Abstract
The toxin-coregulated pilus (TCP) of Vibrio cholerae is required for intestinal colonization and cholera toxin acquisition. Here we report that TCP mediates bacterial interactions required for biofilm differentiation on chitinaceous surfaces. We also show that undifferentiated TCP− biofilms have reduced ecological fitness and, thus, that chitin colonization may represent an ecological setting outside the host in which selection for a host colonization factor may take place.
Vibrio cholerae, the causative agent of the diarrheal disease cholera, is a waterborne bacterium well known for its genome plasticity and clonal diversity (25). Interestingly, only 2 of the more than 200 O-antigen serogroups known to date, O1 and O139, cause epidemic cholera. It is clear that the acquisition of certain genes encoding virulence factors has enabled certain strains of V. cholerae to colonize the human intestine and cause disease (21). The most important of these virulence factors are cholera toxin (CT), which is encoded within the genome of the filamentous bacteriophage CTXφ and thus horizontally transferred (29), and the intestinal colonization factor toxin-coregulated pilus (TCP) (28), which is encoded in the vibrio pathogenicity island (12). The acquisition of the vibrio pathogenicity island, by yet-unclear mechanisms (6), is a critical event in the evolution of epidemic strains of V. cholerae, since TCP also serves as a receptor for CTXφ (29).
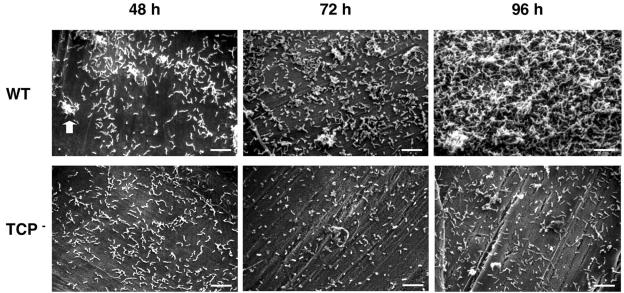
There is enough evidence supporting the concept of an aquatic reservoir of pathogenic V. cholerae organisms (4, 9, 15), and it has become apparent that toxigenic strains have originated from environmental, nontoxigenic clones (2, 11). However, the selective pressures that may act to generate environmental virulent clones remain to be elucidated. Infections upon ingestion of contaminated seafood (13, 25) and V. cholerae's enhanced survival in both the natural environment and the acidic stomach environment when attached to chitinaceous surfaces (1, 20) strongly point to a common link between the colonization of chitin-containing aquatic organisms, such as crustaceans (9), and the persistence of this pathogen in aquatic ecosystems. Chitin, the most abundant natural polymer in aquatic environments, serves as a source of carbon and nitrogen for the growth of a wide range of microbes, including V. cholerae (20), as well as a surface for the formation of surface-attached communities or biofilms (31). In order to gain insight into the genetic basis for biofilm formation on chitin, we performed an unbiased genetic screen to identify Tn10 insertion mutants of a gfp-tagged pandemic V. cholerae El Tor strain (N16961) with defects in the colonization of chitin (the results of this screen will be published elsewhere). Briefly, the screen was performed in 96-well fluorescence black microtiter plates (clear bottom; Corning) whose wells had previously been coated with films of chitin, which were prepared by dissolving finely ground chitin flakes (Sigma) in a solvent of dimethylacetamide and LiCl to a final concentration of 1% (wt/vol) (30, 33) and by using NG2 mineral medium (K2HPO4, 184 mM; KH2PO4, 17 mM; MOPS [morpholinepropanesulfonic acid], 40 mM; NaCl, 86 mM; MgSO4 · 7H2O, 2 mM; FeSO4 · 7H2O, 10 μM; and CaCl2 · 2H2O, 1 mM) (pH 7.8). After 48 h of incubation at 37°C, mutants with colonization defects were identified by measuring the levels of fluorescence associated to the chitin-coated wells. Among the ca. 4,400 Tn10 insertion mutants screened, we identified 126 with colonization defects, including several carrying transposon insertions in genes implicated in the biosynthesis and assembly of TCP, such as tcpB, tcpE, tcpG, and tcpT (8, 18, 23), suggesting a yet-unrecognized role for TCP in the colonization of chitinaceous surfaces. To further investigate this, we used scanning electron microscopy to visualize the development of wild-type (WT) and TCP mutant (a mutant deficient in TcpA, the TCP pilin subunit [7]) biofilms on the surfaces of chitinaceous squid pens (19, 26) (Fig. 1) by growing the bacteria at 37°C in NG2 mineral medium with sterile pieces of squid pen that had been previously washed and autoclaved in phosphate saline buffer. The WT and TCP mutant biofilms were indistinguishable during the first 24 h, both showing a monolayer of cells evenly coating the surfaces of squid pens. By 48 h, the formation of microcolonies was apparent in the WT biofilm, which further differentiated into structured biofilms in the course of the next hours, whereas the TCP mutant biofilms remained unstructured and never formed microcolonies (Fig. 1). Biofilm differentiation in a gfp-tagged WT strain directly correlated with increases in the bacterial biomass associated with the chitinaceous surface, which was detected by measuring the fluorescence from biofilm cells previously detached by sonication (Fig. 2A). However, the numbers of attached TCP− cells remained constant even after prolonged incubation. Differences in biomass inversely correlated to differences in chitinase activity, assayed with 4-methyl-umbelliferyl (4MU)-chitobiose, a 4MU-conjugated substrate analog of chitotriose (22), associated with biofilm cells (Fig. 2B), suggesting that biofilm differentiation on the nutritive surface provides bacteria with a competitive strategy for degrading the recalcitrant substrate. The rates of growth on soluble products of chitin degradation, such as N-acetyl-glucosamine, or rates of growth or degradation of colloidal chitin, an acid-processed form of chitin, were comparable in both strains.
FIG. 1.
Scanning electron microscopy of WT and TCP− mutant biofilms at 48, 72, and 96 h. The arrow points at microcolonies of the WT strain formed during the development of WT biofilms on the surface of the squid pen. TCP− biofilms remained undifferentiated. Bars, 10 μm.
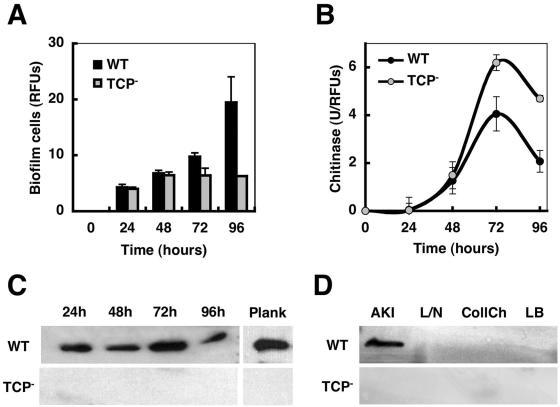
FIG. 2.
Colonization of squid pens by a WT and a TCP− mutant strain of V. cholerae. (A) Biofilm development over a 96-h period. Biofilm biomass was estimated by measuring the fluorescence from gfp-tagged cells (expressed as relative fluorescence units [RFUs]). (B) Chitinase activity associated with biofilm cells of the WT and TCP− mutant strains during the colonization of squid pens. Specific activities of units of chitinase activity (U, described as the amount of enzyme that released 1 pmol of 4MU per min under the assay conditions) per RFU are shown. (C) Immunodetection of TcpA, the TCP pilin subunit, in biofilm cells in the course of 96 h and in planktonic cells at 24 h (Plank). (D) Immunodetection of TcpA in cultures grown under TCP-inducing conditions (AKI medium) or grown for 24 h in the absence of a chitinous surface in NG2 mineral medium with lactate and NH4Cl (L/N) or with colloidal chitin (CollCh), and in LB medium.
Expression of TCP during biofilm formation on chitin surfaces.
As illustrated in Fig. 2C, TcpA, the TCP pilin subunit, was detected using anti-TcpA polyclonal antibodies (27) at comparable levels in both structured and unstructured biofilms and in planktonic cells. However, transduction frequencies of a CTX-Knφ phage, which provide evidence for surface-associated TCP (29), were already detected in WT cells detached with glass beads from 24-h biofilms (transduction frequencies/ml, 0.35 ± 0.15) and were highest in cells from 48-h biofilms (transduction frequencies/ml, 13.80 ± 0.20), when microcolony differentiation occurs. There was a drop in transduction frequencies in later sages of biofilm differentiation (transduction frequencies/ml, <0.01), which may reflect the inaccessibility of the TCP pili within the biofilm matrix. Similar transduction frequencies were detected in WT planktonic cells at the different stages. TcpA expression was not detectable in cells grown in the same medium used for the biofilm assay but with soluble carbon and nitrogen sources (Fig. 2D), nor were TCP filaments apparent in negative stains by transmission electron microscopy, but it was detected when cells were grown under the special in vitro culture conditions (AKI conditions [10]) required by V. cholerae El Tor for expression of CT and TCP. Cells from cultures grown with colloidal chitin, whose colloidal nature prevented the availability of a steady surface for biofilm formation while still serving as a source of carbon and nitrogen, also showed no detectable levels of TcpA expression (Fig. 2D). Therefore, the colonization defect of the TCP mutant was not likely caused by differences in the rates of surface attachment or rates of growth.
Role of TCP in biofilm differentiation.
The lack of observable differences in the numbers of cells attaching to squid pens during the early stages of colonization (Fig. 2A) indicated that TCP does not function as an adhesin. Similarly, TCP does not appear to mediate direct contact with the intestinal epithelium during infection but, rather, mediates bacterial interactions required for microcolony formation and successful colonization (14). To investigate an analogous role for TCP in microcolony formation on chitin, we carried out competition experiments and examined the ability of a gfp-tagged TCP− mutant strain to aggregate and form differentiated biofilms with an unmarked WT strain. Although the mutant showed a reduced competitive index (0.211 ± 0.011, calculated as the ratio of the number of CFU of the mutant strain from a 96-h biofilm on a squid pen to the number of WT and mutant input CFU), coincubation with the WT strain rescued the mutant phenotype, and fluorescence from the TCP mutant cells was present uniformly within the WT biofilm pillars (Fig. 3). Thus, similarly to its role during intestinal colonization, TCP may mediate bacterial interactions during biofilm differentiation on chitinaceous surfaces.
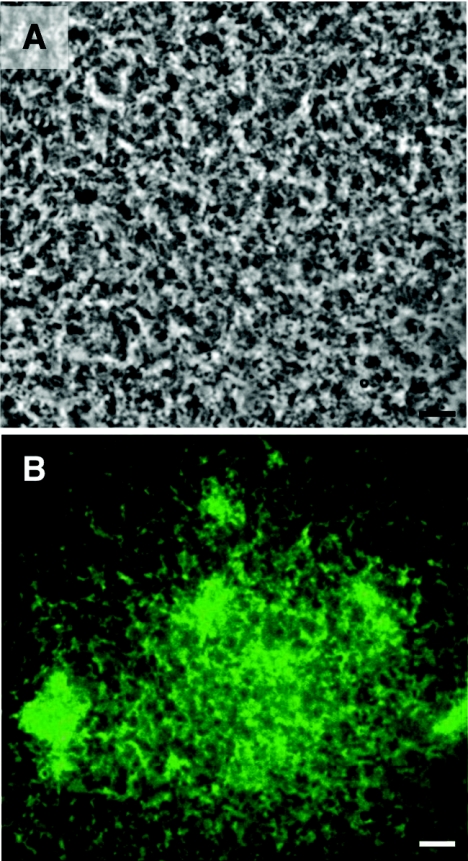
FIG. 3.
Mixed biofilms of WT and TCP− mutant strains. (A) Phase-contrast micrograph showing the differentiated biofilms developed on the surface of a squid pen by a 50:50-mixed culture of a WT and a gfp-tagged TCP− mutant strain over a 96-h period. (B) Fluorescence micrograph placed over the micrograph in panel A showing the uniform distribution of the gfp-tagged TCP− mutant strain within the mixed biofilms. Bars, 10 μm.
Biofilm differentiation and ecological fitness.
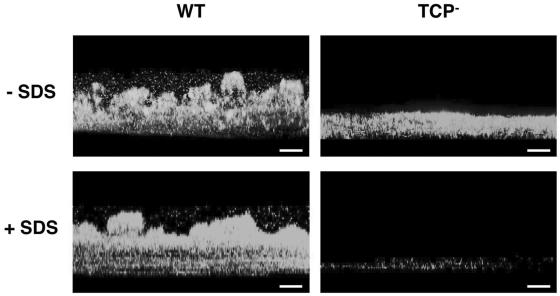
Because undifferentiated biofilms have previously been shown to lack the trademarks of biofilm physiology, such as biocide resistance (5), we examined whether the undifferentiated TCP mutant biofilms of V. cholerae might have increased sensitivity to certain stresses, such as the biocide sodium dodecyl sulfate (SDS) (Fig. 4). Whereas exposure for only 5 min to 0.2% SDS detached most of the cells from the mutant biofilm, no detectable effect was observed in the WT biofilms even after incubation at room temperature for 30 min. Hence, the inability of TCP− strains of V. cholerae to form differentiated biofilms on chitinaceous surfaces is likely to have a significant impact in the environmental fitness of this pathogen. TCP+ biofilms also concentrate a greater number of cells on the nutritive surface, which synergistically act to secrete chitinolytic enzymes to degrade the chitin while providing a mechanical barrier to the diffusion of soluble hydrolysis products, thus maximizing substrate utilization and preventing the release of chemoattractants for bacterial competitors.
FIG. 4.
Effect of SDS treatment on WT and TCP− mutant biofilms on a squid pen. Three-dimensional side views, generated by confocal scanning laser microscopy, of SDS-treated (+ SDS) or untreated (− SDS) biofilms of the WT and TCP mutant strains of V. cholerae showing the detachment of most cells from the undifferentiated TCP− biofilm after SDS treatment are presented; the structured WT biofilms remained unaffected. The substratum (squid pen) is located at the bottom of the images. Bars, 50 μm.
Implications for the evolution of toxigenic strains.
TCP also serves as a receptor for CTXφ; therefore, expression of TCP during the formation of biofilms also makes V. cholerae susceptible to CTXφ transduction. Although the El Tor strain used in this work already carries a chromosomal copy of the CTXφ prophage, incubation of undisrupted biofilms with a CTX-Knφ phage for only 30 min resulted in transduction frequencies of one transductant per 104 cells. Because biofilms provide an ideal environment for the events of horizontal gene transfer thought to be responsible for the acquisition of new genes, including genes coding for virulence factors (32), the CT genes might readily be acquired upon expression of functional TCP within the context of a biofilm.
Conclusions.
While two other pili produced by V. cholerae, the mannose-sensitive hemagglutinin and the chitin-regulated pilus, have been reported to contribute to the colonization of chitin surfaces (3, 17), an environmental role for the TCP pilus, a virulence determinant, had not previously been demonstrated. The data presented here strongly suggest that clones of V. cholerae capable of synthesizing functional TCP are likely to be selected for within chitin-associated biofilms present in aquatic environments. Evidence to date supports the concept of specific symbiotic relationships between V. cholerae and chitinous animals, such as copepods (4, 9, 15), and climate-related zooplankton blooms have been correlated to cholera outbreaks (15, 16, 24), supporting the model of selection of virulence factors such as TCP during V. cholerae's colonization of chitinaceous surfaces. Our findings provide evidence that some of the same virulence factors used by pathogens during infection might have a role in their natural habitat. Hence, the evolution of virulence in bacteria such as V. cholerae might be dictated by factors outside the host, and mechanisms of virulence, as we understand them, are likely to be a reflection of adaptive mechanisms to the environment.
Acknowledgments
We thank John J. Mekalanos and Jun Zhu for providing us with strains and for comments on the manuscript. Also, thanks go to Jun Zhu and the members of the Kolter lab for helpful insights during the development of this work.
This work was supported by grants from the NIH (GM58213), the DOE (DE-FG02-02ER63445), and the Ellison Medical Foundation (ID-SS-0248-02) to R.K. and a postdoctoral fellowship from the Ministerio de Educación, Cultura y Deporte of Spain and the European Social Fund to G.R.
REFERENCES
- 1.Amako, K., S. Shimodori, T. Imoto, S. Miake, and A. Umeda. 1987. Effects of chitin and its soluble derivatives on survival of Vibrio cholerae O1 at low temperature. Appl. Environ. Microbiol. 53:603-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chakraborty, S., A. K. Mukhopadhyay, R. K. Bhadra, A. N. Ghosh, R. Mitra, T. Shimada, S. Yamasaki, S. M. Faruque, Y. Takeda, R. R. Colwell, and G. B. Nair. 2000. Virulence genes in environmental strains of Vibrio cholerae. Appl. Environ. Microbiol. 66:4022-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiavelli, D. A., J. W. Marsh, and R. K. Taylor. 2001. The mannose-sensitive hemagglutinin of Vibrio cholerae promotes adherence to zooplankton. Appl. Environ. Microbiol. 67:3220-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colwell, R. R., J. Kaper, and S. W. Joseph. 1977. Vibrio cholerae, Vibrio parahaemolyticus, and other vibrios: occurrence and distribution in Chesapeake Bay. Science 198:394-396. [PubMed] [Google Scholar]
- 5.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 6.Faruque, A. S. G., J. Zhu, A. M. Kamruzzaman, and J. J. Mekalanos. 2003. Examination of diverse toxin-coregulated pilus-positive Vibrio cholerae strains fail to demonstrate evidence for Vibrio pathogenicity island phage. Infect. Immun. 71:2993-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fullner, K. J., and J. J. Mekalanos. 1999. Genetic characterization of a new type IV-A pilus gene cluster found in both classical and El Tor biotypes of Vibrio cholerae. Infect. Immun. 67:1393-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu, S.-H., J. A. Peek, E. Rattigan, R. K. Taylor, and J. L. Martin. 1997. Structure of TcpG, the DsbA protein folding catalyst from Vibrio cholerae. J. Mol. Biol. 268:137-146. [DOI] [PubMed] [Google Scholar]
- 9.Islam, M. S., B. S. Drasar, and R. B. Sack. 1994. The aquatic flora and fauna as reservoirs of Vibrio cholerae: a review. J. Diarrhoeal Dis. Res. 12:87-96. [PubMed] [Google Scholar]
- 10.Iwanaga, M., K. Yamamoto, N. Higa, Y. Ichinose, N. Nakasone, and M. Tanabe. 1986. Culture conditions for stimulating cholera production by Vibrio cholerae O1 El Tor. Microbiol. Immunol. 30:1075-1083. [DOI] [PubMed] [Google Scholar]
- 11.Jiang, S. C., M. Matte, G. Matte, A. Huq, and R. R. Colwell. 2000. Genetic diversity of clinical and environmental isolates of Vibrio cholerae determined by amplified fragment length polymorphism fingerprinting. Appl. Environ. Microbiol. 66:148-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karaolis, D. K. R., J. A. Johnson, C. C. Bailey, E. C. Boedeker, J. B. Kaper, and P. R. Reeves. 1998. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl. Acad. Sci. USA 95:3134-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaysner, C. A., and W. E. Hill. 1994. Toxigenic Vibrio cholerae O1 in food and water, p. 27-39. In K. I. Wachsmuth, P. A. Blake, and O. Olsvik (ed.), Vibrio cholerae and cholera: molecular to global perspectives. American Society for Microbiology, Washington, D.C.
- 14.Kirn, T. J., M. J. Lafferty, C. M. Sandoe, and R. K. Taylor. 2000. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol. Microbiol. 35:896-910. [DOI] [PubMed] [Google Scholar]
- 15.Lipp, E. K., A. Huq, and R. R. Colwell. 2002. Effects of global climate on infectious disease: the cholera model. Clin. Microbiol. Rev. 15:757-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lobitz, B., L. Beck, A. Huq, B. Wood, G. Fuchs, A. S. Faruque, and R. Colwell. 2000. Climate and infectious disease: use of remote sensing for detection of Vibrio cholerae by indirect measurement. Proc. Natl. Acad. Sci. USA 97:1438-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meibom, K. L., X. B. Li, A. T. Nielsen, C. Y. Wu, S. Roseman, and G. K. Schoolnik. 2004. The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. USA 101:2524-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merrell, D. S., D. L. Hava, and A. Camilli. 2002. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol. Microbiol. 43:1471-1491. [DOI] [PubMed] [Google Scholar]
- 19.Muzzarelli, R. A. A. 1977. Chitin. Pergamon Press, Oxford, United Kingdom.
- 20.Nalin, D. R., V. Daya, A. Reid, and M. M. C. L. Levine. 1979. Adsorption and growth of Vibrio cholerae on chitin. Infect. Immun. 25:768-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nesper, J., A. Kraiβ, S. Schild, J. Blaβ, K. E. Klose, J. Bockemuhl, and J. Reidl. 2002. Comparative and genetic analyses of the putative Vibrio cholerae lipopolysaccharide core oligosaccharide biosynthesis (wav) gene cluster. Infect. Immun. 70:2419-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Brien, M., and R. R. Colwell. 1987. A rapid test for chitinase activity that uses 4-methylumbelliferyl-N-acetyl-β-d-glucosaminide. Appl. Environ. Microbiol. 53:1718-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parsot, C., E. Taxman, and J. J. Mekalanos. 1991. ToxR regulates the production of lipoproteins and the expression of serum resistance in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 88:1641-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pascual, M., X. Rodo, S. P. Ellner, R. Colwell, and M. J. Bouma. 2000. Cholera dynamics and El Nino—southern oscillation. Science 289:1766-1769. [DOI] [PubMed] [Google Scholar]
- 25.Reidl, J., and K. E. Klose. 2002. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol. Rev. 26:125-139. [DOI] [PubMed] [Google Scholar]
- 26.Stretton, S., S. Techkarnjanaruk, A. M. McLennan, and A. E. Goodman. 1998. Use of green fluorescent protein to tag and investigate gene expression in marine bacteria. Appl. Environ. Microbiol. 64:2554-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun, D. X., J. M. Seyer, I. Kovari, R. A. Sumrada, and R. K. Taylor. 1991. Localization of protective epitopes within the pilin subunit of the Vibrio cholerae toxin-coregulated pilus. Infect. Immun. 59:114-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 30.Wan, A. C. A., E. Khor, and G. W. Hastings. 1998. Preparation of a chitin-apatite composite by in situ precipitation onto porous chitin scaffolds. J. Biomed. Mater. Res. 41:541-548. [DOI] [PubMed] [Google Scholar]
- 31.Watnick, P. I., K. J. Fullner, and R. Kolter. 1999. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J. Bacteriol. 181:3606-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson, B. A., and A. A. Salyers. 2003. Is the evolution of bacterial pathogens an out-of-body experience? Trends Microbiol. 11:347-350. [DOI] [PubMed] [Google Scholar]
- 33.Yusof, N. L. B. M., L. Y. Lim, and E. Khor. 2001. Preparation and characterization of chitin beads as a wound dressing precursor. J. Biomed. Mater. Res. 54:59-68. [DOI] [PubMed] [Google Scholar]