to M49-CS101, there were 38 protein spots upregulated in Alt. 1 and 12 protein spots upregulated in Alt. 2. Six of the upregulated Alt. 2 protein spots were quantitated with lower confidence due to inconsistent expression in one of three experiments (Table 2 and data not shown). Including these six spots, 12 protein spots were commonly upregulated in both Alt. 1 and Alt. 2. Some of these proteins were identified as enolase, fatty acid-phospholipid synthesis protein, and NADH oxidase (NOX) (Table 2). Though enolase and the fatty acid/phospholipid synthesis protein were both found in multiple isoforms, only one isoform of each was upregulated (Table 2). The additional 16 protein spots upregulated only in Alt. 1 included two isoforms of enolase, one isoform of fatty acid-phospholipid synthesis protein, a GTP binding protein, cysteine synthase, and biotin carboxylase (Table 2).
When compared to M49-CS101, 19 protein spots were downregulated in Alt. 1 and 28 protein spots were downregulated in Alt. 2. Four of these protein spots from Alt. 1 and one of these protein spots from Alt. 2 showed variability in one experiment. Overall, there were 18 commonly downregulated protein spots in the Alt. 1 and Alt. 2 proteomes. These proteins included cell division protein FtsZ, one of two isoforms of carbamate kinase, the precursor protein to C5a peptidase, heat shock proteins HSP33 and HSP70, phosphotransacetylase, a hypothetical phage protein (M18_1302), and a conserved hypothetical protein (Spy0145) (Table 2). A single protein spot was uniquely downregulated in Alt. 1, while 10 protein spots were uniquely down regulated in Alt. 2. The single protein spot which was uniquely downregulated in Alt. 1 was identified as a cell envelope proteinase (Table 2).
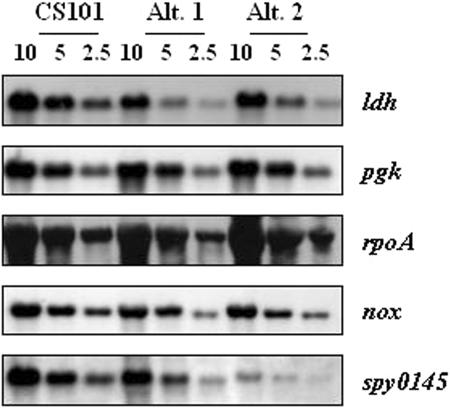
An isoform of the alpha subunit of RNA polymerase (RpoA), which migrated at ∼45 kDa, was detected among the proteins isolated from M49-CS101 and Alt. 1 cultures but not from Alt. 2 cultures. The predicted molecular mass of the subunit is ∼35 kDa. Because RpoA is essential (21) and because an unmodified form was not identified in any of the gels, Northern blotting was done to assess rpoA transcription. Similar levels of transcripts encoding the alpha subunit of RpoA were detected in Alt. 1, Alt. 2, and M49-CS101 cultures (Fig. 4). In addition, an isoform of phosphoglycerate kinase (PGK), which migrated at ∼30 kDa, was detected among proteins isolated from M49-CS101 and Alt. 1 cultures but not from Alt. 2 cultures; PGK is predicted to have a molecular mass of ∼42 kDa. Northern blotting showed that the levels of transcripts encoding PGK are similar in M49-CS101, Alt. 1, and Alt. 2 cultures. It seems likely that additional isoforms of RpoA and PGK are present among the unidentified protein spots or may be below the limit of detection. Nonetheless, the nature, origin, and functional significance of the protein isoforms remain unknown.
FIG. 4.
mRNA expression of typical M49-CS101 colonies and stationary-phase-derived microcolony phenotypes Alt. 1 and Alt. 2. mRNA was isolated from mid-exponential-phase cells (OD600 of 0.50 to 0.65) grown in TH broth. The RNA concentration was determined by spectrophotometric measurement at OD260, appropriate dilutions were made, and RNA from each strain was run on a denaturing agarose gel. Equal loading was confirmed visually by ethidium bromide staining. RNA concentrations are noted above the lanes in each image. These RNA gels were subsequently Northern blotted, and the binding of DNA probes, generated by PCR-mediated digoxigenin incorporation, was determined by CSPD development followed by autoradiography. Specific probes are indicated beside each blot. Fresh RNA was isolated for each experiment, and the results presented here are representative of those from two independently derived M49-CS101, Alt. 1, and Alt. 2 RNA preparations.
LDH, an essential protein in Streptococcus mutans, was not detected in Alt. 2 and was variable in three Alt. 1 cultures (Table 2) (6, 24). Transcripts corresponding to ldh were found in substantially lower quantities in both Alt. 1 and Alt. 2 than in the origin strain M49-CS101 (Fig. This suggested the loss of the LDH protein spot was due to a decrease in transcription or specific mRNA stability. S. pyogenes has a NOX gene (Spy 1150) that may be able to function to recycle NADH in the absence of LDH (20, 40, 42). In agreement with this, a protein spot identified as NOX was upregulated in both Alt. 1 and Alt. 2 (Table 2). NOX upregulation did not appear to be at the level of transcription, since the nox transcript was observed at similar levels for all strains examined (Fig. 4). This suggests that a change had occurred in the posttranscriptional regulation of NADH oxidase in both Alt. 1 and Alt. 2 (Fig. 4).
Spy0145, a protein of unknown function, was not detected in the proteomes of either Alt. 1 or Alt. 2 but was present in the proteome of M49-CS101 (Table 2). Transcription of spy0145 was reduced in Alt. 1 or Alt. 2 compared to M49-CS101 (Fig. 4). This suggests that the absence of a detectable protein spot corresponding to Spy0145 in the Alt. 1 and Alt. 2 proteomes was at least partially a result of a change in regulation at the transcriptional level or in specific mRNA stability.
DISCUSSION
In this paper, we describe the long-term stationary-phase survival of S. pyogenes in batch culture. We found that TH broth, a complex medium containing a carbon source which was exhausted by early stationary phase, supported bacterial survival in excess of 1 year, but CDM did not. The presence of significant amounts of excess sugar after the cessation of growth in cultures containing 1.0% initial glucose concentrations shortened survival in both TH and CDM cultures. In these cultures, entry into stationary phase was not mediated by carbon starvation but instead was probably induced by depletion of another nutrient, cell density, or buildup of toxic products. Two of the most likely explanations for the shortened stationary-phase persistence in these cultures are the accumulation of high levels of lactic acid or the lack of a sugar starvation signal that is necessary to induce a survival response. Since streptococci are classified as lactic acid bacteria, the presence of large amounts of usable glucose at stationary phase represents a carbon source for continued sugar fermentation, lactic acid accumulation, and subsequent pH decrease. The induction of an in vitro survival response resultant from carbon starvation has been previously observed for gram-positive organisms, such as Staphylococcus aureus and Listeria monocytogenes, as well as gram-negative organisms, including E. coli and Salmonella enterica serovar Typhimurium (19, 22, 45, 50). In our studies, the presence of excess (1.0%) glucose in TH broth caused the culture pH to drop to from a range of 5.6 to 6.2 to a range of 4.8 to 4.9, and the bacteria survived for less than 24 h in stationary phase. If either lactic acid (an organic acid) or HCl (an inorganic acid) was used to lower the pH of sugar-starved cultures to pH ∼4.8, the cultures survived for less than 2 days after the addition of exogenous acid. Survival was significantly shortened at pHs of <5.5 regardless of whether lactic acid or HCl was used to reduce the pH. It has been proposed that organic acid stress is different than stresses on cells caused by inorganic acids (1, 9). In gram-negative organisms, such as S. enterica serovar Typhimurium, E. coli, and L. monocytogenes, protection against protonated organic acids and strong acids has been shown to require, at least in part, different regulators (2, 18, 27). Since the addition of both HCl and lactic acid resulted in similar death curves, it is unlikely that the killing is a weak organic acid effect, though it cannot be ruled out. It should be noted that both HCl and lactic acid were added exogenously, whereas in the presence of 1.0% glucose, lactic acid is produced intracellularly, prior to export from the cell, and may have a different effect on intracellular pH. Taken together, these data suggest that control of final culture pH is a prerequisite for survival in stationary phase.
Although a pH level below 5.6 is a limitation for long-term survival of S. pyogenes in stationary phase, maintenance above this value does not ensure survival. In our studies, carbon-starved CDM cultures survived less than 1 week, while carbon-starved TH cultures survived in excess of 1 year. The OD600s of the carbon-starved CDM cultures reached ∼0.3, whereas carbon-starved TH cultures reached an OD600 of ∼1.1 to 1.3. Prolonged survival may depend on a high final culture density at entry into stationary phase. In addition, previous studies performed by Trainor et al. showed that S. pyogenes serotypes M52 and M62 remained culturable in a chemically defined medium for at least 3 weeks in stationary phase under conditions of carbon starvation (46). The total time of persistence in their studies is unknown, since survival was not measured beyond 3 weeks (46). Their CDM formulation differs from the one used in this study in terms of amino acid, vitamin, NADH, and salt concentrations (13). Therefore, the nutrient concentrations and/or NADH/NAD+ ratios may affect the maintenance of a stationary-phase population of S. pyogenes.
The survival response observed in this study is characterized by biphasic survival kinetics. A rapid decrease in overall culturability is followed by the relative stabilization of the surviving bacterial subpopulation. Similar survival kinetics were observed for carbon starvation-induced survival systems described for S. pyogenes, Staphylococcus aureus, and L. monocytogenes (22, 46, 50). Comparable survival responses were observed when both TH and THY broths were used as growth medium, indicating that yeast extract does not influence either survival kinetics or duration of culturability (data not shown). The survival kinetics of the four S. pyogenes strains examined in this study were similar, highlighting that the induction of long-term starvation survival response for S. pyogenes is not limited to M49-CS101.
The formation of VBNC cells among stationary-phase S. pyogenes cells was also investigated. This phenomenon has been previously reported by Trainor et al. (46). They observed a significantly larger number of cells with a functional membrane potential than were able to be cultured by standard streptococcal culture techniques. We observed that cultures of cells killed as a result of prolonged incubation at pH 4.8 (2-week stationary phase in TH with 1.0% glucose condition) yielded no cells retaining intact membranes. However, early-stationary-phase cells which are nonculturable due to pH damage (24-h stationary phase in TH with 1.0% glucose condition) yielded a small percentage of cells retaining membrane integrity. This suggests that there may be a time differential between loss of culturability and loss of membrane integrity for stationary-phase populations of S. pyogenes. Consequently, cells which maintain intact membranes, as measured by fluorescence techniques, may actually represent cells damaged beyond recovery rather than true VBNC cells (34, 35). Nonculturable cells with intact membranes were also found in S. mutans batch and biofilm culture after formaldehyde killing, suggesting dead or damaged cells which stain as “live” are not limited to S. pyogenes (39).
Consistent with this, we were unable to resuscitate the subpopulation of stationary-phase S. pyogenes cells which retained intact membranes but were previously unculturable. Populations of otherwise nonculturable A. hydrophila, V. vulnificus, and E. coli have all been cultured when regrown in the presence of the H2O2-degrading enzyme catalase or the antioxidant pyruvate (3, 30, 49); populations of otherwise unculturable Staphylococcus aureus and V. vulnificus have been shown to require the appropriate types and concentrations of nutrients for recovery (14, 51). Efforts to recover this stationary-phase subpopulation of S. pyogenes by coincubation under analogous conditions resulted in no significant increases in culturability. These data further support the notion that a subpopulation of stationary-phase cells which stain viable with fluorescence techniques may not be VBNC and may instead be damaged beyond their ability to be cultured. Their continued presence in surviving cultures may indicate a slow cell turnover in stationary phase, such as that which has been observed for E. coli and short-term persistence in S. pyogenes (19, 46).
The surviving subpopulations grew with altered phenotypes beginning after approximately 4 weeks, yielding small colonies and atypical large colonies. These phenotypes were stable for greater than 200 generations when they were cultured on TH agar plates. Previous studies have described the ability of S. pyogenes, maintained for greater than 24 h in stationary phase in THY broth, to yield both atypical large and small colonies when regrown on fresh THY plates (28). The colony morphology of these small colonies was associated with the downregulation of capsule and other virulence factors (28). Furthermore, this phenotypic dimorphism was unstable; serial passaging of small colonies on fresh plate media caused reversion to typical large colonies within 100 generations. This is in contrast to the phenotype stability seen in long-term survivors of S. pyogenes M49-CS101 in carbon-depleted TH broth when replated on TH agar plates. Trainor et al. did not observe these altered phenotypes in their stationary-phase cultures; however, the M49-CS101 strain used in these studies produces large amounts of capsule, making any changes in colony size easily recognizable (28, 46). Further studies will need to be done with microcolony-specific markers to determine if all serotypes of S. pyogenes form alternate colony phenotypes during stationary-phase persistence or if this is a strain-specific phenomenon.
The switch to stable atypical large colonies and microcolonies in over 25 independent cultures suggests there is some selective pressure or phase variation favoring the formation of these two phenotypes. The microcolony phenotype seen here is similar to the unstable small-colony phenotype observed in previous studies, which is consistent with phase variation of genes which alter colony morphology (8, 28). The presence of microcolony proteomes (e.g., Alt. 1 and Alt. 2) which differ from each other and the parental strain suggest that the genomes also have accumulated mutations during persistence in stationary phase. Mutation rates are elevated for many bacterial species in stationary phase (as reviewed by Kivisaar [25]), and mutations to the constituents of stationary-phase batch cultures of E. coli have been reported in detail (19, 25, 52, 53). In E. coli cells, these mutations conferred a survival advantage during periods of starvation when cells from aged cultures were competed against their parental strain (19, 52, 53). When independent cultures of the same parental strain were aged in stationary-phase batch culture, they accumulated different mutations when the survivors were compared (19). In Alt. 1 and Alt. 2, it is unclear what advantages, in terms of either persistence or pathogenesis, the changes to the proteomes have conferred. Overall, it seems likely that a combination of cumulative mutation and locked-phase variation of relevant genes may be responsible for the appearance of the alternate colony phenotypes observed in this study.
Changes to the proteomes of both Alt. 1 and Alt. 2 were not limited to a single functional category and could be observed in proteins involved in transcription, translation, division, and other miscellaneous processes; however, the category with the most identified changes was metabolism. There were identifiable proteomic changes in major glycolytic enzymes in both Alt. 1 and Alt. 2, including enolase, PGK, and lactate dehydrogenase (LDH) (Table 2). Enolase showed shifts in its isoform pattern in both Alt. 1 and Alt. 2 (Table 2). Both RpoA and PGK spots were not identified in Alt. 2, while transcript levels remained unchanged (Table 2 and Fig. 4). In M49-CS101 and Alt. 1, both RpoA and PGK had molecular weights significantly different from the predicted molecular weight. The inability to detect PGK and RpoA in Alt. 2 may be the result of changes in protein processing or accumulation of mutations that change the pI or molecular weight of the protein. These observations regarding RpoA and PGK emphasize the general need for caution in interpreting proteomic data. The detection of positional variants is consistent with what has been observed for the comprehensive proteome map for actively dividing Bacillus subtilis cells (5, 16). Up to 183 proteins have been observed to have multiple forms which result from the posttranslational modifications or the processing these proteins have experienced (5, 16). In general, the functional significance of such variants is poorly understood. The variant of RpoA detected in samples obtained from M49-CS101 and Alt. 1 was not detected in samples prepared from Alt. 2 cultures; however, because RpoA is essential, one or more RpoA protein spots is presumably among those proteins which were not identified. This interpretation is supported by results obtained with Northern blotting, which showed wild-type levels of rpoA transcripts in both the Alt. 1 and Alt. 2 strains. Therefore, the results obtained with proteomics revealed clear changes in protein composition between the wild-type and persister strains and identified several proteins for further investigation; however, it remains to be determined which differences are responsible for enhanced persistence of S. pyogenes in the stationary phase of growth.
ldh transcription was reduced, indicating regulation of transcription was responsible for the observed reduction of this enzyme in Alt. 1 (two of three proteomic gels) and Alt. 2 (three of three proteomic gels) (Table 2 and Fig. 4). Changes in these glycolytic enzymes suggest that S. pyogenes may be using an alternate pathway for energy generation. The upregulation of NOX and the reduction in LDH suggest that NOX may play a greater role in recycling NADH in the microcolonies. NOX has been found to be important in aerobic metabolism and is proposed to have a role in an alternative pathway for energy production in the absence of glucose (20, 23, 42). The reliance on NOX to recycle NADH has been noted under conditions of high oxygen tension and under conditions of intermediate oxygen tension and glucose limitation (20).
Although the proteomic analysis of Alt. 1 and Alt. 2 is limited without a fully annotated reference map of the M49-CS101 proteome, it gives an excellent start point for future studies. Taken together, these data show that S. pyogenes can persist for longer than 1 year in stationary phase. This persistence depends on maintenance of pH above 5.6 and the presence of the nutrients in complex medium. Microcolonies with a stable colony morphology are isolated after 4 weeks in stationary phase. Comparison of the proteomes of two microcolonies isolated from two independent colonies revealed a combination of shared and unique changes. Clearly, these changes, as discussed above, can occur by several mechanisms, and further analysis is needed to determine the significance of the changes. Some of the shared changes suggest that microcolonies may use lactate and amino acids as a carbon and energy source. This metabolism may be important for continued slow growth and turnover of cells that allow for persistence and the further accumulation of mutations during stationary phase.
Acknowledgments
We thank P. J. Piggot, Vasant Chary, David Hilbert, John Renye, Shannon Morgan, Valorie Mazeffa, Mauro Meloni, and A. Podbielski for helpful discussions. We thank J. R. Scott and C. P. Moran, Jr., for kindly providing S. pyogenes strains M3-AM3 and M6-JRS4. We thank T. S. Panetti for help with, and the use of, the fluorescent microscope. We thank M. Monestier for use of the camera used to generate colony phenotype pictures.
This work was supported by grant 0160430U from the American Heart Association (to B.A.B.).
REFERENCES
- 1.Audia, J. P., C. C. Webb, and J. W. Foster. 2001. Breaking through the acid barrier: an orchestrated response to proton stress by enteric bacteria. Int. J. Med. Microbiol. 291:97-106. [DOI] [PubMed] [Google Scholar]
- 2.Bearson, B. L., L. Wilson, and J. W. Foster. 1998. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J. Bacteriol. 180:2409-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogosian, G., N. D. Aardema, E. V. Bourneuf, P. J. Morris, and J. P. O'Neil. 2000. Recovery of hydrogen peroxide-sensitive culturable cells of Vibrio vulnificus gives the appearance of resuscitation from a viable but nonculturable state. J. Bacteriol. 182:5070-5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandt, C. M., F. Allerberger, B. Spellerberg, R. Holland, R. Lutticken, and G. Haase. 2001. Characterization of consecutive Streptococcus pyogenes isolates from patients with pharyngitis and bacteriological treatment failure: special reference to prtF1 and sic / drs. J. Infect. Dis. 183:670-674. [DOI] [PubMed] [Google Scholar]
- 5.Buttner, K., J. Bernhardt, C. Scharf, R. Schmid, U. Mader, C. Eymann, H. Antelmann, A. Volker, U. Volker, and M. Hecker. 2001. A comprehensive two-dimensional map of cytosolic proteins of Bacillus subtilis. Electrophoresis 22:2908-2935. [DOI] [PubMed] [Google Scholar]
- 6.Chen, A., J. D. Hillman, and M. Duncan. 1994. l-(+)-lactate dehydrogenase deficiency is lethal in Streptococcus mutans. J. Bacteriol. 176:1542-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cleary, P. P., and D. Cue. 2000. High frequency invasion of mammalian cells by beta hemolytic streptococci. Subcell. Biochem. 33:137-166. [DOI] [PubMed] [Google Scholar]
- 8.Cleary, P. P., L. McLandsborough, L. Ikeda, D. Cue, J. Krawczak, and H. Lam. 1998. High-frequency intracellular infection and erythrogenic toxin A expression undergo phase variation in M1 group A streptococci. Mol. Microbiol. 28:157-167. [DOI] [PubMed] [Google Scholar]
- 9.Cotter, P. D., and C. Hill. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67:429-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cottrell, M. T., and D. L. Kirchman. 2000. Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl. Environ. Microbiol. 66:5116-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courtney, H. S., D. L. Hasty, and J. B. Dale. 2002. Molecular mechanisms of adhesion, colonization, and invasion of group A streptococci. Ann. Med. 34:77-87. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dassy, B., and J. E. Alouf. 1983. Growth of Streptococcus pyogenes and streptolysin O production in complex and synthetic media. J. Gen. Microbiol. 129:643-651. [DOI] [PubMed] [Google Scholar]
- 14.Diaper, J. P., and C. Edwards. 1994. Survival of Staphylococcus aureus in lakewater monitored by flow cytometry. Microbiology 140:35-42. [DOI] [PubMed] [Google Scholar]
- 15.Dinkla, K., M. Rohde, W. T. Jansen, J. R. Carapetis, G. S. Chhatwal, and S. R. Talay. 2003. Streptococcus pyogenes recruits collagen via surface-bound fibronectin: a novel colonization and immune evasion mechanism. Mol. Microbiol. 47:861-869. [DOI] [PubMed] [Google Scholar]
- 16.Eymann, C., A. Dreisbach, D. Albrecht, J. Bernhardt, D. Becher, S. Gentner, T. Tam le, K. Buttner, G. Buurman, C. Scharf, S. Venz, U. Volker, and M. Hecker. 2004. A comprehensive proteome map of growing Bacillus subtilis cells. Proteomics 4:2849-2876. [DOI] [PubMed] [Google Scholar]
- 17.Facinelli, B., C. Spinaci, G. Magi, E. Giovanetti, and P. E. Varaldo. 2001. Association between erythromycin resistance and ability to enter human respiratory cells in group A streptococci. Lancet 358:30-33. [DOI] [PubMed] [Google Scholar]
- 18.Ferreira, A., D. Sue, C. P. O'Byrne, and K. J. Boor. 2003. Role of Listeria monocytogenes σB in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl. Environ. Microbiol. 69:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finkel, S. E., and R. Kolter. 1999. Evolution of microbial diversity during prolonged starvation. Proc. Natl. Acad. Sci. USA 96:4023-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson, C. M., T. C. Mallett, A. Claiborne, and M. G. Caparon. 2000. Contribution of NADH oxidase to aerobic metabolism of Streptococcus pyogenes. J. Bacteriol. 182:448-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayward, R. S., K. Igarashi, and A. Ishihama. 1991. Functional specialization within the alpha-subunit of Escherichia coli RNA polymerase. J. Mol. Biol. 221:23-29. [DOI] [PubMed] [Google Scholar]
- 22.Herbert, K. C., and S. J. Foster. 2001. Starvation survival in Listeria monocytogenes: characterization of the response and the role of known and novel components. Microbiology 147:2275-2284. [DOI] [PubMed] [Google Scholar]
- 23.Higuchi, M., Y. Yamamoto, L. B. Poole, M. Shimada, Y. Sato, N. Takahashi, and Y. Kamio. 1999. Functions of two types of NADH oxidases in energy metabolism and oxidative stress of Streptococcus mutans. J. Bacteriol. 181:5940-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hillman, J. D., A. Chen, M. Duncan, and S. W. Lee. 1994. Evidence that l-(+)-lactate dehydrogenase deficiency is lethal in Streptococcus mutans. Infect. Immun. 62:60-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kivisaar, M. 2003. Stationary phase mutagenesis: mechanisms that accelerate adaptation of microbial populations under environmental stress. Environ. Microbiol. 5:814-827. [DOI] [PubMed] [Google Scholar]
- 26.Kuo, C. F., J. J. Wu, P. J. Tsai, F. J. Kao, H. Y. Lei, M. T. Lin, and Y. S. Lin. 1999. Streptococcal pyrogenic exotoxin B induces apoptosis and reduces phagocytic activity in U937 cells. Infect. Immun. 67:126-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lambert, L. A., K. Abshire, D. Blankenhorn, and J. L. Slonczewski. 1997. Proteins induced in Escherichia coli by benzoic acid. J. Bacteriol. 179:7595-7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leonard, B. A., M. Woischnik, and A. Podbielski. 1998. Production of stabilized virulence factor-negative variants by group A streptococci during stationary phase. Infect. Immun. 66:3841-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michailova, L., N. Markova, T. Radoucheva, S. Stoitsova, V. Kussovski, and M. Jordanova. 2000. Atypical behaviour and survival of Streptococcus pyogenes L forms during intraperitoneal infection in rats. FEMS Immunol. Med. Microbiol. 28:55-65. [DOI] [PubMed] [Google Scholar]
- 30.Mizunoe, Y., S. N. Wai, A. Takade, and S. Yoshida. 1999. Restoration of culturability of starvation-stressed and low-temperature-stressed Escherichia coli O157 cells by using H2O2-degrading compounds. Arch. Microbiol. 172:63-67. [DOI] [PubMed] [Google Scholar]
- 31.Molinari, G., and G. S. Chhatwal. 1999. Streptococcal invasion. Curr. Opin. Microbiol. 2:56-61. [DOI] [PubMed] [Google Scholar]
- 32.Molinari, G., M. Rohde, C. A. Guzman, and G. S. Chhatwal. 2000. Two distinct pathways for the invasion of Streptococcus pyogenes in non-phagocytic cells. Cell. Microbiol. 2:145-154. [DOI] [PubMed] [Google Scholar]
- 33.Neeman, R., N. Keller, A. Barzilai, Z. Korenman, and S. Sela. 1998. Prevalence of internalisation-associated gene, prtF1, among persisting group-A streptococcus strains isolated from asymptomatic carriers. Lancet 352:1974-1977. [DOI] [PubMed] [Google Scholar]
- 34.Nystrom, T. 2003. Nonculturable bacteria: programmed survival forms or cells at death's door? Bioessays 25:204-211. [DOI] [PubMed] [Google Scholar]
- 35.Nystrom, T. 2001. Not quite dead enough: on bacterial life, culturability, senescence, and death. Arch. Microbiol. 176:159-164. [DOI] [PubMed] [Google Scholar]
- 36.Osterlund, A., and L. Engstrand. 1997. An intracellular sanctuary for Streptococcus pyogenes in human tonsillar epithelium—studies of asymptomatic carriers and in vitro cultured biopsies. Acta Otolaryngol. 117:883-888. [DOI] [PubMed] [Google Scholar]
- 37.Osterlund, A., R. Popa, T. Nikkila, A. Scheynius, and L. Engstrand. 1997. Intracellular reservoir of Streptococcus pyogenes in vivo: a possible explanation for recurrent pharyngotonsillitis. Laryngoscope 107:640-647. [DOI] [PubMed] [Google Scholar]
- 38.Podbielski, A., A. Flosdorff, and J. Weber-Heynemann. 1995. The group A streptococcal virR49 gene controls expression of four structural vir regulon genes. Infect. Immun. 63:9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renye, J. A., Jr., P. J. Piggot, L. Daneo-Moore, and B. A. Buttaro. 2004. Persistence of Streptococcus mutans in stationary-phase batch cultures and biofilms. Appl. Environ. Microbiol. 70:6181-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito, M., S. Ohga, M. Endoh, H. Nakayama, Y. Mizunoe, T. Hara, and S. Yoshida. 2001. H2O2-nonproducing Streptococcus pyogenes strains: survival in stationary phase and virulence in chronic granulomatous disease. Microbiology 147:2469-2477. [DOI] [PubMed] [Google Scholar]
- 41.Schmitt-Slomska, J., A. Boué, and R. Caravano. 1972. Induction of L-variants in human diploid cells infected by group A streptococci. Infect. Immun. 5:389-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seki, M., K. Iida, M. Saito, H. Nakayama, and S. Yoshida. 2004. Hydrogen peroxide production in Streptococcus pyogenes: involvement of lactate oxidase and coupling with aerobic utilization of lactate. J. Bacteriol. 186:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sela, S., R. Neeman, N. Keller, and A. Barzilai. 2000. Relationship between asymptomatic carriage of Streptococcus pyogenes and the ability of the strains to adhere to and be internalised by cultured epithelial cells. J. Med. Microbiol. 49:499-502. [DOI] [PubMed] [Google Scholar]
- 44.Shaw, J. H., and D. B. Clewell. 1985. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J. Bacteriol. 164:782-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spector, M. P. 1998. The starvation-stress response (SSR) of Salmonella. Adv. Microb. Physiol. 40:233-279. [DOI] [PubMed] [Google Scholar]
- 46.Trainor, V. C., R. K. Udy, P. J. Bremer, and G. M. Cook. 1999. Survival of Streptococcus pyogenes under stress and starvation. FEMS Microbiol. Lett. 176:421-428. [DOI] [PubMed] [Google Scholar]
- 47.Tsai, P.-J., Y.-S. Lin, C.-F. Kuo, H.-Y. Lei, and J.-J. Wu. 1999. Group A streptococcus induces apoptosis in human epithelial cells. Infect. Immun. 67:4334-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van de Rijn, I., and R. E. Kessler. 1980. Growth characteristics of group A streptococci in a new chemically defined medium. Infect. Immun. 27:444-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wai, S. N., Y. Mizunoe, A. Takade, and S. Yoshida. 2000. A comparison of solid and liquid media for resuscitation of starvation- and low-temperature-induced nonculturable cells of Aeromonas hydrophila. Arch. Microbiol. 173:307-310. [DOI] [PubMed] [Google Scholar]
- 50.Watson, S. P., M. O. Clements, and S. J. Foster. 1998. Characterization of the starvation-survival response of Staphylococcus aureus. J. Bacteriol. 180:1750-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whitesides, M. D., and J. D. Oliver. 1997. Resuscitation of Vibrio vulnificus from the viable but nonculturable state. Appl. Environ. Microbiol. 63:1002-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zinser, E. R., and R. Kolter. 1999. Mutations enhancing amino acid catabolism confer a growth advantage in stationary phase. J. Bacteriol. 181:5800-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zinser, E. R., and R. Kolter. 2000. Prolonged stationary-phase incubation selects for lrp mutations in Escherichia coli K-12. J. Bacteriol. 182:4361-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]