Abstract
Klebsiella aerogenes UreE, a metallochaperone that delivers nickel ions during urease activation, consists of distinct “peptide-binding” and “metal-binding” domains and a His-rich C terminus. Deletion analyses revealed that the metal-binding domain alone is sufficient to facilitate urease activation. This domain was purified and shown to exhibit metal-binding properties similar to those of UreE lacking only the His-rich tail.
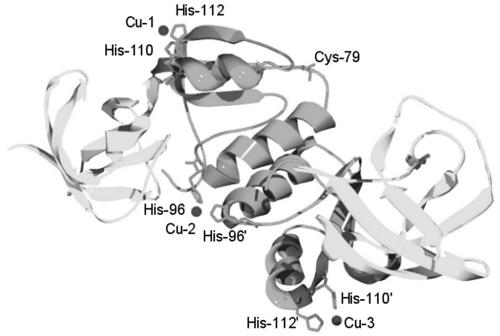
Urease is a Ni enzyme that catalyzes the hydrolysis of urea, thus contributing to the volatilization of ammonia from urea-based fertilizers and to the virulence of several human pathogens (9, 14). From studies with the archetypal bacterial urease from Klebsiella aerogenes, Ni is inserted into the apoprotein (UreABC) in a GTP-dependent process that requires the action of UreD, UreF, and UreG and is facilitated by UreE—a putative metallochaperone that delivers Ni (8, 11, 15). Homologues of ureE are conserved in almost all urease-producing microbes (14, 17), and cells containing partial ureE deletions exhibit reduced urease specific activities and yield purified enzyme with reduced Ni stoichiometry (11). K. aerogenes UreE binds up to 6 Ni molecules per dimer (12), but a truncated form lacking the 15 C-terminal His-rich residues (H144*UreE) still binds up to three Ni molecules per dimer and is fully competent in facilitating Ni-dependent activation of urease in vivo and in an in vitro reconstituted system (1, 20). H144*UreE has been studied in detail by spectroscopic (2), kinetic (4), site-directed mutagenic (3), and crystallographic (19) methods. Of significance to the present work, the protein contains a region that has been termed the peptide-binding domain (residues 1 to 70) and a metal-binding domain (residues 71 to 143, which also comprise the dimer interface region) (Fig. 1). This work examines the functional roles of the two domains of UreE by analysis of deletions and by characterization of the isolated metal-binding domain.
FIG. 1.
Structure of dimeric H144*UreE. The peptide-binding domain (residues 1 to 69) is shown in light grey, and the metal-binding domain (residues 70 to 143) is shown in dark grey. The histidine metal ligands (side chains from the second monomer of the dimeric UreE are denoted with a prime) as well as the location of one Cys79 are shown. Cu is displayed at three metal-binding sites.
Constructions.
Oligonucleotide primers were designed to create specific deletions of ureE within pTBEFH144* (1) by using PCR methods. Reactions were carried out with the QuikChange mutagenesis kit (Stratagene) according to the manufacturer's instructions, and the blunt-ended products were ligated to recircularize the linear fragments containing the desired deletions (Table 1). Plasmid DNAs from deletion constructs were verified by sequencing. Residues 2 to 136 were deleted by using a similar strategy, eliminating potential complications of the His-rich C-terminal region in the interpretation of metal-binding experiments while leaving the ureF start site undisturbed. To study ureE deletions while leaving the other six urease genes unaltered, restriction fragments containing the Δ2-158, Δ2-69, Δ70-158, and Δ2-136f ureE deletions were substituted for the corresponding fragment in pACT-KKWT, which contains the entire urease operon cloned into pACT3 (6). This expression plasmid contains a p15A ori gene which allows for complementation experiments using two compatible plasmids. An expression plasmid for production of the metal-binding domain was constructed by replacing ureE of pETWTΔG (analogous to pETH144*ΔG as described in reference 3, except it carries full-length ureE) with a similar fragment encoding only the metal-binding domain, to yield plasmid pETΔ2-69ΔG-20.
TABLE 1.
Plasmids used
| Plasmid | Relevant characteristics | Reference |
|---|---|---|
| pTBEFH144* | BamHI-AvrII fragment of urease operon containing ureEF cloned into pUC19; the ureE gene contains a stop at codon 144 | 1 |
| pETWTΔG | Expression plasmid pET21 containing ureEF | 3 |
| pETH144*ΔG | Expression plasmid pET21 containing ureEF with a deletion of the His-rich C terminus | 3 |
| pETΔ2-69ΔG-20 | pETH144*ΔG with a deletion of the ureE peptide-binding domain | This study |
| pACT3 | E. coli expression vector with a p15A ori gene | 6 |
| pACT-KKWT | Plasmid pACT3 containing ureDABCEFG with wild-type ureE | This study |
| pACT-KKH144* | Same as pACT-KKWT but with a truncated ureE gene lacking the C-terminal 15-residue His-rich region | This study |
| pACT-KKΔ2-158 | Plasmid pACT3 containing ureDABCFG with a total deletion of the ureE coding region | This study |
| pACT-KKΔ2-69 | Plasmid pACT3 containing ureDABCEFG with a deletion of ureE residues 2 to 69; also has H144* | This study |
| pACT-KKΔ70-158 | Plasmid pACT3 containing ureDABCEFG with a deletion of ureE residues 70 to 158 | This study |
| pACT-KKΔ2-136f | Plasmid pKK223-3 containing ureDABCEFG with a deletion of ureE residues 2 to 136; frame shift produces a small peptide, MSRRLRNREPRSSSCSS-COOH | This study |
Effects of ureE deletions on in vivo urease activation.
Escherichia coli DH5α cells containing plasmids carrying the K. aerogenes urease gene cluster with deletions of ureE were grown in Luria-Bertani medium (30 to 40 ml in 125-ml flasks) containing 30 μg/ml chloramphenicol with shaking at 37°C. After 30 min, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to 0.1 mM, and shaking was continued. Due to sequestering of metal ions by components in this medium, no significant urease activity was observed, even in cells containing the wild-type K. aerogenes urease operon (11). When turbidity reached an A600 of ≈0.1 to 0.25, spectinomycin was added to 100 μg/ml to stop protein synthesis (so only preformed urease apoprotein and accessory proteins would participate in activation, Ni was added, and shaking was continued for 20 to 40 min.
For Ni dependence experiments (Fig. 2A), 10-ml aliquots were transferred into sterile flasks containing premeasured NiSO4 concentrations, shaken for 4 h at 37°C, harvested by centrifugation, washed in 20 mM phosphate buffer (pH 7.4) containing 0.5 mM EDTA and 1 mM β-mercaptoethanol, disrupted by sonication, and assayed for urease activity. Cells carrying pACT-KKWT or pACT-KKH144* exhibited the highest activities with increasing Ni concentrations. Cells harboring pACT-KKΔ2-69 had lower activities, especially below 1.0 mM Ni2+. Cells carrying the pACT-KKΔ2-136f plasmid with a totalureE deletion showed the smallest amount of activation overall, but this was complemented with H144*UreE from a second plasmid, resulting in activities nearly equivalent to that from pACT-KKH144*-containing cells. These results are qualitatively comparable to those of earlier studies (1, 11) in which the urease gene cluster was cloned into higher-copy plasmids, cultures were grown overnight rather than for 4 h, and the plasmid constructs contained unsequenced deletions within ureE. Thus, full-length or H144* forms of UreE facilitate activation of urease, while deletions of ureE do not eliminate urease activation. Significantly, UreE missing both the peptide-binding domain and the His-rich region supported an intermediate level of urease activation consistent with a critical role of the metal-binding domain.
FIG. 2.
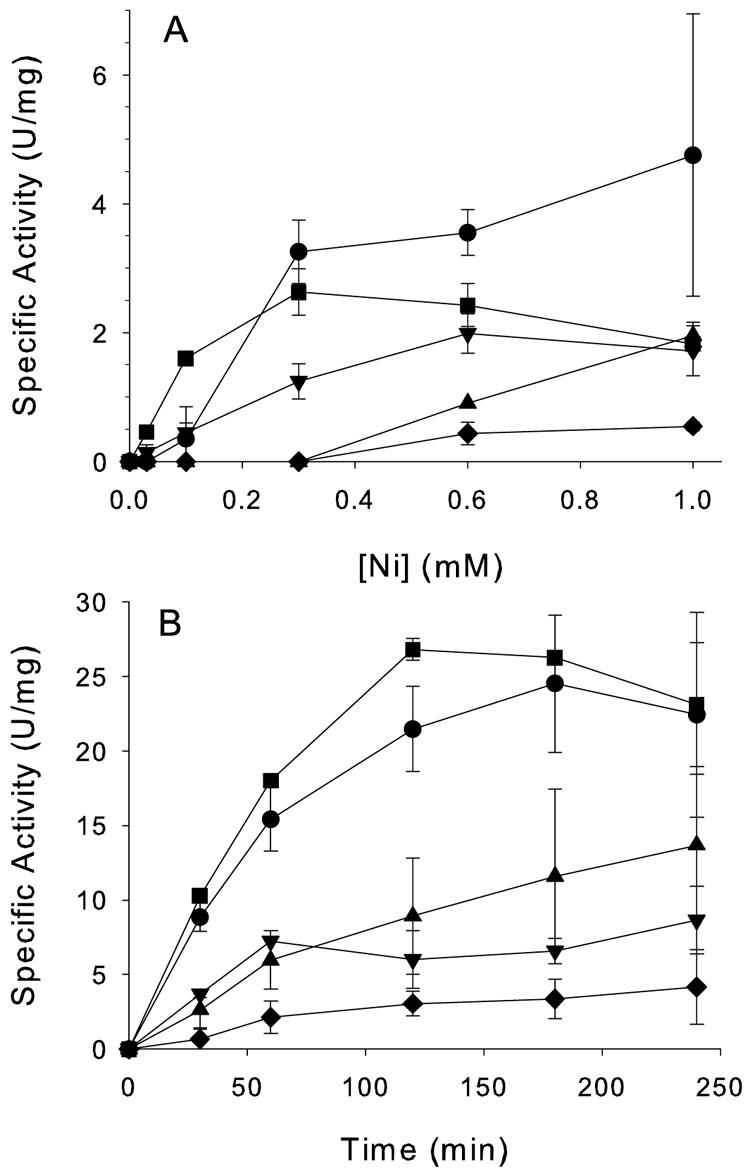
Effects of urease deletions on in vivo urease activation. Spectinomycin-treated cells were (A) exposed to various concentrations of Ni2+ and incubated for 4 h or (B) treated with 1 mM NiSO4 and sampled over time. Aliquots were analyzed for urease specific activity. Cultures contained pACT-KKWT (containing wild-type ureE [•]), pACT-KKH144* (containing truncated ureE lacking the His-rich C-terminal 15 amino acids [▪]), pACT-KKΔ2-69 (containing only the ureE metal-binding domain [▴]), pACT-KKΔ2-136f (with the entire ureE deletion [♦]), or pACT-KKΔ2-136f and pTBEFH144* (with the total ureE deletion plasmid cotransformed with a second compatible plasmid encoding ureEF [▾]).
For time dependence studies (Fig. 2B), cultures were supplemented with 1 mM NiSO4 and assayed at various time intervals. Cells with wild-type and H144* forms of UreE showed the highest specific activities throughout the time course, while cells containing pACT-KKΔ2-69 had intermediate rates of activation and cells harboring pACT-KKΔ2-136f exhibited the lowest activation rates. The final specific activities differ from those shown in Fig. 2A, reflecting the wide variation obtained with these in vivo experiments; nevertheless, the trends shown were quite reproducible. As with the Ni dependence studies, pTBEFH144* partially complemented the complete ureE deletion, though not to wild-type levels. Western blot analyses of culture samples with anti-UreE antibodies (12) detected UreE in cells containing the two plasmids (data not shown). Similar results showing lower-than-wild-type specific activities have been seen in other complementation experiments with urease accessory protein genes (11, 16).
Characterization of the UreE metal-binding domain.
H144*UreE was purified from cultures containing pETH144*ΔG (3) as previously described, except that E. coli C41(DE3) (13) was used as the host. The UreE metal-binding domain was purified by the same procedure using E. coli C41(DE3) carrying plasmid pETΔ2-69ΔG-20. Protein concentration was determined by using the bicinchoninic acid protein assay kit (Pierce) with bovine serum albumin as the standard. The purity of the sample was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10) using 18% running and 4.5% stacking gels with Bio-Rad broad-range molecular weight standards; the metal-binding domain produced a single band of the expected size (Mr, 8,372). The yield of metal-binding domain was 1.9 mg from 2 liters of cells, compared to approximately 47 mg of H144*UreE from 1 liter of culture. The low yield of the single-domain protein may result from its decreased stabilization in the absence of the N-terminal domain.
Native molecular weight estimations of the metal-binding domain utilized gel permeation chromatography with a Protein-Pak 125 column (7.8 by 300 mm; Waters) connected to a Waters Breeze chromatography system and a buffer of 20 mM Tris-0.2 M NaCl-10 μM NiSO4 (pH 7.8). To test the effects of Ni on the native molecular weight of the UreE metal-binding domain, some samples were chromatographed in 20 mM Tris-0.2 M NaCl-0.1 mM EDTA (pH 7.8). Mixtures of protein molecular weight standards (Bio-Rad and Calbiochem) were chromatographed under identical conditions. Major and minor species of metal-binding domain (36,000 ± 5,000 kDa and 17,600 ± 3,000 Da) were consistent with tetrameric and dimeric structures. The presence of 10 μM Ni2+ or 0.1 mM EDTA did not affect the quaternary structure.
Metal ion-binding properties of H144*UreE and the metal-binding domain.
H144*UreE and the metal-binding domain (10 μM subunit in 50 mM Tris, pH 7.9, containing 85 mM NaCl) were subjected to equilibrium dialysis as described previously (3) using a 3,500-kDa cutoff membrane (Spectrapor), and the data were analyzed by fitting to a two-cooperative-site Adair equation (3). H144*UreE bound 3.1 ± 0.1 mol of Ni per mole of dimer with a Kd1 of 16.7 ± 3.1 μM and a Kd2 of 4.3 ± 1.3 μM. This stoichiometry is in excellent agreement with the crystal structure (19), although it exceeds the 1.9 mol of Ni/mole of dimer determined in an earlier equilibrium dialysis study (1). The binding constants are close to the previously published numbers. The isolated metal-binding domain bound 2.3 ± 0.1 mol of Ni/mole of dimer with a Kd1 of 16.7 ± 6.5 μM and a Kd2 of 2.5 ± 1.2 μM. If the metal-binding domain were to form a dimeric structure in the same way as H144*UreE, a maximum of three mol of Ni would be expected to be bound. Since this domain behaves mostly as a tetramer, it is conceivable that some metal-binding sites are inaccessible in the larger aggregate (i.e., 4 to 5 mol of Ni/mole of tetramer). Another possibility is that a small fraction of the metal-binding domain does not fold properly in the absence of its N-terminal peptide-binding domain.
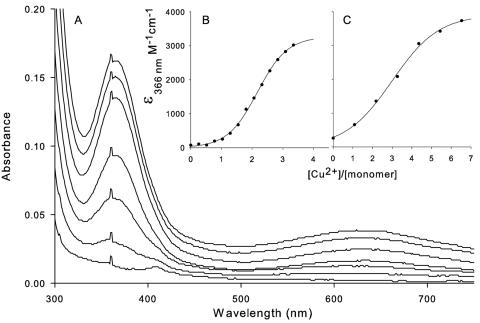
The binding of Cu2+ or Ni2+ to H144*UreE and UreE metal-binding domain proteins was monitored in a Shimadzu 2401PC spectrophotometer with a 160-μl cuvette (Starna) in 50 mM Tris buffer, pH 7.9, containing 85 mM NaCl. Binding of Cu2+ to H144*UreE results in a significant absorbance increase centered at 366 nm (2) attributed to a thiolate-to-Cu2+ charge transfer transition involving Cys79, since mutation or chemical modification of this residue eliminated the spectral changes. This same feature was observed for the isolated metal-binding domain (Fig. 3A). Plots of the absorbance changes at 366 nm versus Cu2+ concentration revealed sigmoid effects for both samples (Fig. 3B and C), with lower midpoint Cu concentrations (2.48 ± 0.03 mol of Cu2+/mole of monomer) for H144*UreE than for the metal-binding domain (3.0 ± 0.1 mol of Cu2+/mole of monomer).
FIG. 3.
UV-visible spectroscopic changes resulting from titration of CuSO4 into H144*UreE and the metal-binding domain. (A) Raw data from titration of 46 μM UreEΔ2-69 with CuSO4 in 50 mM Tris (pH 8)-85 mM NaCl. (B) Data from a similar titration of 85.2 μM H144*UreE with CuSO4 in identical buffer conditions and corrected for dilution effects to calculate extinction coefficients at 366 nm. (C) Data from the titration shown in panel A, corrected for dilution effects to calculate extinction coefficients.
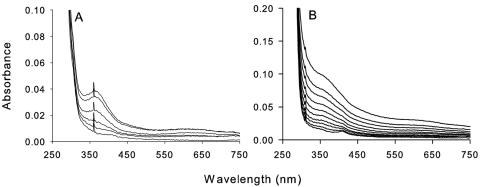
Ni titration of H144*UreE and the metal-binding domain resulted in less-intense absorbance changes than for Cu (Fig. 4). The spectral changes of the control sample were somewhat more pronounced than what was observed in earlier studies (2) and led to the development of a defined peak at 366 nm. This Ni-induced absorbance is consistent with a weak thiolate-to-Ni2+ charge transfer transition, as reported in other Ni-containing proteins (7, 22) and model compounds (5), and suggests that Ni2+ can coordinate to Cys79 when present at a high concentration. Maximal absorption of this chromophore required six equivalents of Ni2+ per H144* dimer, suggesting that Ni binds to Cys79 only after the first three binding sites approach full occupancy. Addition of Ni to the isolated metal-binding domain produced a general increase in absorbance at 320 to 400 nm with no distinct peak observed. The Ni concentrations resulting in a half-maximal absorbance change for the metal-binding domain (14.4 ± 0.6 mol of Ni2+/mole of dimer) is significantly greater than that for the control H144*UreE sample (3.8 ± 0.4 mol of Ni2+/mole of dimer). These results suggest that aggregation of the metal-binding protein hinders access of Ni to the thiol associated with this chromophore.
FIG. 4.
UV-visible spectra of H144*UreE and the UreE metal-binding domain titrated with Ni. (A) Uncorrected spectra of 85.2 μM H144*UreE after successive additions of NiSO4. (B) Uncorrected spectra for 46 μM UreE metal-binding domain titrated with NiSO4.
Conclusions.
The presence of UreE facilitates urease activation in cells that are grown in media with low Ni availability; however, deletion of the entire ureE gene did not eliminate urease activity. Similar results were noted in the case of a Helicobacter pylori ureE chromosomal knockout that produced urease at about 1% of wild-type levels (21). Cells containing the urease operon with only the UreE metal-binding domain possessed higher activities than cells with complete ureE deletion, demonstrating that the single domain protein can deliver Ni to urease apoprotein and the N-terminal domain is not required. The isolated metal-binding domain is primarily a tetramer, in contrast to the dimeric wild-type UreE or H144*UreE. It is worth noting that a concentration- and metal ion-dependent dimer-tetramer equilibrium has been observed for the full-length Bacillus pasteurii UreE (18, 23). The K. aerogenes UreE metal-binding domain exhibits Kd values nearly identical to those of the H144*UreE control sample, consistent with retention of the three high-affinity Ni-binding sites observed in the crystal structure (Fig. 1). The absorbance changes revealed by UV/visible spectroscopy upon Cu2+ and Ni2+ binding involve a distinct metal-binding site associated with Cys79. Differences in the two samples are consistent with decreased access of metals to this binding site in the tetrameric protein. Our overall results indicate that the fold of the purified metal-binding domain closely resembles that of H144*UreE, allowing it to function in urease activation.
Acknowledgments
We thank Merhawit Negussie for assistance with the initial phases of these studies.
This study was supported by National Institutes of Health grant DK45686.
REFERENCES
- 1.Brayman, T. G., and R. P. Hausinger. 1996. Purification, characterization, and functional analysis of a truncated Klebsiella aerogenes UreE urease accessory protein lacking the histidine-rich carboxyl terminus. J. Bacteriol. 178:5410-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colpas, G. J., T. G. Brayman, J. McCracken, M. A. Pressler, G. T. Babcock, L. J. Ming, C. M. Colangelo, R. A. Scott, and R. P. Hausinger. 1998. Spectroscopic characterization of metal binding by Klebsiella aerogenes UreE urease accessory protein. J. Biol. Inorg. Chem. 3:150-160. [Google Scholar]
- 3.Colpas, G. J., T. G. Brayman, L. J. Ming, and R. P. Hausinger. 1999. Identification of metal-binding residues in the Klebsiella aerogenes urease nickel metallochaperone, UreE. Biochemistry 38:4078-4088. [DOI] [PubMed] [Google Scholar]
- 4.Colpas, G. J., and R. P. Hausinger. 2000. In vivo and in vitro kinetics of metal transfer by the Klebsiella aerogenes urease nickel metallochaperone, UreE. J. Biol. Chem. 275:10731-10737. [DOI] [PubMed] [Google Scholar]
- 5.Desrochers, P. J., R. W. Cutts, P. K. Rice, M. L. Golden, J. B. Graham, T. M. Barclay, and A. W. Cordes. 1999. Characteristics of five-coordinate nickel-cysteine centers. Inorg. Chem. 38:5690-5694. [Google Scholar]
- 6.Dykxhoorn, D. M., R. St Pierre, and T. Linn. 1996. A set of compatible tac promoter expression vectors. Gene 177:133-136. [DOI] [PubMed] [Google Scholar]
- 7.Gencic, S., and D. A. Grahame. 2003. Nickel in subunit beta of the acetyl-CoA decarbonylase/synthase multienzyme complex in methanogens. J. Biol. Chem. 278:6101-6110. [DOI] [PubMed] [Google Scholar]
- 8.Hausinger, R. P., G. J. Colpas, and A. Soriano. 2001. Urease: a paradigm for protein-assisted metallocenter assembly. ASM News 67:78-84. [Google Scholar]
- 9.Hausinger, R. P., and P. A. Karplus. 2001. Urease, p. 867-879. In A. Messerschmidt (ed.), Handbook of metalloproteins. John Wiley & Sons, Ltd., West Sussex, United Kingdom.
- 10.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 11.Lee, M. H., S. B. Mulrooney, M. J. Renner, Y. Markowicz, and R. P. Hausinger. 1992. Klebsiella aerogenes urease gene cluster: sequence of UreD and demonstration that four accessory genes (ureD, ureE, ureF, and ureG) are involved in nickel metallocenter biosynthesis. J. Bacteriol. 174:4324-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, M. H., H. S. Pankratz, S. Wang, R. A. Scott, M. G. Finnegan, M. K. Johnson, J. A. Ippolito, D. W. Christianson, and R. P. Hausinger. 1993. Purification and characterization of Klebsiella aerogenes UreE protein: a nickel-binding protein that functions in urease metallocenter assembly. Protein Sci. 2:1042-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289-298. [DOI] [PubMed] [Google Scholar]
- 14.Mobley, H. L. T., M. D. Island, and R. P. Hausinger. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59:451-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mulrooney, S. B., and R. P. Hausinger. 2003. Nickel uptake and utilization by microorganisms. FEMS Microbiol. Rev. 27:239-261. [DOI] [PubMed] [Google Scholar]
- 16.Mulrooney, S. B., and R. P. Hausinger. 1990. Sequence of the Klebsiella aerogenes urease genes and evidence for accessory proteins facilitating nickel incorporation. J. Bacteriol. 172:5837-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musiani, F., B. Zambelli, M. Stola, and S. Ciurli. 2004. Nickel trafficking: insights into the fold and function of UreE, a urease metallochaperone. J. Inorg. Biochem. 98:803-813. [DOI] [PubMed] [Google Scholar]
- 18.Remaut, H., N. Safarov, S. Ciurli, and J. Van Beeumen. 2001. Structural basis for Ni2+ transport and assembly of the urease active site by the metallochaperone UreE from Bacillus pasteurii. J. Biol. Chem. 276:49365-49370. [DOI] [PubMed] [Google Scholar]
- 19.Song, H. K., S. B. Mulrooney, R. Huber, and R. P. Hausinger. 2001. Crystal structure of Klebsiella aerogenes UreE, a nickel-binding metallochaperone for urease activation. J. Biol. Chem. 276:49359-49364. [DOI] [PubMed] [Google Scholar]
- 20.Soriano, A., G. J. Colpas, and R. P. Hausinger. 2000. UreE stimulation of GTP-dependent urease activation in the UreD-UreF-UreG-urease apoprotein complex. Biochemistry 39:12435-12440. [DOI] [PubMed] [Google Scholar]
- 21.Voland, P., D. L. Weeks, E. A. Marcus, C. Prinz, G. Sachs, and D. Scott. 2003. Interactions among the seven Helicobacter pylori proteins encoded by the urease gene cluster. Am. J. Physiol. Gastrointest. Liver Physiol. 284:G96-G106. [DOI] [PubMed] [Google Scholar]
- 22.Wang, S. C., A. V. Dias, S. L. Bloom, and D. B. Zamble. 2004. Selectivity of metal binding and metal-induced stability of Escherichia coli NikR. Biochemistry 43:10018-10028. [DOI] [PubMed] [Google Scholar]
- 23.Won, H. S., Y. H. Lee, J. H. Kim, I. S. Shin, M. H. Lee, and B. J. Lee. 2004. Structural characterization of the nickel-binding properties of Bacillus pasteurii urease accessory protein (Ure)E in solution. J. Biol. Chem. 279:17466-17472. [DOI] [PubMed] [Google Scholar]