Abstract
P1par family members promote the active segregation of a variety of plasmids and plasmid prophages in gram-negative bacteria. Each has genes for ParA and ParB proteins, followed by a parS partition site. The large virulence plasmid pWR100 of Shigella flexneri contains a new P1par family member: pWR100par. Although typical parA and parB genes are present, the putative pWR100parS site is atypical in sequence and organization. However, pWR100parS promoted accurate plasmid partition in Escherichia coli when the pWR100 Par proteins were supplied. Unique BoxB hexamer motifs within parS define species specificities among previously described family members. Although substantially different from P1parS from the P1 plasmid prophage of E. coli, pWR100parS has the same BoxB sequence. As predicted, the species specificity of the two types proved identical. They also shared partition-mediated incompatibility, consistent with the proposed mechanistic link between incompatibility and species specificity. Among several informative sequence differences between pWR100parS and P1parS is the presence of a 21-bp insert at the center of the pWR100parS site. Deletion of this insert left much of the parS activity intact. Tolerance of central inserts with integral numbers of helical DNA turns reflects the critical topology of these sites, which are bent by binding the host IHF protein.
The enteric bacterium Shigella flexneri is the causative agent of shigellosis, a severe human enteric disease. It is one of the leading causes of death in Third World countries. Pathogenic strains of S. flexneri contain a large (200-kb) plasmid, pWR100, which encodes many of the factors essential for virulence (17). The sequence of this plasmid has been determined (3) (accession number AL391753). It revealed a homolog of the partition region of the plasmid prophage of bacteriophage P1.
The P1par (partition) region consists of an operon containing parA and parB genes and a centromere analog site, parS, that lies downstream (1). Like similar systems found in other low-copy-number plasmids, it assures the faithful distribution of the plasmids between dividing cells by an active process akin to mitosis (10, 19). The parA gene encodes a Walker-type ATPase essential for plasmid movement during partition (6, 15). The parB gene encodes a protein that can bind tightly to the partition site, parS (5, 8). It is required for capture of the plasmid at the cell center prior to partition (15).
There are now six identified members of the P1par family. New loci have been identified in the large virulence plasmids of Salmonella and Shigella species and the Rts1 plasmid of Proteus vulgaris (A. Dabrazhynetskaya, K. Sergueev, and S. Austin, unpublished data). In addition to P1par, two others have been previously studied: those found in bacteriophage P7 from Escherichia coli, and the virulence plasmid pMT1 from Yersinia pestis (25; Dabrazhynetskaya et al., unpublished). These two have a similar structure and organization to P1par. Each has a unique species specificity. The Par protein of each species works only with the cognate parS site from the same species (12, 25; Dabrazhynetskaya et al., unpublished). The three previously studied species also have unique specificities for partition-mediated incompatibility. Two plasmids with identical par systems are incompatible because they displace each other from the cell. This is due to competition for selection of the plasmid as a substrate for partition (2). In the cases studied so far, incompatibility and species specificity appear to correlate: each family member exerts incompatibility against plasmids with its own type of par element but does not interfere with the activity of the other par family members (12, 25; Dabrazhynetskaya et al., unpublished). The P1parS site consists of a central integration host factor (IHF) binding region flanked by two arms that bind ParB. Within the arms lie a specifically arranged set of motifs, the BoxA and BoxB sequences (Fig. 1B). BoxA sequences are the principle binding sites for ParB (5, 9). The BoxB sequences also contact ParB. They are essential for partition and control the species specificity by allowing P1 ParB to bind and by excluding the binding of P7 ParB (20, 23). P7parS is organized similarly to P1parS but has a different BoxB motif. Substitution of the P7 BoxB sequences into an otherwise-intact P1 parS site switches the specificity, so that the site then works exclusively with the P7 Par proteins (12, 13). Similarly, the unique BoxB sequences of the pMT1parS site also determine species specificity (Dabrazhynetskaya et al., unpublished). There are two hexamer BoxB sequences in each parS site. They generally differ from those of another species by only one or two bases. The species-specific information in the cognate ParB protein also appears to be limited to a small region: a run of less than 11 amino acids near the carboxy terminus in the case of P7 ParB (20). Changes in a small amount of information in BoxB and its contact point on the ParB protein appear to be capable of defining a number of species specificities for the system.
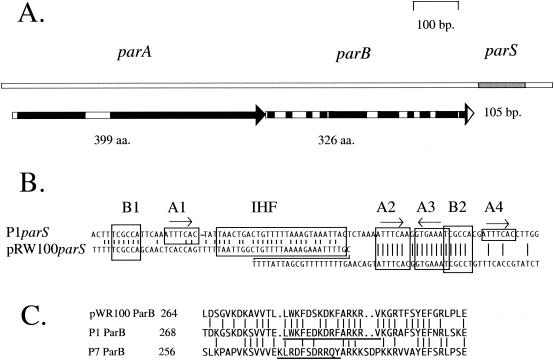
FIG. 1.
The pWR100 par region. A. Map of the pWR100par region showing the parA and parB open reading frames and the parS site. Black boxes on the open reading frame arrows show regions of very high similarity to the amino acid sequences of the equivalent P1 Par proteins. Blocks of >10 amino acids with >90% similarity to the P1 proteins are included. B. Alignment of the parS sequences of pWR100 and P1. The pWR100parS site is displayed in a staggered configuration in order to accommodate an additional 21-bp sequence that is absent from P1parS. Vertical lines join identities. The boxed sequences B1 and B2 are the discriminator hexamers whose variants determine the species specificity differences for parS between various P1par family members. The boxes A1 through A4 are repeats of the heptamer BoxA sequence with the consensus ATTTCAC. They are the principle binding motifs for the ParB protein. Boxes A1 and A4 are conserved in previously characterized P1par family members but appear to be absent in pWR100parS. C. Alignment of amino acid sequences from near the C termini of the ParB proteins of pWR100, P1, and P7. Vertical lines join identities. The underlined P1 and P7 sequences are the regions within which resides the information thought to be involved in the specific recognition of the respective P1 and P7parS BoxB motifs (20).
Here, we characterize the pWR100par element from S. flexneri and show that its sequence and specificity confirm the importance of the BoxB-ParB contact in species specificity. The results provide further support for the hypothesis that species specificity governs partition-mediated incompatibility and constitutes a special mechanism for plasmid speciation and evolution. The results also confirm that two prominent sequence features that are conserved in the previously characterized family members are either unnecessary or redundant.
MATERIALS AND METHODS
General methods and materials.
General methods and materials were as previously described (20).
Bacterial strains.
Strain DH5α was used for general DNA manipulations and for tests of operon regulation in β-galactosidase assays (22). Strain CC2056 recA56 trpam thi lacZam λW82 (24) was used for the colony color assay and for incompatibility tests.
Plasmids.
Plasmids pALA1513 and pALA1514, which carry the P1 and P7 par operons, respectively, were produced by taking the purified EcoRI-XbaI fragment of pALA1413 or pALA1414 (20) and inserting it between the EcoRI and BamHI sites of the pGB2 vector (4). Ligation was carried out in the presence of a BamHI-XbaI linker formed by annealing the oligonucleotides 5′-GATCCGAACCCCTTCG and 5′-CTAGCGAAGGGGTTCG. Plasmid pALA2601, which contains the pWR100 parA-parB operon, consists of the pWR100 sequence bp 28818 to 31240 (accession number AL391753). The par operon was obtained as a PCR product with BamHI and SalI extensions at the left and right ends, cut with SalI and BamHI, and inserted between the homologous sites in the vector pBR322. The PCR primers used were 5′-CCTGTGGATCCTCTACGCTGAACTGGATTATTCGGGGGAA and 5′-CATCGGTCGACGCTCTCCAATTAAAAACTGGTGAGTTGC; the underlined sequences are complementary to pWR100.
Plasmid pALA2602 contains a 255-bp pWR100 fragment (bp 31200 to 31454) that includes the parS site. The parS site was obtained as a PCR product with BamHI extensions on both ends. The PCR used the primers 5′-TGTCCGGATCCAAATTCAAGTTTTTCGCCAGC and 5′-CGAGGATCCGGTATCCCGCTTACTTTTGTTG, where the underlined sequences are complementary to pWR100. The PCR product was then cut with BamHI and inserted in the homologous sites of pALA1991 (25). Plasmid pALA2602 was recombined with λ-P1:5RΔ1005 to give λ-P1:5RΔ1005::pALA2602 as previously described (24).
Plasmid pALA2609 was constructed by excising the BamHI fragment of pALA2602 and inserting it into the BclI site of pACYC184. The smaller PvuII fragment of plasmid pALA2609 was removed, and the remainder was religated to produce pALA2611. This deleted the C-terminal portion of the chloramphenicol resistance cassette.
A PCR fragment containing the promoter of the pWR100 par operon was inserted as an EcoRI-BamHI fragment between the equivalent sites of the lacZ reporter plasmid pALA1426 (14) to give the plasmid pALA2615. The promoter region was obtained as a PCR product using the primers 5′-GATCGGAATTCCCGGGGTGAACTGGATTATTCGGGGG and 5′-TGTCCGGATCCCCGGGTAGGGCGAGCAACATTTTAT, where the underlined sequences are complementary to the pWR100 template.
Plasmid pALA2620 is the result of cloning of the BamHI-SalI fragment from pALA2601 into the same sites in the pGB2 vector (4). A 21-bp deletion within the parS site of plasmid pALA2602 was made to give pALA2621. This was generated using the GeneTailor site-directed mutagenesis system (Invitrogen) using 5′-TAAAAGAAATTTTGCTTTTAATTTCACGGTGAAAT and 5′-TAAAAGCAAAATTTCTTTTAAAACAGCCAATTA as mutagenic primers. The deletion starts at position 31268 and extends to position 31288 of the pWR100 sequence.
Colony color partition assay.
The colony color partition assays were performed in strain CC2056 (20) using high-titer lysates as described elsewhere (24). The appropriate mini-P1 plasmid containing the respective parS site was recombined into a λ-P1:5RΔpar phage vector and introduced by infection into cells supplying the P1, P7, or pWR100 Par proteins. The recombined elements replicate as low-copy-number plasmids driven by the P1 replicon. Their maintenance stability was measured after 25 generations of unselected growth, scoring for the ability of the supF marker that they carry to suppress the lacZ amber mutation in the strain and hence give a red colony on lactose MacConkey indicator plates (20).
Incompatibility tests.
The ability of supernumerary parS sites to exert incompatibility against plasmids maintained by a par system was carried out as follows. Colony color partition tests were carried out as described above, except that each strain carried an additional plasmid derived from the vector pACYC184 that carried the parS site from P1 (pALA1849), P7 (pALA1850), or pWR100 (pALA2611). Tetracycline (5 μg/ml) was added to the medium to ensure retention of pACYC184 derivatives.
Assays of β-galactosidase activity.
Assays of β-galactosidase activity were performed on cells permeabilized with sodium dodecyl sulfate (18). The DH5α cells contained the plasmid pALA2615, which has the lacZ gene under control of the pWR100par promoter-operator region, and plasmid pBR322 or its derivative pALA2601, which produced the pWR100 Par proteins. The β-galactosidase values presented are the averages from assays done in triplicate.
RESULTS
pWR100 par region.
The putative ParA and ParB proteins of S. flexneri pWR100 are similar in size and sequence to their bacteriophage P1 counterparts (3). ParA has 399 residues and shows 86% similarity to P1 ParA. ParB has 326 residues and is 76% similar to P1 ParB (Fig. 1A). The stop codon of the parA open reading frame overlaps the start codon of parB by 1 nucleotide, a feature seen in P7 and pMT1 par operons, but not in P1par (16, 25).
The organization of the putative pWR100parS sequence differs considerably from those of the previously characterized family members (Dabrazhynetskaya et al., unpublished) (Fig. 1B). It begins 13 bp downstream of the parB open reading frame. The right arm of the site is P1 like, with boxes A2, A3, and B2 clearly present (3; Dabrazhynetskaya et al., unpublished). However, there is no recognizable BoxA4 sequence, and the central region contains a 21-bp sequence, not present in P1parS, that acts as a spacer between the right-arm sequences and a sequence clearly homologous to the P1 IHF binding site. The left arm has no recognizable BoxA1 sequence but has a BoxB1 that is a perfect repeat of BoxB2. The distance from the BoxB1 sequence to the start of the IHF site is similar to those of other family members (Fig. 1B).
The pWR100par system is functional in E. coli.
We used the colony color partition test to study the activity of the pWR100par region in E. coli. This assay measures the stability of the maintenance of a λ-mini-P1 plasmid carrying the parS partition site when Par proteins are supplied in trans. The proteins were produced from a pGB2 plasmid vector carrying the par genes under control of their endogenous promoter. The putative pWR100par operon and parS partition site were separately amplified by PCR, using primers that introduced new restriction sites at the ends of the pWR100 sequences. The parS site and the par operon were then cloned into λ-mini-P1 and pGB2 plasmid vectors, respectively, as described in Materials and Methods. Table 1 shows that the λ-mini-P1pWR100parS test plasmid was faithfully maintained, but only when the pWR100 Par proteins were supplied. The presence of the pWR100parS site on the target plasmid was required for this effect. We conclude that the pWR100par system is functional in E. coli. Its efficiency in this assay was slightly less than those of the P1 and P7par systems assayed under similar conditions (Table 1).
TABLE 1.
Colony color partition assaysa
| Plasmid | par operon carried on plasmid | Test plasmidbparS site | % Retention (25 generations) |
|---|---|---|---|
| pGB2 | None | pWR100parS | <2 |
| pALA2620 | pWR100 parA-parB | None | <2 |
| pALA2620 | pWR100 parA-parB | pWR100parS | 70 |
| pALA2620 | pWR100 parA-parB | P1parS | 30 |
| pALA2620 | pWR100 parA-parB | P7parS | <2 |
| pALA1513 | P1parA-parB | P1parS | 97 |
| pALA1513 | P1parA-parB | pWR100parS | 92 |
| pALA1514 | P7parA-parB | P7parS | 93 |
| pALA1514 | P7parA-parB | pWR100parS | <2 |
Retention of a test plasmid carrying the appropriate parS site was measured when a second, pGB2-based plasmid supplying the Par proteins from the appropriate par operon was present.
The test plasmids were λ-P1:5RΔ1005::pALA1952 (P1parS), λ-P1:5RΔ1005::pALA1991 (no parS site), λ-P1:5RΔ1005::pALA1993 (P7parS), or λ-P15R:5RΔ1005::pALA2602 (pWR100 parS).
The pWR100par genes are autoregulated at the level of transcription.
We constructed a transcriptional fusion consisting of 264 bp of the pWR100par operon linked to the lacZ gene of the vector pRS415 (pALA2615). The sequence begins 201 bp upstream from the start of the parA open reading frame. This pWR100parA-lacZ fusion expressed high levels of β-galactosidase activity in the absence of Par proteins but was repressed eightfold when both pWR100 ParA and ParB proteins were supplied in trans from a compatible plasmid (Table 2). Thus, expression of the pWR100par operon is autoregulated at the level of transcription by interaction of the Par proteins with control sequences located near the start of the ParA open reading frame, as is the case with other members of the P1par family (7).
TABLE 2.
Autoregulation of the pWR100par operona
| Supplying plasmid | Par proteins supplied | β-Galactosidase activity (Miller units) |
|---|---|---|
| pBR322 | None | 12,074 +/− 407 |
| pALA2601 | pWR100 ParA, ParB | 1,575 +/− 305 |
The DH5α cells contained the plasmid pALA2615 that has the lacZ gene under control of the pWR100par promoter-operator region.
The pWR100 and P1 par systems have the same species specificity for the action of the Par proteins at parS.
The partition assay used here has the parS site and the par operon on two different plasmids. On substituting one or the other plasmid with its equivalent from different plasmid species, the species specificities of the site and proteins can be assessed. Table 1 shows that the P1 Par proteins were able to function efficiently with the pWR100parS site. Also, the test plasmid carrying the P1parS site was partitioned when the pWR100 Par proteins were supplied, although the efficiency was significantly less than when the native P1 proteins were used. There was no apparent interaction between pWR100 Par components and those of the P7 plasmid (Table 1). We conclude that the Par proteins and parS sites of pWR100 and P1 are substantially interchangeable. They share the same species specificity for this interaction, despite considerable sequence differences.
The pWR100 and P1par systems are mutually incompatible.
When two identical par regions are present on two different plasmids in the same cell, the plasmids become incompatible with each other. This partition-mediated incompatibility is caused by like parS sites competing with each other during partition. It is a major factor in determining whether two plasmids can coexist in the same cell (2). A convenient assay for this effect consists of the introduction of supernumerary parS sites on an otherwise-compatible cloning vector into cells containing the relevant partitioning plasmid. Partition-mediated incompatibility is seen when the resident plasmid is displaced from the cell. Using this assay, we have previously shown that the P1, P7, and pMT1par systems each show unique incompatibility properties: their parS sites exerted incompatibility against par plasmids of their own species, but not against those of two other species (2, 12, 25).
The pWR100parS site was inserted into a pACYC184 vector (see Materials and Methods), and the resulting plasmid was transformed into the partition assay strain containing a test plasmid maintained by the pWR100par system. On selecting only for the incoming plasmid, the test plasmid was rapidly lost from the cells (Table 3). Thus, pWR100parS exerts an incompatibility effect against its own par system similar to that exerted by P1, P7, or pMT1 parS sites against their respective par systems. We then tested the ability of other parS species to exert incompatibility against the pWR100par system. The P1parS site was able to exert incompatibility against pWR100par. Likewise, the pWR100parS site exerted incompatibility against the P1par system (Table 3). There was no comparable effect exerted between P7 and pWR100 par components, although the P7parS site may have a small destabilizing effect on the pWR100par system (a reduction from 70% to 60% retention) (Tables 1 and 4). Thus, the pWR100 and P1 systems exhibit the same incompatibility specificity as well as sharing species specificity for parS recognition by the Par proteins. This further supports the theory that species specificity and incompatibility have a common mechanistic basis in members of the P1par family of partition elements (Dabrazhynetskaya et al., unpublished).
TABLE 3.
Incompatibility assays
| parS site on resident target plasmida | Par protein(s) suppliedb | Supernumerary parS sites carried by incoming pACYC184 derivativec | % Retention of target plasmid (25 generations) |
|---|---|---|---|
| P7 | P7 | P7 | 3 |
| P1 | P1 | P1 | 3 |
| pWR100 | pWR100 | pWR100 | 5 |
| P1 | P1 | pWR100 | 10 |
| P7 | P7 | pWR100 | 95 |
| pWR100 | pWR100 | P1 | 5 |
| pWR100 | pWR100 | P7 | 60 |
| P1 | P1 | P7 | 95 |
| P7 | P7 | P1 | 95 |
The target plasmids were λ-P1::5RΔ1005::pALA1993 (P7parS), λ-P1::5RΔ1005::pALA1952 (P1parS), or λ-P1::5RΔ1005::pALA2602 (pWR100parS).
The Par proteins were supplied from pALA1414 (P7 ParA and ParB), pALA1413 (P1 ParA and ParB), or pALA2620 (pMT1 ParA and ParB).
Supernumerary parS sites were provided by the presence of the following pACYC184 derivatives: pALA1850 (P7parS), pALA1849 (P1parS), or pALA2609 (pWR100parS).
TABLE 4.
Effect of deleting the noncanonical 21-bp sequence from the pWR100parS site
| Plasmid | par operon carried | Test plasmid parSa site | % Retentionb (25 generations) |
|---|---|---|---|
| pGB2 | None | pWR100parSΔ21 | <2 |
| pALA2620 | pWR100parA-parB | pWR100parS | 72 |
| pALA1413 | P1parA-parB | pWR100parS | 96 |
| pGB2 | None | pWR100parSΔ21 | <2 |
| pALA2620 | pWR100parA-parB | pWR100parSΔ21 | 28 |
| pALA1413 | P1parA-parB | pWR100parSΔ21 | 78 |
The test plasmids were λ-P1::5RΔ1005::pALA2602 (pWR100parS) or λ-P1::5RΔ1005::pALA2621 (pWR100parSΔ21).
All values were obtained from a single experiment. Duplicate experiments gave similar results. The controls using the wild-type pWR100parS are duplications of tests reported in Table 1, where similar but not identical values were obtained.
Deletion of the noncanonical 21-bp sequence in the pWR100parS site.
The most prominent difference between pWR100 and P1 parS sites is the presence in pWR100parS of a 21-bp extension between the IHF binding site and BoxA2 (Fig. 1). In order to investigate the significance of this sequence, the 21 nucleotides were deleted. The mutated parS site was inserted in the λ mini-P1 test plasmid, and its stability was checked when pWR100 or P1 ParA and ParB proteins were supplied in trans. As shown in Table 4, the deletion retained significant function, although the maintenance efficiency of the plasmid was reduced almost threefold. The deleted pWR100parS site also functioned when the P1 ParA and ParB proteins were supplied. Notably, the degree of the stabilization by the P1 proteins remained high when the deletion was introduced. The mutant site worked considerably better with the P1 proteins than with its native pWR100 proteins (Table 4). Thus, although the specificities of the pWR100 and P1 sites substantially overlap, the pWR100 proteins appear to have adapted to function with the longer parS site so that they work less well without this extra sequence element.
DISCUSSION
The P1par family of partition loci is a group of versatile elements that are employed in a wide variety of plasmid types. They have presumably been disseminated to these plasmids by lateral exchange. Lateral exchanges of DNA elements (cassettes) containing various plasmid functions are common (19). The currently described P1par family members are from plasmid prophages (P1 and P7), large virulence plasmids (pMT1, pSLT, and pWR100), and antibiotic resistance plasmids (Rts1). These general plasmid types have little else in common. They are found in five different bacterial genera: Escherichia, Salmonella, Yersinia, Shigella, and Proteus (Dabrazhynetskaya et al., unpublished). Thus, by associating with different cassettes containing a variety of different replication systems, virulence factors, phage genes, etc., the P1par family members have achieved a broad distribution in nature as elements in diverse plasmids and hosts.
The pWR100 ParA and ParB proteins are typical of the P1par family, with the highest degree of similarity to the ParA and ParB proteins of P1par and pSLTpar (Dabrazhynetskaya et al., unpublished). However, the pWR100parS site is atypical, with a 21-bp inclusion between the IHF and A2 boxes and the absence of any obvious A1 or A4 box (Fig. 1). The inclusion is not essential for function, as it can be deleted. The P1parS site tolerates inserts with integral numbers of helical turns in a similar position (11). Shorter or longer inserts are not tolerated. This was interpreted as a requirement for the two arms of the site to have specific faces of the helix aligned with each other when brought together by the severe bending imposed by IHF binding (11). The 21-bp inclusion in the pWR100parS site constitutes two turns of the helix. By analogy with the P1 case, its existence and dispensability can therefore be understood.
The P1 Par proteins work efficiently with the pWR100parS site that lacks the A1 and A4 boxes, even though their cognate P1parS site has them. Also, the pWR100 Par proteins work with the P1parS site that has these features. This is consistent with the A1 and A4 boxes being redundant or nonfunctional in P1 and also, perhaps, in the other P1par family members that have them. The A4 boxes of P1and P7parS can be deleted without affecting partition (11, 12). Multiple mutations in BoxA1 reduced, but did not eliminate, the partition activity of P7parS (13).
The P1 species specificity difference from P7 is determined by the interaction of a short tract of amino acids in ParB with a unique BoxB motif (21). The species specificity of pMT1 also proved to reside in a unique BoxB motif. Changing just a single base in one of the pMT1parS BoxB repeats was sufficient to change the species specificity of the pMT1 site to that of P1 (Dabrazhynetskaya et al., unpublished). Based on these findings, it appears that the BoxB-ParB contact is a special mechanism for defining several unique specificities. Minor variations in the BoxB motif, in conjunction with minor differences in a small region of the cognate ParB protein, can generate several different exclusive specificities for ParB action at parS. This appears to lend the system considerable flexibility for rapid speciation during plasmid evolution.
The properties of pWR100par extend support for BoxB as a unique determinant of species specificity. The pWR100par system showed the same species specificity as P1par. Although pWR100parS is an atypical parS, with considerable divergence from the form and sequence of other family members, it has recognizable BoxB sequences that are identical to those of P1. The ParB regions thought to contain the contact with BoxB are also very similar in the two species (Fig. 1C). In contrast, the equivalent P7 ParB, which fails to function with pWR100 or P1 parS, shows little similarity (Fig. 1C).
For the P1, P7, and pMT1 systems, it has been shown that species specificity correlates with partition-mediated incompatibility. Also, BoxB alterations that change species specificity can change the incompatibility specificity correspondingly (Dabrazhynetskaya et al., unpublished). This correlation is also extended here: pWR100parS and P1parS have identical BoxB sequences and the same species specificity and show the same incompatibility type, despite considerable differences in sequence and organization of other features. Thus, species specificity for Par protein action at parS and partition-mediated incompatibility appear to have a common mechanistic basis. Two plasmids with like ParB-parS complexes compete with each other during partition, leading to incompatibility. Unlike ones fail to compete and thus are compatible. The basis of this competition might be the ability to form mixed plasmid pairs via the bound ParB proteins prior to segregation (2).
REFERENCES
- 1.Austin, S., and A. Abeles. 1983. Partition of unit-copy miniplasmids to daughter cells. II. The partition region of miniplasmid P1 encodes an essential protein and a centromere-like site at which it acts. J. Mol. Biol. 169:373-387. [DOI] [PubMed] [Google Scholar]
- 2.Austin, S. J., and K. Nordstrom. 1990. Partition-mediated incompatibility of bacterial plasmids. Cell 60:351-354. [DOI] [PubMed] [Google Scholar]
- 3.Buchrieser, C., P. Glaser, C. Rusniok, H. Nedjari, H. D'Hauteville, F. Kunst, P. Sansonetti, and C. Parsot. 2000. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38:760-771. [DOI] [PubMed] [Google Scholar]
- 4.Churchward, G., D. Belin, and Y. Nagamine. 1984. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 31:165-171. [DOI] [PubMed] [Google Scholar]
- 5.Davis, M. A., and S. J. Austin. 1988. Recognition of the P1 plasmid centromere analog involves binding of the ParB protein and is modified by a specific host factor. EMBO J. 7:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis, M. A., L. Radnedge, K. A. Martin, F. Hayes, B. Youngren, and S. J. Austin. 1996. The P1 ParA protein and its ATPase activity play a direct role in the segregation of plasmid copies to daughter cells. Mol. Microbiol. 21:1029-1036. [DOI] [PubMed] [Google Scholar]
- 7.Friedman, S. A., and S. J. Austin. 1988. The P1 plasmid-partition system synthesizes two essential proteins from an autoregulated operon. Plasmid 19:103-112. [DOI] [PubMed] [Google Scholar]
- 8.Funnell, B. E. 1988. Mini-P1 plasmid partitioning: excess ParB protein destabilizes plasmids containing the centromere parS. J. Bacteriol. 170:954-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Funnell, B. E., and L. Gagnier. 1993. The P1 plasmid partition complex at parS. II. Analysis of ParB protein binding activity and specificity. J. Biol. Chem. 268:3616-3624. [PubMed] [Google Scholar]
- 10.Gerdes, K., J. Moller-Jensen, and R. Bugge Jensen. 2000. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 37:455-466. [DOI] [PubMed] [Google Scholar]
- 11.Hayes, F., and S. Austin. 1994. Topological scanning of the P1 plasmid partition site. J. Mol. Biol. 243:190-198. [DOI] [PubMed] [Google Scholar]
- 12.Hayes, F., and S. J. Austin. 1993. Specificity determinants of the P1 and P7 plasmid centromere analogs. Proc. Natl. Acad. Sci. USA 90:9228-9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayes, F., M. A. Davis, and S. J. Austin. 1993. Fine-structure analysis of the P7 plasmid partition site. J. Bacteriol. 175:3443-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayes, F., L. Radnedge, M. A. Davis, and S. J. Austin. 1994. The homologous operons for P1 and P7 plasmid partition are autoregulated from dissimilar operator sites. Mol. Microbiol. 11:249-260. [DOI] [PubMed] [Google Scholar]
- 15.Li, Y., A. Dabrazhynetskaya, B. Youngren, and S. Austin. 2004. The role of Par proteins in the active segregation of the P1 plasmid. Mol. Microbiol. 53:93-102. [DOI] [PubMed] [Google Scholar]
- 16.Ludtke, D. N., B. G. Eichorn, and S. J. Austin. 1989. Plasmid-partition functions of the P7 prophage. J. Mol. Biol. 209:393-406. [DOI] [PubMed] [Google Scholar]
- 17.Makino, S.-I., C. Sasakawa, and M. Yoshikawa. 1988. Genetic relatedness of the basic replicon of the virulence plasmid in shigellae and enteroinvasive Escherichia coli. Microb. Pathog. 5:267-274. [DOI] [PubMed] [Google Scholar]
- 18.Miller, J. R. 1992. A short course in bacterial genetics: a laboratory manual for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Nordstrom, K., and S. J. Austin. 1989. Mechanisms that contribute to the stable segregation of plasmids. Annu. Rev. Genet. 23:37-69. [DOI] [PubMed] [Google Scholar]
- 20.Radnedge, L., M. A. Davis, and S. J. Austin. 1996. P1 and P7 plasmid partition: ParB protein bound to its partition site makes a separate discriminator contact with the DNA that determines species specificity. EMBO J. 15:1155-1162. [PMC free article] [PubMed] [Google Scholar]
- 21.Radnedge, L., B. Youngren, M. Davis, and S. Austin. 1998. Probing the structure of complex macromolecular interactions by homolog specificity scanning: the P1 and P7 plasmid partition systems. EMBO J. 17:6076-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Surtees, J. A., and B. E. Funnell. 2001. The DNA binding domains of P1 ParB and the architecture of the P1 plasmid partition complex. J. Biol. Chem. 276:12385-12394. [DOI] [PubMed] [Google Scholar]
- 24.Youngren, B., and S. Austin. 1997. Altered ParA partition proteins of plasmid P1 act via the partition site to block plasmid propagation. Mol. Microbiol. 25:1023-1030. [DOI] [PubMed] [Google Scholar]
- 25.Youngren, B., L. Radnedge, P. Hu, E. Garcia, and S. Austin. 2000. A plasmid partition system of the P1-P7par family from the pMT1 virulence plasmid of Yersinia pestis. J. Bacteriol. 182:3924-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]