Abstract
Adaptation of Lactococcus lactis towards progressive carbon starvation is mediated by three different types of transcriptomic responses: (i) global responses, i.e., general decreases of functions linked to bacterial growth and lack of induction of the general stress response; (ii) specific responses functionally related to glucose exhaustion, i.e., underexpression of central metabolism genes, induction of alternative sugar transport and metabolism, and induction of the arginine deiminase pathway; and (iii) other responses never described previously during carbon starvation.
Fundamental knowledge concerning the adaptation of Lactococcus lactis to adverse environments and more particularly to carbon starvation is still very fragmented. Studies have generally been focused on glycolysis and bacterial survival (15, 19, 23, 24, 26), providing only parceled results. In order to provide a more exhaustive picture of the adaptation, carbon starvation in L. lactis was investigated with whole-transcriptome analyses. Transcriptomic data were integrated in a more global physiological study based on metabolic flux determination, amino acid consumption, and measurement of certain enzyme activities. Modifications in the transcriptional profile were analyzed during progressive adaptation, allowing carbon starvation stimulons, a prerequisite of regulation network analysis, to be identified.
Overview and dynamic analysis.
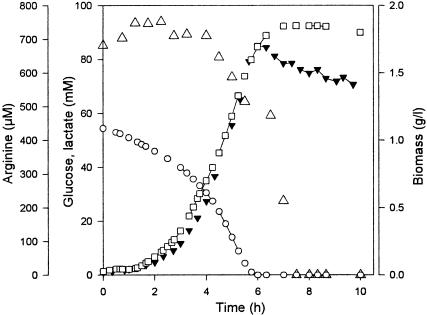
The growth of L. lactis IL-1403 and its adaptation towards glucose starvation were investigated with a pH-regulated fermentor and in the chemically defined medium CDM (22), commonly used for L. lactis cultivation (Fig. 1). After an exponential phase associated with nutrient excess (0 to 5 h) and a short deceleration phase due to decreasing glucose concentration, a nongrowth stationary phase characterized by glucose exhaustion succeeded after 6 h of fermentation. The first 1.5 h of stationary phase corresponded to simple carbon starvation, since glucose was totally consumed but other nutrients, and particularly amino acids, were still available. Arginine was significantly consumed by nongrowing cells and was also exhausted from the medium after 1.5 h of starvation. The metabolism remained homolactic all along the fermentation since no fermentation products other than lactate accumulated significantly.
FIG. 1.
Growth (▾), lactate production (□), glucose consumption (○), and arginine consumption (Δ) during the progressive adaptation of L. lactis IL-1403 to carbon starvation.
Whole-genome transcripts were measured on nylon membranes after hybridization of cDNA labeled by retro-transcription with [P33]dCTP in the different phases of the culture: exponential phase, deceleration phase, onset of carbon starvation (1.25 h), and after 3.5 h of starvation when arginine was no longer available. Three independent measurements were studied, and abundance ratios were calculated with the exponential phase as the reference in order to characterize expression pattern evolution. Only 30% of the genome was involved in the response to carbon starvation, as 704 genes showed at least one significant expression variation during the fermentation (Student's t test with a P value below 0.05). Globally, two times more genes were underexpressed than overexpressed, and this transcriptomic response, related to transcription or degradation rate changes, was triggered before glucose exhaustion. Indeed, the first expression reorganization took place as soon as the glucose concentration reached a threshold value of 14 mM. Further modification of the expression pattern was observed at the onset of carbon starvation (1.25 h), whereas expression varied very slightly once carbon starvation was established. Genes overexpressed during deceleration phase (226 genes) generally remained overexpressed during carbon starvation. Inversely, genes underexpressed during deceleration phase (411 genes) remained underexpressed. However, within each of these two groups, different kinetic profiles were observed. Last, few genes (67 genes) were transiently induced during the deceleration phase or from the onset of carbon starvation.
Global response.
For most of the genes involved in general processes associated with cellular growth, a general decline of expression was observed during the experiment. This is consistent with growth arrest due to carbon starvation but not necessarily linked to protein decrease. Indeed, the various RNA polymerase subunit-encoding genes were underexpressed (rpoA, -B, -C, and -E). Since the amount of transcripts and the total RNA concentration in the cells decreased during deceleration and starvation phases (Table 1), the rate of transcription should have been considerably reduced. The translation rate should also have decreased, as 47% of the translation apparatus genes, including 35 ribosomal protein-, 8 tRNA synthase-, 8 translation factor-, and 4 GTPase-encoding genes were underexpressed during the three phases studied. Furthermore, a 15-fold reduction of the global translation rate was previously observed after a 2-h carbon starvation period in L. lactis by Kunji et al. (19). The rate of nucleotide biosynthesis should also have decelerated, as the genes involved in nucleotide metabolism were all underexpressed during the deceleration and starvation phases. This was reinforced by underexpression of genes encoding nucleotidic base uptake (pyrP and pbuX). Similarly, most of the genes involved in cell envelope metabolism, i.e., genes for fatty acid and phospholipid metabolism (8 genes), membrane biosynthesis (5 genes), and cell wall biosynthesis (17 genes), and in the cell division process (8 genes) were underexpressed. At the opposite extreme, replication seemed not so drastically affected by carbon starvation, since 10 genes were underexpressed while 7 genes were over- or transiently overexpressed. This global expression decrease of genes involved in processes typical of growing cells (transcription, translation, nucleotide biosynthesis, cell envelope metabolism, and cell division) can be compared to a stringent response, previously described for Escherichia coli or Bacillus subtilis during amino acid starvation (3, 9, 28). The stringent response had already been observed in Staphylococcus aureus during carbon starvation (5) and in B. subtilis (1). Thus, the stringent response appears to be involved in various stresses and should occupy a more general function of coordination of the global transcriptional pattern with growth conditions as previously postulated for E. coli by Chatterji and Ojha (3).
TABLE 1.
Relative mRNA proportions and total RNA concentrations during progressive adaptation of L. lactis IL1403 to carbon starvation
| Parameter | Mean ± SD
|
|||
|---|---|---|---|---|
| Exponential (4 h) | Deceleration (5.75 h) | 1.25 h of starvation (7.25 h) | 3.5 h of starvation (9.5 h) | |
| Relative proportion of mRNA in total RNAa | 100 | 58 ± 1 | 43 ± 4 | 34 ± 22 |
| Total RNA concn (g · 100 g dried cells−1) | 11.7 ± 1.3 | 7.6 ± 0.8 | 8.2 ± 1.4 | 7.3 ± 0.8 |
Calculated as ratios of whole intensities of the membranes, with the exponential phase as reference.
Carbon starvation is known to confer an increased resistance towards various stresses, such as heat, osmosis, and acid, ethanol, and oxidative stresses (15), suggesting the induction in L. lactis, as in other bacteria, of the general stress response. However, in our experiment, chaperone genes were not induced, contrary to what was already observed during heat shock or acid or salt stress (2, 12, 18). Similarly, Clp proteases, regulated by ctsR (7, 27), are generally induced at high temperature or low pH to degrade misfolded proteins (11). In this study, clpP and clpE were underexpressed and clpB, -C, and -X were constitutive, in agreement with the constitutive expression of ctsR. Moreover, htrA, encoding a surface protease essential in defense against stress conditions (10), was strongly underexpressed during starvation. In addition, cold shock protein genes (cspD and -E) and stress protein genes (grpE, dinF, yjbE, and a ytgH, homologue of the gls24 gene in Enterococcus faecalis [13]) did not show significant expression increases. Last, the two-component system C, encoded by llrC and kinC, postulated to regulate the general stress response (21), was not shown to be induced in our study. Thus, unlike in B. subtilis (1, 16), the general stress response was not observed in L. lactis during carbon starvation, suggesting that previously described cross-protection should be linked to a different mechanism.
Responses specific to glucose exhaustion.
Cells also developed a set of responses typical of carbon starvation. Abundances of most of the mRNAs encoding enzymes of central metabolism (ackA1, adhE, aldB, enoA, fbaA, frdC, mae, pfl, pflA, pgiA, pmg, pta, and tpiA) diminished from the deceleration phase. However, concentrations of glycolytic and pyruvate utilization enzymes, determined by in vitro enzyme activity measurements, were maintained throughout the fermentation. These results, in contradiction with the drop of certain enzyme activities previously observed in L. lactis ML3 (19, 24), are consistent with a carbon flux control located at the level of the sugar transport in strain IL-1403 (8). Uncorrelated profiles of enzymes and transcripts indicated that the in vivo regulation of central metabolism during carbon starvation was located rather at the enzyme activity level by allosteric control than at the gene level by transcriptional control.
Alternative-carbon-source utilization was favored during deceleration phase and after glucose exhaustion. Induction was observed at three different levels: utilization pathways, transport, and regulation. Genes specifically involved in galactose, lactose, maltose, ribose, and other sugar utilization (galM, lacZ, malQ, rbsK, uxaC, xylX, ygjD, and yidC) or polysaccharide degradation (apu and yucG) were overexpressed throughout the fermentation, while genes encoding transporters for ribose, xyloside, or uncharacterized sugars were induced transiently (msmK, rbsA, rbsC, xynT, yngF, ypbD, and ypdA). Induction of sugar utilization was supported by underexpression of repressors involved in sugar operon regulation (fruR, gntR, and kdgR). Induction of three genes encoding citrate lyase (citC, -E, and -F), probably related to the underexpression of the citR repressor from deceleration phase, indicated that the citrate utilization pathway was activated during carbon starvation. This induction was consistent with the late consumption of citrate (present in the CDM medium at very low concentrations) during carbon starvation. Last, glycerol metabolism seemed to play a crucial role in the carbon starvation response, since five genes involved in this pathway were induced at high levels from deceleration phase: dhaL and -M, encoding dihydroxyacetone kinases (respective ratios of 7.1 and 3.9), already shown to be induced during carbon starvation in E. faecalis (20); glpD and -K, encoding, respectively, glycerol-3-P dehydrogenase and glycerol kinase; and glpF1, encoding a glycerol uptake enzyme (ratio of 6 at the onset of carbon starvation). Such induction of the metabolism of various alternative carbon sources, already observed in B. subtilis during carbon starvation (1), may be a general response to counteract carbon starvation.
The culture medium contained most of the amino acids since L. lactis IL-1403, like other lactic acid bacteria, requires numerous amino acids to grow (4). Amino acid consumption rates were reduced during deceleration phase and generally stopped at growth arrest. The expression of amino acid biosynthesis genes did not show a predominant profile (many genes over- or underexpressed), while amino acid carrier-encoding genes were generally underexpressed from deceleration phase (11 genes). Unlike other amino acids, arginine was consumed in the stationary phase through the arginine deiminase pathway, leading to equimolar productions of ornithine. The switching on of the arginine deaminase (ADI) pathway in L. lactis, in agreement with the results of a previous study of continuous cultures (6), enabled the cells to be supplied with maintenance energy during carbon starvation since one ATP per arginine is produced. ADI pathway regulation was shown to occur at the transcriptional level, since genes arcA, -B, and -C1 and argF, encoding all the enzymes involved in this pathway (arginine deiminase, ornithine carbamoyltransferase, and carbamate kinase), were strongly induced from deceleration phase and remained overexpressed during starvation phases. Surprisingly, genes encoding the arginine/ornithine carrier, arcD1 and D2, presented a constitutive profile during carbon starvation. Very little information is available about ADI pathway transcriptional regulation. llrA was supposed to regulate the ADI pathway during acid stress conditions (21), but its role during carbon starvation was not firmly established since it was underexpressed during starvation. However, llrH, hypothetically involved in ADI pathway regulation (14), should be a better candidate, since it was highly overexpressed throughout the fermentation.
Other responses.
Five competence genes among the 13 genes involved in the transformation pathway, namely, radA, comEA, comGA, comGB, and the alternative sigma comX, were progressively overexpressed. nucA, involved in DNA cleavage before its internalization during the transformation process in B. subtilis (25), was also overexpressed throughout the fermentation. L. lactis is generally considered a noncompetent microorganism, and this natural competence induction at the transcriptional level has not previously been described. Competence induction was already observed in B. subtilis during nitrogen starvation (17) but not during carbon starvation (1), probably indicating that specific nutritional starvation is required to induce competence in each microorganism.
The induction of 48 phage- and prophage-related function genes was also observed in response to glucose diminution and exhaustion. Despite this induction, no bacterial lysis was observed (upon 15 repetitions of the culture), indicating that the expression of a gene(s) essential for phage development is still missing.
Various ion uptake genes (amtB, yafB, ydaE, yddA, yfgQ, yndG, and ypbB) and multidrug resistance transporters genes (cydD, napC, ycdH, ydiC, yniG, and yxbD) were also overexpressed. Last, two genes, vacB1 and vacB2, encoding uncharacterized ribonucleases were highly induced during carbon starvation, though no RNA degradation was observed throughout the culture.
In this study, carbon starvation stimulons were identified and some responses, specific to L. lactis, were highlighted (the arginine deaminase pathway and the lack of a general stress response). Since no alternative stress sigma factor has been identified in L. lactis, more information on transcriptional regulatory mechanisms is needed to better understand this pleiotropic response.
Acknowledgments
Membrane spotting and analysis support were provided by Plateforme Génomique (Toulouse, France).
This work was supported by a grant obtained from the Toulouse Genopole.
REFERENCES
- 1.Bernhardt, J., J. Weibezahn, C. Scharf, and M. Hecker. 2003. Bacillus subtilis during feast and famine: visualization of the overall regulation of protein synthesis during glucose starvation by proteome analysis. Genome Res. 13:224-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Champomier-Verges, M. C., E. Maguin, M. Y. Mistou, P. Anglade, and J. F. Chich. 2002. Lactic acid bacteria and proteomics: current knowledge and perspectives. J. Chromatogr. B 771:329-342. [DOI] [PubMed] [Google Scholar]
- 3.Chatterji, D., and A. K. Ojha. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 4:160-165. [DOI] [PubMed] [Google Scholar]
- 4.Cocaign-Bousquet, M., C. Garrigues, L. Novak, N. D. Lindley, and P. Loubiere. 1995. Rational development of a simple synthetic medium for the sustained growth of Lactococcus lactis. J. Appl. Bacteriol. 79:108-116. [Google Scholar]
- 5.Crosse, A. M., D. L. Greenway, and R. R. England. 2000. Accumulation of ppGpp and ppGp in Staphylococcus aureus 8325-4 following nutrient starvation. Lett. Appl. Microbiol. 31:332-337. [DOI] [PubMed] [Google Scholar]
- 6.Crow, V. L., and T. D. Thomas. 1982. Arginine metabolism in lactic streptococci. J. Bacteriol. 150:1024-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derre, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol. Microbiol. 31:117-131. [DOI] [PubMed] [Google Scholar]
- 8.Even, S., N. D. Lindley, and M. Cocaign-Bousquet. 2001. Molecular physiology of sugar catabolism in Lactococcus lactis IL1403. J. Bacteriol. 183:3817-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eymann, C., G. Homuth, C. Scharf, and M. Hecker. 2002. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J. Bacteriol. 184:2500-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foucaud-Scheunemann, C., and I. Poquet. 2003. HtrA is a key factor in the response to specific stress conditions in Lactococcus lactis. FEMS Microbiol. Lett. 224:53-59. [DOI] [PubMed] [Google Scholar]
- 11.Frees, D., and H. Ingmer. 1999. ClpP participates in the degradation of misfolded protein in Lactococcus lactis. Mol. Microbiol. 31:79-87. [DOI] [PubMed] [Google Scholar]
- 12.Frees, D., F. K. Vogensen, and H. Ingmer. 2003. Identification of proteins induced at low pH in Lactococcus lactis. Int. J. Food Microbiol. 87:293-300. [DOI] [PubMed] [Google Scholar]
- 13.Giard, J. C., N. Verneuil, Y. Auffray, and A. Hartke. 2002. Characterization of genes homologous to the general stress-inducible gene gls24 in Enterococcus faecalis and Lactococcus lactis. FEMS Microbiol. Lett. 206:235-239. [DOI] [PubMed] [Google Scholar]
- 14.Guedon, E., E. Jamet, and P. Renault. 2002. Gene regulation in Lactococcus lactis: the gap between predicted and characterized regulators. Antonie Leeuwenhoek 82:93-112. [PubMed] [Google Scholar]
- 15.Hartke, A., S. Bouche, X. Gansel, P. Boutibonnes, and Y. Auffray. 1994. Starvation-induced stress resistance in Lactococcus lactis subsp. lactis IL1403. Appl. Environ. Microbiol. 60:3474-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hecker, M., and U. Volker. 2001. General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44:35-91. [DOI] [PubMed] [Google Scholar]
- 17.Jarmer, H., R. Berka, S. Knudsen, and H. H. Saxild. 2002. Transcriptome analysis documents induced competence of Bacillus subtilis during nitrogen limiting conditions. FEMS Microbiol. Lett. 206:197-200. [DOI] [PubMed] [Google Scholar]
- 18.Kilstrup, M., S. Jacobsen, K. Hammer, and F. K. Vogensen. 1997. Induction of heat shock proteins DnaK, GroEL, and GroES by salt stress in Lactococcus lactis. Appl. Environ. Microbiol. 63:1826-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunji, E. R. S., T. Ubbink, A. Matin, B. Poolman, and W. N. Konings. 1993. Physiological responses of Lactococcus lactis ML3 to alternating conditions of growth and starvation. Arch. Microbiol. 159:372-379. [Google Scholar]
- 20.Leboeuf, C., L. Leblanc, Y. Auffray, and A. Hartke. 2000. Characterization of the ccpA gene of Enterococcus faecalis: identification of starvation-inducible proteins regulated by ccpA. J. Bacteriol. 182:5799-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Connell-Motherway, M., D. van Sinderen, F. Morel-Deville, G. F. Fitzgerald, S. D. Ehrlich, and P. Morel. 2000. Six putative two-component regula-tory systems isolated from Lactococcus lactis subsp. cremoris MG1363. Microbiology 146:935-947. [DOI] [PubMed] [Google Scholar]
- 22.Poolman, B., and W. N. Konings. 1988. Relation of growth of Streptococcus lactis and Streptococcus cremoris to amino acid transport. J. Bacteriol. 170:700-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poolman, B., E. J. Smid, H. Veldkamp, and W. N. Konings. 1987. Bioenergetic consequences of lactose starvation for continuously cultured Streptococcus cremoris. J. Bacteriol. 169:1460-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poolman, B., B. Bosman, J. Kiers, and W. N. Konings. 1987. Control of glycolysis by glyceraldehyde-3-phosphate dehydrogenase in Streptococcus cremoris and Streptococcus lactis. J. Bacteriol. 169:5887-5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Provvedi, R., I. Chen, and D. Dubnau. 2001. NucA is required for DNA cleavage during transformation of Bacillus subtilis. Mol. Microbiol. 40:634-644. [DOI] [PubMed] [Google Scholar]
- 26.Thompson, J., and T. D. Thomas. 1977. Phosphoenolpyruvate and 2-phosphoglycerate: endogenous energy sources for sugar accumulation by starved cells of Streptococcus lactis. J. Bacteriol. 130:583-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varmanen, P., H. Ingmer, and F. K. Vogensen. 2000. ctsR of Lactococcus lactis encodes a negative regulator of clp gene expression. Microbiology 146:1447-1455. [DOI] [PubMed] [Google Scholar]
- 28.Vogel, U., M., Sorensen, S. Pedersen, K. F. Jensen, and M. Kilstrup. 1992. Decreasing transcription elongation rate in Escherichia coli exposed to amino acid starvation. Mol. Microbiol. 6:2191-2200. [DOI] [PubMed] [Google Scholar]