Abstract
The Yersinia enterocolitica prophage PY54 replicates as a linear DNA molecule with covalently closed ends. For replication of a circular PY54 minimal replicon that has been derived from a linear minireplicon, two phage-encoded loci are essential in Escherichia coli: (i) the reading frame of the replication initiation gene repA and (ii) its 212-bp origin located within the 3′ portion of repA. The RepA protein acts in trans on the origin since we have physically separated the PY54 origin and repA onto a two-plasmid origin test system. For this trans action, the repA 3′ end carrying the origin is dispensable. Mutagenesis by alanine scan demonstrated that the motifs for primase and for nucleotide binding present in the protein are essential for RepA activity. The replication initiation functions of RepA are replicon specific. The replication initiation proteins DnaA, DnaG, and DnaB of the host are unable to promote origin replication in the presence of mutant RepA proteins that carry single residue exchanges in these motifs. The proposed origins of the known related hairpin prophages PY54, N15, and PKO2 are all located toward the 3′ end of the corresponding repA genes, where several structure elements are conserved. Origin function depends on the integrity of these elements.
Three temperate bacterial viruses of different organisms generating linear prophages are known, PY54, N15, and PKO2. The DNA carries covalently closed hairpins at their ends. Hence we propose the name “hairpin prophage.” Two of the phages, PY54 of Yersinia enterocolitica and PKO2 of Klebsiella oxytoca, have been discovered very recently (4, 27). However, the paradigm of the linear prophages, N15 of Escherichia coli, was described already in 1967 by Victor Ravin (13). The linear DNA form is also known from brown algae viruses (7, 8) and in the pathogens Borrelia burgdorferi and Agrobacterium tumefaciens (2, 14). Although a straightforward mechanistic scheme has been suggested by Rybchin and Svarchevsky (31), the replication of the linear DNA is still poorly understood. The replication model is still valid today. The crucial components are (i) an origin of replication of the theta type proposing bidirectional DNA synthesis, (ii) a multifunctional phage-encoded replication protein guiding the initiation of replication, and (iii) a protelomerase essential for the conversion of circular precursor DNA into the linear form at a sequence called the telomere resolution site. The initial work on the phage systems apparently stimulated studies on the Borrelia system (5).
The protelomerase is a tyrosine integrase-like enzyme that catalyzes the intermolecular strand transfer in the double-stranded DNA molecule by cleavage and rejoining of phosphodiester bonds to yield the linear prophage with closed hairpin ends (9, 10). The same principle of action of protelomerases has been confirmed on several enzymes of different origins (4, 16, 21). It was rather simple to predict and locate the telomere resolution sites because they map at the ends of the linear DNA. In the circular substrates these sites consist of palindromic sequences of 40 to 70 bp arranged diametrically opposite to cos.
The prediction for the gene of the replication protein was originally assisted by computer search. Two motifs present in the multifunctional replication protein α of the E. coli satellite phage P4 were recognized in the potential product of the open reading frame for repA of N15 (31) and later on in the corresponding RepA sequences of PY54 (17) and PKO2 (4). The motif of the catalytic center of prokaryotic primases—EGYATA—and a type A nucleotide binding site in the P4 α protein are responsible for primase and helicase activity, respectively, (20, 42). Similar motifs are present in the same order as in P4 α in the RepA proteins of N15, PY54, and PKO2. The nucleotide binding motif including flanking residues is identical in N15 and PKO2, whereas in PY54 just the two residues following the central GKT deviate from the other sequences. The primase motif in the RepA sequence is slightly different from that in P4 α (EGFATG versus EGYATA). As in other primase sequences already known (40), the position of the tyrosine is occupied by a phenylalanine in the corresponding RepA motif. We had shown previously that the change of Y to F enhances primase activity in vitro of P4 α and of the primase of the conjugative plasmid RP4 (33).
The P4 α protein has origin binding ability (39) and interacts with the copy number regulation protein Cnr (34, 38). The α gene has been reported to contain the sequence for a secondary replication origin for P4 prophage replication (35). Therefore, the replication properties of the circular prophage P4, i.e., the α protein, provided a realistic model for unraveling the replication properties of the circular minireplicons of the linear hairpin prophages. The experiments to localize the PY54 origin were further stimulated by the existence of autonomously replicating linear and circular miniplasmids of N15 and of PY54 (16, 29). Recently, data supporting the hypothesis that ori might be located within the repA gene have been reported for N15 (30).
The Yersinia prophage PY54 replicates also in E. coli (16), demonstrating that all host-encoded enzymes required for phage propagation are interchangeable between Y. enterocolitica and E. coli. Hence, in all our studies we made use of the advantages this genetically well-characterized host bacterium offers. Here we describe the definition of the circular minimal replicon derived from the linear PY54 prophage. This includes the mapping of RNA polymerase binding sites of the minireplicon to determine the position of the repA promoter and the preparation of an anti-RepA serum for subsequent RepA identification. We locate the repA-dependent origin within the coding region of repA toward the 3′ end. To resolve the role of PY54 RepA in replication, we devised and implemented a direct genetic selection for (i) repA alleles that encode full-length proteins and (ii) origin variants; we subjected both to phenotypic characterization. We demonstrate that RepA acts in trans on the PY54 origin sequence located on a separate plasmid. Furthermore, we show that in vivo (i) for RepA function the motifs for primase and nucleotide binding are essential, (ii) the coding capacity of the origin is dispensable for repA dependent replication, and (iii) defined elements within the origin play a role in replication.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli strain SCS1 [recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1] (Stratagene) was used as host for plasmids, overexpression, and in trans complementation experiments. The PY54 library was constructed in E. coli strain GeneHogs [F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara-leu)7697 galU galK rpsL(Smr) nupG, fhuA::IS2] (Invitrogen). DH5α (15) served as host for the pSH plasmids. Media were as described previously (41). When appropriate, antibiotics were added to the following concentrations: ampicillin (sodium salt; 100 μg/ml), chloramphenicol (10 μg/ml), tetracycline · HCl (10 μg/ml), or neomycin (30 μg/ml). Plasmids are listed in Table 1. The construction of plasmids containing PY54-derived DNA is described in the supplemental material.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | PY54 position (bp) | Reference or origin |
|---|---|---|---|
| pBR329 | Cloning vector, pMB1 replicon, Apr, Cmr, Tcr | 6 | |
| pMS119EH | Cloning vector; pMB1 replicon, Ptac/lacI; Apr | 1 | |
| pMS470Δ8 | Cloning vector; pMB1 replicon, Ptac/lacI; T7 gene 10 Shine-Dalgarno sequence; Apr | 1 | |
| pGZ119EH/HE | Cloning vector; ColD replicon, Ptac/lacI; Cmr | 25 | |
| pGZ119EH/HEX | pGZ119EH/HE, ColD replicon excisable by XhoI | This work | |
| pGZ119EH/HEXA | pGZ119EH/HE, ColD replicon excisable by XhoI or AscI | This work | |
| pKKlux | Promoter probe vector; pMB1 replicon; promoterless luxAB genes from Vibrio harveyi; T1T2 of rrnB; Apr | 19 | |
| pKKL700lux | pKKlux with Solanum tuberosum ST-LS promoter | 19 | |
| pSH101 | pMS119EH Δ[BamHI-HindIII] Ω[PCR PY54 tel 1,990 bp, BamHI-HindIII] | 24592-26571 | 16 |
| pSH101a | pGZ119HEXA Δ[BamHI-HindIII] Ω[pSH101 BamHI-HindIII, 1,996 bp] | 24592-26571 | This work |
| pSH120 | Circular PY54 minireplicon; Kmr | 27398-31809 | 16, 17 |
| pSH121 | Circular PY54 minireplicon; Kmr, Tcr | 31681-27483::Kmra and 27492-27398 | This work |
| pSH122 | Circular PY54 minireplicon; Tcr | 31809-31673::Tcra and 31681-27652 | This work |
| pJH101a | pGZ119EHXA Ω[PY54, 7,945 bp, NheI] | 11171-19115 | This work |
| pJH102a | pGZ119EHXA Ω[PY54, 7,746 bp, NheI] | 3425-11170 | This work |
| pJH103a | pGZ119EHXA Ω[PY54, 6,577 bp, NheI] | 25433-32009 | This work |
| pJH103-1a | pGZ119EHXA Δ[XbaI-SmaI] Ω[PY54 2,281 bp, NheI-EcoRV] | 25433-27713 | This work |
| pJH104a | pGZ119EHXA Ω[PY54, 6,317 bp, NheI] | 19116-25432 | This work |
| pJH105a | pGZ119EHXA Ω[PY54, 4,688 bp, NheI] | 32010-36697 | This work |
| pJH106a | pGZ119EHXA Ω[PY54, 3,206 bp, NheI] | 42305-45510 | This work |
| pJH107a | pGZ119EHXA Ω[PY54, 2,550 bp, NheI] | 39755-42304 | This work |
| pJH108a | pGZ119EHXA Ω[PY54, 2,547 bp, NheI] | 37208-39754 | This work |
| pJH201a | pGZ119EHXA Ω[PY54, 9,992 bp, EcoRV] | 11126-21117 | This work |
| pJH202a | pGZ119EHXA Ω[PY54, 7,762 bp, EcoRV] | 36420-44181 | This work |
| pJH204a | pGZ119EHXA Ω[PY54, 5,058 bp, EcoRV] | 6115-11725 | This work |
| pJH206a | pGZ119EHXA Ω[PY54, 2,688 bp, EcoRV] | 27713-30400 | This work |
| pJH207a | pGZ119EHXA Ω[PY54, 1,785 bp, EcoRV] | 30401-32185 | This work |
| pJH210a | pGZ119EHXA Ω[PY54, 8,272 bp, EcoRV] | 1-6014 and 44182-46399 | This work |
| pGB154 | pMS470Δ8 Δ[NdeI-HindIII] Ω[PCR PY54 repA 3,993 bp, NdeI-HindIII] | 31623-27631 | This work |
| pGB154X000Y | pGB154 encoding the amino acid exchange at the RepA position indicated | 31623-27631 | This work |
| pGB154-2 | pMS470Δ8 [NdeI-HindIII] Ω[PCR PY54 repAΔ2 1,347 bp, NdeI-HindIII] | 31623-30277 | This work |
| pGB154-3 | pMS470Δ8 Δ[NdeI-HindIII] Ω[PCR PY54 repAΔ3 1,360 bp, NdeI-HindIII] | 30292-28933 | This work |
| pGB154-4 | pMS470Δ8 Δ[NdeI-HindIII] Ω[PCR PY54 repAΔ4 1,311 bp, NdeI-HindIII] | 28941-27631 | This work |
| pGB154-6 | pMS470Δ8 Δ[NdeI-HindIII] Ω[PCR PY54 repAΔ6, 2,691 bp, NdeI-HindIII] | 31623-28933 | This work |
| pGB154-7 | pMS470Δ8 Δ[NdeI-HindIII] Ω[PCR PY54 repAΔ7 2,661 bp, NdeI-HindIII] | 30292-27631 | This work |
| pAM154 | pGZ119EHX Δ[BamHI-HindIII] Ω [pGB154 BamHI-HindIII, 4,042 bp] | 31623-27631 | This work |
| pAM154HE | pGZ119HEX Δ[BamHI-HindIII] Ω[pGB154 BamHI-HindIII, 4,042 bp] | 27631-31623 | This work |
| pAM155 | pAM154 Δ[XhoI-XhoI, 988 bp] | 31623-27631 | This work |
| pAM155HE | pAM154HE Δ[XhoI-XhoI, 988 bp], not viable | 27631-31623 | This work |
| pAM155-4 | pAM155 Δ[BamHI-HindIII, 4042 bp] Ω [pGB154-4 BamHI-HindIII, 1,363 bp] | 28032-27631 | This work |
| pAM155-9 | pGZ119EHX Δ[BamHI/T4 DNA polymerase-HindIII] Ω[pGB154 StyI/T4 DNA polymerase-HindIII, 407 bp] | 28035-27631 | This work |
| pNT154-10 | pGZ119EHX Δ[BamHI-HindIII] Ω [PCR PY54 repA 212 bp, BamHI-HindIII] | 27973-27762 | This work |
| pNT154-11 | pGZ119EHX Δ[BamHI-HindIII] Ω [oligonucleotides, repA 54 bp, BamHI-HindIII] | 27912-27859 | This work |
| pNT155-10 | pNT154-10 Δ[XhoI-XhoI, 988 bp] | 27973-27762 | This work |
| pNT155-11 | pNT154-11 Δ[XhoI-XhoI, 988 bp], not viable | 27912-27859 | This work |
| pNT154-12 | pGB154 Δ[StyI-HindIII, 399 bp] Ω[StyI-HindIII adapter providing a stop codon] | 31623-28031 | This work |
| pNT153-14 | pGB154 Δ[EcoRI-HindIII, 798 bp] Ω[PCR PY54 500 bp, EcoRI-HindIII] | 31623-27935 | This work |
| pNT153-15 | pGB154 Δ[StyI-HindIII, 399 bp] Ω[PCR PY54 230 bp, StyI-HindIII] | 31623-27806 | This work |
| pKB153-16 | pGB154 Δ[NdeI-CpoI, 487 bp] Ω[PCR PY54 401 bp, NdeI-CpoI] | 31536-27631 | This work |
| pKB153-19 | pGB154 Δ[NdeI-CpoI, 487 bp] Ω[NdeI-CpoI adapter, PY54 repA 11 bp] | 31584-27631 | This work |
Transposon insertions resulted in the generation of 10-bp (Kmr) and 9-bp (Tcr) target site sequence duplications. See supplemental material for details.
Construction of the replicon/origin isolation plasmids pGZ119EHX/HEX and pGZ119EHXA/HEXA.
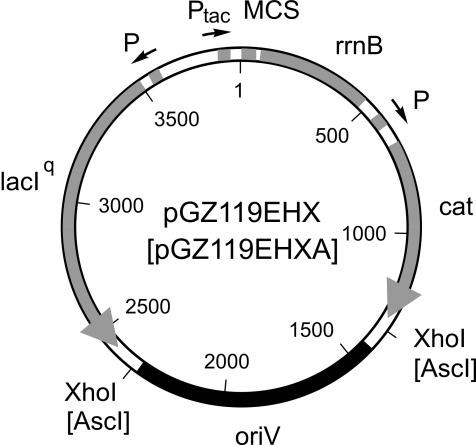
For DNA manipulation standard techniques were used (32). The Ptac/lacIq-regulated expression vectors pGZ119EH/HE carry the ColD replication origin and the chloramphenicol resistance gene cat (25). A unique XhoI site is located at position 2341. To construct pGZ119EHX/HEX (Fig. 1), a second XhoI recognition sequence was generated at position 1353 by a site-directed mutagenesis approach based on PCR (36). This excisable 988-bp XhoI fragment encodes the complete RNA I and RNA II and comprises the corresponding promoters and ori, the site where the primer RNA II is elongated by DNA polymerase I. To construct pGZ119EHXA/HEXA, an AscI recognition sequence was introduced at each XhoI site of pGZ119EHX/HEX via a PCR-based approach (Fig. 1).
FIG. 1.
pGZ119EHX/HEX and pGZ119EHXA/HEXA as vector systems for controlled gene expression and replicon/ori isolation. The expression unit is composed of the tac promoter, the lacO operator sequence, the lacI repressor gene, and the rrnB operon containing the Rho-independent terminators T1 and T2. The antibiotic resistance gene is cat of Tn9, the replicon stems from ColD-CA23, and the multicloning site (MCS) is that of M13mp18 or mp19 for the EH or HE version, respectively. For the purpose of replicon/ori isolation, the ColD origin was made excisable as an XhoI fragment or an AscI fragment.
Construction of a PY54 DNA fragment library.
To construct a library of overlapping restriction fragments of PY54, a high-titer phage lysate was purified by cesium chloride gradient centrifugation (32). Phage DNA was isolated as described previously (18). Circular DNA was obtained by ligating the cohesive ends of the viral DNA with T4 DNA ligase. The DNA was digested with the restriction endonucleases EcoRV and NheI, which cut the PY54 genome into 10 and 11 fragments, respectively. The fragments were inserted into the corresponding sites of tet of pBR329 (Apr, Cmr, and Tcr) (6). Upon transformation of E. coli strain GeneHogs, recombinant plasmids of tetracycline-sensitive colonies were characterized by restriction analysis. Plasmids containing NheI restriction fragments resulted in the pJH100 series, and those harboring EcoRV restriction fragments gave the pJH200 series (Table 1). To construct the pJH100a and pJH200a series, the NheI and EcoRV fragments of the library were inserted into pGZ119EHXA (Fig. 1) digested with XbaI and SmaI, respectively. The plasmids obtained carry the insert in either orientation. To assay for replicon activity, the ColD origin was removed by cleavage with AscI and subsequent gel electrophoresis. Following religation of the remaining fragments (pJH100b and pJH200b series), SCS1 was transformed.
Site-directed mutagenesis.
Point mutations were introduced into repA directly using a PCR-based method (36) with pGB154 as template. The primers used were designed by changing as few bases as possible. Following mutagenesis, the nucleotide sequence of each repA allele was determined.
Analysis of promoter activity in vivo.
RepA promoter activity was studied by using the promoter search vector pKKlux (19). pKKlux is a derivative of pKK232-8 that carries the promoterless luxAB genes of the bioluminescent bacterium Vibrio harveyi. PCR products to be studied for repA promoter activity contained a SmaI site at the 5′ end and a XbaI site at the 3′ end and were inserted into the corresponding sites of pKKlux. After transformation of E. coli DH5α, the nucleotide sequences of suitable constructs were verified by DNA sequencing. The luminescence measurements were carried out as previously described (19). Briefly, cultures of plasmid-containing DH5α were grown at 28°C to an A600 of 1.0 and diluted to contain 106 cells per ml. A total of 100 μl of the diluted cultures was transferred to a microtiter plate. Fifty microliters of 2% decylaldehyde in 50 mM sodium phosphate buffer, pH 7.0, was added, and luminescence was measured at 28°C for 10 sec in a Microlumat LB96P (EG & G Berthold, Bad Wildbad, Germany). To determine the background of the assay, a DH5α strain containing the promoter search vector pKKlux was used. To normalize the results, DH5α(pKKL700lux) was used (19). pKKL700lux carries the ST-LS1 promoter from Solanum tuberosum which is fully active in bacteria.
RESULTS
Replicon search on the PY54 genome.
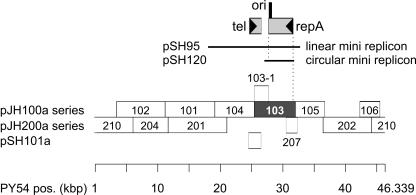
The pair of replicon isolation vectors pGZ119EHXA/HEXA allows for removing easily the vector origin ColD by digestion with AscI or XhoI (Fig. 1). Plasmids of the pJH100a and pJH200a series carry the NheI and EcoRV restriction fragments of the circularized DNA of phage PY54 (see Material and Methods). To ensure that the fragmentation did not disrupt the phage-encoded replicon, fragments were chosen to overlap each other for at least 176 bp (pJH207/pJH105) (Fig. 2 and Table 1). The fragments were present in each of both orientations with respect to the tac promoter of pGZ119EHXA. To assay for replicon activity, the ColD origin of the pJH100a and pJH200a series was removed, and SCS1 cells were transformed (see Material and Methods). The sole fragment conferring replicon activity was the 6,577-bp NheI fragment of plasmid pJH103a. The removal of the ColD origin resulted in the autonomously replicating plasmid pJH103b. The conclusion is that it carries a replication origin and possibly a phage replication gene. According to published sequence data, pJH103b encodes the entire repA gene, i.e., the potential replication gene of PY54 (17). Thus, in the following we concentrated on the identification of the replicon on pJH103b by making use of already existing derivatives, which carry PY54 sequences contained within the NheI fragment of pJH103b (Table 1 and Fig. 2).
FIG. 2.
PY54 fragment libraries used for the identification of the phage-encoded replicon and for the repA-dependent origin (ori). The pJH100a and pJH200a series of plasmids based on the vector pGZ119EHXA contain the NheI and EcoRV fragments of PY54, respectively. pSH101a carries essentially tel. Only fragments overlapping each other are shown. The gray bar represents the PY54-derived NheI fragment acting as a replicon. None of the other fragments carries a repA-dependent ori that functions in trans (see text for details). The localization and orientation of tel and repA are shown. Within the repA reading frame, ori is marked in black. The PY54 portions of the linear minireplicon pSH95 and of the circular minireplicon pSH120 are indicated. The scale for the PY54 viral genome is given at the bottom, beginning and ending with the cohesive ends (17).
The PY54 repA gene supports autonomous plasmid replication.
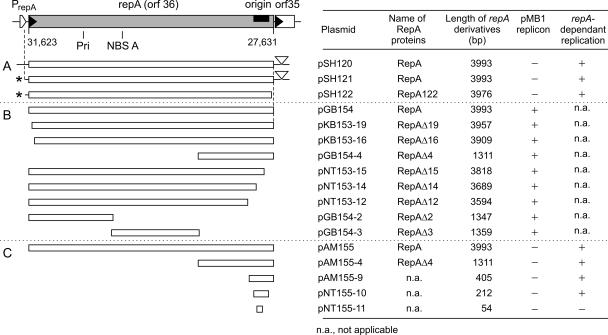
Plasmid pSH120 consists of a kanamycin resistance gene and the repA region of prophage PY54. This region comprises the complete repA gene, a 186-bp intergenic region upstream of repA, and the 5′ region (218 bp) of orf35, which is split in two halves by the kanamycin resistance gene (Table 1 and Fig. 2 and 3). Derivatization yielded pSH121 and pSH122 (Table 1; see supplemental material). The existence of these minireplicons suggested that repA functions as the replicon for the PY54 prophage replication because of three reasons: (i) there exists the prophage-derived linear minireplicon pSH95 (Fig. 2) mainly comprising repA, tel, and par (16); (ii) the circular minireplicon pSH120 was derived from this linear minireplicon; and (iii) pSH122 carries just a truncated repA with a short intergenic region (Fig. 3). This intergenic region did not seem to confer properties of a replication origin. To prove the minireplicon hypothesis, only the coding region of repA devoid of any flanking PY54 nucleotide sequences has been inserted into the vector for replicon and ori isolation.
FIG. 3.
Replicon identification and isolation of the replication origin of the PY54 prophage. The repA region is drawn at the top of the figure (left panel) as a portion of the PY54 genome. The sequence positions of repA (31623 to 27631) are according to EMBL accession no. AJ564013. The direction of transcription is from left to right with the repA promoter PrepA marked by an open arrowhead. The repA gene (orf36) and orf35 are included. Motifs in the deduced amino acid sequence are indicated: NBS A and the primase motif (Pri) as part of the catalytic site of the proposed RepA primase activity. The black bar marks the position of the replication origin. For a description of the construction of the plasmids, see the supplemental material. The open bars represent the repA portions of the plasmids given. The table (right panel) summarizes the names of the plasmids and the lengths of repA portions contained. The plasmids that carry the pMB1 replicon or that replicate in a repA-dependent manner are indicated. Section A (both panels) includes autonomously replicating PY54 plasmids. The asterisks in the left panel indicate the tetC/tetR divergent promoters; the wedges symbolize the insertion site of the aphA-Tn903 cassette within orf35. In pSH122, the truncated repA gene ends with some non-repA codons (for an explanation, see the supplemental material). Section B (both panels) includes plasmids carrying repA deletion derivatives. Each of these constructs contains the pMB1 origin in the vector portion. Section C (both panels) includes plasmids used to locate the minimal PY54 origin within repA. These constructs are devoid of the vector origin. pNT155-11 does not replicate in the presence of repA.
The minireplicon consists of repA and a promoter.
Since we anticipated that repA expression is needed for replicon function, we chose the chemically inducible expression vectors pGZ119EHX and pGZ119HEX carrying the ColD replicon (Fig. 1) for the isolation of the repA gene. Based on these vectors, pAM154 and pAM154HE contain the entire repA coding region including the T7 gene 10 Shine-Dalgarno sequence in the transcriptive and antitranscriptive orientation with respect to the tac promoter (Fig. 3; see supplemental material). After removal of the ColD origin, only pAM155 that carries repA in the transcriptive orientation gave viable colonies upon transformation of SCS1 (Table 2). Hence, pAM155 replicates autonomously in E. coli and thus contains a functional PY54 minimal replicon. Although the tac promoter is controlled by LacI, it is known that the leakiness of the promoter under noninduced conditions is high enough to allow for a basal level of transcription. The data obtained demonstrate that sufficient repA is transcribed for replicon function. Since pAM155HE carrying repA in antitranscriptive orientation does not exist, we conclude that replication via the origin within repA depends on the presence of RepA protein (see below). As pAM155 replicates, the alternative repA start GTG, nine codons upstream of the proposed ATG start codon, is at least dispensable for RepA-driven replication in E. coli. Hence, in the following we ignored the GTG start codon and used the repA version beginning at ATG.
TABLE 2.
In vivo replication of PY54 ori plasmidsa
| Ori plasmid | pAM155 | pAM155HE | pAM155-4 | pAM155-4 | pAM155-4 | pAM155-4 | pAM155-9 | pNT155-10 | pNT155-11 |
| Helper plasmid | − | − | − | pGB154 | pKB153-19 | pNT153-14 | pGB154 | pGB154 | pGB154 |
| Replication | + | − | − | + | − | + | + | + | − |
| RepA action | cis | NA | NA | trans | NA | trans | trans | trans | NA |
The series of pAM155 and pNT155 plasmids (Cmr) lack the original replication origin (ColD) of the vector pGZ119EHX. The pMB1 replicon-based pMS119EH serves as the vector for pGB154, pKB154-19, and pNT153-14 (all Apr). NA, not applicable.
PY54-dependent replication requires repA transcription.
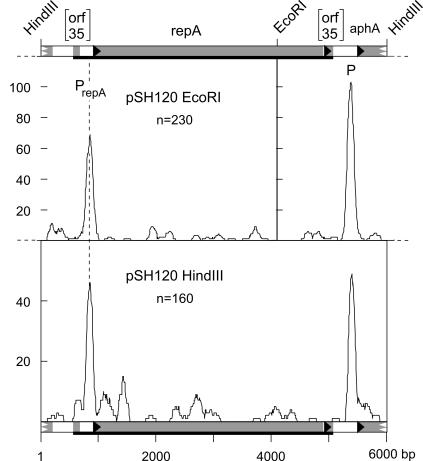
The existence of pAM155 (see above) implies that in pSH120 transcription of repA must occur, maybe from its original promoter, because pSH120 carries a 186-bp intergenic region upstream of repA that could easily provide promoter activity. Since computer predictions failed to identify any promoter sequence in this region (17), we examined this hypothesis by RNA polymerase binding studies. We have applied E. coli RNA polymerase because the pSH series of plasmids replicate in E. coli and because there is a close phylogenetic relationship between E. coli and Y. enterocolitica. The binding studies on RNA polymerase/DNA complexes carried out by electron microscopy demonstrated that two pronounced binding sites exist in pSH120. One accounted for the aphA promoter; the other one was observed in the intergenic region upstream of repA (Fig. 4). Thus, the histograms showing RNA polymerase binding sites on pSH120 demonstrate the presence of a promoter upstream of repA. In pSH122 part of the repA upstream region has been deleted. Only one peak mapping toward the junction tetC-PY54 DNA was observed (data not shown) which is caused by the divergent tandem promoter for tetC and for the tetracycline repressor gene tetR. This tetR promoter firing toward repA is present on the EZ::TN insertion cassette used and is responsible for transcription of repA.
FIG. 4.
RNA polymerase binding sites on PY54 minireplicon pSH120 (6,008 bp) linearized with EcoRI or HindIII. Covalent complexes of E. coli σ70 RNA polymerase and plasmid DNA were used to localize the position of the RNA polymerase binding sites on the DNA by electron microscopy (10). The gene organization of the plasmid is shown at the top of the histograms. The gray boxes represent genes; the 5′ ends are highlighted by triangles. Orf35 is split in two portions by the transposon inserted. The bold black lines mark the extension of PY54-derived DNA. The peaks of bound RNA polymerase demonstrate the presence of a promoter, P. The positions of relevant restriction sites are given. In separate experiments the plasmid was digested with two restriction enzymes, each cutting the DNA once. The histograms were generated by determining the frequency of distribution of RNA polymerase bound over 1,000 equidistant points on the target DNA, counting all protein complexes bound within a 2% window (in bp) centered at each point. The total number of complexes measured is given within the histograms. The abscissa gives the number of complexes measured with protein bound at the respective position.
Even though the significant RNA polymerase binding site upstream of repA clearly demonstrates the presence of a promoter signal (Fig. 4), the corresponding region results in only minute transcription activity in vivo (Table 3), indicating that transcription at the repA promoter is initiated probably quite inefficiently. It is important to notice that the promoter test vector by itself provides a basal transcription activity (19). However, the tetR promoter of pSH122 resulted in efficient transcription. This enhanced transcription level might be key for a copy number increase of repA minireplicons.
TABLE 3.
Luminescence of E. coli DH5α strains harboring plasmidsa
| Plasmid | Insert | Luminescence (RLU/10 sec) | Source |
|---|---|---|---|
| None (LB) | 161 ± 90 | ||
| pKKlux | None | 914 ± 194 | |
| pKK700lux | ST-LS1 promoter | 11,477 ± 920 | S. tuberosum |
| pSH109 | PY54 position 31737 to 31624 | 542 ± 14 | pSH120 |
| pSH110 | PY54 position 31805 to 31624 | 843 ± 57 | pSH120 |
| pSH111 | PY54 position 31681 to 31624 + tetR promoter | 79,951 ± 3188 | pSH122 |
Nucleotide sequences upstream of repA as indicated were inserted into the promoter search vector pKKlux (see Material and Methods). The data shown were averaged from at least three independent experiments done in triplicate. The plasmids pKKlux and pKK700lux were used as controls. RLU, relative light units produced during the first 10s after the addition of the substrate.
Does efficient repA transcription increase the copy number of the minireplicon?
Upon transposon mutagenesis (tetC) of pSH120, the copy number of the resulting plasmids increased considerably. The insertion of the tetC cassette into pSH120 yielding pSH121 caused a more than 30-fold increase in copy number compared to that of pSH120 (data not shown). The plasmid amount isolated from cells harboring pSH120 is low, whereas cells carrying pSH121 or pSH122 contain high but comparable amounts of plasmid DNA. Since the insertion occurred in the upstream region of repA, the increase in copy number might well have something to do with the alteration of the original repA promoter or the removal of a regulatory sequence for protein binding. The next question we asked was whether RepA acts in cis or in trans to its origin of replication. In other words, is it possible to separate part of repA and identify an origin?
RepA acts in trans at ori.
The fragment library in pGZ119EHXA was also used to ensure that except for the repA gene no other region of the phage genome carries a repA-dependent replication origin (Fig. 2). Upon removal of the ColD origin of the pJH plasmid series and transformation of SCS1(pGB154) that is RepA+, no colonies were obtained, suggesting that additional repA-dependent origins are absent from the PY54 genome. Plasmid pJH103b, which carries repA and flanking regions (Fig. 2), is compatible with the repA-encoding helper pGB154 because the transformation under selective conditions yielded the anticipated number of colonies with the expected plasmid content.
To identify the potential origin, three overlapping fragments of the repA gene, the 5′ portion starting with the predicted initiation codon ATG, its middle part, and the 3′ end were prepared from pGB154-2, pGB154-3, and pGB154-4 (Fig. 3), respectively, and inserted into pGZ119EHX. Following removal of the vector ColD origin, colonies have been obtained only in the presence of a complete repA gene in trans for pAM155-4 containing the 3′ repA portion (Fig. 3 and Table 2). The other two possible derivatives with either the 5′ portion or the middle part of repA cannot be replicated in the presence of an intact repA gene in trans. Thus, the 3′ end of repA contains the replication origin. RepA acts in trans to this origin.
The origin localizes toward the 3′ end of repA.
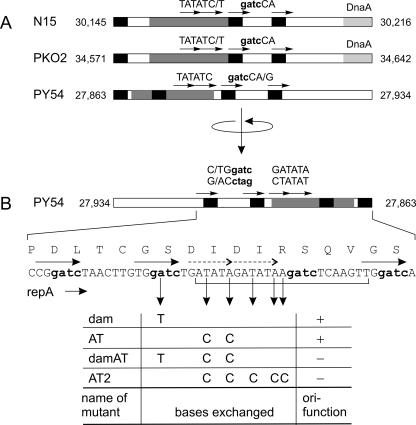
To narrow down the origin moiety, two additional experiments have been carried out. The same procedure just described for origin isolation yielded pAM155-9 and pNT155-10, carrying considerably smaller repA fragments of 405 bp and 212 bp, respectively (Fig. 3 and Table 2). Thus, the very 3′ end of repA embraces the origin of PY54 prophage replication. pNT155-11 carries a 54-bp segment composed of four dam methylation sites, three of which are part of 6-bp sequence repetitions, and an AT-rich region containing 6-bp direct repeats. These structural elements are conserved among PY54, N15, and PKO2 (Fig. 5A). Since pNT155-11 is not viable in the presence of the helper plasmid pGB154 providing repA (Table 2), these elements alone are insufficient to act as a repA-dependent origin.
FIG. 5.
(A) Structural elements conserved in the proposed origins of N15, PKO2, and PY54. The numbers at both sides of the bars are the nucleotide positions within the respective genome. The GenBank/EMBL accession numbers are as follows: N15, AF064539; PKO2, AY374448; PY54, AJ564013. The black boxes and the light gray box represent the dam-methylation sites GATC and the DnaA consensus site, respectively. The AT-rich region is given in dark gray. The arrows mark the positions of direct repeats conserved in all three origins. (B) The bar representing the PY54 segment of panel A has been mirrored to show the relevant portion of the repA nucleotide sequence and the RepA primary structure in the convenient direction from left to right. Above the coding strand, the deduced amino acid sequence is shown. The dam methylation sites are marked in boldface lowercase letters. The bracket highlights the AT-rich region. Arrows mark the inverted repeats containing dam sites and the inverted repeats within the AT-rich segment. Nucleotides exchanged by mutagenesis are given.
Mutagenesis of defined origin elements abolishes origin function.
To study the importance of the conserved elements described above, site-directed mutagenesis was applied. Four mutants were generated (Fig. 5B) as follows: (i) the alteration of one dam site that is part of a 6-bp repeat, (ii) the alteration of one arm of the direct repeat within the AT-rich segment, (iii) the combination of the first and second alterations; and (iv) the combination of the second alteration and further changes in the AT-rich segment generating a direct repeat that differs from the original one and reduces the AT content of the segment. The alterations were designed to conserve the wild-type amino acid sequence of the RepA protein; i.e., in most cases the wobble position of a codon was changed (Fig. 5B). The mutations were introduced into pAM155-9 to be tested for origin function in trans and into pAM155 to be tested for replicon function. Furthermore, the respective full-length repA variants were tested for RepA overproduction.
The alteration of a single element had little or no effect on replication of the respective mutant PY54-ori plasmid pAM155-9 and of the corresponding replicon derivatives pAM155 (Fig. 3 and 5B). If two elements were destroyed, the origin function was completely lost, and accordingly the replicon was inactive. Hence, the set of repeats containing the dam sites or the dam site within the AT-rich region is essential for a functional origin. However, the alteration of one arm keeps the origin active, whereas the combination of two mutations abolished replication.
The existence of an nonfunctional PY54 origin within the full-length repA gene resulted in a significant increase of the RepA amount detected in extracts of induced cells. However, under inducing conditions in the presence of a functional RepA-dependent origin in trans, the RepA amount decreases again to the limit of detection (data not shown). In both cases limited degradation of RepA was detected (not shown).
The coding capacity of the ori region is dispensable for RepA function.
To answer the question whether or not the entire repA gene is needed to drive replication from the minimal origin, deletion derivatives of repA from the 5′ and 3′ ends were generated and used in the in vivo complementation assay for the replication of the ori plasmid pAM155-9 (Fig. 3). The ori plasmid did not replicate in the presence of pKB153-19 or pKB153-16 (Fig. 3 and Table 2). Hence, apparently the entire 5′ end of repA is required to promote origin replication. Not even 36 bp are dispensable.
We knew already from pSH122 that short deletions at the 3′ end by the removal of 18 bp did not abolish the replication activity of the encoded RepA variant (Fig. 3). Similarly, removal of 175 bp and 307 bp yielded pNT153-15 and pNT153-14, respectively. The corresponding proteins RepAΔ15 and RepAΔ14 were still replication proficient. Only RepAΔ12 encoded by pNT153-12 that lacks 399 bp of repA was replication deficient (Fig. 3). The data demonstrate that the region upstream of the origin encodes the replication activity of RepA (Fig. 3). The coding capacity of the origin region within repA apparently is dispensable to drive replication from an origin in trans. The following deals with the identification of the proposed 1330-residue RepA protein.
C terminal modification facilitates the identification of RepA.
Expression vector cloning of repA gave minute amounts of RepA protein undetectable in gels stained with Coomassie blue. Independent of whether the original Shine-Dalgarno sequence or the one of T7 gene 10 was used, there was no improvement. Similarly the use of the predicted initiation codons ATG or the GTG nine codons upstream was applied without the expected success. Additionally, a rare codon effect could not be observed. However, overexpression of repA deletion derivatives (Fig. 3) resulted in prominent amounts of product (data to be published elsewhere). Microsequencing of the various RepA deletion variants confirmed their N-terminal integrity. By far the best of these derivatives for protein overproduction was RepAΔ3 encoded by pGB154-3. Following solubilization in urea, the corresponding truncated product RepAΔ3 was purified and applied as an antigen to raise anti-RepAΔ3 serum in rabbits.
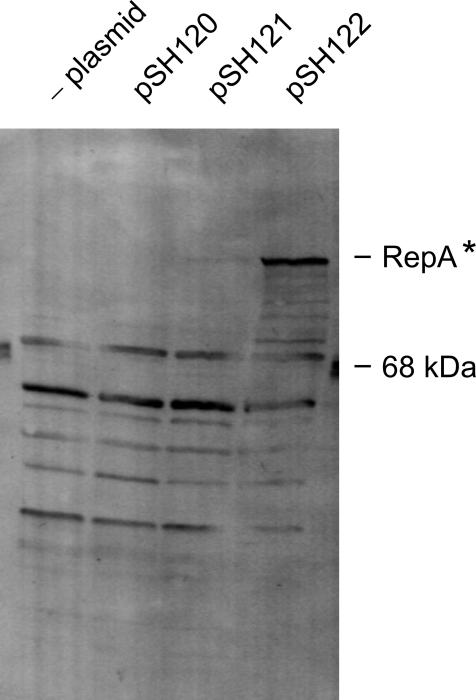
The RepAΔ3 antiserum was used to probe extracts of cells harboring the series of pSH plasmids (Fig. 6). Extracts of DH5α(pSH122) gave the best signal, whereas the signal for pSH121 was very weak, and for pSH120 the signal was not detectable at all. Although there are nonspecific signals seen in the extract of plasmid-free host strain DH5α, the RepA signal is clearly visible because this part of the gel is essentially free of nonspecific signals. Accordingly, only in DH5α(pSH122) extracts was RepA observed in Coomassie blue-stained gels (data not shown). The alteration at the repA 3′ end seems to allow for accumulation of the pSH122-encoded RepA variant in the cell.
FIG. 6.
Identification of the repA gene product. Standardized amounts (measured as A600 of the cultures) of stationary DH5α cells with and without plasmids were used for preparation of sodium dodecyl sulfate cell extracts (12). An equal volume of each extract was electrophoresed on a 15% polyacrylamide gel containing 0.1% sodium dodecyl sulfate. After gel electrophoresis, the proteins were electroblotted onto a nitrocellulose membrane using a semidry transfer device, followed by reaction with 1:500 diluted anti-RepAΔ3 serum and subsequently with dichlorotriazinylaminofluorescein-conjugated goat antirabbit immunoglobulin G. The fluorescence signal was visualized using the FluorImager 575 and ImageQuant software version 5.1. RepA* is the RepA fusion protein encoded by pSH122 (see Material and Methods). The position of bovine serum albumin (68 kDa) is given.
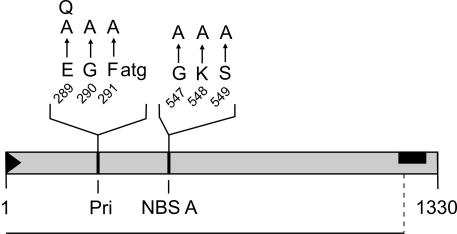
Intact primase and nucleotide binding motifs are essential for RepA-dependent replication.
The primase motif is part of the catalytic center of DnaG and other primases, and the nucleotide binding motif type A (NBS A) interacts with the NTP triphosphate group of NTP-consuming enzymes like helicases. To investigate the influence of these motifs on RepA function(s), a series of point mutations was generated (Fig. 7). The in vivo complementation assay for RepA activity demonstrated that none of the seven mutations showed any activity. Thus, both motifs prove to be essential for RepA-dependent replication of the ori plasmid pAM155-9 (Fig. 3). In addition, we conclude that the DnaG protein of the host cannot substitute for the action of RepA. The same conclusion holds true for the NBS A motif that could belong to a postulated helicase activity of RepA. Again, the motif is essential; a host-encoded helicase seems to be unable to substitute for the RepA activity associated with the NBS A motif. Since the primase and the helicase functions of the host do not support PY54 ori replication, the data suggest the absolute replicon specificity of RepA.
FIG. 7.
Mutations generated in prominent amino acid motifs in RepA. The gray bar represents the RepA protein. The positions of the primase motif (Pri) and of NBS A are indicated. Highly conserved residues shown in capital letters were changed to alanine. Additionally, the glutamate was replaced by glutamine. In the primase motif, the residues weakly conserved are in lowercase letters. The RepA amino acid positions of the residues exchanged are given. The line denotes the RepA portion essential for replication of the RepA-dependent origin in trans. The black bar marks the protein domain that corresponds to the origin region in the repA gene.
DISCUSSION
The circular minimal replicon of PY54 consists of repA and a promoter region driving repA transcription. The original PY54 repA promoter is interchangeable with other promoters like that of tetR or tac without loss of the replication ability of the minireplicon. Since a few potential −10 regions were spotted upstream of the repA gene, they provide several possibilities to propose a promoter sequence. Only for one of the regions has a suitable −35 region (TCAACA) been extracted that fits in the distance of 17 bp to an appropriate −10 region (GTTGAT; PY54 positions 31681 to 31686) 60 bp upstream of repA. Since the proposed promoter sequence coincides with the position of the peak in the histogram displaying the RNA polymerase binding sites (Fig. 4), it likely functions as the initiation signal for repA transcription in the PY54 prophage. The lack of any RNA polymerase binding sites in the repA coding sequence indicates that transcription, except for repA expression, probably is dispensable for replicon activity. Transcriptional activation for replication initiation is unlikely since in our in vivo replication test system the origin functions independent of the orientation relative to a promoter; also the insertion of a strong terminator between the promoter and the origin has no significant effect on repA-dependent replication (data not shown). The copy number effect in the pSH plasmid series is likely to be related to the efficiency of the promoter, because in pSH120 the original promoter is weak, whereas in pSH121 and pSH122, the tetR promoter is strong (Table 3). However, we cannot exclude that the repA promoter is regulated. The only available element for regulation in pSH120 apparently is the product of repA itself, probably acting as a repressor on sequences upstream of the gene. Thus, autoregulation by RepA is still a possible scenario for copy number regulation of pSH120 and, hence, also of PY54 prophage replication.
A closer look at the sequence of the PY54 repA gene offered the in-frame initiation codon GTG nine codons upstream of the originally proposed ATG. Our data demonstrate that the additional nine codons are dispensable for minireplicon activity (Fig. 3 and Table 2). The use of either GTG or ATG resulted in the same phenotype and the same minute RepA amount detected by immunoblotting (data not shown). Hence, RepA encoded by the gene starting with ATG is suitable for RepA replication functions.
From the data obtained for the set of pSH plasmids, we learned that the C-terminal end of RepA seems to play an important role for repA expression and/or protein stability. The recombinant RepA* protein of pSH122 is devoid of six original residues and ends with 29 residues of incidental origin. Nevertheless, this almost full-length RepA protein, which is fully active in vivo, represents one of our most efficient derivatives for overexpression of the repA gene (Fig. 6). We already know that proteases Lon and OmpF are not responsible for a potential RepA degradation (data not shown). Expression of repA portions yielded reasonable protein amounts, whereas the full-length gene led to a drop in appreciable expression. If RepA-dependent replication in the cell was prevented, the amount of RepA detected in extracts of induced cells increased significantly (data not shown). This observation was made for inactive mutant RepA proteins carrying single amino acid exchanges and even for wild-type RepA encoded by a repA gene that carries an inactive PY54 origin (Fig. 5B). In either case, limited RepA degradation took place. Hence, the amount of RepA present in the cell is downregulated if RepA-dependent replication occurs. The mechanism(s) of this regulation remains unknown. Regulation may occur at the level of repA transcription and translation. Also RepA degradation may play a role.
It has been reported that by the use of conditional lethal E. coli strains, the phages N15 and P4 replicate independently of host-encoded initiation factors DnaA, DnaB, and DnaG (3, 31). In P4 replication, this autonomy has been explained by the action of the multifunctional replication protein α (42). The specificity of α and of the host initiation factors must differ significantly because they cannot replace α. It is also important to note that the α gene cannot suppress the phenotype of the conditionally lethal dna mutations of the host as it is known for the P1 ban gene in a dnaB-null strain (24) or for the primase genes of conjugative plasmids in a dnaG(Ts) background (23).
The striking similarity of the arrangement of motifs in P4 α protein and RepA proteins of the hairpin phages may provide the explanation for the PY54 replication property. As in P4 α, the functions proposed for RepA, replicative helicase, primase, and possibly origin binding would make the host initiation factors dispensable. The point mutations generated in motifs for primase and nucleotide binding (Fig. 7) strongly support this notion, since DnaA, DnaB, and DnaG cannot substitute for RepA. Thus, the situation is similar to that in phage P4. In both cases the replication properties of the α protein and of RepA confer replicon specificity.
Whereas P4 α protein consists of 777 amino acid residues, the proposed number of residues for the RepA proteins of the three related hairpin phages is 1330 (PY54), 1324 (N15), and 1328 (PKO2), almost twice the size of the α protein. The α protein plays four roles: it acts as a replicative helicase, a primase, and an origin recognition protein (37, 42) and interacts with the P4 Cnr protein (copy number regulation (34, 38). Therefore, the RepA proteins may exert additional activities to the proposed ones that do not become readily obvious by sequence comparisons.
We conclude that the integrity of the whole protein is needed for RepA activity, since (i) complementation between two separately expressed portions of repA, i.e., pGB154-5/pAM155-4, does not occur and (ii) repAΔ7 is unable to promote replication (data not shown). Additionally, the removal of the proposed primase domain (pGB154-7) is deleterious for replication, indicating that DnaG cannot complement the defect in RepA (see above). As in P4, the PY54 replication gene contains the replication origin. The origin in P4 α is, in fact, a secondary origin (oriII), the purpose of which is not yet understood (35). The P4 primary origin (oriI) and also oriII are composed of two components, the crr (cis replication region) portion and the ori portion. Origin function is assured only if both parts are present. In vivo it has been shown that the P4 α protein functions in trans at oriII (35). In an in vitro P4 DNA replication system α acts also in trans at oriI (11).
The single, one-component origin of PY54 has been downsized to 212 bp toward the 3′ end of repA (Fig. 3 and Table 2). Data from the deletion analyses indicated that the size of the fully functional trans-acting RepA protein consists of 1,215 amino acid residues. This finding demonstrates that the coding capacity of the repA 3′ region is dispensable for RepA function.
For N15, it has been claimed that RepA functions in cis only (28). Cis action of replication proteins has been discussed by McFall (26). This article warns of a trivial explanation for data obtained by the cis-trans complementation assay. In the classical case of the gene A of phage φX174, it has been suggested that the A* protein is responsible for the cis effect because of competition with the full-length A protein for target sites (22). Thus, in vivo the A protein may act preferentially in cis. However, in realitas it acts in trans, as has been undoubtedly demonstrated by the use of the purified A protein in reconstituted in vitro replication systems. As the core sequences of the origins of the hairpin prophages PY54, N15, and PKO2 are well conserved (Fig. 5A), it is unlikely that the action of the corresponding RepA proteins should be fundamentally different. Since we have physically separated the PY54 origin and repA onto two plasmids, there is no doubt that the 212-bp repA fragment contains the functional origin and that RepA functions in trans.
For an in vivo replication test system, it is of invaluable advantage to have the origin and repA on two discrete plasmids, because this allows the evaluation of mutagenesis data on each of the items separately. Of primary concern is origin mutagenesis since the changes made in the origin retain the native protein sequence of the trans-acting product, in our case RepA. The reverse question could also be asked: do amino acid changes in the corresponding ori portion alter the protein's function?
Sequences of the active PY54 ori and those suggested to function in N15 as the prophage origin (28) are conserved in the arrangement of sites in a range of about 50 bp of PY54, N15, and PKO2 (Fig. 5A). The nucleotide sequence of this core is nearly identical in N15 and PKO2, whereas in PY54 it differs slightly from the other two. Altering one dam site or one arm of the TATATC repeat (Fig. 5B) leaves the PY54 origin functional, whereas the combination of both mutations results in an origin completely inactive in our two-plasmid replication test system. Mutations in both arms of the repeat also abolished replication. These base exchanges led to a significant reduction of the AT content from 78% to 57% within the AT-rich region. The enhanced local stability of the double-stranded DNA might hamper the initial melting of both strands required for replication initiation. From our data we conclude that the PY54 origin we identified functions as the initiation site for prophage replication. Whereas the origin seems to be quite resistant to certain sequence alterations without loosing activity, the overall structure has to be maintained for replication function. In PY54 the DnaA box present in both N15 and PKO2 is missing. Since the PY54 minimal replicon replicates perfectly well in E. coli, the DnaA box in N15 and PKO2 might normally also be dispensable.
Supplementary Material
Acknowledgments
The expert technical assistance of Marianne Schlicht and Iris Klein is greatly appreciated. We thank Hans Lehrach for generous support and Gerhild Lüder for preparing the electron micrographs.
This work was supported by a grant of the Deutsche Forschungsgemeinschaft to E.L.
REFERENCES
- 1.Balzer, D., G. Ziegelin, W. Pansegrau, V. Kruft, and E. Lanka. 1992. KorB protein of promiscuous plasmid RP4 recognizes inverted sequence repetitions in regions essential for conjugative plasmid transfer. Nucleic Acids Res. 20:1851-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbour, A. G., and C. F. Garon. 1987. Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science 237:409-411. [DOI] [PubMed] [Google Scholar]
- 3.Briani, F., G. Dehò, F. Forti, and D. Ghisotti. 2001. The plasmid status of satellite bacteriophage P4. Plasmid 45:1-17. [DOI] [PubMed] [Google Scholar]
- 4.Casjens, S. R., E. B. Gilcrease, W. M. Huang, K. L. Bunny, M. L. Pedulla, M. E. Ford, J. M. Houtz, G. F. Hatfull, and R. W. Hendrix. 2004. The pKO2 linear plasmid prophage of Klebsiella oxytoca. J. Bacteriol. 186:1818-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaconas, G., P. E. Stewart, K. Tilly, J. L. Bono, and P. Rosa. 2001. Telomere resolution in the Lyme disease spirochete. EMBO J. 20:3229-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Covarrubias, L., and F. Bolivar. 1982. Construction and characterization of new cloning vehicles. VI. Plasmid pBR329, a new derivative of pBR328 lacking the 482-base-pair inverted duplication. Gene 17:79-89. [DOI] [PubMed] [Google Scholar]
- 7.Delaroque, N., W. Boland, D. G. Müller, and R. Knippers. 2003. Comparisons of two large phaeoviral genomes and evolutionary implications. J. Mol. Evol. 57:613-622. [DOI] [PubMed] [Google Scholar]
- 8.Delaroque, N., D. G. Müller, G. Bothe, T. Pohl, R. Knippers, and W. Boland. 2001. The complete DNA sequence of the Ectocarpus siliculosus virus EsV-1 genome. Virology 287:112-132. [DOI] [PubMed] [Google Scholar]
- 9.Deneke, J., G. Ziegelin, R. Lurz, and E. Lanka. 2000. The protelomerase of temperate Escherichia coli phage N15 has cleaving-joining activity. Proc. Natl. Acad. Sci. USA 97:7721-7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deneke, J., G. Ziegelin, R. Lurz, and E. Lanka. 2002. Phage N15 telomere resolution. Target requirements for recognition and processing by the protelomerase. J. Biol. Chem. 277:10410-10419. [DOI] [PubMed] [Google Scholar]
- 11.Diaz Orejas, R., G. Ziegelin, R. Lurz, and E. Lanka. 1994. Phage P4 DNA replication in vitro. Nucleic Acids Res. 22:2065-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenbrandt, R., M. Kalkum, R. Lurz, and E. Lanka. 2000. Maturation of IncP pilin precursors resembles the catalytic dyad-like mechanism of leader peptidases. J. Bacteriol. 182:6751-6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Golub, E. I., and V. K. Ravin. 1967. A new system of phage conversion. Dokl. Akad. Nauk SSSR. 174:465-467. (In Russian.) [PubMed] [Google Scholar]
- 14.Goodner, B., G. Hinkle, S. Gattung, N. Miller, M. Blanchard, B. Qurollo, B. S. Goldman, Y. Cao, M. Askenazi, C. Halling, L. Mullin, K. Houmiel, J. Gordon, M. Vaudin, O. Iartchouk, A. Epp, F. Liu, C. Wollam, M. Allinger, D. Doughty, C. Scott, C. Lappas, B. Markelz, C. Flanagan, C. Crowell, J. Gurson, C. Lomo, C. Sear, G. Strub, C. Cielo, and S. Slater. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323-2328. [DOI] [PubMed] [Google Scholar]
- 15.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 16.Hertwig, S., I. Klein, R. Lurz, E. Lanka, and B. Appel. 2003. PY54, a linear plasmid prophage of Yersinia enterocolitica with covalently closed ends. Mol. Microbiol. 48:989-1003. [DOI] [PubMed] [Google Scholar]
- 17.Hertwig, S., I. Klein, V. Schmidt, S. Beck, J. A. Hammerl, and B. Appel. 2003. Sequence analysis of the genome of the temperate Yersinia enterocolitica phage PY54. J. Mol. Biol. 331:605-622. [DOI] [PubMed] [Google Scholar]
- 18.Hertwig, S., A. Popp, B. Freytag, R. Lurz, and B. Appel. 1999. Generalized transduction of small Yersinia enterocolitica plasmids. Appl. Environ. Microbiol. 65:3862-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacob, D., A. Lewin, B. Meister, and B. Appel. 2002. Plant-specific promoter sequences carry elements that are recognised by the eubacterial transcription machinery. Transgenic Res. 11:291-303. [DOI] [PubMed] [Google Scholar]
- 20.Keck, J. L., D. D. Roche, A. S. Lynch, and J. M. Berger. 2000. Structure of the RNA polymerase domain of E. coli primase. Science 287:2482-2486. [DOI] [PubMed] [Google Scholar]
- 21.Kobryn, K., and G. Chaconas. 2002. ResT, a telomere resolvase encoded by the Lyme disease spirochete. Mol. Cell 9:195-201. [DOI] [PubMed] [Google Scholar]
- 22.Kornberg, A., and T. A. Baker. 1992. DNA replication. W. H. Freeman and Company, New York, N.Y.
- 23.Lanka, E., and P. T. Barth. 1981. Plasmid RP4 specifies a deoxyribonucleic acid primase involved in its conjugal transfer and maintenance. J. Bacteriol. 148:769-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemonnier, M., G. Ziegelin, T. Reick, A. M. Gomez, R. Diaz-Orejas, and E. Lanka. 2003. Bacteriophage P1 Ban protein is a hexameric DNA helicase that interacts with and substitutes for Escherichia coli DnaB. Nucleic Acids Res. 31:3918-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lessl, M., D. Balzer, R. Lurz, V. L. Waters, D. G. Guiney, and E. Lanka. 1992. Dissection of IncP conjugative plasmid transfer: definition of the transfer region Tra2 by mobilization of the Tra1 region in trans. J. Bacteriol. 174:2493-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McFall, E. 1986. Cis-acting proteins. J. Bacteriol. 167:429-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Popp, A., S. Hertwig, R. Lurz, and B. Appel. 2000. Comparative study of temperate bacteriophages isolated from Yersinia. Syst. Appl. Microbiol. 23:469-478. [DOI] [PubMed] [Google Scholar]
- 28.Ravin, N. V., V. V. Kuprianov, E. B. Gilcrease, and S. R. Casjens. 2003. Bidirectional replication from an internal ori site of the linear N15 plasmid prophage. Nucleic Acids Res. 31:6552-6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ravin, N. V., and V. K. Ravin. 1999. Use of a linear multicopy vector based on the mini-replicon of temperate coliphage N15 for cloning DNA with abnormal secondary structures. Nucleic Acids Res. 27:e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravin, V., N. Ravin, S. Casjens, M. E. Ford, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequence and analysis of the atypical temperate bacteriophage N15. J. Mol. Biol. 299:53-73. [DOI] [PubMed] [Google Scholar]
- 31.Rybchin, V. N., and A. N. Svarchevsky. 1999. The plasmid prophage N15: a linear DNA with covalently closed ends. Mol. Microbiol. 33:895-903. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. T. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Strack, B., M. Lessl, R. Calendar, and E. Lanka. 1992. A common sequence motif,-E-G-Y-A-T-A-, identified within the primase domains of plasmid-encoded I-and P-type DNA primases and the α protein of the Escherichia coli satellite phage P4. J. Biol. Chem. 267:13062-13072. [PubMed] [Google Scholar]
- 34.Terzano, S., R. Christian, F. H. Espinoza, R. Calendar, G. Dehò, and D. Ghisotti. 1994. A new gene of bacteriophage P4 that controls DNA replication. J. Bacteriol. 176:6059-6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tocchetti, A., G. Galimberti, G. Dehò, and D. Ghisotti. 1999. Characterization of the oriI and oriII origins of replication in phage-plasmid P4. J. Virol. 73:7308-7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiner, M. P., L. G. Costa, W. Schoettlin, J. Cline, E. Mathur, and J. T. Bauer. 1994. Site-directed mutagenesis of double-stranded DNA by polymerase chain reaction. Gene 151:119-123. [DOI] [PubMed] [Google Scholar]
- 37.Yeo, H. J., G. Ziegelin, S. Korolev, R. Calendar, E. Lanka, and G. Waksman. 2002. Phage P4 origin-binding domain structure reveals a mechanism for regulation of DNA-binding activity by homo- and heterodimerization of winged helix proteins. Mol. Microbiol. 43:855-867. [DOI] [PubMed] [Google Scholar]
- 38.Ziegelin, G., R. Calendar, D. Ghisotti, S. Terzano, and E. Lanka. 1997. Cnr protein, the negative regulator of bacteriophage P4 replication, stimulates specific DNA binding of its initiator protein α. J. Bacteriol. 179:2817-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ziegelin, G., R. Calendar, R. Lurz, and E. Lanka. 1997. The helicase domain of phage P4 α protein overlaps the specific DNA binding domain. J. Bacteriol. 179:4087-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ziegelin, G., and E. Lanka. 1995. Bacteriophage P4 DNA replication. FEMS Microbiol. Rev. 17:99-107. [DOI] [PubMed] [Google Scholar]
- 41.Ziegelin, G., N. A. Linderoth, R. Calendar, and E. Lanka. 1995. Domain structure of phage P4 α protein deduced by mutational analysis. J. Bacteriol. 177:4333-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ziegelin, G., E. Scherzinger, R. Lurz, and E. Lanka. 1993. Phage P4 α protein is multifunctional with origin recognition, helicase and primase activities. EMBO J. 12:3703-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.