Abstract
Nuclear magnetic resonance (NMR) data are rarely deposited in open databases, leading to loss of critical scientific knowledge. Existing data reporting methods (images, tables, lists of values) contain less information than raw data, and are poorly standardized. Together, these issues limit FAIR (findable, accessible, interoperable, reusable) access to these data, which in turn creates barriers for compound dereplication and the development of new data-driven discovery tools. Existing NMR databases are either not designed for natural products data, or employ complex deposition interfaces that disincentivize deposition. Journals, including the Journal of Natural Products (JNP), are now requiring data submission as part of the publication process, creating the need for a streamlined, user-friendly mechanism to deposit and distribute NMR data.
Recently, our team reported the development of the Natural Products Magnetic Resonance Database (NP-MRD; www.np-mrd.org). In this paper we present a new data deposition platform for the NP-MRD project that is designed to enable users to deposit NMR data for published or submitted manuscripts in under five minutes. This platform includes a suite of automated data extraction and standardization tools, together with a simple-to-use web-based interface and detailed error reporting to simplify the data deposition process and is available at https://depositions.np-mrd.org/.
Graphical Abstract
The physical sciences are moving towards an open data model where raw data are routinely released as part of the publication process.1–3 This change has been enabled by improvements in the information technology sector that have lowered the barrier to entry for creating and maintaining public repositories, and by increased scrutiny on data dissemination by funders and journals.
Despite these advances, NMR data deposition rates currently lag behind those of other data types.4 This is in part because NMR data are complicated compared to other biological or chemical data formats (e.g. sequence data or chemical structures). However it is also due in part to the culture of data reporting for small molecule characterization which has historically favored NMR assignment tables and images of NMR spectra in supporting information files over raw data deposition. Although several established platforms exist for NMR data deposition,5,6 uptake has been low among the natural products community. The effect of this is that while some data types such as biosynthetic gene cluster sequence and mass spectrometry fragmentation data are routinely deposited into subject-specific databases,7,8 other data types such as NMR and bioassay data are typically lost (Figure 1).
Figure 1.
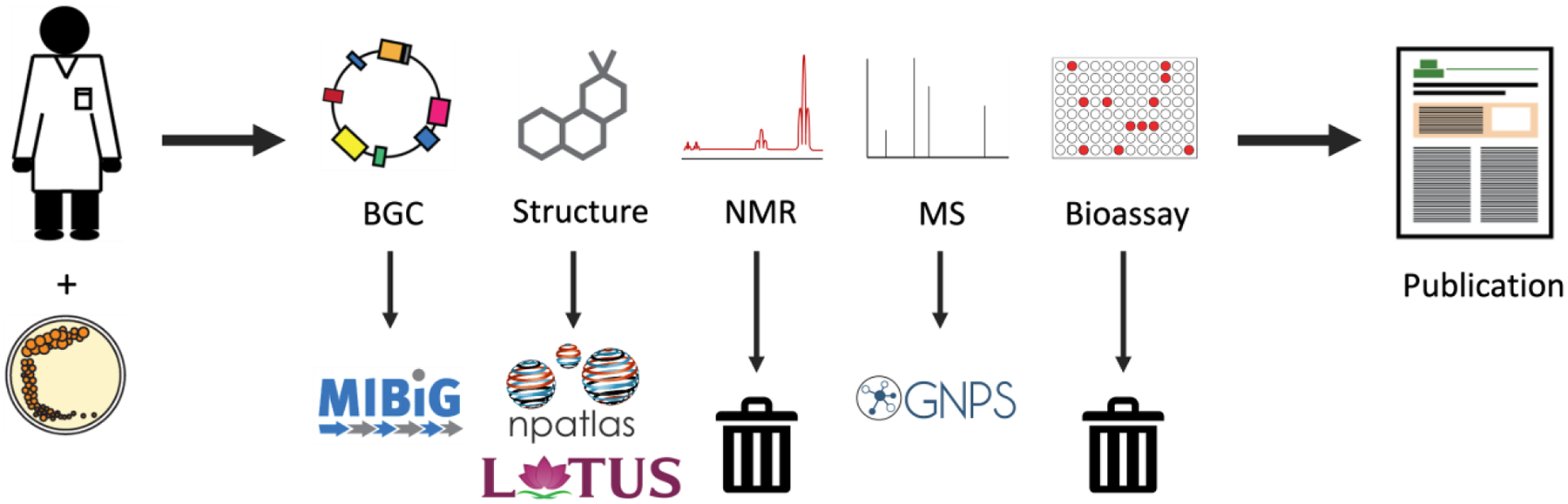
Workflow illustrating the common data types and example destinations during the data lifecycle for natural products discovery. Notably, most NMR and bioassay data are not currently deposited in open repositories, precluding their reuse by the scientific community.
Recognizing the value of raw NMR data for the natural products community, in 2020 the National Institutes of Health National Center for Complementary and Integrative Health (NCCIH) and the Office of Dietary Supplements (ODS) jointly funded the creation of a new data repository for NMR data for natural products; the Natural Products Magnetic Resonance Database (NP-MRD). The NP-MRD is designed to house raw NMR data, chemical shift assignments, and calculated chemical shifts and coupling constants for all available natural products. Details about the design and functionality of the NP-MRD are available in the paper reporting its initial release.9 This database has grown rapidly since its initial release in 2020, and now contains over 280,000 natural product structures and more than 5.4 million experimental, simulated and predicted NMR spectra.
More recently, some journals are beginning to require data deposition of raw NMR data as part of the publication process. The Journal of Natural Products (JNP) has led the way in this area among the ACS journals by requiring raw NMR data deposition for all papers reporting new natural product structures published after June 1 of this year.10 This requirement will have a direct effect on data deposition rates. In the six months leading up to the deposition requirement JNP published 92 articles reporting new NPs (from a total of 156), of which just 11 (12%) included raw NMR data in the data availability statement, illustrating the need for such an initiative.
To facilitate data deposition from the community and reduce the barrier to entry for sharing raw NMR data the NP-MRD team has developed a new data deposition platform that simplifies the deposition process and provides a seamless mechanism for submitting, managing, and releasing NMR data to the NP-MRD database.
Results and Discussion
To develop the new deposition platform we first defined the desired attributes of the system, which included:
Ease of use (<5 minute deposition)
Built in extraction, verification, and standardization of spectral metadata
Detailed error reporting and user feedback
Secure infrastructure
Flexible data model (able to accommodate data from published articles, presubmissions, and private collections)
Embargo options for presubmission articles
Capacity to generate private links to share with reviewers
Ability to accept multiple data types (raw fids, processed data and peak lists)
Compatibility with all major NMR spectrometer vendors (Bruker, Agilent/Varian, JEOL)
Compatibility with JCAMP-DX open exchange format for integration with external NMR software including MNova and other commercial and open-source tools
Automated generation and assignment of NP-MRD accession numbers to depositions and delivery to depositors
To accomplish these objectives we developed a ‘full stack’ application built in Python and Next.js using the Django framework. This application includes a ‘frontend’ web server that allows users to interact with the system, a ‘backend’ server where most of the data processing is performed, and a database and data store where the data are housed. This stand-alone system is connected to the main NP-MRD database via an automatic programming interface (API) that enables transfer of completed submissions for final validation and insertion. To simplify access for users we have embedded this service as part of the NP-MRD website (https://depositions.np-mrd.org/).
The data model for the deposition interface centers on article-based submissions, meaning that all data for a given article can be deposited at the same time, rather than compound by compound. The model can either accept data from existing publications or from ‘presubmission’ articles that are entering the peer review process. In addition it can accommodate data from private repositories for which no article is available, making it suitable for most sources of NMR data.
Currently, the deposition platform accepts raw NMR data for all types of 1D and 2D NMR experiments, and can accommodate the full range of NMR-active nuclei. In addition the system will accept peak lists for proton and carbon chemical shifts where available. Fortunately, raw NMR data from all three of the major NMR instrument vendors (Bruker, Agilent/ Varian, and JEOL) includes a broad set of associated metadata (NMR solvent, observed nuclei, spectrometer frequency, temperature etc.). This greatly simplifies the deposition process because key metadata can be extracted from the uploaded files without requiring manual data entry by users. Leveraging this existing spectral metadata allowed us to write code to automatically populate many fields in the database, and to simplify the deposition process so that users are required to provide only those key data (e.g. compound name and structure) not present in the NMR data file.
After logging in to the system the workflow for data deposition includes five key steps (Figure 2, blue boxes):
Select deposition type. Users provide information about the article or repository from which the data derive. If the article is already published then users supply only the digital object identifier (DOI) of the paper, and our system automatically searches three literature databases (PubMed, Scopus and CrossRef) to return standardized citation information. If the paper is not yet published then users supply the working title and author list from the draft manuscript and the deposition platform automatically appends the DOI and citation information once the paper is published. Finally, if the data are from a private repository users enter some key details about that repository so that the provenance of the NMR data are recorded.
Input structures. Users supply the names, structures (as SMILES strings), and source organisms (genus and species) for each compound they wish to deposit. All compounds can be added on the same page, which includes buttons to dynamically add or remove compound entries.
Upload raw NMR data. Users prepare all the NMR experiments for a given compound as a single zipped file. No specific folder naming convention is required, and all 1D and 2D experiment types are accepted. Users drag and drop the zipped file for each compound into the area next to each compound and wait for upload to complete (typically a few seconds).
Input peak lists. Users can optionally provide peak lists for proton and/or carbon signals. Peak lists are valuable for training automated spectral processing tools, and help users to deconvolute complex spectra. Both peak list and raw data deposition are optional, meaning that users can submit raw data, or peak lists, or both, for any compound provided that at least one of these data types is included.
Approve submission. Finally, users are offered the opportunity to review the submitted data and correct any errors before completing the submission.
Figure 2:
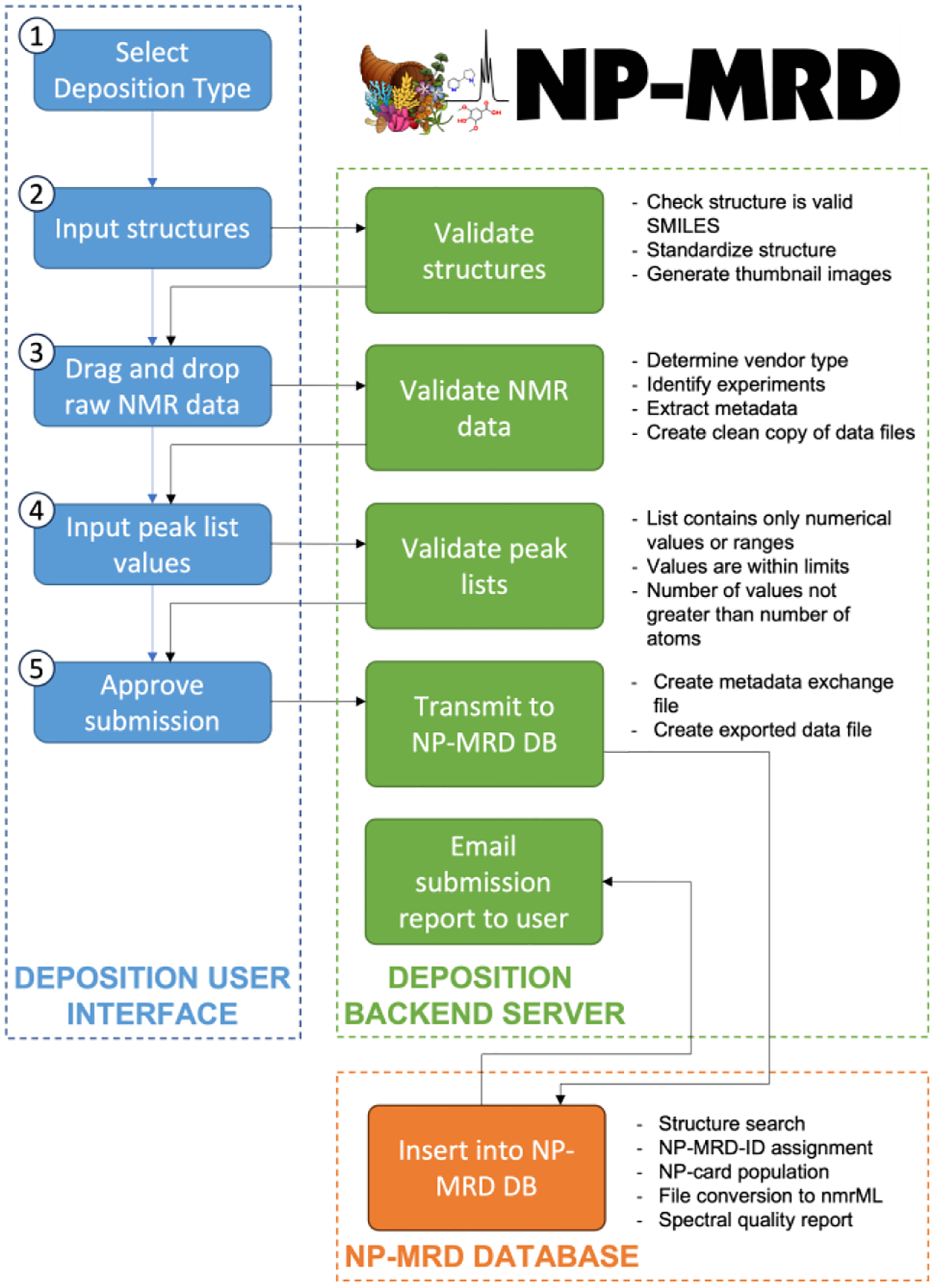
Schematic of data deposition workflow. Blue boxes represent steps in the deposition process on the frontend webserver. Green boxes represent actions taken on the backend server to validate and standardize submitted data. The orange box represents the main NP-MRD database, to which deposited data are transmitted and from which the insertion report is returned.
Data Standardization
A core principle of database development is the idea of data standardization.11,12 If data derive from a range of sources, then the same information may be provided in different formats for different entries. For example, a journal may be listed as ‘Journal of Natural Products’ from one source, but ‘J. Nat. Prod.’ in another. This creates redundant terms in the database and leads to fragmentation of the underlying data, which in turn causes incomplete search results and other errors.
To address this issue, the data deposition system performs a suite of metadata extraction, validation, and standardization checks at each step in the deposition process. After each step in the deposition process (Figure 2, blue boxes), deposited and extracted data are automatically reviewed to ensure that they meet the required standards for each field (Figure 2, green boxes). For example, SMILES strings for chemical structures are inspected to make sure that they can be parsed by chemoinformatics software and that they describe a single molecule. Any field that fails validation is returned to the front end and highlighted to the user, along with a detailed error message that describes the error and how to fix it. The development of tools for metadata extraction and detailed error reporting were both essential steps in meeting our design goal of five-minute depositions.
The core data validation and standardization steps include:
Citation data. We have developed a microservice that uses either the Digital Object Identifier (DOI; most journals) or the Publisher Item Identifier (PII; Elsevier journals) to sequentially search Pubmed, Scopus and Crossref for complete citation information for published articles. This approach means that users only have to supply a single field (DOI/PII) for each paper, and that the returned citation data are pre-standardized by these services. If the article returns an abstract then this is analyzed using a machine-learning-based natural language processing (NLP) module that has been trained to identify compound names and the genus and species of producing organisms. This module examines the relationship between compounds and organisms to determine the origin of each natural product. For example, if the abstract contains the sentence “Examplamides A - C (1 – 3) were isolated from the marine-derived bacterium Nocardia sp.” then the NLP module will return three compounds (examplamide A, examplamide B, and examplamide C) and connect each to the source organism Nocardia sp. The module is designed to recognize molecules that derive from endosymbionts and the assign them to the correct producing organism. For example, if the abstract text states “Examplamides A - C (1 – 3) were derived from the symbiotic bacterium Nocardia sp. isolated from the marine sponge Dysidea herbacea” then the NLP module will correctly assign the producing organism as Nocardia sp., rather than the sponge host. If available, compound details are prepopulated into the compound submission page to simplify the submission process.
Compound data. Compound data are accepted as SMILES strings, which can be easily copied from ChemDraw and other chemical drawing programs. As described above, SMILES strings are first validated using the Python package RDKit13 to ensure they are valid molecular representations. Strings that pass validation are standardized and converted to thumbnail images, which are displayed to users. Each compound must be assigned a producing organism (if known). Genus names are validated to ensure that they contain only characters from the Roman alphabet, and that they are at least five characters long to prevent submission of ambiguous abbreviations like S. erythraea. Species names may contain characters from the Roman and Greek alphabets, numbers, and a range of punctuation characters to permit the submission of strain codes if desired. Species names must also be at least five characters long. If either genus or species names are shorter than 5 characters then the submission is flagged for manual review by the NP-MRD data curation team prior to data release.
-
Raw NMR data. Processing raw NMR data is one of the most complicated steps in the submission process. Unlike databases of structures (e.g., the Natural Products Atlas or LOTUS)14,15 or biosynthetic gene clusters (e.g., MIBiG)7 that distribute data as text strings, the NP-MRD must distribute both text-based information and directories of raw NMR data. Raw data must therefore be reviewed carefully to ensure that they do not contain malicious code or irrelevant files (images, word processor documents etc.). Fortunately, data from each of the NMR instrument vendors are generated in a single format for each vendor, making it straightforward to scan, filter, and copy submitted data to create standardized files for distribution.
In addition, the platform uses the submitted data to extract key metadata for each experiment. The system first inspects the file structure to determine which instrument manufacturer the data derive from. Next, manufacturer-specific scripts are used to determine the experiment type, observed nuclei, frequencies, solvent, and temperature for each spectrum. This approach has two advantages over manual data entry. Firstly, it ensure that all entries conform to data standards, reducing error in the underlying database (e.g. Chloroform vs. chloroform, vs. CDCl3, vs chloroform-d etc.). Secondly, it reduces the amount of information that users must supply, which eliminates data entry errors and significantly simplifies the data deposition process.
Peak lists. Each peak list is submitted as a single comma separated string. To validate peak lists we first examine each entry in the list to ensure that it is a single value or a range. Values that are outside the accepted range for each nucleus (−2 to 20 ppm for proton, −10 to 250 ppm for carbon) raise an error that is reported to the user. These values must be corrected before submission can proceed. Values that are unusual but within the allowed ranges (−2 to −1 and 16 to 20 ppm for proton, −10 to 0 and 230 to 250 ppm for carbon) raise warnings that are reported to the user, but do not prevent submission. The number of values provided in the peak list is also compared to the atom counts for the structure. Carbon and proton peak lists cannot contain more values than the carbon and proton atom counts from the SMILES structure. Users can submit either raw data or peak lists, or both but must submit at least one NMR data type for each compound to complete the submission.
Data Insertion
Following data validation, users are presented with a summary page that lists each of the compounds they have submitted, along with a list of the data available for each compound. Approved submissions are transmitted via the API to the NP-MRD database for insertion which involves the following four steps:
Compound search. The NP-MRD database model is ‘compound-centric’, with data organized into NP cards, one card for each structure. New depositions are searched against the database to determine whether or not an NP card exists for that structure. If the compound is not in the database, then a new NP card is created, and a new NP-MRD ID number assigned to the card.
Data insertion. Once the correct NP card has been identified the NMR data for each compound are inserted into the database using the metadata extracted from the raw files during the deposition process. Each insertion is attributed to the depositor, with attribution for each deposition clearly displayed next to the data on the NP card. Because NP-MRD accepts data from both new and known natural products this means that some compounds can include multiple examples of a given experiment type (e.g. proton spectrum) deposited by different users from different studies. This offers users multiple examples of data against which to dereplicate isolated compounds, and provides tool developers with real-world examples of spectrum variability between laboratories; a valuable resource for creating robust informatics tools. As part of the data insertion step raw data are converted to the nmrML open data format which is offered as a download option for each spectrum.
Quality report generation. A long-term goal of the NP-MRD project is to generate quality reports for all spectra deposited to the database. These quality metrics help users to select data for applications such as dereplication, and provide a mechanism for the database administrators to identify, review, and, if necessary, remove spectra of very low quality. Currently quality reports are generated for 1D spectra from Bruker instruments. Development of tools for other experiment types and vendor formats is ongoing.
Insertion summary. Finally, the NP-MRD database sends an insertion report summary back to the deposition platform to confirm successful insertion, identify any errors or issues raised during the insertion process, provide the quality report (if available), and define the accession code (NP-MRD ID) for each compound.
In the final step a summary of the insertion status for each compound in the deposition dataset is sent to the depositor by email. This summary includes hyperlinks to the deposited data, the accession codes for each compound, quality report values for relevant spectra, and preformatted text for inclusion in the data availability statement of the manuscript. If relevant, the summary also includes information about the embargo period (see below) and instructions about managing public release of embargoed data.
Data Sources
The deposition system accepts data from three sources: published articles, presubmission articles, and private collections. Presubmission articles are defined as those papers that are in the publication process but have not yet been published or assigned a DOI. Private collections are defined as those datasets for which no associated article is available. A core design feature of the NP-MRD database is that the provenance of every data point should be defined. Linking NMR data to publications provides a wealth of additional information about the methods used to collect, isolate, and identify each compound. However, many laboratories possess internal reference libraries of spectra that will never be reported in publications, including data for frequently encountered known compounds that are of particular value for dereplication. Enabling depositions that are not connected to the scientific literature is therefore important for broadening the scope and coverage of the database.
For published articles the only information that is required is the article DOI. As described above, the DOI is used to retrieve all other data about the article in a standard format. Because the article and chemical structures are already in the public domain there is no need for an embargo period on data from published articles, so these data are released immediately upon submission.
For presubmission articles both the title and the author list are required. Because these papers are unpublished it is not always appropriate to release the raw data immediately upon submission. Instead, users select one of three embargo options: ‘release immediately’, ‘release on a given date’, or ‘release upon publication’. Data that are subject to embargo are transferred to the NP-MRD database and assigned an NP-MRD ID number which is returned to the depositor. However, the data are not made public until either the embargo date is reached, or the user manually changes the dataset to ‘public’ in their account settings.
If users select ‘release upon publication’ then the deposition system uses the title and author list to scan the published literature daily to identify the corresponding published article. Because titles and author lists can sometimes change between initial submission and publication we use a ‘fuzzy’ string match that can accommodate moderate variations in the text. Candidate matches are manually reviewed by a member of the deposition team prior to data release. If a match is identified then data are set to public and the submission is updated to include the new DOI. Occasionally titles or author lists change significantly, precluding automated recognition of published articles. If data from a presubmission article have not been published after six months then a follow up email is sent to the depositor requesting an update on the publication status.
For private collections additional metadata are required. The structure of this metadata is dependent on the source of each molecule. Users can select from ‘purified in-house’, ‘commercial’, ‘compound library’ or ‘other’ as source types. The website dynamically changes the required fields based on source type to ensure that each dataset includes basic information about compound provenance. These data are displayed with the submitted data on the NP card after submission.
Literature Tracking
Scientific databases are most valuable when they include a broad cross section of the available data in a given subject area. The Protein Data Bank16 and the X-ray structures held by the Cambridge Crystallographic Data Center17 are two examples of repositories that have matured into rich sources of open access standardized data for scientific discovery and the development of new tools. However, achieving high rates of compliance for data submission is challenging. Fields such as X-ray crystallography that have succeeded in making deposition an expected part of the scientific process have done so over many years through a combination of incentives and enforcement.
Raw data availability is currently weak in the field of natural products. Few publications provide raw NMR, mass spectrometry, or screening data as supplementary information, instead presenting these data as figures and tables in the main manuscript or images in the supporting information. The reasons for this lack of data availability are complex, but two factors are clear contributors to the issue. Firstly, until recently there were few available repositories for data sharing, and most journals would only accept pdf files as supporting information. This made it functionally difficult to store raw data in permanent locations that were open to the general public. Secondly, there is a concern among some members of the community that data sharing can expose research groups to significant downsides (possibility of being ‘scooped’, encroachment on research areas by others, greater scrutiny on published work) with little upside return, as discussed in a recent review on this topic.12
To increase the rate of data deposition to the NP-MRD we created a system for real-time tracking of the scientific literature that creates customized deposition pages for all new articles reporting natural products discovery (Figure 3). First, we perform a daily download of article titles and abstracts from the RSS feeds of 50 journals known to publish papers about natural products discovery. Next, we identify articles about natural products discovery from this pool of text using a support vector model machine learning classifier trained on ~20,000 published articles. Articles classified as reporting natural products isolation are processed using the natural language processing (NLP) package described above that identifies compound names and source organisms and determines the relationships between these categories using named entity recognition (NER). This step returns a list of isolated compounds and the organisms from which they derive. After manual review by a member of the deposition team to correct any errors from the NLP step a custom deposition page is created that is pre-populated with all the information required for submission except the chemical structures and the NMR data. An email invitation is sent to the corresponding author(s) inviting them to use this preformatted page to deposit their data to the NP-MRD. Currently uptake from these emails is ~10%, suggesting that while some groups see the value of sharing raw data, this has yet to become an established activity in our field.
Figure 3.
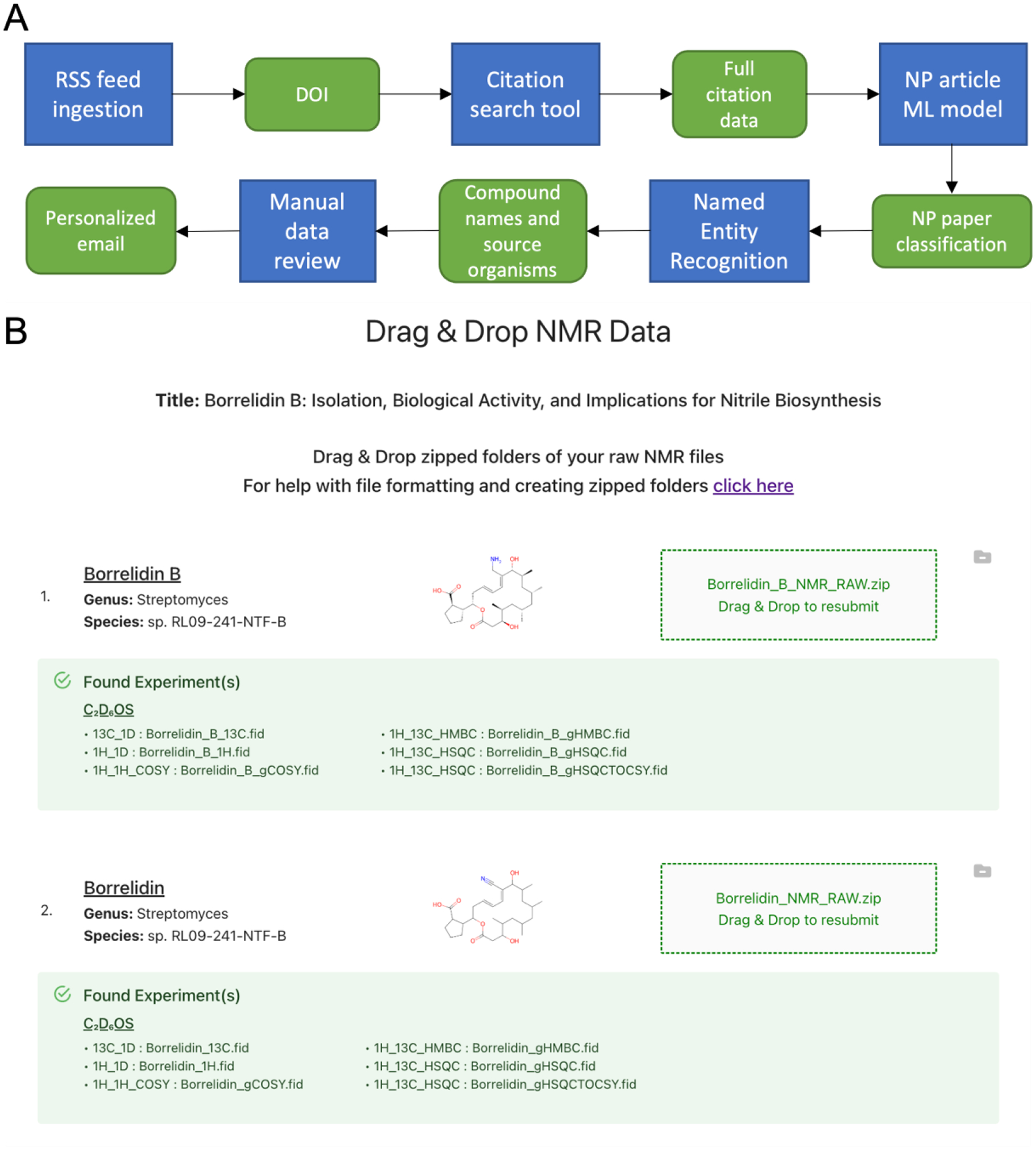
A) Workflow of literature tracking module. Blue boxes denote steps in the automated literature tracking pipeline. Green boxes denote information passed from one step in the process to the next. B) Screenshot example of customized NMR data submission page.
The same pre-population system is used for unsolicited depositions of data from published articles (discussed above). When a user enters the DOI of the article they wish to submit, the deposition system uses the literature service and the NLP/NER tool to generate a prepopulated compound page for the article. A caveat with this approach is that these data are not subject to review by a subject expert. This means that not all articles return prepopulated fields (something that is fixed manually for literature solicitations), and that prepopulated data must be reviewed carefully for accuracy.
Together these features provide a streamlined resource for raw data dissemination for natural products-based NMR data. Although the NP-MRD is a new resource, these data are already being integrated into a number of innovative new initiatives. For example, the Natural Products Atlas14 is engaged in an ongoing project to create cross-links to the NP-MRD,9 GNPS,8 and MIBiG,7 providing a central hub for integrating structural, taxonomic, biosynthetic and spectroscopic data for microbial natural products. Separately the NP-MRD is being recognized as a valuable resource for future applications in metabolomics and small molecule discovery, including computer-aided structure elucidation.18
Conclusion
The new NP-MRD deposition platform has been live since mid-2022. To date (October 16th 2023) we have received data for 1,532 compounds, including 8,168 unique NMR spectra. This has increased the raw data content of the NP-MRD by a factor of six, and established a steady source of data for this growing resource. The interface for the deposition platform and the underlying data structure have both been carefully designed with user experience in mind. The website is supported by a rich suite of data inspection and standardization tools, and is accompanied by detailed error reporting and feedback to help users successfully complete data depositions.
Notwithstanding the ease of use, rates of data deposition to the NP-MRD remain relatively low. We hope that new initiatives such as JNP’s recent requirement for NMR data deposition for new natural products will increase the volume of data being placed in the public domain. The benefits to our community are obvious if we look to other fields where this is the norm. With better data availability comes the opportunity to develop new tools that can revolutionize and simplify the ways in which science is performed. For example, the availability of protein structures in the Protein Data Bank was critical to the development of AlphaFold 2,19 which has revolutionized the field of structural biology. One can envision many new opportunities that could be provided to the natural products community by the creation of a large NMR data repository. Dereplication, de novo structure elucidation, and even bioactivity target prediction could all be subjects for the development of new discovery tools. We hope that this new deposition interface will encourage greater user participation in the broader NP-MRD initiative, and provide JNP readers with a facile mechanism by which they can comply with new data deposition requirements.
Funding Sources
This project was supported through funding from the U.S. National Institutes of Health (NCCIH/ODS) under grant number U24AT010811 to J.R.C., L.W.S., D.S.W. and R.G.L.
REFERENCES
- (1).Wilkinson MD; Dumontier M; Aalbersberg IJ; Appleton G; Axton M; Baak A; Blomberg N; Boiten J-W; da Silva Santos LB; Bourne PE; Bouwman J; Brookes AJ; Clark T; Crosas M; Dillo I; Dumon O; Edmunds S; Evelo CT; Finkers R; Gonzalez-Beltran A; Gray AJG; Groth P; Goble C; Grethe JS; Heringa J; ‘t Hoen PAC; Hooft R; Kuhn T; Kok R; Kok J; Lusher SJ; Martone ME; Mons A; Packer AL; Persson B; Rocca-Serra P; Roos M; van Schaik R; Sansone S-A; Schultes E; Sengstag T; Slater T; Strawn G; Swertz MA; Thompson M; van der Lei J; van Mulligen E; Velterop J; Waagmeester A; Wittenburg P; Wolstencroft K; Zhao J; Mons B The FAIR Guiding Principles for Scientific Data Management and Stewardship. Scientific Data 2016, 3 (1), 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Rutz A; Sorokina M; Galgonek J; Mietchen D; Willighagen E; Gaudry A; Graham JG; Stephan R; Page R; Vondrášek J; Steinbeck C; Pauli GF; Wolfender J-L; Bisson J; Allard P-M The LOTUS Initiative for Open Knowledge Management in Natural Products Research. Elife 2022, 11. 10.7554/eLife.70780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Else H A Guide to Plan S: The Open-Access Initiative Shaking up Science Publishing. Nature 2021. 10.1038/d41586-021-00883-6. [DOI] [PubMed] [Google Scholar]
- (4).McAlpine JB; Chen SN; Kutateladze A; Macmillan JB; Appendino G; Barison A; Beniddir MA; Biavatti MW; Bluml S; Boufridi A; Butler MS; Capon RJ; Choi YH; Coppage D; Crews P; Crimmins MT; Csete M; Dewapriya P; Egan JM; Garson MJ; Genta-Jouve G; Gerwick WH; Gross H; Harper MK; Hermanto P; Hook JM; Hunter L; Jeannerat D; Ji NY; Johnson TA; Kingston DGI; Koshino H; Lee HW; Lewin G; Li J; Linington RG; Liu M; McPhail KL; Molinski TF; Moore BS; Nam JW; Neupane RP; Niemitz M; Nuzillard JM; Oberlies NH; Ocampos FMM; Pan G; Quinn RJ; Reddy DS; Renault JH; Rivera-Chávez J; Robien W; Saunders CM; Schmidt TJ; Seger C; Shen B; Steinbeck C; Stuppner H; Sturm S; Taglialatela-Scafati O; Tantillo DJ; Verpoorte R; Wang BG; Williams CM; Williams PG; Wist J; Yue JM; Zhang C; Xu Z; Simmler C; Lankin DC; Bisson J; Pauli GF The Value of Universally Available Raw NMR Data for Transparency, Reproducibility, and Integrity in Natural Product Research. Natural Product Reports. 2019, pp 35–107. 10.1039/c7np00064b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Hoch JC; Baskaran K; Burr H; Chin J; Eghbalnia HR; Fujiwara T; Gryk MR; Iwata T; Kojima C; Kurisu G; Maziuk D; Miyanoiri Y; Wedell JR; Wilburn C; Yao H; Yokochi M Biological Magnetic Resonance Data Bank. Nucleic Acids Res. 2023, 51 (D1), D368–D376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Steinbeck C; Kuhn S NMRShiftDB -- Compound Identification and Structure Elucidation Support through a Free Community-Built Web Database. Phytochemistry 2004, 65 (19), 2711–2717. [DOI] [PubMed] [Google Scholar]
- (7).Terlouw BR; Blin K; Navarro-Muñoz JC; Avalon NE; Chevrette MG; Egbert S; Lee S; Meijer D; Recchia MJJ; Reitz ZL; van Santen JA; Selem-Mojica N; Tørring T; Zaroubi L; Alanjary M; Aleti G; Aguilar C; Al-Salihi SAA; Augustijn HE; Avelar-Rivas JA; Avitia-Domínguez LA; Barona-Gómez F; Bernaldo-Agüero J; Bielinski VA; Biermann F; Booth TJ; Carrion Bravo VJ; Castelo-Branco R; Chagas FO; Cruz-Morales P; Du C; Duncan KR; Gavriilidou A; Gayrard D; Gutiérrez-García K; Haslinger K; Helfrich EJN; van der Hooft JJJ; Jati AP; Kalkreuter E; Kalyvas N; Kang KB; Kautsar S; Kim W; Kunjapur AM; Li Y-X; Lin G-M; Loureiro C; Louwen JJR; Louwen NLL; Lund G; Parra J; Philmus B; Pourmohsenin B; Pronk LJU; Rego A; Rex DAB; Robinson S; Rosas-Becerra LR; Roxborough ET; Schorn MA; Scobie DJ; Singh KS; Sokolova N; Tang X; Udwary D; Vigneshwari A; Vind K; Vromans SPJM; Waschulin V; Williams SE; Winter JM; Witte TE; Xie H; Yang D; Yu J; Zdouc M; Zhong Z; Collemare J; Linington RG; Weber T; Medema MH MIBiG 3.0: A Community-Driven Effort to Annotate Experimentally Validated Biosynthetic Gene Clusters. Nucleic Acids Res. 2023, 51 (D1), D603–D610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Wang M; Carver JJ; Phelan VV; Sanchez LM; Garg N; Peng Y; Nguyen DD; Watrous J; Kapono CA; Luzzatto-Knaan T; Porto C; Bouslimani A; Melnik AV; Meehan MJ; Liu W-T; Crüsemann M; Boudreau PD; Esquenazi E; Sandoval-Calderón M; Kersten RD; Pace LA; Quinn RA; Duncan KR; Hsu C-C; Floros DJ; Gavilan RG; Kleigrewe K; Northen T; Dutton RJ; Parrot D; Carlson EE; Aigle B; Michelsen CF; Jelsbak L; Sohlenkamp C; Pevzner P; Edlund A; McLean J; Piel J; Murphy BT; Gerwick L; Liaw C-C; Yang Y-L; Humpf H-U; Maansson M; Keyzers RA; Sims AC; Johnson AR; Sidebottom AM; Sedio BE; Klitgaard A; Larson CB; Boya PCA; Torres-Mendoza D; Gonzalez DJ; Silva DB; Marques LM; Demarque DP; Pociute E; O’Neill EC; Briand E; Helfrich EJN; Granatosky EA; Glukhov E; Ryffel F; Houson H; Mohimani H; Kharbush JJ; Zeng Y; Vorholt JA; Kurita KL; Charusanti P; McPhail KL; Nielsen KF; Vuong L; Elfeki M; Traxler MF; Engene N; Koyama N; Vining OB; Baric R; Silva RR; Mascuch SJ; Tomasi S; Jenkins S; Macherla V; Hoffman T; Agarwal V; Williams PG; Dai J; Neupane R; Gurr J; Rodríguez AMC; Lamsa A; Zhang C; Dorrestein K; Duggan BM; Almaliti J; Allard P-M; Phapale P; Nothias L-F; Alexandrov T; Litaudon M; Wolfender J-L; Kyle JE; Metz TO; Peryea T; Nguyen D-T; VanLeer D; Shinn P; Jadhav A; Müller R; Waters KM; Shi W; Liu X; Zhang L; Knight R; Jensen PR; Palsson BØ; Pogliano K; Linington RG; Gutiérrez M; Lopes NP; Gerwick WH; Moore BS; Dorrestein PC; Bandeira N Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol 2016, 34 (8), 828–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Wishart DS; Sayeeda Z; Budinski Z; Guo A; Lee BL; Berjanskii M; Rout M; Peters H; Dizon R; Mah R; Torres-Calzada C; Hiebert-Giesbrecht M; Varshavi D; Varshavi D; Oler E; Allen D; Cao X; Gautam V; Maras A; Poynton EF; Tavangar P; Yang V; van Santen JA; Ghosh R; Sarma S; Knutson E; Sullivan V; Jystad AM; Renslow R; Sumner LW; Linington RG; Cort JR NP-MRD: The Natural Products Magnetic Resonance Database. Nucleic Acids Res. 2022, 50 (D1), D665–D677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Proteau PJ Journal of Natural Products 2023 - New NMR Data Requirements and Editor Changes. J. Nat. Prod 2023, 86 (4), 653–654. [DOI] [PubMed] [Google Scholar]
- (11).Sivade M; Alonso-López D; Ammari M; Bradley G; Campbell NH; Ceol A; Cesareni G; Combe C; De Las Rivas J; del-Toro N; Heimbach J; Hermjakob H; Jurisica I; Koch M; Licata L; Lovering RC; Lynn DJ; Meldal BHM; Micklem G; Panni S; Porras P; Ricard-Blum S; Roechert B; Salwinski L; Shrivastava A; Sullivan J; Thierry-Mieg N; Yehudi Y; Van Roey K; Orchard S Encompassing New Use Cases - Level 3.0 of the HUPO-PSI Format for Molecular Interactions. BMC Bioinformatics 2018, 19 (1). 10.1186/s12859-018-2118-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).van Santen JA; Kautsar SA; Medema MH; Linington RG Microbial Natural Product Databases: Moving Forward in the Multi-Omics Era. Nat. Prod. Rep 2021, 38 (1), 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).RDKit: Open-Source Cheminformatics. Https://www.rdkit.org.
- (14).van Santen JA; Poynton EF; Iskakova D; McMann E; Alsup TA; Clark TN; Fergusson CH; Fewer DP; Hughes AH; McCadden CA; Parra J; Soldatou S; Rudolf JD; Janssen EM-L; Duncan KR; Linington RG The Natural Products Atlas 2.0: A Database of Microbially-Derived Natural Products. Nucleic Acids Res. 2022, 50 (D1), D1317–D1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Rutz A; Sorokina M; Galgonek J; Mietchen D; Willighagen E; Gaudry A; Graham JG; Stephan R; Page R; Vondrášek J; Steinbeck C; Pauli GF; Wolfender J-L; Bisson J; Allard P-M The LOTUS Initiative for Open Knowledge Management in Natural Products Research. Elife 2022, 11. 10.7554/elife.70780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Burley SK; Bhikadiya C; Bi C; Bittrich S; Chao H; Chen L; Craig PA; Crichlow GV; Dalenberg K; Duarte JM; Dutta S; Fayazi M; Feng Z; Flatt JW; Ganesan S; Ghosh S; Goodsell DS; Green RK; Guranovic V; Henry J; Hudson BP; Khokhriakov I; Lawson CL; Liang Y; Lowe R; Peisach E; Persikova I; Piehl DW; Rose Y; Sali A; Segura J; Sekharan M; Shao C; Vallat B; Voigt M; Webb B; Westbrook JD; Whetstone S; Young JY; Zalevsky A; Zardecki C RCSB Protein Data Bank (Rcsb.org): Delivery of Experimentally-Determined PDB Structures alongside One Million Computed Structure Models of Proteins from Artificial Intelligence/Machine Learning. Nucleic Acids Res. 2023, 51 (D1), D488–D508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Groom CR; Bruno IJ; Lightfoot MP; Ward SC The Cambridge Structural Database. Acta Crystallogr. B Struct. Sci. Cryst. Eng. Mater 2016, 72 (Pt 2), 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Sahayasheela VJ; Lankadasari MB; Dan VM; Dastager SG; Pandian GN; Sugiyama H Artificial Intelligence in Microbial Natural Product Drug Discovery: Current and Emerging Role. Nat. Prod. Rep 2022, 39 (12), 2215–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Jumper J; Evans R; Pritzel A; Green T; Figurnov M; Ronneberger O; Tunyasuvunakool K; Bates R; Žídek A; Potapenko A; Bridgland A; Meyer C; Kohl SAA; Ballard AJ; Cowie A; Romera-Paredes B; Nikolov S; Jain R; Adler J; Back T; Petersen S; Reiman D; Clancy E; Zielinski M; Steinegger M; Pacholska M; Berghammer T; Bodenstein S; Silver D; Vinyals O; Senior AW; Kavukcuoglu K; Kohli P; Hassabis D Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596 (7873), 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]


