Abstract
Agrobacterium tumefaciens translocates DNA and protein substrates between cells via a type IV secretion system (T4SS) whose channel subunits include the VirD4 coupling protein, VirB11 ATPase, VirB6, VirB8, VirB2, and VirB9. In this study, we used linker insertion mutagenesis to characterize the contribution of the outer-membrane-associated VirB9 to assembly and function of the VirB/D4 T4SS. Twenty-five dipeptide insertion mutations were classified as permissive for intercellular substrate transfer (Tra+), completely transfer defective (Tra−), or substrate discriminating, e.g., selectively permissive for transfer only of the oncogenic transfer DNA and the VirE2 protein substrates or of a mobilizable IncQ plasmid substrate. Mutations inhibiting transfer of DNA substrates did not affect formation of close contacts of the substrate with inner membrane channel subunits but blocked formation of contacts with the VirB2 and VirB9 channel subunits, which is indicative of a defect in assembly or function of the distal portion of the secretion channel. Several mutations in the N- and C-terminal regions disrupted VirB9 complex formation with the outer-membrane-associated lipoprotein VirB7 or the inner membrane energy sensor VirB10. Several VirB9.i2-producing Tra+ strains failed to elaborate T pilus at detectable levels (Pil−), and three such Tra+ Pil− mutant strains were rendered Tra− upon deletion of virB2, indicating that the cellular form of pilin protein is essential for substrate translocation. Our findings, together with computer-based analyses, support a model in which distinct domains of VirB9 contribute to substrate selection and translocation, establishment of channel subunit contacts, and T-pilus biogenesis.
Agrobacterium tumefaciens uses the VirB/D4 type IV secretion system (T4SS) to translocate oncogenic transfer DNA (T-DNA) and effector proteins to plant cells (11, 15). Recently, by use of a coupled formaldehyde cross-linking immunoprecipitation assay, termed transfer DNA immunoprecipitation (TrIP), we obtained evidence that the T-DNA substrate forms close contacts with 6 of the 12 VirB/D4 subunits (13). Further TrIP studies of T4SS mutant strains led to formulation of a DNA translocation pathway in which VirD4, a member of a large family of ATP-binding receptors termed coupling proteins (T4CPs) (20, 26, 37), recruits the DNA substrate to the secretion machine. Next, VirD4 transfers the substrate to the inner-membrane-associated ATPase VirB11, which in turn mediates transfer to the integral inner membrane subunits VirB6 and VirB8. VirB6 and VirB8 function together to transfer the substrate to VirB2 and VirB9, presumably for passage through the periplasm to the cell surface (13). Very recently, we determined that the first two steps of this translocation pathway, T-DNA substrate docking at the VirD4 T4CP and transfer to VirB11, proceed independently of ATP energy utilization by either of these subunits. In contrast, delivery of substrate to the channel components VirB6 and VirB8 requires ATP binding or hydrolytic activities of VirD4 and VirB11 as well as VirB4, a third energetic component of the VirB/D4 T4SS (2).
The TrIP assay and other biochemical, structural, and genetic approaches are advancing a mechanistic understanding of how ATP energy, the three ATP-binding subunits (VirD4, VirB4, and VirB11), and VirB6 and VirB8 contribute to DNA substrate transfer across the inner membrane (2, 12, 13, 26, 31, 52). Presently, however, there is very little information about how VirB2 and VirB9 coordinate passage of DNA and, presumably, protein substrates through the periplasm and across the outer membrane. VirB2 and VirB9 possess cleavable signal sequences, and both subunits are exported across the inner membrane (6, 19, 23, 43, 48). Both subunits partition predominantly with the outer membrane of wild-type cells during fractionation, although inner-membrane-associated forms of both proteins are also detected (43, 48). A current working model is that VirB2 and VirB9 form the portion of the secretion channel that extends from the inner membrane to and across the outer membrane. VirB2 also polymerizes as the extracellular T pilus by a mechanism dependent on the 10 other VirB proteins (34, 35), but at this time it is not known how the secretion channel and the T pilus physically or functionally interact.
Here, we initiated a structure-function study of VirB9 by characterizing the phenotypic consequences of dipeptide insertions introduced along the length of the protein. We report the isolation of substrate-discriminating mutations located mainly in the N-terminal third of the protein that enable the selective export of the oncogenic T-DNA and VirE2 to plant cells or a mobilizable IncQ plasmid to bacterial cells. We further identify mutations that affect VirB9 stability and that disrupt interactions with the outer-membrane-associated VirB7 lipoprotein (23) and inner membrane VirB10 shown recently to function as an energy sensor subunit (12). Finally, we identify “uncoupling” mutations that block biogenesis of the T pilus but not substrate transfer to recipient cells. Results of these mutational studies together with computer analyses establish that VirB9 is composed of distinct regions or domains that contribute in various ways to secretion channel activity and T-pilus biogenesis.
MATERIALS AND METHODS
Bacterial strains and induction conditions.
A. tumefaciens A348 served as the wild-type strain. PC1009 and PC1011 are A348 derivatives deleted of virB9 and virB11, respectively (7). A nonpolar virB2 deletion mutation was introduced into PC1009 and PC1011 by marker exchange-eviction mutagenesis as described previously (7). Briefly, the sacB suicide plasmid pBB120 (7) carrying the ΔvirB2 mutation with approximately 750 bp of upstream and downstream flanking DNA sequence was introduced by electroporation into strains PC1009 and PC1011 with selection for kanamycin-resistant (Kanr) transformants. Following sucrose counterselection, Sucr Kans tranformants were analyzed by PCR amplification across the virB1-virB3 junction to identify the desired ΔvirB2ΔvirB9 and ΔvirB2ΔvirB11 mutant strains, designated PC1209 and PC1211, respectively. The ΔvirB2 mutations were nonpolar as shown by genetic complementation. Conditions for growth of A. tumefaciens and Escherichia coli and for vir gene induction with 200 μM acetosyringone (AS) in induction medium (IM) have been described previously (7). Plasmids were maintained in A. tumefaciens and E. coli by the addition of carbenicillin (100 μg/ml), kanamycin (100 μg/ml), tetracycline (5 μg/ml), gentamicin sulfate (100 μg/ml), or spectinomycin (600 μg/ml).
virB9 gene constructions.
Plasmid pLL373 served as the template for construction of virB9 insertion mutations. First, the unique XhoI restriction site in the polylinker of pJW373 (51) was eliminated by XhoI digestion, blunt-ending of the linearized plasmid with the Klenow fragment of DNA polymerase I, and religation to yield plasmid pLL373. Next, XhoI restriction sites were introduced in frame at approximately 30-bp intervals along the length of the virB9 gene of pLL373 by oligonucleotide-directed mutagenesis (33). The mutagenic oligonucleotides carried the XhoI restriction site with at least 15 bp of 5′ and 3′ sequence complementary to the regions of virB9 flanking the insertion site. The oligonucleotide sequences are available upon request. Mutant alleles carrying the XhoI site, which specifies codons for the Leu-Glu dipeptide, were identified by restriction enzyme digestion and confirmed by sequencing of the entire virB9 gene. The resulting plasmids were designated pLL9xxx, where xxx denotes the codon immediately preceding the XhoI insertion site. These ColE1 plasmids were introduced into A. tumefaciens by ligation to the broad-host-range plasmid pSW172 (IncP) (14) or a Kanr derivative of pSW172 designated as pXZ151 (54).
Plasmid pKVD46 expressing Ptac-GST-virB9 was constructed as follows. First, an ∼2.0-kb BamHI fragment from pUI1637 (22) containing an nptII gene and additional unique restriction sites including XbaI was introduced into the BamHI site of pGEX-2T (Pharmacia), resulting in plasmid pKVD10. Next, an NdeI site carried on an XbaI-HindIII fragment from pZD1398 (21) was introduced into pKVD10, resulting in plasmid pKVD44. Finally, full-length virB9 was introduced as an NdeI-XhoI fragment from pPC994 (7) into pKVD44 to yield plasmid pKVD46.
TrIP.
The TrIP assay was performed as previously described (13). Briefly, A. tumefaciens cells were treated with formaldehyde and solubilized with detergent. Anti-VirB antibodies were used to precipitate the respective VirB proteins from the detergent-soluble fraction, and the presence of DNA in the immunoprecipitates was assessed by PCR amplification. Primers for amplification of the T-DNA region corresponding to gene 7 of the T-DNA were 5′-GGGCGATTATGGCATCCAGAAAGCC-3′ and 5′-GTCGGCGGCCCACTTGGCACACAG-3′. Primers for the nontransferred ophDC locus (a region of the Ti plasmid ∼25 kb from the transferred T-DNA) were 5′-CCTGCGGATGTCAGGGCTCTCGT-3′ and 5′-TGTCCGTGCTTGCCAATCCCCG-3′. PCR products were separated by electrophoresis through a 1.2% gel and were visualized by ethidium bromide staining.
To compare levels of T-strand by quantitative TrIP (QTrIP), immunoprecipitates were subjected to 20 cycles of PCR amplification using the primers specific for gene 7 of the T-DNA. On the 21st cycle, within the logarithmic phase of the amplification cycle (reference 13 and data not shown) a single round of PCR amplification was performed with the addition of 1.0 μCi of [32P]dGTP (American Biosciences). PCR products were column purified with a Qiaquick PCR purification kit (QIAGEN) to remove unincorporated nucleotides. Aliquots of the eluted material were mixed with 3.5 ml of scintillation liquid (Ecolite ICN) and counted with a Beckman Coulter instrument. QTrIP experiments to quantify levels of the pML122 transfer intermediate were performed as described previously (13) but with primers specific for mobA: 5′-CTCAGTGGTTCAAGCGGTACA-3′ and 5′-TGATAGTTCTTCGGGCTGGTT-3′. TrIP studies monitoring translocation of pML122 through the VirB/D4 T4SS will be described elsewhere (E. Cascales et al., unpublished data). The entire QTrIP protocol was repeated two times in duplicate, and the average value from a single experiment was reported.
Coimmunoprecipitation.
Antibodies to VirB10 were used to assay for a VirB9-VirB10 interaction by coimmunoprecipitation, as previously described (12).
Protein analyses and immunodetection.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using glycine or tricine buffer systems, Western blotting, and immunostaining with anti-Vir antibodies was performed as previously described (32). Proteins were loaded on SDS-polyacrylamide gels on a per cell equivalent basis for comparisons between strains of steady-state levels of T4SS subunits. Anti-VirB9 antibodies were raised by overproduction and purification of a glutathione S-transferase (GST)-VirB9 fusion protein from E. coli BL21(DE3, pKVD46). A 100-ml culture was grown to an optical density at 600 nm (OD600) of 0.3, IPTG (isopropyl-β-d-thiogalactopyranoside; 1 mM final concentration) was added to induce GST-virB9 expression, and cells were incubated with shaking for 4 h at 37°C. GST-VirB9 present in the soluble fraction of cell lysates was purified by passage through a glutathione Sepharose column and eluted according to the manufacturer's instructions (Pharmacia). Purified GST-VirB9 was sent to Cocalico Biologicals, Inc. (Reamstown, Pa.), for injection into New Zealand White rabbits. Anti-VirB9 antibodies were enriched from the polyclonal antiserum by adsorption to purified GST-VirB9 immobilized on nitrocellulose as previously described (3).
Cellular fractionation and T-pilus isolation.
T pili were isolated by growth of A. tumefaciens strains to an OD600 of 0.5 in MG/L (mannitol glutamate-Luria salts) medium at 28°C. Cells were pelleted, diluted fivefold in IM, and incubated for 6 h at 22°C. Next, 200 μl of an AS-induced culture was spread on IM agar plates, and the plates were incubated for 3 days at 18°C. Cells were then gently scraped off the plates in 1 ml of 50 mM KPO4 buffer (pH 5.5) and pelleted by centrifugation at 14,000 × g for 15 min at room temperature. The supernatant was removed, and the cell pellet was resuspended in 1 ml of 50 mM KPO4, pH 5.5. This suspension was passed through a 25-gauge needle 10 times to collect flagella, pili, and surface proteins. The sheared bacterial cells were pelleted by centrifugation at 14,000 × g for 30 min at 4°C. The remaining supernatant was filtered through a 0.22-μm-pore-size cellulose acetate membrane to remove unpelleted cells. T pili were harvested by centrifugation of filtered exocellular material at 100,000 × g for 1 h at 4°C. Pelleted material was analyzed by SDS-PAGE and immunostaining (39).
Conjugation assays.
The RSF1010 derivative pML122 was introduced into various A. tumefaciens donor strains by diparental mating with E. coli strain S17-1(pML122) (39). A. tumefaciens strains carrying pML122 were mated with A348Spcr recipient cells, as previously described (32, 39). Transfer frequencies are reported as the number of transconjugants recovered per donor cell. Experiments were repeated in triplicate, and results are reported for a representative experiment.
Virulence and mixed infection assays.
A. tumefaciens strains were tested for virulence by inoculating wound sites of Kalanchoe daigremontiana leaves. As controls, all leaves were coinoculated with wild-type (WT) strain A348 and the isogenic ΔvirB9 mutant (7). Relative virulence was assessed by inoculation of serially diluted cultures adjusted to the same OD600 on plant wound sites (7). Tumor formation was monitored over a period of 6 weeks and scored on a scale of 0 (−, avirulent) to 4 pluses (+, WT virulence). VirB9.i2-producing strains were also tested for the ability to export VirE2 or T-DNA to plant cells by mixed infection assays (38). Fresh plate cultures of avirulent virB9 mutant strains were mixed with the avirulent virE2 mutant 358mx (45) or T-DNA deletion mutant LBA4404 (38) and the resulting mixture was inoculated on K. daigremontiana wound sites to assay for tumor formation. Each experiment was repeated at least three times for each strain on separate leaves.
RESULTS
VirB9 insertion mutations alter T4SS function.
Twenty-five VirB9 mutant proteins bearing dipeptide insertion mutations (i2) were assayed for effects on substrate transfer and T-pilus biogenesis. Initial studies showed that most of the VirB9.i2 proteins migrated at the same position and accumulated at comparable steady-state levels as native VirB9 in SDS-polyacrylamide gels (Fig. 1A). However, four mutant proteins (96.i2, 226.i2 256.i2, and 276.i2) reproducibly accumulated at aberrantly low levels, and two (196.i2 and 236.i2) could not be detected in whole-cell extracts by immunostaining. Below, we present results suggesting that these mutations might destabilize VirB9 through disruption of partner protein contacts.
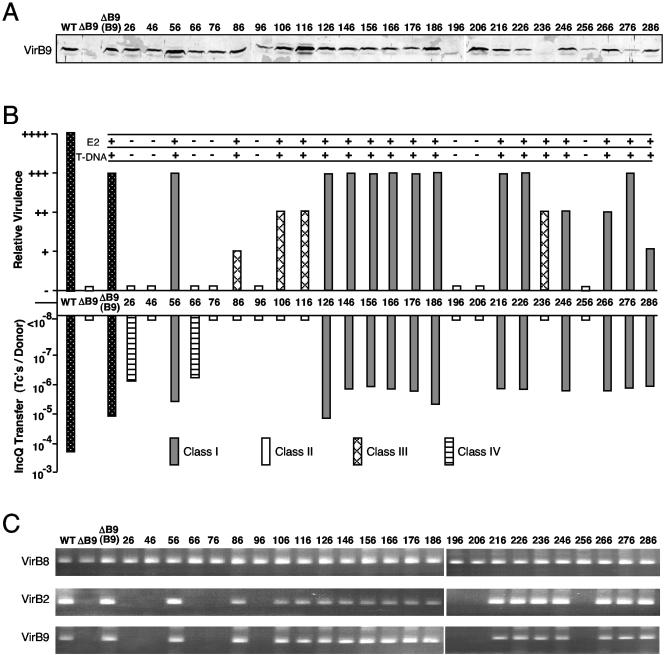
FIG. 1.
Effects of VirB9 insertion mutations on substrate transfer. (A) Total cellular proteins from AS-induced A348 (WT), PC1009 (ΔB9), and PC1009 producing the VirB9.i2 mutant proteins (identified by residue at i2 insertion site) were analyzed by SDS-PAGE and development of immunoblots with antibodies specific for VirB9. Protein samples were loaded on a per cell equivalent basis.
(B) Effects of i2 mutations on virulence and pML122 and VirE2 substrate transfer. Inoculated K. daigremontiana leaves were scored for tumor formation on a scale of no tumors (−) to tumors incited by WT A348 (++++). PC1009(pLL9xx, pML122) donors were mated with A348Spcr recipients, and transfer efficiencies are reported as the number of transconjugants per donor cell for a representative experiment. VirE2 and T-DNA transfer were monitored by coinoculation of PC1009(pLL9xxx) strains, respectively, with the virE2 mutant strain Mx358 or the T-DNA deletion strain LBA4404. Pluses denote VirE2 or T-DNA transfer. Dotted bars indicate WT A348 and PC1009(pLL373) producing native VirB9. PC1009(pLL9xxx) producing VirB9.i2 mutants are indicated as follows: gray bars, class I (Tra+); white bars, class II (Tra−); hatched bars, class III (T-DNA+ IncQ−); striped bars, class IV (T-DNA− IncQ+). (C) T-DNA substrate interactions with VirB8, VirB2, and VirB9 channel subunits assessed by TrIP. The T-DNA substrate was detected in material precipitated with antibodies to the VirB proteins listed at left by PCR amplification and agarose gel electrophoresis. PC1009 strains producing native VirB9 and the class I (Tra+) and III (T-DNA+ IncQ−) mutants displayed WT patterns of substrate contacts; PC1009 producing class II (Tra−) and IV (T-DNA− IncQ+) mutants showed no substrate contacts with VirB2 or the VirB9.i2 mutants.
Strain PC1009 (ΔvirB9) is avirulent, and WT virB9 expression from an IncP replicon restores virulence on K. daigremontiana leaves, albeit not completely to WT levels (Fig. 1B). Alleles for 17 of the 25 VirB9.i2 derivatives also complemented the ΔvirB9 mutation as shown by restoration of virulence, establishing that the corresponding i2 mutations did not disrupt assembly of a functional secretion channel (Fig. 1B). The VirB/D4 T4SS separately translocates the T-DNA and protein substrates (8, 46, 49), which can be demonstrated by mixed infection assays with strains deleted of the T-DNA or a gene encoding an exported effector protein, e.g., the VirE2 single-stranded DNA-binding protein that binds the T-DNA in the plant cell and facilitates its movement to the plant nuclear pore complex (16, 25). The eight VirB9.i2-producing strains that failed to incite tumor formation also failed to export VirE2, as shown by mixed infections with strain Mx358 (virE2 mutant), or T-DNA, as shown by mixed infections with LBA4404 (ΔT-DNA). These eight VirB9.i2 mutant proteins thus block intercellular translocation of both T-DNA and the VirE2 substrates (Fig. 1B).
In mating experiments, most of the VirB9.i2-producing strains also mobilized the transfer of the IncQ plasmid pML122 to A. tumefaciens recipient cells (Fig. 1B). Interestingly, however, four strains competent for T-DNA transfer (86.i2, 106.i2, 116.i2, and 236.i2) failed to translocate pML122 at detectable levels, whereas two strains defective for T-DNA transfer (26.i2 and 66.i2) fairly efficiently translocated the IncQ plasmid. We thus assigned the mutations to four classes according to their effects on intercellular substrate transfer. Class I mutations are permissive for transfer of the tested DNA and protein substrates (Tra+), and class II mutations completely inhibit substrate transfer (Tra−). The class III and IV mutations confer substrate-selective transfer such that the class III mutations translocate only the T-DNA and VirE2 to plant cells. Conversely, the class IV mutations translocate only the IncQ plasmid to agrobacterial recipients. As shown in Fig. 1B, most class I permissive mutations are located in the central and C-terminal regions of VirB9. The class III and IV substrate-discriminating mutations are located almost exclusively in the first third of the protein. The class II Tra− mutations are distributed throughout the protein, but the results of computer analyses showed that most of these mutations reside in clusters of conserved residues among VirB9 family members (see Discussion).
VirB9 i2 mutations arrest T-DNA translocation through the VirB/D4 T4SS.
In WT strain A348, the T-DNA transfer intermediate forms a series of sequentially and spatially ordered close contacts with the inner membrane channel subunits VirD4, VirB11, VirB6, and VirB8 and then with the periplasmic and outer-membrane-associated subunits VirB2 and VirB9, as shown by TrIP (13). As expected, strains producing the class I (Tra+) and III (T-DNA Tra+, IncQ Tra−) mutant proteins displayed a WT pattern of T-DNA contacts with all six of these channel subunits (Fig. 1C and data not shown). By contrast, strains producing the class II (Tra−) and IV (T-DNA Tra−, IncQ Tra+) mutant proteins displayed a transfer arrest such that the T-DNA substrate formed WT contacts with the four inner membrane channel subunits, as represented with VirB8, but no detectable contacts with VirB2 or the VirB9.i2 derivatives (Fig. 1C). The class II and IV mutations thus appear to block progression of the T-DNA substrate beyond the inner membrane portion of the secretion channel.
An N-terminal region confers substrate discrimination.
TrIP studies have supplied evidence that the translocation pathway for the IncQ plasmid pML122 is identical to that of the T-DNA substrate (E. Cascales et al., unpublished data). Here, we quantitated the effects of the i2 mutations on T-DNA and pML122 substrate translocation by use of the highly sensitive QTrIP assay (13), with a particular interest in the substrate-discriminating mutations (Fig. 2).
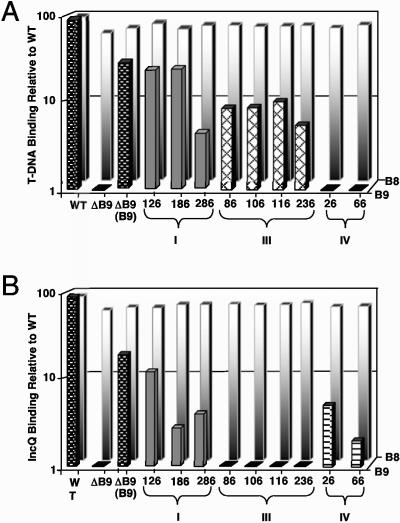
FIG. 2.
Effects of i2 mutations on the efficiency of T-DNA and pML122 translocation through the VirB/D4 T4SS. (A) T-DNA substrate precipitated with antibodies to VirB8 (background white-shaded bars) and VirB9 (foreground bars) from cell extracts of strain A348 (WT), PC1009 (ΔB9), and PC1009 strains producing native VirB9 (B9) or the VirB9.i2 mutants. (B) pML122 substrate precipitated with antibodies to VirB8 (background white-shaded bars) and VirB9 (foreground bars) from cell extracts of strains depicted in panel A engineered to carry pML122. For both panels, levels of the precipitated DNA substrates are presented as counts per minute of incorporated radionucleotide during one cycle in the logarithmic phase of PCR amplification. Values for the mutant strains are represented as percentages of the values for the WT strain A348 in a log scale. Standard deviations between experiments were <1 to 2% for a given immunoprecipitation.
PC1009 (ΔvirB9) synthesizing native VirB9 or several tested class I (Tra+) VirB9.i2 mutants (126.i2, 186.i2, and 286.i2) from an IncP replicon formed WT patterns of T-DNA and pML122 substrate contacts with the VirD4, VirB11, VirB6, VirB8, VirB2, and VirB9.i2 subunits. Quantitatively, however, these strains displayed WT levels of substrate binding to VirB8 but appreciable reductions in levels of substrate binding to the VirB9.i2 derivatives (Fig. 2A and B). The levels of T-DNA binding to these channel subunits ranged from 10 to 40% of WT values, whereas the levels of pML122 substrate binding ranged from 5 to 15% of WT values. These levels of substrate binding, however, were well above background levels and very reproducible between experiments. Moreover, for a given strain, the levels of substrate binding to the VirB9 mutants detected by QTrIP correlated with the capacity to incite plant tumor formation and conjugally transfer plasmid pML122 to agrobacterial recipients (compare Fig. 1B and 2).
Strains producing the class III (T-DNA Tra+, IncQ Tra−) and IV (T-DNA Tra−, IncQ Tra+) mutant proteins also formed WT patterns of T-DNA and pML122 substrate contacts with the inner membrane channel subunits, as represented with VirB8 (Fig. 2A and B). Very strikingly, however, the class III mutations (86.i2, 106.i2, and 236.i2) permitted formation of T-DNA substrate contacts with the VirB9.i2 derivatives but completely blocked formation of the corresponding pML122 substrate contacts (Fig. 2A and B). Conversely, the class IV mutations (26.i2 and 66.i2) permitted formation of pML122 substrate contacts, albeit at low levels, with the VirB9.i2 derivatives but completely blocked formation of the corresponding T-DNA substrate contacts. As with the class I mutations, the levels of substrate binding to the class III and IV VirB9.i2 derivatives correlated with the virulence and conjugation data (compare Fig. 1B and 2). Taken together, results of the TrIP and QTrIP studies strongly indicate that the class III and IV mutations selectively block passage of the IncQ plasmid and T-DNA substrates, respectively, through the distal portion of the secretion channel composed of VirB2 and VirB9 (see Discussion).
VirB9 partner interactions with VirB7 and VirB10.
VirB9 forms an intermolecular disulfide bridge via reactive Cys residue 262 with lipoprotein VirB7 (1, 4, 5, 24, 44). Assembly of the heterodimer strongly stabilizes VirB9, as shown by instability of native VirB9 in a ΔvirB7 mutant (24) and of VirB9Cys262Ser in a ΔvirB9 mutant producing native VirB7 (44). The WT strain A348, PC1009(pLL373) producing native VirB9, and most PC1009(pLL9xxx) strains producing the VirB9.i2 mutants accumulated the VirB9 species almost exclusively as disulfide-cross-linked VirB7-VirB9 heterodimers, as shown by electrophoresis of cell extracts through SDS-polyacrylamide gels under nonreducing conditions (Fig. 3A).
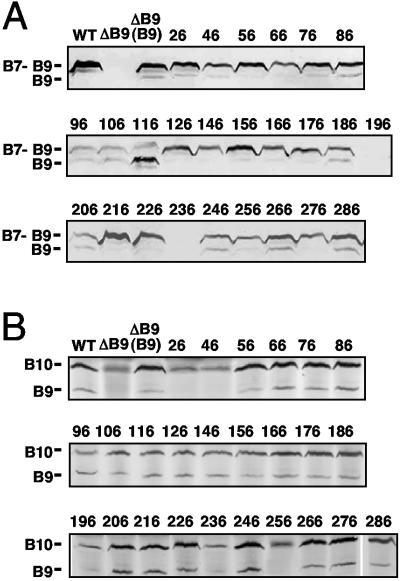
FIG. 3.
Effects of i2 mutations on VirB9 partner interactions with VirB7 and VirB10. (A) Detection of disulfide-cross-linked VirB7-VirB9 dimers (∼36-kDa) and VirB9 monomers (29-kDa) in total cell extracts by nonreducing SDS-PAGE and immunoblot analysis with anti-VirB9 antibodies. (B) Detection of the putative VirB9-VirB10 complexes by immunoprecipitation with anti-VirB10 antibodies. Immunoprecipitates were analyzed for VirB9 and VirB10 by SDS-PAGE and development of immunoblots with antibodies to both proteins. WT, strain A348; ΔB9, strains PC1009 and PC1009 producing native VirB9 (B9) and the VirB9.i2 mutants (i2 insertion sites shown).
However, several VirB9.i2-producing strains inefficiently formed the heterodimers, as shown by low levels of the 36-kDa cross-linked species and a corresponding increase in amounts of the 29-kDa VirB9 monomer species (Fig. 3A). Interestingly, the mutations affecting dimer formation were juxtaposed to those shown in Fig. 1A (96.i2, 196.i2, and 236.i2) to exert strong destabilizing effects on VirB9. The mutations affecting dimer formation included 96.i2 and flanking mutations (86.i2, 106.i2, and 116.i2), mutations flanking residue 196 (186.i2 and 206.i2), and mutations located distal to residue 236 (246.i2, 266.i2, and 286.i2). The 96.i2 mutant protein accumulated at low but detectable levels, whereas the 196.i2 and 236.i2 mutant proteins were undetectable in whole-cell extracts, reminiscent of the instability of native VirB9 in a ΔvirB7 mutant (24). These three i2 mutations thus exert correlative effects on disulfide cross-linking, VirB9 instability, and substrate translocation (Fig. 1A). Several mutations near these residues that affected dimer formation also exerted corresponding but less severe effects on T4SS function (Fig. 1B). While we had anticipated that mutations near Cys262 would disrupt disulfide bridge formation, clearly mutations elsewhere in the protein also directly or indirectly affect the VirB7-VirB9 contact. Interestingly, residues 96, 196, and 236 are located in regions of sequence conservation among VirB9 family members, underscoring the general importance of these motifs for VirB9 function (see Discussion).
The VirB7-VirB9 heterodimer interacts with the bitopic inner membrane subunit VirB10 (5, 12, 18, 50), as can be shown by immunoprecipitation with anti-VirB10 antibodies in the absence of added reductant (12). If reducing agent is added to extracts prior to immunoprecipitation, the VirB7-VirB9 disulfide bridge is broken, and the anti-VirB10 antibodies coprecipitate a stable VirB9-VirB10 complex, demonstrating that the VirB7-VirB9-VirB10 complex forms via a VirB9-VirB10 binary interaction (Fig. 3B) (12). Formation of the VirB7-VirB9 dimer contributes to stabilization of VirB10, as evidenced by a low abundance of VirB10 in PC1007 (ΔvirB7) and PC1009 (ΔvirB9) mutants (5, 7, 24). Correspondingly, anti-VirB10 antibodies immunoprecipitated a low level of VirB10 from PC1009 extracts (Fig. 2B). Formation of the VirB9-VirB10 complex is essential for T-DNA substrate translocation through the distal portion of the secretion channel (12, 13) (see Discussion). As expected, therefore, the anti-VirB10 antibodies coprecipitated presumptive VirB10-VirB9.i2 complexes in all strains competent for translocating the T-DNA (Fig. 1B and 3B). Although difficult to detect in the blot shown in Fig. 3B, the anti-VirB10 antibodies even coprecipitated a presumptive VirB10-VirB9.i2-236 complex despite the instability of this class III mutant protein. Similarly, the anti-VirB10 antibodies coprecipitated detectable levels of VirB10 and several class II Tra− mutants, including the 76.i2 mutant protein and the low-abundance 96.i2 and 196.i2 mutant proteins (Fig. 3B).
In striking contrast, the anti-VirB10 antibodies coprecipitated low levels of VirB10 but no detectable amounts of the 26.i2, 46.i2, and 256.i2 mutant proteins, which is suggestive of a lack of interactions. The 256.i2 mutation is near the reactive Cys262 residue and might disrupt the VirB9-VirB10 interaction indirectly through unidentified effects on assembly of the VirB7-VirB9 dimers or higher-order multimers. By contrast, results of yeast two-hybrid (18) and substitution mutagenesis studies (5) have led to proposals that the N-terminal region of VirB9 mediates complex formation with VirB10. Hence, the 26.i2 and 66.i2 mutations might exert their effects directly through disruption of this intermolecular contact. Of final interest, the class III 26.i2 mutation selectively blocks T-DNA but not IncQ plasmid transfer (Fig. 1B). An inability of the 26.i2 mutant protein to interact productively with VirB10 contact might be related to this substrate discrimination phenotype (see Discussion).
N- and C-terminal insertion mutations disrupt T-pilus assembly.
Certain mutations in the VirB11 ATPase (39) and the polytopic membrane component VirB6 (32) block T-pilus production without preventing intercellular substrate transfer. These mutations are postulated to “uncouple” two parallel biochemical pathways mediating assembly of the T pilus and the secretion channel (32, 39). As expected, most class I VirB9.i2-producing strains (Tra+) possessed abundant levels of exocellular VirB2 pilin, which is suggestive of T-pilus assembly (Fig. 4A). In contrast, the sheared fraction from the 286.i2-producing strain did not possess detectable levels of VirB2, even when concentrated 100-fold or subjected to ultracentrifugation at 100,000 × g to recover the high-molecular-weight T pilus (Fig. 4). This mutation therefore blocks pilus production (Pil−), without affecting substrate transfer (Tra+) (Fig. 1B and 2A and B). Similarly, we were unable to detect VirB2 in the exocellular fractions of strains producing two class III mutations, 116.i2 and 236.i2 (Fig. 4). These mutations also block pilus production but not a functional secretion channel (Fig. 1B and 3). We discuss below how the 286.i2, 116.i2, and 235.i2 “uncoupling” mutations might exert their effects.
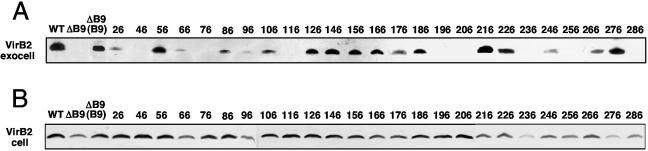
FIG. 4.
Effects of VirB9.i2 mutations on exocellular and cellular forms of VirB2 pilin. (A) Exocellular VirB2 as a marker for the T pilus detected in material recovered by mechanical shearing of equivalent numbers of AS-induced cells. (B) Cellular form of VirB2 in the corresponding shear-resistant fraction. Protein samples were subjected to SDS-PAGE and development of immunoblots with anti-VirB2 antibodies. WT, strain A348; ΔB9, strains PC1009 and PC1009 producing native VirB9 (B9) and the VirB9.i2 mutant proteins.
Finally, several PC1009(pL9xxx) strains, mainly those producing VirB9 derivatives with i2 mutations in the C-terminal region, accumulated low levels of the cellular form of VirB2 pilin (Fig. 4). This form of pilin is defined as that which remains cell associated following removal of surface proteins, pili, and other organelles from the cell surface by extensive mechanical shearing. Taken together with evidence from the TrIP studies that VirB2 and VirB9 functionally interact (13), these findings suggest that the C-terminal region of VirB9 might mediate a stabilizing interaction with the cellular form of VirB2 pilin.
Requirement for cellular VirB2, not the T pilus, for substrate transfer.
The isolation of VirB9 “uncoupling” mutations adds to an accumulating body of evidence that the WT T pilus is not essential for conjugal DNA transfer (32, 39). We sought to determine whether the “uncoupling” mutations also bypass the requirement for synthesis of the cellular form of VirB2. We constructed two double mutant strains, PC1209 (ΔvirB2ΔvirB9) and PC1211 (ΔvirB2ΔvirB11), and then introduced alleles for the “uncoupling” mutants of VirB9 (116.i2, 236.i2, and 286.i2) or VirB11 (I265T) (39). PC1009 strains producing the 116.i2, 236.i2, and 286.i2 mutant proteins are Tra+ Pil−, as is PC1011 (ΔvirB11) producing the VirB11.I265T mutant (Table 1; Fig. 2 and 4) (39). In striking contrast, the isogenic double mutant strains PC1209 and PC1211 producing the “uncoupling” mutant proteins but no VirB2 remained avirulent (Table 1). Therefore, while the “uncoupling” mutant proteins support substrate transfer in the absence of WT T-pilus production, they do not bypass the requirement for cellular VirB2. These genetic data complement the TrIP findings (13) indicating that the cellular form of VirB2 pilin functions together with VirB9 to mediate substrate passage through the periplasm to the cell surface.
TABLE 1.
Uncoupling mutations do not bypass VirB2 pilin requirement for DNA substrate translocation
| Host strain | Complementing construct | Virulence |
|---|---|---|
| A348 (WT) | +++ | |
| ΔvirB9 | − | |
| ΔvirB9 | PvirB-virB9 | ++ |
| ΔvirB11 | PvirB-virB11 | ++ |
| ΔvirB9 | PvirB-virB9.i2-116 | ++ |
| ΔvirB9 | PvirB-virB9.i2-236 | ++ |
| ΔvirB9 | PvirB-virB9.i2-286 | ++ |
| ΔvirB11 | PvirB-virB11-I265T | ++ |
| ΔvirB2ΔvirB9 | PvirB-virB2, PvirB-virB9 | ++ |
| ΔvirB2ΔvirB11 | PvirB-virB2, PvirB-virB11 | ++ |
| ΔvirB2ΔvirB9 | PvirB-virB9.i2-116 | − |
| ΔvirB2ΔvirB9 | PvirB-virB9.i2-236 | − |
| ΔvirB2ΔvirB9 | PvirB-virB9.i2-286 | − |
| ΔvirB2ΔvirB11 | PvirB-virB11-I265T | − |
DISCUSSION
We initiated a structure-function analysis of VirB9, an outer-membrane-associated component of the A. tumefaciens VirB/D4 secretory apparatus, by characterizing effects of two-residue insertion mutations on machine function. Results of these mutational studies, coupled with computer-based analyses, suggest that VirB9 is composed of three functional domains, as depicted in Fig. 5. Domains corresponding roughly to the N- and C-terminal thirds of VirB9 are highly conserved among family members and contribute to establishment of intersubunit contacts, selection and trafficking of DNA substrates, protein stability, and biogenesis of the T pilus. A central, nonconserved region tolerates i2 mutations and might adopt a structure required for substrate passage across the outer membrane.
FIG. 5.
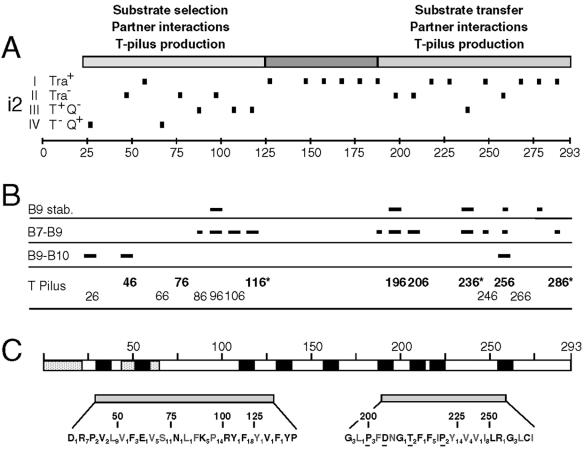
VirB9 domains identified by mutational and computer analyses. (A) VirB9 domains identified by i2 mutagenesis are depicted in the schematic at the top, with residue numbers identifying the approximate boundaries. The i2 mutations with positions indicated with short vertical dashes are classified according to the effects (in parentheses) on substrate translocation by host strains as follows: I (Tra+), no effect on substrate transfer; II (Tra−), transfer defective; III (T+ Q−), T-DNA Tra+, pML122 (IncQ) Tra−; IV (T− Q+), T-DNA Tra−, pML122 (IncQ) Tra+. Scale in 25-residue intervals. (B) Effects of i2 mutations on VirB9 stability (B9 stab.) and assembly of the disulfide-cross-linked VirB7-VirB9 dimers (B7-B9), the VirB9-VirB10 complex (B9-B10), and the T pilus. Short and long dashes, respectively, correspond to modest and severe deviations from WT patterns. For the T pilus, the sites of i2 mutations that completely abolished (bold typeface) or significantly diminished (normal typeface) T pilus production are identified. Residues marked with an asterisk correspond to sites of the “uncoupling” (Tra+ Pil−) mutations. No marks or residue numbers indicate a WT pattern. (C) Computer predictions of features of interest, domain structures, and sequence conservation of VirB9 family members. Schematic shows the locations of two stretches of hydrophobicity (speckled bars), the first corresponding to the signal sequence and nine possible β-barrel transmembrane segments (black bars). Two conserved domains, corresponding to the N- and C-terminal domains identified by the i2 mutational studies, are separated by a nonconserved central region. Conserved residues are distributed throughout the N- and C-terminal domains among VirB9-related proteins: bold residues, conserved among over 70 VirB9 family members; gray residues, additionally conserved residues among 17 closely related VirB9 proteins identified mainly in α-proteobacterial species. Glu631 and Gly206 residues are invariant among all VirB9 homologs; underlined residues are invariant among the 17 close homologs; subscripts represent the numbers of intervening nonconserved residues.
N-terminal substrate-specifying domain.
The N-terminal domain is postulated to extend from the signal sequence cleavage site at residue 21 through residue 116. Complementing our experimental support (Fig. 5B) for this domain assignment, a ProDom (http://www.toulouse.inra.fr/prodom.html) (17) analysis identifies this as a conserved region among over 70 VirB9-like proteins (Fig. 5C). This region possesses numerous conserved residues and an invariant Glu corresponding to residue 63 of A. tumefaciens VirB9. As expected, the number of sequence similarities and identities increase when 17 closely related VirB9 proteins identified mainly in α-proteobacterial species (Fig. 5C) are analyzed by Clustal W multiple alignment (http://npsa-pbil.ibcp.fr/cgi-bin/align_clustalw.pl) (47). The complete sequence alignments of VirB9-related proteins in the database are available through our laboratory website (http://mmg.uth.tmc.edu.webpages/faculty/pchristie/html). The N-terminal region is generally hydrophilic with the exception of a stretch of hydrophobicity between residues 44 to 64. The single class I Tra+ mutation (56.i2) mapped within this hydrophobic stretch, whereas all other N-terminal mutations disrupted VirB9 function in some way.
Remarkably, several mutations in the N-terminal domain selectively blocked translocation of the IncQ plasmid (class III) or T-DNA (class IV) substrates. These mutations did not affect early transfer reactions leading to formation of substrate contacts with the inner membrane channel subunits VirD4, VirB11, VirB6, and VirB8, whereas they prevented formation of substrate contacts with the periplasmic and outer-membrane-associated channel subunits VirB2 and VirB9 (Fig. 1C and 2). The capacity of VirB9 mutants to mediate recognition and selective transfer of two DNA substrates through the distal portion of the secretion channel is of considerable interest because, until now, biochemical and genetic evidence suggested that the T4CP, e.g., VirD4, fulfills the task of substrate recognition and recruitment to the secretion channel (3, 13, 20, 26, 37). Our findings indicate that VirB9, through its N terminus, contributes a second substrate selection mechanism, acting as a checkpoint to regulate substrate passage through the periplasm or the outer membrane.
How might the N-terminal domain of VirB9 confer substrate discrimination? The VirB/D4 T4SS translocates T-DNA as a nucleoprotein particle composed of the VirD2 relaxase covalently bound to a single strand (T-strand) of T-DNA (41). This T4SS also translocates the IncQ plasmid as a MobA relaxase-R-strand intermediate (46). In view of a demonstrated 5′ to 3′ directional movement of the T-strand during conjugation, the relaxase is thought to “pilot” the T-strand through the secretion channel (15, 16). VirB9 thus might directly interact with secretion signals carried by relaxase proteins. Accordingly, the class II mutations might abolish relaxase signal contacts altogether, whereas the class III and IV mutations could disrupt recognition of novel motifs carried by MobA or VirD2, respectively.
Alternatively, VirB9 might mediate substrate recognition indirectly through its partner subunit interactions. For example, the N-terminal region of VirB9 probably interacts directly with VirB10, as indicated by our coimmunoprecipitation data (Fig. 4) and results of a substitution mutagenesis study (5). In that study, Gly52Glu and, to a slightly lesser extent, Val68Iso and Ala78Thr substitution mutations disrupted intercellular DNA transfer as well as the VirB9-dependent assembly of higher-order VirB10 complexes detectable by chemical cross-linking (5). A complementary yeast two-hybrid screen also detected an interaction between a VirB9 N-terminal fragment (residues 17 to 122) and VirB10 (18). Recently, we supplied evidence that, in A. tumefaciens, the VirB9-VirB10 interaction is dynamic and driven by a conformational change in VirB10 that occurs upon sensing of ATP utilization by the VirD4 and VirB11 energetic components (12). Moreover, VirB9-VirB10 complex formation is required for movement of the T-DNA substrate through the distal portion of the secretion channel (12, 13). The N-terminal region of VirB9 therefore might regulate substrate passage through contacts with the VirB10 energy sensor subunit. Accordingly, the VirB9 substrate-discriminating mutations might exert their effects through disruption of the VirB9-VirB10 contact. It is interesting that the 26.i2 mutation abolished detectable VirB9-VirB10 complex formation and T-DNA substrate transfer but not IncQ plasmid transfer (Fig. 1B). Further studies of this mutation might unveil differences in structural or mechanistic requirements for passage of T-DNA versus IncQ plasmid substrates through the distal portion of the secretion channel.
Finally, it is interesting that the class III and IV mutations selectively blocked substrate transfer to agrobacterial and plant recipient cells, respectively. Though these mutations likely exert their effects at the periplasmic side of the inner membrane as suggested by the TrIP data (Fig. 1C and 2), it is possible that the capacity of VirB9 to regulate substrate transfer is subject to control by secretion channel surface interactions with recipient cell receptors. In the E. coli F plasmid transfer system, early genetic studies provided evidence for recipient contact-dependent triggering of donor conjugal transfer, yet the mechanism by which the recipient cell communicates its presence to the donor cell remains unknown (36). In A. tumefaciens, the importance of recipient-specific signals for channel gating is illustrated by the fact that, to date, interbacterial transmission of T-DNA-based substrates has not been shown, even though we have demonstrated that this substrate forms close contacts with all six channel subunits in AS-induced cultures (13).
Central structural and C-terminal stabilization domains.
The central domain, proposed to span residues ∼126 to 186, tolerated all i2 mutations (Fig. 5). With the exception of a cluster of related residues between 132 to 139 (Fig. 5C), this region is not conserved among the VirB9-related proteins. Of possible significance, the central domain possesses several potential β-strands according to algorithms of Schirmer and Cowan (42; S. Buchanan, personal communication) and Zhai and Saier (53). These findings led Cao and Saier (10) to suggest that this region of VirB9 spans the outer membrane. At this point, this hypothesis remains untested, but it is interesting that none of the i2 mutations throughout this region affected VirB9 function (Fig. 5A and B). Studies have shown that membrane β-barrel proteins often tolerate small insertion mutations, particularly if the mutations map in extramembranous loops. The Bordetella pertussis Fha transporter retains the capacity to secrete hemagglutinin when bearing a two-residue insertion at 35 of 48 positions (28). Most derivatives of the E. coli BtuB transporter bearing six-residue insertions also retain the capacity to transport at least a subset of its substrates (27). Outer membrane secretins XcpQ and XpsD tolerate small insertions of less than five residues at multiple positions (9), whereas larger insertions tend to inactivate secretins (29, 30). Most of the i2 mutations in the central region of VirB9 also reside in predicted extramembranous loops (Fig. 5A and C). Presently, we are testing a model that the central domain of VirB9 forms part of the outer membrane channel for this T4SS.
The C-terminal region of VirB9 possesses several distinctive features. The ProDom and Clustal W alignment algorithms also identify this as a conserved region among VirB9-related proteins (Fig. 5 C) (see our laboratory website at http://mmg.uth.tmc.edu.webpages/faculty/pchristie/html). The computer analyses identify several potential β-strand transmembrane segments suggesting this region might also associate with the outer membrane. As with the N-terminal mutations, insertions mapping within or adjacent to highly conserved sequence motifs (196.i2, 206.i2, and 256.i2) abolished protein function, demonstrating the importance of these conserved motifs (Fig. 5C). Mutations in these and flanking sites also correlated with diminished steady-state abundance of the VirB9 mutant (Fig. 1A) as well as the cellular form of VirB2 pilin, suggesting that the C terminus of VirB9 might adopt a structure or critical partner contacts for stabilization of both subunits. Indeed, several C-terminal mutations disrupted complex formation with the lipoprotein VirB7 and one (256.i2) disrupted complex formation with VirB10. Effects of the C-terminal mutations on partner contacts thus might account for the observed instabilities of VirB9, VirB2, and VirB10.
Contribution of cell-associated VirB2 pilin to substrate translocation.
A central question surrounding type IV secretion is the role of the T pilus and the pilin protein in translocation. The isolation of “uncoupling” mutations in this (Fig. 5C) and previous studies (32, 40) suggests minimally that a WT T pilus is dispensable for intercellular substrate transfer. Here, we further determined that the “uncoupling” mutations do not obviate the requirement for the cellular form of VirB2 pilin for transfer (Table 1). These findings, the correlative effects of C-terminal mutations on VirB9 and VirB2 stabilities (Fig. 1A and 4), and results of the TrIP studies of T4SS mutants (Fig. 1B and 2) (13) together strongly argue that the cellular form of VirB2 functionally and probably physically interacts with VirB9 to deliver substrates to the cell surface. Intriguingly, no direct interactions between VirB2 and VirB9 have been reported, although VirB9 and VirB7 form a precipitable complex with inner membrane VirB6 (32), and there is also evidence for an interaction between VirB7 and the exocellular form of VirB2 (40). Deciphering the complexity of the channel subunit interactions in the periplasm and the overall architecture of this portion of the secretion channel remains a fascinating area for future study.
Acknowledgments
We thank Lawrence Lee, Vitaliya Sagulenko, and Johnny Fernandez for strain constructions. We thank members of the lab for valuable discussions and critiques of the manuscript.
This work was supported by NIH grant GM48746.
REFERENCES
- 1.Anderson, L. B., A. V. Hertzel, and A. Das. 1996. Agrobacterium tumefaciens VirB7 and VirB9 form a disulfide-linked protein complex. Proc. Natl. Acad. Sci. USA 93:8889-8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atmakuri, K., E. Cascales, and P. J. Christie. 2004. Energetic components VirD4, VirB11, and VirD4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol. Microbiol. 54:1199-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atmakuri, K., Z. Ding, and P. J. Christie. 2003. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Mol. Microbiol. 49:1699-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baron, C., Y. R. Thorstenson, and P. C. Zambryski. 1997. The lipoprotein VirB7 interacts with VirB9 in the membranes of Agrobacterium tumefaciens. J. Bacteriol. 179:1211-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaupré, C. E., J. Bohne, E. M. Dale, and A. N. Binns. 1997. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J. Bacteriol. 179:78-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beijersbergen, A., S. J. Smith, and P. J. Hooykaas. 1994. Localization and topology of VirB proteins of Agrobacterium tumefaciens. Plasmid 32:212-218. [DOI] [PubMed] [Google Scholar]
- 7.Berger, B. R., and P. J. Christie. 1994. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 176:3646-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binns, A., C. Beaupré, and E. Dale. 1995. Inhibition of VirB-mediated transfer of diverse substrates from Agrobacterium tumefaciens by the IncQ plasmid RSF1010. J. Bacteriol. 177:4890-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bitter, W., M. Koster, M. Latijnhouwers, H. de Cock, and J. Tommassen. 1998. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol. Microbiol. 27:209-219. [DOI] [PubMed] [Google Scholar]
- 10.Cao, T. B., and M. H. Saier, Jr. 2001. Conjugal type IV macromolecular transfer systems of Gram-negative bacteria: organismal distribution, structural constraints and evolutionary conclusions. Microbiology 147:3201-3214. [DOI] [PubMed] [Google Scholar]
- 11.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cascales, E., and P. J. Christie. 2004. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc. Natl. Acad. Sci. USA 101:17228-17233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cascales, E., and P. J. Christie. 2004. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304:1170-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, C. Y., and S. C. Winans. 1991. Controlled expression of the transcriptional activator gene virG in Agrobacterium tumefaciens by using the Escherichia coli lac promoter. J. Bacteriol. 173:1139-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christie, P. J. 2004. Bacterial type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems. Biochem. Biophys. Acta 1694:219-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Citovsky, V., and P. Zambryski. 1993. Transport of nucleic acids through membrane channels: Snaking through small holes. Annu. Rev. Microbiol. 47:167-197. [DOI] [PubMed] [Google Scholar]
- 17.Corpet, F., F. Servant, J. Gouzy, and D. Kahn. 2000. ProDom and ProDom-GC: tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res. 28:267-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das, A., and Y.-H. Xie. 2000. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J. Bacteriol. 182:758-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das, A., and Y. H. Xie. 1998. Construction of transposon Tn3phoA: its application in defining the membrane topology of the Agrobacterium tumefaciens DNA transfer proteins. Mol. Microbiol. 27:405-414. [DOI] [PubMed] [Google Scholar]
- 20.Ding, Z., K. Atmakuri, and P. J. Christie. 2003. The outs and ins of bacterial type IV secretion substrates. Trends Microbiol. 11:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding, Z., Z. Zhao, S. J. Jakubowski, A. Krishnamohan, W. Margolin, and P. J. Christie. 2002. A novel cytology-based, two-hybrid screen for bacteria applied to protein-protein interaction studies of a type IV secretion system. J. Bacteriol. 184:5572-5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eraso, J. M., and S. Kaplan. 1996. Complex regulatory activities associated with the histidine kinase PrrB in expression of photosynthesis genes in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 178:7037-7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez, D., T. A. T. Dang, G. M. Spudich, X.-R. Zhou, B. R. Berger, and P. J. Christie. 1996. The Agrobacterium tumefaciens virB7 gene product, a proposed component of the T-complex transport apparatus, is a membrane-associated lipoprotein exposed at the periplasmic surface. J. Bacteriol. 178:3156-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez, D., G. M. Spudich, X.-R. Zhou, and P. J. Christie. 1996. The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J. Bacteriol. 178:3168-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gelvin, S. B. 2003. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 67:16-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomis-Ruth, F. X., M. Sola, F. de la Cruz, and M. Coll. 2004. Coupling factors in macromolecular type-IV secretion machineries. Curr. Pharm. Des. 10:1551-1565. [DOI] [PubMed] [Google Scholar]
- 27.Gudmundsdottir, A., C. Bradbeer, and R. J. Kadner. 1988. Altered binding and transport of vitamin B12 resulting from insertion mutations in the Escherichia coli btuB gene. J. Biol. Chem. 263:14224-14230. [PubMed] [Google Scholar]
- 28.Guedin, S., E. Willery, J. Tommassen, E. Fort, H. Drobecq, C. Locht, and F. Jacob-Dubuisson. 2000. Novel topological features of FhaC, the outer membrane transporter involved in the secretion of the Bordetella pertussis filamentous hemagglutinin. J. Biol. Chem. 275:30202-30210. [DOI] [PubMed] [Google Scholar]
- 29.Guilvout, I., K. R. Hardie, N. Sauvonnet, and A. P. Pugsley. 1999. Genetic dissection of the outer membrane secretin PulD: are there distinct domains for multimerization and secretion specificity? J. Bacteriol. 181:7212-7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu, N. T., M. N. Hung, D. C. Chen, and R. T. Tsai. 1998. Insertion mutagenesis of XpsD, an outer-membrane protein involved in extracellular protein secretion in Xanthomonas campestris pv.campestris. Microbiology 144:1479-1486. [DOI] [PubMed] [Google Scholar]
- 31.Jakubowski, S. J., V. Krishnamoorthy, E. Cascales, and P. J. Christie. 2004. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion system. J. Mol. Biol. 341:961-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jakubowski, S. J., V. Krishnamoorthy, and P. J. Christie. 2003. Agrobacterium tumefaciens VirB6 protein participates in formation of VirB7 and VirB9 complexes required for type IV secretion. J. Bacteriol. 185:2867-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunkel, T. A., K. Bebenek, and J. McClary. 1991. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 204:125-139. [DOI] [PubMed] [Google Scholar]
- 34.Lai, E. M., and C. I. Kado. 1998. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J. Bacteriol. 180:2711-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai, E. M., and C. I. Kado. 2000. The T-pilus of Agrobacterium tumefaciens. Trends Microbiol. 8:361-369. [DOI] [PubMed] [Google Scholar]
- 36.Lawley, T. D., W. A. Klimke, M. J. Gubbins, and L. S. Frost. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224:1-15. [DOI] [PubMed] [Google Scholar]
- 37.Llosa, M., F. X. Gomis-Ruth, M. Coll, and F. de la Cruz. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 38.Otten, L., H. De Greve, J. Leemans, R. Hain, P. Hooykaas, and J. Schell. 1984. Restoration of virulence of vir region mutants of Agrobacterium tumefaciens strain B6S3 by coinfection with normal and mutant Agrobacterium strains. Mol. Gen. Genet. 195:159-163. [Google Scholar]
- 39.Sagulenko, E., V. Sagulenko, J. Chen, and P. J. Christie. 2001. Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J. Bacteriol. 183:5813-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sagulenko, V., E. Sagulenko, S. Jakubowski, E. Spudich, and P. J. Christie. 2001. VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus. J. Bacteriol. 183:3642-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheiffele, P., W. Pansegrau, and E. Lanka. 1995. Initiation of Agrobacterium tumefaciens T-DNA processing. Purified proteins VirD1 and VirD2 catalyze site- and strand-specific cleavage of superhelical T-border DNA in vitro. J. Biol. Chem. 270:1269-1276. [DOI] [PubMed] [Google Scholar]
- 42.Schirmer, T., and S. W. Cowan. 1993. Prediction of membrane-spanning β-strands and its application to maltoporin. Protein Sci. 2:1361-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shirasu, K., and C. Kado. 1993. Membrane location of the Ti plasmid VirB proteins involved in the biosynthesis of a pilin-like conjugative structure on Agrobacterium tumefaciens. FEMS Microbiol. Lett. 111:287-294. [DOI] [PubMed] [Google Scholar]
- 44.Spudich, G. M., D. Fernandez, X.-R. Zhou, and P. J. Christie. 1996. Intermolecular disulfide bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport apparatus. Proc. Natl. Acad. Sci. USA 93:7512-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stachel, S. E., and E. W. Nester. 1986. The genetic and transcriptional organization of the vir region of the A6 Ti. EMBO J. 5:1445-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stahl, L. E., A. Jacobs, and A. N. Binns. 1998. The conjugal intermediate of plasmid RSF1010 inhibits Agrobacterium tumefaciens virulence and VirB-dependent export of VirE2. J. Bacteriol. 180:3933-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thorstenson, Y., G. Kuldau, and P. Zambryski. 1993. Subcellular localization of seven VirB proteins of Agrobacterium tumefaciens: implications for the formation of a T-DNA transport structure. J. Bacteriol. 175:5233-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vergunst, A. C., B. Schrammeijer, A. den Dulk-Ras, C. M. de Vlaam, T. J. Regensburg-Tuink, and P. J. Hooykaas. 2000. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290:979-982. [DOI] [PubMed] [Google Scholar]
- 50.Ward, D. V., Draper, O., and P. C. Zambryski. 2002. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc. Natl. Acad. Sci. USA 99:11493-11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ward, J. E., Jr., E. M. Dale, E. W. Nester, and A. N. Binns. 1990. Identification of a VirB10 protein aggregate in the inner membrane of Agrobacterium tumefaciens. J. Bacteriol. 172:5200-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yeo, H. J., and G. Waksman. 2004. Unveiling molecular scaffolds of the type IV secretion system. J. Bacteriol. 186:1919-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhai, Y., and M. H. Saier, Jr. 2002. The beta-barrel finder (BBF) program, allowing identification of outer membrane beta-barrel proteins encoded within prokaryotic genomes. Protein Sci. 11:2196-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou, X.-R., and P. J. Christie. 1999. Mutagenesis of Agrobacterium VirE2 single-stranded DNA-binding protein identifies regions required for self-association and interaction with VirE1 and a permissive site for hybrid protein construction. J. Bacteriol. 181:4342-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]