Abstract
Naphthalene dioxygenase (NDO) from Ralstonia sp. strain U2 has not been reported to oxidize nitroaromatic compounds. Here, saturation mutagenesis of NDO at position F350 of the α-subunit (NagAc) created variant F350T that produced 3-methyl-4-nitrocatechol from 2,6-dinitrotoluene (26DNT), that released nitrite from 23DNT sixfold faster than wild-type NDO, and that produced 3-amino-4-methyl-5-nitrocatechol and 2-amino-4,6-dinitrobenzyl alcohol from 2-amino-4,6-dinitrotoluene (2A46DNT) (wild-type NDO has no detectable activity on 26DNT and 2A46DNT). DNA shuffling identified the beneficial NagAc mutation G407S, which when combined with the F350T substitution, increased the rate of NDO oxidation of 26DNT, 23DNT, and 2A46DNT threefold relative to variant F350T. DNA shuffling of NDO nagAcAd also generated the NagAc variant G50S/L225R/A269T with an increased rate of 4-amino-2-nitrotoluene (4A2NT; reduction product of 2,4-dinitrotoluene) oxidation; from 4A2NT, this variant produced both the previously uncharacterized oxidation product 4-amino-2-nitrocresol (enhanced 11-fold relative to wild-type NDO) as well as 4-amino-2-nitrobenzyl alcohol (4A2NBA; wild-type NDO does not generate this product). G50S/L225R/A269T also had increased nitrite release from 23DNT (14-fold relative to wild-type NDO) and generated 2,3-dinitrobenzyl alcohol (23DNBA) fourfold relative to wild-type NDO. The importance of position L225 for catalysis was confirmed through saturation mutagenesis; relative to wild-type NDO, NDO variant L225R had 12-fold faster generation of 4-amino-2-nitrocresol and production of 4A2NBA from 4A2NT as well as 24-fold faster generation of nitrite and 15-fold faster generation of 23DNBA from 23DNT. Hence, random mutagenesis discovered two new residues, G407 and L225, that influence the regiospecificity of Rieske non-heme-iron dioxygenases.
2,4-Dinitrotoluene (24DNT) and 26DNT are priority pollutants (17) generated during the synthesis of the explosive 2,4,6-trinitrotoluene (TNT), polyurethane foams, toluene, diisocyanate, and dyestuffs (10, 33a). These DNT isomers are genotoxic in Wistar rats (23). Other DNT isomers such as 23DNT, 25DNT, and 34DNT are found in industrial effluent streams (7) and are reported to be human carcinogens (29). Aminonitrotoluenes are generated biologically from the reduction of DNTs (35), add to the complexity of sites contaminated with nitroaromatic compounds, and provide motivation for engineering oxygenase enzymes to initiate their biodegradation. In the environment, TNT is also readily reduced to the aminodinitrotoluene isomers 2-amino-4,6-dinitrotoluene (2A46DNT) and 4A26DNT (5, 37), and these reduction products are more toxic than the parent compound TNT (14).
Recombinant Escherichia coli strains expressing 24DNT dioxygenase (DDO) from the Burkholderia cepacia R34 and Burkholderia sp. strain DNT (DNT DDO) oxidize 2A46DNT at the 3,4 position, releasing nitrite to form 3-amino-4-methyl-5-nitrocatechol (3A4M5NC), and hydroxylation of the methyl group forms 2-amino-4,6-dinitrobenzyl alcohol (2A46DNBA) (Fig. 1) (13). The recombinant nitrobenzene dioxygenase from Comamonas sp. strain JS765 has been shown to oxidize 4A26DNT at the 2,3 position, releasing nitrite and to generate 3A6M5NC (13). The wild-type naphthalene dioxygenase (NDO) from Ralstonia sp. strain U2 has not been reported to degrade these aminodinitrotoluenes or any other nitroaromatic compounds (6). Since few nitroaromatic compounds are generated through biological activity (5), the instances of microbiological degradation of nitroaromatic compounds are relatively sparse; therefore, directed evolution of enzymes to create organisms capable of transforming or mineralizing these compounds may benefit bioremediation efforts.
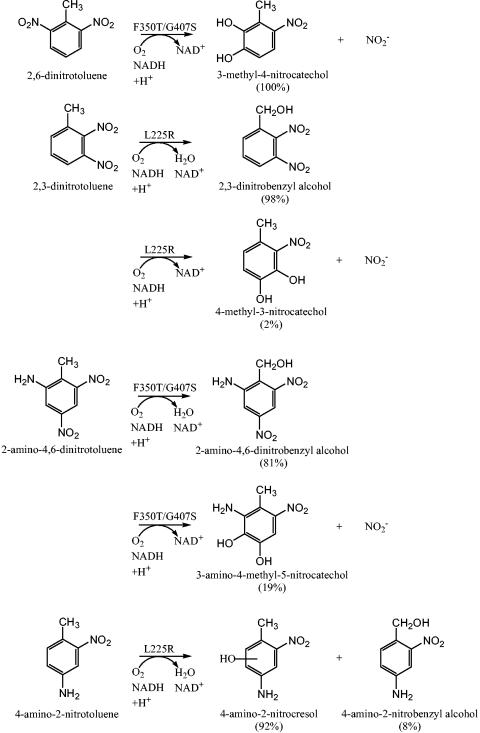
FIG. 1.
Wild-type and variant NDO dioxygenase reactions with 2,6-dinitrotoluene, 2,3-dinitrotoluene, and 2-amino-4,6-dinitrotoluene, and the monooxygenase-like reactions toward 2,3-dinitrotoluene, 2-amino-4,6-dinitrotoluene, and 4-amino-2-nitrotoluene. The NagAc variant with the highest rate of product formation is indicated above each reaction arrow, and product percentages are shown in parentheses for that variant.
The initial step in the utilization of naphthalene as a carbon source by Ralstonia sp. strain U2 is catalyzed by NDO, which is encoded by nagAaAbAcAd. NDO is a non-heme-iron, ring-hydroxylating dioxygenase (6) similar to the naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4 (NCIB NDO). Unlike NCIB NDO, NDO from U2 contains an active salicylate 5-hydroxylase (encoded by nagGH) within the nagAaGHAbAcAd operon (6). The NagG (salicylate 5-hydroxylase large-subunit) and NagH (salicylate 5-hydroxylase small-subunit) terminal hydroxylase subunits utilize the same electron transfer subunits, NagAa and NagAb, as the terminal dioxygenase (6) to catabolize naphthalene through gentisate rather than through catechol (6). Truncated homologs of nagG are present in all four known nitroarene dioxygenases from strains B. cepacia R34 (DDO), Burkholderia sp. strain DNT (DNT DDO), Acidovorax sp. strain JS42 (2-nitrotoluene dioxygenase) (25), and Comamonas sp. strain JS765 (nitrobenzene dioxygenase) (20). Hence, the U2 nagAaGHAbAcAd operon may be the naphthalene dioxygenase that evolved into the nitroarene dioxygenases (6).
Saturation mutagenesis at the DDO DntAc alpha-subunit position V350 was shown by us to increase the rate of DDO for the addition of a single-oxygen atom to the benzene ring of substituted phenols (e.g., o-methoxyphenol and o-nitrophenol) (16). The NDO terminal oxygenase (alpha) subunit, NagAc (448 amino acids), has 89% amino acid sequence identity with NahAc of NCIB NDO (12) and 91% identity with DntAc of DDO (12).
Our goal was to engineer the NDO NagAc for activity toward nitroaromatic compounds through saturation mutagenesis (at position F350) and through DNA shuffling of the U2 NDO nagAcAd terminal oxygenase genes. We reasoned that by starting with a dioxygenase with no detectable activity towards nitroaromatic compounds, we would be able to determine key residues that influence NDO activity, and these residues might provide insight into the structure/function relationships of this family of Rieske non-heme-iron dioxygenases. Here, through protein engineering, we generated NDO activity toward nitroaromatic compounds and discovered that alpha-subunit residues L225, F350, and G407 influence NDO activity with nitroaromatic compounds (alpha-subunit residues analogous to L225 and G407 have not been linked previously to the catalytic activity of any Rieske non-heme-iron dioxygenases). This appears to be the first report of the biological synthesis 4-amino-2-nitrocresol and 2,3-dinitrobenzyl alcohol (23DNBA), of the use of the Ralstonia sp. strain U2 NDO for the oxidation of nitroaromatic compounds, and the first directed evolution of a dioxygenase for nitroaromatic compounds.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
E. coli strain TG1 (32) was used as the host for the recombinant dioxygenase constructs during cloning, solid-phase screening, and whole-cell transformations of 26DNT. E. coli JVQ2 with two nitroreductase mutations (nfsAB) (36) was used during whole-cell transformations of 2A46DNT, 23DNT, and 4A2NT to minimize the undesired reduction of the nitro groups.
For whole-cell dioxygenase transformations, single colonies were initially inoculated into 25 ml of LB medium (32) containing 1.0% glucose and kanamycin (100 μg/ml) and grown overnight at 37°C at 250 rpm. Glucose maintained plasmid segregational stability by suppressing dioxygenase expression until enzyme activity was desired. Ten milliliters of the overnight culture was inoculated into 250 ml of LB medium containing 1.0 mM isopropyl-β-d-thiogalactopyranoside and kanamycin (100 μg/ml) and grown at 37°C at 250 rpm from an initial optical density at 600 nm (OD600) of 0.3 to 2.0 (∼3 h of incubation). The 24DNT dioxygenases from B. cepacia R34 and Burkholderia sp. strain DNT were expressed in both E. coli TG1 and E. coli JVQ2 from the plasmids pBS(Kan)R34 (16) and pBS(Kan)DNT (21), respectively. The relative expression of the nagAaAbAcAd loci from E. coli TG1/pBS(Kan)NDO− and E. coli JVQ2/pBS(Kan)NDO− was evaluated using SDS-PAGE (32) with a 12% acrylamide gel.
Chemicals.
Dimethyl formamide and chloroform were purchased from Fisher Scientific Co. (Fairlawn, N.J.). 24DNT, 26DNT, 23DNT, 2-amino-4-nitrotoluene (2A4NT), 4A2NT, 4A26DNT, sulfanilamide, and N-(1-naphthyl)ethyldiamine were purchased from Sigma Chemical Co. (St. Louis, MO). 2A46DNT was purchased from AccuStandard Inc. (New Haven, CT). 3-Methyl-4-nitrocatechol (3M4NC), 4M5NC, and 3A4M5NC were supplied by Jim Spain of the U.S. Air Force. 4-Amino-2-nitrobenzyl alcohol (4A2NBA) and 2,4-dinitrobenzyl alcohol (24DNBA) were supplied by Masa-aki Mori (23).
Construction of pBS(Kan)NDO.
The expression vector pBS(Kan)NDO was constructed by PCR amplification of the Ralstonia sp. strain U2 nagAaGHAbAcAd loci from plasmid pWWF6 (6) using standard PCR conditions and Pfu high-fidelity DNA polymerase (Stratagene, La Jolla, CA). The front primer BSIIKpnNag-Fwd (Table 1) contains the restriction site KpnI, and the rear primer BSIIBamNag-Rvs (Table 1) contains the restriction site BamHI; these primers allowed the directional cloning of the 5.2-kb nagAaGHAbAcAd loci downstream of the lac promoter of the expression vector pBS(Kan) (high-copy-number plasmid derived from pBluescript II KS−) (2) and generated the 9,284-bp plasmid pBS(Kan)NDO. E. coli TG1 cells were transformed with the plasmid constructs using Bio-Rad Gene Pulser (Hercules, CA) at 1.5 V, 25 μF, and 200 Ω; correctly constructed plasmids were selected based on the production of indigoid compounds from recombinant TG1 colonies.
TABLE 1.
Primers used for the cloning the U2 nagAaGHAbAcAd loci of Ralstonia sp. strain U2 into pBS(Kan), for the saturation mutagenesis of F350, G407, and L225 of NDO NagAc, and for the sequencing of the wild-type and mutant insert regions nagAc and nagAd
| Primer | Sequence |
|---|---|
| nagAaGHAbAcAd loci cloninga | |
| BSIIKpnNag Fwd | 5′-GGCGGAGAGGTACCAACTGAAAAAAGAGCTTGCATGG-3′ |
| BSIIBamNag Rvs | 5′-GGATGACCGGATCCGCCATCAAGGCAGGCATCCCCAGG-3′ |
| Saturation mutagenesis and shufflingb | |
| nagAb Fwd | 5′-CCCTGAAGGCGATGTGGTCG-3′ |
| dntAcXba Rvs | 5′-AGGGTTTTCCCAGTCACG-3′ |
| nag Back PCR Fwd | 5′-GCCCTCTGCACCAAGGTCG-3′ |
| pBSii Back PCR Rvs | 5′-GGGCGAATTGGAGCTCC-3′ |
| nagAc F350 In Fwd | 5′-GCTGACGCGGTTCAGCGCACTNNNGGACCAGCAGG-3′ |
| nagAc F350 in Rvs | 5′-CCCAGAATCCTGCTGGTCCNNNAGTGCGCTGAACC-3′ |
| U2 G407 In Fwd | 5′-CGTCGTTGCCAAATCGGCAATCNNNGAAACCAGCTATCG-3′ |
| U2 G407 In Rvs | 5′-GGTAGAATCCGCGATAGCTGGTTTCNNNGATTGCCGATTTGG-3′ |
| U2 L225 In Fwd | 5′-GGTCAGTCGATATTTACCCCTNNNGCGGGCAACGC-3′ |
| U2 L225 In Rvs | 5′-GGAAGCATAGCGTTGCCCGCNNNAGGGGTAAATATCG-3′ |
Restriction sites used in the cloning of the U2 nagAaGHAbAcAd loci are represented in italics and are underlined.
Degenerate positions used in the randomization of positions F350, G407, and L225 are denoted as N (underlined).
To avoid the possibility of interference from an active salicylate hydroxylase, genes nagG and nagH were removed from the original pBS(Kan)NDO construct using two naturally occurring BsrGI restriction sites (generating a 1,208-bp fragment); the remaining 8,076-bp pBS(Kan)NDO vector was ligated on itself to generate the new U2 nagAaAbAcAd expression vector pBS(Kan)NDO− which was electroporated into E. coli TG1 cells. Deletion of nagGH from pBS(Kan)NDO was verified through restriction enzyme digests using BsrGI, EcoRI, and BamHI.
Saturation mutagenesis of NDO nagAc.
Saturation mutagenesis at position F350 of the alpha subunit (nagAc) of NDO was performed as described previously (16) using the degenerate primers (Table 1) nagAc-F350-In-Fwd (forward) and nagAc-F350-In-Rvs (reverse) as well as primers nagAb-Fwd (upstream of the unique EcoRI site and the start codon for NagAc) and dntAcXba-Rvs (downstream of the stop codon for NagAd and the unique XbaI site). A degenerate 1,281-bp PCR fragment was amplified using primers nagAb-Fwd and nagAc-F350-In-Rvs, and a 1,293-bp degenerate PCR fragment was amplified using primers nagAc-F350-In-Fwd and dntAcXba-Rvs. The two fragments were combined during the final reassembly PCR using the nested outer primers nag-Back-PCR-Fwd and pBSii-Back-PCR-Rvs (Table 1) as described previously (16). The resulting randomized 2,258-bp PCR product was ligated into pBS(Kan)NDO−, after the double digestion of both vector and insert with EcoRI and XbaI, replacing the wild-type region. The resulting plasmid library was electroporated into E. coli TG1.
For saturation mutagenesis of NagAc position G407, a 2,258-bp reassembly PCR product containing NNN for the 407 codon was obtained from the degenerate 1,592-bp PCR product generated by the primers nagAb-Fwd and U2-G407-In-Rvs (Table 1) and the degenerate 916-bp PCR product generated by U2-G407-In-Fwd and dntAcXba-Rvs. For NagAc position L225, a 2,258-bp reassembly PCR product containing NNN for the 225 codon was obtained from the degenerate 1,041-bp PCR product generated by the primers nagAb-Fwd and U2-L225-In-Rvs (Table 1) and the degenerate 1,416-bp PCR product generated by U2-L225-In-Fwd and dntAcXba-Rvs. The final degenerate G407 and L225 PCR fragments were combined during the final reassembly PCR using the nested outer primers nag-Back-PCR-Fwd and pBSii-Back-PCR-Rvs (Table 1).
DNA shuffling of NDO nagAcAd.
DNA shuffling of 100% of nagAc and 100% of nagAd of NDO was performed using DNaseI to generate 50-bp fragments as described previously for toluene ortho-monooxygenase (2). The reassembly PCR primers (Table 1) nag-Back-PCR-Fwd and pBSii-Back-Rvs anneal upstream and downstream of the natural restriction site EcoRI and restriction site XbaI (encoded in the multiple-cloning site), respectively, allowing cloning of only the mutagenized region. The shuffled reassembly product was then ligated into plasmid pBS(Kan)NDO−, replacing the 2,058-bp nagAcAd region.
Subcloning of NDO NagAc mutants.
The variant F350T fragment (1,235 bp) was ligated into the G407S plasmid backbone fragment (6,841 bp) via the naturally occurring restriction sites EcoRI (5′ restriction site) and AccI (3′ restriction site), generating the variant F350T/G407S. The G50S/L225R/A269T triple mutant fragment (821 bp) was ligated into the F350T plasmid backbone fragment (7,255 bp) via the naturally occurring restriction sites EcoRI (5′ restriction site) and Bsu36I (3′ restriction site), generating the quadruple mutant G50S/L225R/A269T/F350T. After subcloning, both the F350T/G407S and G50S/L225R/A269T/F350T mutations were verified by sequencing the variant nagAc gene from the reconstructed pBS(Kan)NDO−.
Colony screening.
The randomized libraries generated through saturation mutagenesis and DNA shuffling were screened for the production of NDO-mediated oxidized products using a solid-phase assay as described previously (16); these oxidation products were detected after extracellular auto-oxidation. Each screening plate contained 300 to 1,000 μM concentrations of the substrates 24DNT, 26DNT, 2A46DNT, 4A26DNT, o-cresol, or p-cresol.
Product identification and rates of formation via HPLC.
After two rounds of solid-phase screening, the positive variants were grown in liquid culture for analysis using reverse-phase high-pressure liquid chromatography (HPLC) as described previously (16). For all the nitroaromatic substrates, 10 to 20 ml of exponentially grown cells resuspended in 50 mM Tris-Cl buffer, pH 7.6, were incubated in 250-ml glass shake flasks with 0.5 mM substrate (dimethyl formamide was the diluent and 10 to 20 μl of substrate stock solution was added to the cell suspension). After 0 to 60 min, 1.5 ml of the incubating cell suspension was removed and centrifuged at 14,000 rpm for 1 to 2 min in a Spectrafuge 16 M Microcentrifuge (Labnet Inc., Woodbridge, N.J.). The substrates and products were separated using a 100- by 4.6-mm Chromolith Performance RP-18e column (Merck KGaA, Darmstadt, Germany). For the analysis of 26DNT and 2A46DNT oxidation, a gradient elution was performed with H2O-0.1% formic acid and acetonitrile (95:5 for 0 to 3.5 min, 85:15 for 5 to 8 min, 60:40 for 9 to 12 min, and returned to 95:5 at 15 min) as the mobile phases at a flow rate of 1 ml/min for the first 1.5 min, at 3 ml/min for 3.5 min, and remaining at 1 ml/min for 10 min. For the analysis of 23DNT and 4A2NT oxidation, a gradient elution was performed with H2O-0.1% formic acid and acetonitrile (100:0 for 0 to 15 min, 0:100 for 15 to 20 min, and returned to 100:0 after 30 min) as the mobile phases at a flow rate of 2 ml/min.
The products were identified by comparing their HPLC retention times and UV-visible spectra to standard chemicals and were corroborated through coelution (equal concentration of an authentic standard added to the newly identified product to verify its identify via the formation of a single peak). To ensure the accuracy of the retention times, the HPLC columns were equilibrated with the appropriate elution buffer prior to evaluation of various transformations. The authentic product standards 3M4NC, 4A2NBA, and 3A4M5NC were also evaluated for their stability to determine appropriate sampling times.
Initial product formation rates were determined by collecting supernatant samples at 5-min intervals for the first 20 min and at 10- to 30-min intervals thereafter. Product formation rates were quantified in nmol/min/mg of protein by converting product peak areas to concentrations using standard curves prepared at the specific absorbance wavelength for each product formed. The initial rates of product formation for the wild-type NDO and NagAc variants were determined from at least two independent whole-cell transformation experiments. Protein content was 0.22 mg of protein/ml/OD unit for recombinant E. coli TG1 and 0.18 mg of protein/ml/OD unit for recombinant E. coli JVQ2 as determined using the Protein Assay Kit (Sigma Diagnostics Inc., St. Louis, Mo.). Authentic standards were not available for the products 23DNBA, 4-amino-2-nitrocresol, and 2A46DNBA; hence, for these products, approximate relative rates were determined using the initial linear plot of product peak area (at the maximum absorbance wavelength) versus time and were normalized using the cell suspension optical density to determine the relative performance of the wild-type NDO and the NagAc variants.
LC-MS analysis.
To identify 2A46DNBA generated by the NDO NagAc variant F350T/G407S from 2A46DNT (0.5 mM) after 45 min of incubation (incubating cell suspension had an optical density of 20), HPLC-mass spectrometry (LC-MS) was performed with a Hewlett-Packard (Palo Alto, CA) 1090 series II Liquid Chromatograph with a diode array detector coupled to a Micromass Q-TOF2 (Beverly, MA) mass spectrometer. Separation was achieved using a 100- by 4.6-mm Chromolith Performance RP-18e column with a mobile phase consisting of H2O-0.1% formic acid and acetonitrile and a gradient elution at 2.0 ml/min starting from 100% H2O-0.1% formic acid to 0% in 12 min, with a 3-min hold at 100% acetonitrile and a return to 100% H2O-0.1% formic acid in 15 min. The Q-TOF2 was operated in positive-ion electrospray mode with 3.0 kV applied to the inlet capillary and 30 V applied to the extraction cone. During the 2A46DNBA analyses, LC-MS was also used to confirm the synthesis of 3A4M5NC from 2A46DNT by F350T/G407S. The products generated during the incubation of the NDO variant F350T/G407S with 2A46DNT were compared against the retention times and molecular mass of the authentic standard compound 3A4M5NC and the expected molecular mass of 2A46DNBA under the conditions used for the LC-MS analysis.
To identify 4M3NC and 23DNBA generated by the NagAc variant L225R from 23DNT (0.5 mM) after 60 min of incubation with a cell suspension of optical density of 15 to 20, LC-MS analyses were performed with a Finnigan MAT P4000 (Thermo Separation Products Inc., San Jose, CA) and Finnigan MAT UV6000LP diode array detector coupled to a LCQ (Thermo Separation Products Inc., San Jose, CA) mass spectrometer. Separation was achieved using the Chromolith Performance RP-18e column with a mobile phase consisting of H2O-0.1% formic acid and acetonitrile and a gradient elution at 1.0 ml/min starting from 100% H2O-0.1% formic acid to 0% in 20 min, with a 5-min hold at 100% acetonitrile and a return to 100% H2O-0.1% formic acid in 20 min. The LCQ was operated in negative-ion electrospray mode with 5.1 kV applied to the inlet capillary and 3.5 V applied to the extraction cone. To determine that an additional MS peak was either an acetonitrile or formic acid adduct product formed during the MS procedure (in addition to the 4M3NC and 23DNBA generated by the NagAc variant L225R from 23DNT) (4, 11), the MS characteristics of the analogous compounds 4M5NC and 24DNBA were determined including m/z fragment patterns, the tendency to form acetonitrile adducts or formate adducts, and UV-visible spectra.
To identify 4-amino-2-nitrocresol generated by the wild-type NDO and NagAc variant L225R from 4A2NT, LC-MS was performed using the identical conditions as described during the analysis of the 23DNT oxidation products; however, the LCQ was operated in positive-ion electrospray mode with 4.5 kV applied to the inlet capillary and 3.5 V applied to the extraction cone. During the 4-amino-2-nitrocresol analyses, LC-MS was also used to confirm the synthesis of 4A2NBA from 4A2NT by L225R.
Modeling of NagAc, nitrite assay, and DNA sequencing.
The altered residues were visualized using the DeepView program (Swiss-Pdb Viewer) (33) after the wild-type NDO NagAc α-subunit was determined using the SWISS-MODEL server (33) based on the NCIB NDO NahAc template 1O7G (15) (polymer chain A). The nitrite generated from the oxidation of all nitroaromatic compounds was detected spectrophotometrically as described previously (16) based on the formation of an azo dye complex between nitrite, sulfanilamide, and N-(1-naphthyl)ethyldiamine; the azo dye has an absorbance maximum at 543 nm. DNA sequencing of the nagAc and nagAd genes was performed as described previously (16) using the primers nagAb-Fwd and pBSii-Back-PCR-Rvs (Table 1), U2ctgB (5′-CGAAAGGCTTTGTGTGCAG-3′), U2ctgC (5′-GCGGGCTTACAAATGACC-3′), U2ctgD (5′-CAACATGGAGACGGAGTCG-3′), and U2ctgE (5′-AACGCCTGGCTTGAACAC-3′).
RESULTS
Saturation mutagenesis of NagAc F350.
To convert wild-type NDO into an enzyme which has activity on nitroaromatic compounds, a saturation mutagenesis library of 500 variants for NDO NagAc F350 was screened for increased activity toward 24DNT, 26DNT, 2A46DNT, 4A26DNT, and p-cresol using the solid-phase assay. Five hundred variants were chosen at random (some produced indigoid compounds on LB agar transformation plates) from an initial pool of 2,000 transformation colonies based on our calculation that there is a 99% probability that each of the 64 codons is sampled for a population of this size (30). Five NDO variants of the F350 mutagenesis library generated oxidation products that diffused from the streaked colonies into the agar media and that auto-oxidized to form colored high-molecular-weight compounds (22) that were consistent with novel oxidation activity toward the substrates 26DNT and 2A46DNT: F350T, F350Q, F350L, F350S, and F350N. Sequencing the complete nagAcAd genes for the NagAc F350T variant showed there were no additional amino acid changes. No dioxygenases with altered activity toward 24DNT, 4A26DNT, or p-cresol were found.
DNA shuffling of NDO nagAcAd.
To identify new amino acid positions that influence NDO catalytic activity, a DNA shuffling mutant library totaling 1,500 random NDO nagAcAd variants was screened for enhanced activity toward 24DNT, 4A26DNT, and p-cresol using the solid-phase assay. Variant NagAc G407S with three nucleotide changes (two silent) over the entire 2,058-bp shuffled region was selected due to the production of a uniquely colored p-cresol oxidation product, and NagAc G50S/L225R/A269T with three nucleotide changes (all coding) over the entire 2,058-bp shuffled region was identified due to the production of a uniquely colored, 24DNT-derived, oxidation product. Screening of the NDO nagAcAd shuffling library failed to identify dioxygenases with altered activity toward 4A26DNT. Sequencing the shuffled nagAcAd region revealed one silent mutation and one amino acid change, F71L, in the nagAd gene (GenBank accession no. AF036940); these nucleotide changes were not introduced during cloning since they were confirmed through sequencing of the original nagAd gene from the plasmid pWWF-6 (6).
Saturation mutagenesis of NagAc L225 and G407.
To confirm the influence of the L225R substitution in the DNA shuffling variant NagAc G50S/L225R/A269T, a saturation mutagenesis library of 500 variants for NDO NagAc L225 was screened for enhanced activity toward 24DNT, 26DNT, 2A46DNT, and 4A26DNT using the solid-phase assay. Position L225 was chosen for saturation mutagenesis due to its close proximity to the active-site mononuclear iron (10 Å); positions G50 (38 Å) and A269 (25 Å) are located on the outer shell of the alpha subunit and were not believed to influence the catalytic activity of NagAc. Three different L225R variants (each encoding a different R225 codon) generated a colored 24DNT oxidation product that was similar to the reaction identified for the triple mutant G50S/L225R/A269T. Sequencing the nagAcAd genes indicated that no other amino acids were changed.
To investigate the role of NagAc G407 on NDO catalytic activity, a saturation mutagenesis library of 500 variants for NDO NagAc G407 was screened for enhanced activity toward 24DNT, o-cresol, and p-cresol using the solid-phase assay. Variants G407S and G407C were identified from this library as having similar activity towards o-cresol and p-cresol relative to the initial NagAc G407S DNA shuffling mutant. Variant G407T was also found that generated a high level of indigoid-like products during growth on the LB media agar plates.
Whole-cell transformation of 26DNT and 23DNT.
The product generated from the NDO NagAc variant F350T oxidation of 26DNT was determined via HPLC analysis and coelution to be 3M4NC, which releases nitrite (Fig. 1 and Table 2). Hence, the F350T mutation created an NDO variant able to oxidize 26DNT to 3M4NC (only product peak), whereas wild-type NDO has no activity on this substrate. NagAc F350T released nitrite and produced 3M4NC at the same rate as wild-type DDO of B. cepacia R34 (0.4 ± 0.2 nmol NO2−/min/mg of protein and 0.33 ± 0.06 nmol 3M4NC/min/mg of protein) and DNT DDO of Burkholderia sp. strain DNT (0.39 ± 0.08 nmol NO2−/min/mg of protein and 0.29 ± 0.09 nmol 3M4NC/min/mg of protein); therefore, the newly acquired rate of oxidation by the variant NDO was comparable to enzymes evolved for growth on 24DNT. NagAc variants G50S/L225R/A269T and L225R did not oxidize 26DNT.
TABLE 2.
Oxidation rates of 26DNT, 2A46DNT, and 23DNT by wild-type NDO, NDO NagAc saturation variant F350T, and NagAc saturation and DNA shuffling variant F350T/G407Sa
| Substrate | Product | Wild-type NDO
|
F350T
|
F350T/G407S
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Nitrite release rate | Product formation rate | Nitrite release rate | Product formation rate | Relative to NDO | Nitrite release rate | Product formation rate | Relative to NDO | Relative to F350T | ||
| 26DNTb | 3-methyl-4-nitrocatechol | 0 | 0 | 0.36 ± 0.07 | 0.3 ± 0.1c (100%) | ∞ | 1.1 ± 0.3 | 0.8 ± 0.1c (100%) | ∞ | 3d |
| 2A46DNTe | 2-amino-4,6-dinitrobenzyl alcohol | NA | 0 | NA | 70 ± 20f (73%)g | ∞ | NA | 170 ± 10f (81%)g | ∞ | 2 |
| 3-amino-4-methyl-5-nitrocatechol | 0 | 0 | 0.032 ± 0.006 | 0.043 ± 0.008c (27%)g | ∞ | 0.12 ± 0.04 | 0.12 ± 0.03c (19%)g | ∞ | 3d | |
| 23DNTe | 2,3-dintrobenzyl alcohol | NA | 115h (100%)i | NA | 36h (100%)i | 0.3 | NA | 104h (100%)i | 0.9 | 3 |
| 4-methyl-3-nitrocatechol | 0.014 ± 0.003 | NDj | 0.08 ± 0.02 | ND | 6 | 0.15 ± 0.01 | ND | 11 | 2 | |
Nitrite release rate was determined in nmol NO2−/min/mg protein. Product percentages are shown in parentheses with the product formation rate. NA, not available.
Transformations of 26DNT were performed using E. coli TG1 with 0.5 mM substrate.
Rate determined in nmol product/min/mg protein.
Relative performance is average of nitrite release rate and product formation rate calculated from identical time course experiments.
Transformations of 2A46DNT and 23DNT were performed using E. coli JVQ2 with 0.5 mM substrate.
Approximate relative activity determined by comparison of slope for product peak area at 300 nm/min/incubating suspension OD 600 nm.
Percentages of each product generated are relative to the total product area formed, and are based on peak area (not concentration) at 300 nm.
Approximate relative activity determined by comparison of slope for product peak area at 260 nm/min/incubating suspension OD 600 nm.
Percentages of each product generated are relative to the total product area formed, and are based on peak area (not concentration) at 260 nm.
4-methyl-3-nitrocatechol peak not detected (ND) through HPLC.
Variant G50S/L225R/A269T did not have increased activity on 24DNT even though it was identified from 24DNT agar plates. However, G50S/L225R/A269T had enhanced activity on the host-formed reduction product from 24DNT, 4A2NT (Fig. 1), and this is why it was identified using the agar plate screening (Table 3).
TABLE 3.
Oxidation rates of 4A2NT and 23DNT by wild-type NDO, NDO NagAc saturation variant L225R, and DNA shuffling variant G50S/L225R/A269Ta
| Substrate | Product | Wild-Type NDO
|
L225R
|
G50S/L225R/A269T
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Nitrite release rate | Product formation rate | Nitrite release rate | Product formation rate | Relative to NDO | Relative to G50S/L225R/A269T | Nitrite release rate | Product formation rate | Relative to NDO | ||
| 4A2NTb | 4-amino-2-nitrocresol | NA | 90 ± 60c (100%)d | NA | 1100 ± 140c (92%)d | 12 | 1 | NA | 1000 ± 40c (93%)d | 11 |
| 4-amino-2-nitrobenzyl alcohol | NA | 0 | NA | 0.7 ± 0.1e (8%)d | ∞ | 1 | NA | 0.53 ± 0.08e (7%)d | ∞ | |
| 23DNTb | 2,3-dintrobenzyl alcohol | NA | 115f (100%)g | NA | 1709f (98%)g | 15 | 3 | NA | 498f (100%)g | 4 |
| 4-methyl-3-nitrocatechol | 0.014 ± 0.003 | NDh | 0.3 ± 0.1 | −i (2%)g | 24 | 2 | 0.19 ± 0.05 | ND | 14 | |
Rate determined in nmol NO2−/min/mg protein. Product percentages are shown in parentheses with product formation rate. NA, not available.
Transformations of 4A2NT and 23DNT were performed using E. coli JVQ2 with 0.5 mM substrate.
Approximate relative activity determined by comparison of slope for product peak area at 300 nm/min/incubating suspension OD 600 nm.
Percentages of each product generated are relative to the total product area formed, and are based on peak area (not concentration) at 300 nm.
Rate determined in nmol product/min/mg protein.
Approximate relative activity determined by comparison of slope for product peak area at 260 nm/min/incubating suspension OD 600 nm.
Percentages of each product generated are relative to the total product area formed, and are based on peak area (not concentration) at 260 nm.
4-methyl-3-nitrocatechol peak not detected (ND).
4-methyl-3-nitrocatechol peak detected only after 60 minutes of incubation with 23DNT, relative activity could not be quantified through HPLC analyses.
LC-MS identified 23DNBA (m/z = 197) as the primary product generated from the oxidation of 23DNT by NagAc L225R (Fig. 1 and Table 3). During the identification of 23DNBA, an aberrant m/z fragment of 242 was found and was most likely due to the formation of a 23DNBA formic acid adduct within the MS ionization trap. The formation of the m/z fragment of 242 was also identified during MS analysis with the authentic standard for the analogous nitroaromatic compound 24DNBA (m/z = 197). Using HPLC, it was determined that 23DNBA synthesis from 23DNT was enhanced fourfold by G50S/L225R/A269T and 15-fold by L225R relative to wild-type NDO (Fig. 1 and Table 3). NagAc F350T generated 23DNBA from 23DNT at one-third the rate of wild-type NDO (Table 2).
The secondary product from the oxidation of 23DNT by variant L225R was identified through LC-MS analyses as 4M3NC (m/z = 168), which releases nitrite (Fig. 1 and Table 3). During the identification of 4M3NC, the aberrant m/z fragment of 213 that was formed was most likely due to the formation of a 4M3NC formic acid adduct within the MS ionization trap. The formation of the m/z fragment of 213 was also identified during MS analysis with the analogous authentic standard for 4M5NC (m/z = 168). Only L225R generated 4M3NC at a high enough concentration to be detected during the LC-MS and HPLC analyses. The identification of the 4M3NC product generated by variant L225R corroborated the release of nitrite from 23DNT detected during the whole-cell transformations. The initial rate of nitrite formation (Tables 2 and 3) shows that 23DNT oxidation was enhanced sixfold for F350T, 14-fold for G50S/L225R/A269T, and 24-fold for L225R relative to the wild-type NDO. The negative control E. coli TG1/pBS(Kan) did not generate 3M4NC from 26DNT, and E. coli JVQ2/pBS(Kan) did not generate 23DNBA or 4M3NC during incubation with 23DNT; hence, the enhanced oxidation of these substrates was from the NDO mutations.
Whole-cell transformation of 2A46DNT.
LC-MS identified 2A46DNBA as the primary product generated by NagAc variant F350T from the oxidation of 2A46DNT (Fig. 1); wild-type NDO had no activity on this substrate (Table 2). Variant F350T generated 2A46DNBA at 20% of the rate of wild-type DDO of B. cepacia R34 (13).
Through comparisons of HPLC retention times, UV-visible spectra, and coelution with the authentic standard, the secondary product generated from the oxidation of 2A46DNT by F350T was determined to be 3A4M5NC, which releases nitrite (Fig. 1 and Table 2); again, wild-type NDO had no activity on this substrate (Table 2). NagAc F350T has a rate of nitrite release and 3A4M5NC formation that is 1/10 the initial rate of the DDO of B. cepacia R34 (0.4 ± 0.1 nmol NO2−/min/mg of protein) and 1/30 the initial rate of the DNT DDO of Burkholderia sp. strain DNT (1.5 ± 0.5 nmol NO2−/min/mg of protein). The negative control, E. coli JVQ2/pBS(Kan), did not generate 2A46DNBA from 2A46DNT or 3A4M5NC from 2A46DNT, so the enhanced oxidations were from the altered NDO.
Whole-cell transformation of 4A2NT.
Aminonitrotoluene isomers were considered as substrates for the wild-type NDO and the NagAc variants since it was determined that the E. coli TG1 host cells were reducing 24DNT to two reduction products, 2A4NT and 4A2NT, and it was the enhanced activity of the G50S/L225R/A269T variant on 4A2NT that led it to its selection from 24DNT-containing agar plates. To minimize this undesired reduction reaction, NDO was expressed in E. coli JVQ2 to allow the quantification of the enhanced activity of the NagAc variants on 4A2NT. No nitrite was measured during the incubation of wild-type NDO and G50S/L225R/A269T with 24DNT, 4A2NT, or 2A4NT.
Through HPLC time course analyses, 4-amino-2-nitrocresol was generated from 4A2NT 11-fold faster by G50S/L225R/A269T and 12-fold faster by L225R (Table 3) relative to wild-type NDO. 2A4NT was not oxidized by the wild-type NDO, G50S/L225R/A269T, or L225R. NagAc variant F350T generated trace amounts of 4-amino-2-nitrocresol from 4A2NT. The negative control, E. coli JVQ2/pBS(Kan), did not generate 4-amino-2-nitrocresol from 4A2NT, so the enhanced oxidations were from the altered NDO.
Using LC-MS, the synthesis of 4-amino-2-nitrocresol (m/z = 169) was identified as the primary product generated during the oxidation of 4A2NT by variant L225R (Fig. 1 and Table 3). During the identification of 4-amino-2-nitrocresol, the aberrant m/z fragment of 209 that was formed was most likely due to the formation of a 4-amino-2-nitrocresol acetonitrile adduct within the MS ionization trap.
Through comparisons of HPLC retention times, UV-visible spectra, and coelution with the authentic standard, the variants G50S/L225R/A269T and L225R were determined to generate 4A2NBA as the secondary product from 4A2NT (wild-type NDO has no activity on this substrate) (Fig. 1 and Table 3). LC-MS also confirmed the identification of the product 4A2NBA (m/z = 169), and the aberrant m/z fragment of 209 was also identified with the authentic standard for 4A2NBA.
Combination of beneficial NagAc mutations.
The mutation F350T was combined with both the G50S/L225R/A269T and G407S variations to determine the impact of multiple beneficial mutations on catalytic activity toward the substrates 26DNT and 2A46DNT. NagAc variant F350T/G407S oxidized 26DNT threefold faster relative to NagAc F350T (Table 2), which exceeds by threefold the rate of 26DNT oxidation by both wild-type 24DNT dioxygenase strains; hence, the beneficial mutations could be combined to improve activity. During contact with 23DNT, the F350T/G407S variant also generated twofold greater nitrite relative to F350T and enhanced the relative rate of 23DNBA formation threefold relative to F350T (Table 2).
The primary product generated by NagAc variant F350T/G407S from 2A46DNT was identified through LC-MS as 2A46DNBA (m/z = 213). F350T/G407S oxidized 2A46DNT to 2A46DNBA with a twofold enhancement relative to NagAc F350T (Table 2). The formation of 3A4M5NC as the secondary product from 2A46DNT by F350T/G407S was confirmed through LC-MS by comparing the expected LC retention times and molecular masses (m/z = 184) of the product peak and of the authentic 3A4M5NC standard. During incubation with 2A46DNT, NagAc F350T/G407S released nitrite and generated 3A4M5NC threefold relative to F350T (Table 2). The NagAc variant G50S/L225R/A269T/F350T did not generate any products from 23DNT, 26DNT or 2A46DNT during whole-cell transformations.
Protein expression analysis.
Using SDS-PAGE, similar concentrations were found for the NagAa (35.2 kDa), NagAc (49.6 kDa), and NagAd (23.1 kDa) subunits from E. coli TG1/pBS(Kan)NDO− for wild-type NDO, variant G407S, variant F350T, and variant F350T/G407S as well as from E. coli JVQ2/pBS(Kan)NDO− for wild-type NDO, variant G50S/L225R/A269T, and variant L225R. Hence, the changes in catalytic activity of the NDO variants (changed regiospecificity and rates) were not due to changes in enzyme expression.
NagAc modeling.
The root mean square deviations of the modeled NDO was determined to be 0.07 Å, using 445 Cα atoms of the NahAc and NagAc alpha subunits, and the spatial configurations of the amino acids that coordinate the iron atoms of the Rieske Fe2-S2 center were conserved. The F350T substitution increases the distance between this residue and the three amino acids that coordinate the active-site mononuclear iron by approximately 2.4 Å, and the F350T substitution allows the formation of a new hydrogen bond to occur between the hydroxyl R-group of the threonine at position 350 and carboxyl group of peptide backbone for V346 (2.6 Å separation). Similarly, the L225R substitution allows the formation of a new hydrogen bond between the R-group amine moiety of R225 and the carboxyl moiety of the Y205 peptide backbone (2.8 Å separation). The R225 R-group becomes tucked inside the alpha-carbon backbone, potentially creating a void near the active-site mononuclear iron that was not permitted by the double methyl groups of the L225 R-group.
The G407S and G407T substitutions also allow the formation of a new hydrogen bond between the R-group hydroxyl group present in serine and threonine and the hydroxyl group on the R-group of S404 and the backbone carboxyl group of I406 (2.6 Å and 2.3 Å separation, respectively). The sulfur atom in the R-group for the G407C substitution generates a similar hydrogen bond between positions S404 and I406 (2.5 Å and 2.2 Å separation, respectively).
DISCUSSION
This is the first report to use protein engineering to increase the activity of a naphthalene dioxygenase toward nitroaromatic compounds. It is shown clearly in this paper that substitution of the F350 residue of the alpha subunit (NagAc) of NDO with T350 creates an enzyme with enhanced reaction rates toward the dinitrotoluene isomers, 26DNT and 23DNT, and 2A46DNT and that combining the F350T mutation with the G407S amino acid substitution enhances further the activity of the variant NDO toward nitroaromatic compounds. In addition, the L225R substitution generates a variant NagAc with enhanced catalytic activity toward 23DNT and 4A2NT (synthesizing the previously uncharacterized products 23DNBA and 4-amino-2-nitrocresol, respectively).
The alpha-subunit residues L225 and G407 have not been previously identified as influencing the catalytic activity of Rieske-dioxygenases (including biphenyl, toluene, and naphthalene dioxygenases) using DNA shuffling, molecular breeding, error-prone PCR, and site-directed mutagenesis (1, 18, 19, 27, 28, 31, 34, 38, 39, 41). Based on primary amino acid sequence alignments, NagAc L225 corresponds to L253 in the biphenyl dioxygenase alpha subunit from both Burkholderia xenovorans LB400 and Pseudomonas pseudoalcaligenes KF707, to M242 in the toluene dioxygenase alpha subunit from P. putida F1, and to L227 from NCIB NDO. NagAc G407 corresponds to A434 in the biphenyl dioxygenase alpha subunit from both B. xenovorans LB400 and P. pseudoalcaligenes KF707, to S423 in the toluene dioxygenase alpha subunit from P. putida F1, and to G409 from NCIB NDO. Both alpha subunits of the nitroarene dioxygenases from R34 DDO and DNT DDO encode L225 and S407 as well as the residues V350 and T350, respectively. The NagAc F350V variation was not found while screening the NagAc F350 saturation mutagenesis library.
Given the high rate of activity of NagAc F350T toward 26DNT, it is interesting that this variant does not recognize the substrate 4A26DNT, since both of these nitroaromatic compounds share a six-position nitro group. This lack of a relationship between activity and nitro group position is also apparent by comparing the activity of F350T toward 2A46DNT but not 24DNT, even though these compounds share a four-position nitro group. These differences in activity suggest that more than one of the functional groups on the benzene ring is dictating the orientation of the substrate inside of the active-site pocket. This also suggests that more than one amino acid substitution is necessary to convert a dioxygenase with a narrow range of nitroaromatic activity to one with a broad substrate range.
The importance of position L225 in the mutant G50S/L225R/A269T was proven through saturation mutagenesis and subsequent rounds of solid-phase screening. Variant L225R, without the G50S and A269T mutations, exhibited a higher rate of nitrite release from 23DNT than the parent NagAc triple mutant (Table 3). This enhancement in activity is possibly due to the removal of the slightly detrimental mutations G50S and A269T. Conversely, the combination of the beneficial F350T mutation with NagAc G50S/L225R/A269T appeared to deactivate the catalytic properties of NDO. In this quadruple mutant the proximity of both the L225R and F350T substitutions to the active site pocket may have caused an adverse conformational change of the active site pocket.
Though any assumption is highly speculative without extensive docking simulations, the observation that the NagAc F350T substitution permits a high rate of activity toward 26DNT and 2A46DNT implies that the smaller size of the threonine R-group versus the larger phenylalanine R-group may dictate the entrance of substrates into the NagAc active site. The results of this study also show that amino acids not nearby the NagAc active site can impact the regiospecific preference of the terminal oxygenase. This is clearly seen for the mutations L225R and G407S, which are greater than 10 Å and 15 Å, respectively, away from the active site mononuclear iron. It seems counterintuitive in the case of the NagAc variant L225R that an amino acid substitution in which the new amino acid has a larger R-group would promote increased reaction rates. However, the introduction of the positively charged arginine R-group may favor an electronic interaction with the negatively charged nitroaromatic compounds. Other than the formation of new hydrogen bonds, it is not clear how the G407S substitution influences the conformation of the NagAc active site or introduces new contacts with the various substrates.
Due to limitations in substrate specificity of the downstream degradation enzymes involved in naphthalene (40) and 24DNT (24) degradation, it is not our intention to suggest that nitroaromatic compounds may be completely mineralized by Ralstonia sp. strain U2 expressing the NDO variants described here. Beyond bioremediation, isomers of the hydroxylated products that we have synthesized may be used in reactions to produce pharmaceutically relevant compounds. For example, 24DNBA has been evaluated as a precursor for carbamates used in the synthesis of tumor treatments (3) and antibiotic prodrugs (9); hence, the 23DNBA product synthesized as described in this report from 23DNT, which is not commercially available, may be useful as a precursor. In addition, isomers of 4-amino-2-nitrocresol (generated here from the oxidation of 4A2NT) such as 6-amino-4-nitro-o-cresol and 2-amino-5-nitro-p-cresol have been evaluated as a precursors for fungicides (42) and antiparasitic agents (8), respectively.
An attempt to convert the naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4 to a nitroarene dioxygenase through the NahAc F352T mutation did not generate an NCIB NDO variant with activity towards 24DNT (26). Hence, the results shown in this current study may provide further evidence that the Ralstonia sp. strain U2 NDO is the archetypal nitroarene dioxygenase.
Acknowledgments
This study was supported by the National Science Foundation (BES-0114126) and by the U.S. Department of Education through the Graduate Assistance in Areas of National Need Award in Environmental Biotechnology (P200A000821).
We thank P. A. Williams for plasmid pWWF6, I. B. Lambert for E. coli JVQ2, A. Fishman for her help with the HPLC analyses, and A. Kind and D. Hill for their assistance with the LC-MS analyses.
REFERENCES
- 1.Brühlmann, F., and W. Chen. 1999. Tuning biphenyl dioxygenase for extended substrate specificity. Biotechnol. Bioeng. 63:544-551. [DOI] [PubMed] [Google Scholar]
- 2.Canada, K. A., S. Iwashita, H. Shim, and T. K. Wood. 2002. Directed evolution of toluene ortho-monooxygenase for enhanced 1-naphthol synthesis and chlorinated ethene degradation. J. Bacteriol. 184:344-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Culbert, P. A., A. V. Wearing, M. J. Chamberlain, and D. H. Hunter. 1993. Radioiodinated 2-nitrobenzyl carbamates as bioreductive alkylating agents for tissue hypoxia. Nucl. Med. Biol. 20:477-485. [DOI] [PubMed] [Google Scholar]
- 4.Danikiewicz, W., T. Bienkowski, and D. Poddebniak. 2004. Generation and reactions of anionic sigma-adducts of 1,3-dinitrobenzene and 1,3,5-trinitrobenzene with carbanions is a gas phase, using an electrospray ion source as the chemical reactor. J. Am. Soc. Mass. Spectrom. 15:927-933. [DOI] [PubMed] [Google Scholar]
- 5.Esteve-Núñez, A., A. Caballero, and J. L. Ramos. 2001. Biological degradation of 2,4,6-trinitrotoluene. Microbiol. Mol. Biol. Rev. 65:335-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuenmayor, S. L., M. Wild, A. L. Boyes, and P. A. Williams. 1998. A gene cluster encoding steps in conversion of naphthalene to gentisate in Pseudomonas sp. strain U2. J. Bacteriol. 180:2522-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashimoto, A., H. Sakino, T. Kojima, E. Yamagami, S. Tateishi, and T. Akiyama. 1982. Sources and behaviour of dinitrotoluene isomers in sea water. Water Res. 16:891-897. [Google Scholar]
- 8.Haugwitz, R. D., R. G. Angel, G. A. Jacobs, B. V. Maurer, V. L. Narayanan, L. R. Cruthers, and J. Szanto. 1982. Antiparasitic agents. 5. Synthesis and anthelmintic activities of novel 2-heteroaromatic-substituted isothiocyanatobenzoxazoles and benzothiazoles. J. Med. Chem. 25:969-974. [DOI] [PubMed] [Google Scholar]
- 9.Hay, M. P., B. M. Sykes, W. A. Denny, and C. J. O'Connor. 1999. Substituent effects on the kinetics of reductively initiated fragmentation of nitrobenzyl carbamates designed as triggers for bioreductive prodrugs. J. Chem. Soc. Perkin Trans. I 19:2759-2770. [Google Scholar]
- 10.Howe-Grant, M. 1991. Nitrobenzene and nitrotoluenes, p. 133-152. In M. Howe-Grant (ed.), Kirk-Othmer encyclopedia of chemical technology, 4th ed., vol. 17. Wiley-Interscience Publishers, New York, N.Y.
- 11.Jemal, M., and D. J. Hawthorne. 1999. Effect of high performance liquid chromatography mobile phase (methanol versus acetonitrile) on the positive and negative ion electrospray response of a compound that contains both an unsaturated cactone and a methyl sulfone group. Rapid Commun. Mass Spectrom. 13:61-66. [Google Scholar]
- 12.Johnson, G. R., R. K. Jain, and J. C. Spain. 2002. Origins of the 2,4-dinitrotoluene pathway. J. Bacteriol. 184:4219-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, G. R., B. F. Smets, and J. C. Spain. 2001. Oxidative transformation of aminodinitrotoluene isomers by multicomponent dioxygenases. Appl. Environ. Microbiol. 67:5460-5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, L. R., R. Davenport, H. Balbach, and D. J. Schaeffer. 1994. Phototoxicity. 3. Comparative toxicity of trinitrotoluene and aminodinitrotoluenes to Daphnia magna, Dugesia dorotocephala, and sheep erythrocytes. Ecotoxicol. Environ. Safe. 27:34-49. [DOI] [PubMed] [Google Scholar]
- 15.Karlsson, A., J. V. Parales, R. E. Parales, D. T. Gibson, H. Eklund, and S. Ramaswamy. 2003. Crystal structure of naphthalene dioxygenase: side-on binding of dioxygen to iron. Science 299:1039-1042. [DOI] [PubMed] [Google Scholar]
- 16.Keenan, B. G., T. Leungsakul, B. F. Smets, and T. K. Wood. 2004. Saturation mutagenesis of Burkholderia cepacia R34 2,4-dinitrotoluene dioxygenase at DntAc valine 350 for synthesizing nitrohydroquinone, methylhydroquinone, and methoxyhydroquinone. Appl. Environ. Microbiol. 70:3222-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keith, L. H., and W. A. Telliard. 1979. Priority pollutants. Environ. Sci. Technol. 13:416-423. [Google Scholar]
- 18.Kimura, N., A. Nishi, M. Goto, and K. Furukawa. 1997. Functional analysis of a variety of chimeric dioxygenases constructed from two biphenyl dioxygenases that are similar structurally but different functionally. J. Bacteriol. 179:3936-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumamaru, T., H. Suenaga, M. Mitsuoka, T. Watanabe, and K. Furukawa. 1998. Enhanced degradation of polychlorinated biphenyls by directed evolution of biphenyl dioxygenase. Nat. Biotechnol. 16:663-666. [DOI] [PubMed] [Google Scholar]
- 20.Lessner, D. J., G. R. Johnson, R. E. Parales, J. C. Spain, and D. T. Gibson. 2002. Molecular characterization and substrate specificity of nitrobenzene dioxygenase from Comamonas sp. strain JS765. Appl. Environ. Microbiol. 68:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leungsakul, T., B. G. Keenan, H. Yin, B. F. Smets, and T. K. Wood. Saturation mutagenesis of 2,4-DNT dioxygenase of Burkholderia sp. strain DNT for enhanced dinitrotoluene degradation. Biotechnol. Bioeng., in press. [DOI] [PubMed]
- 22.Meyer, A., A. Schmid, M. Held, A. H. Westphal, M. Rothlisberger, H.-P. E. Kohler, W. J. H. van Berkel, and B. Witholt. 2002. Changing the substrate reactivity of 2-hydroxybiphenyl 3-monooxygenase from Pseudomonas azelaica HBP1 by directed evolution. J. Biol. Chem. 277:5575-5582. [DOI] [PubMed] [Google Scholar]
- 23.Mori, M., M. Shoji, M. Sayama, T. Kondo, M. Inoue, and K. Kodaira. 2000. Secondary metabolism of dinitrobenzyl glucuronide related to production of genotoxic compounds of dinitrotoluene in male Wistar rat. J. Health Sci. 46:329-335. [Google Scholar]
- 24.Nishino, S. F., G. C. Paoli, and J. C. Spain. 2000. Aerobic degradation of dinitrotoluenes and pathway for bacterial degradation of 2,6-dinitrotoluene. Appl. Environ. Microbiol. 66:2139-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parales, J. V., A. Kumar, R. E. Parales, and D. T. Gibson. 1996. Cloning and sequencing of the genes encoding 2-nitrotoluene dioxygenase from Pseudomonas sp. JS42. Gene 181:57-61. [DOI] [PubMed] [Google Scholar]
- 26.Parales, R. E., K. Lee, S. M. Resnick, H. Jiang, D. J. Lessner, and D. T. Gibson. 2000. Substrate specificity of naphthalene dioxygenase: effect of specific amino acids at the active site of the enzyme. J. Bacteriol. 182:1641-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parales, R. E., J. V. Parales, and D. T. Gibson. 1999. Aspartate 205 in the catalytic domain of naphthalene dioxygenase is essential for activity. J. Bacteriol. 181:1831-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parales, R. E., S. M. Resnick, C.-L. Yu, D. R. Boyd, N. D. Sharma, and D. T. Gibson. 2000. Regioselectivity and enantioselectivity of naphthalene dioxygenase during arene cis-dihydroxylation: control by phenylalanine 352 in the α subunit. J. Bacteriol. 182:5495-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rickert, D. E., B. E. Butterworth, and J. A. Popp. 1984. Dinitrotoluene: acute toxicity, oncongenicity, genotoxicity, and metabolism. Crit. Rev. Toxicol. 13:217-234. [DOI] [PubMed] [Google Scholar]
- 30.Rui, L., Y. M. Kwon, A. Fishman, K. F. Reardon, and T. K. Wood. 2003. Saturation mutagenesis of toluene ortho-monooxygenase for enhanced 1-naphthol synthesis and chloroform degradation. Appl. Environ. Microbiol. 70:3246-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakamoto, T., J. M. Joern, A. Arisawa, and F. H. Arnold. 2001. Laboratory evolution of toluene dioxygenase to accept 4-picoline as a substrate. Appl. Environ. Microbiol. 67:3882-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2 ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Spanggord, R. J., J. C. Spain, S. F. Nishino, and K. E. Mortelmans. 1991. Biodegradation of 2,4-dinitrotoluene by a Pseudomonas sp. Appl. Environ. Microbiol. 57:3200-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suenaga, H., M. Mitsuoka, Y. Ura, T. Watanabe, and K. Furukawa. 2001. Directed evolution of biphenyl dioxygenase: emergence of enhanced degradation capacity for benzene, toluene, and alkylbenzene. J. Bacteriol. 183:5441-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valli, K., B. J. Brock, D. K. Joshi, and M. H. Gold. 1992. Degradation of 2,4-dinitrotoluene by the lignin-degrading fungus Phanerochaete chrysosporium. Appl. Environ. Microbiol. 58:221-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whiteway, J., P. Koziarz, J. Veall, N. Sandhu, P. Kumar, B. Hoecher, and I. B. Lambert. 1998. Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J. Bacteriol. 180:5529-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin, H., T. K. Wood, and B. F. Smets. 2005. Reductive transformation of TNT by Escherichia coli: pathway description. Appl. Microbiol. Biotechnol. 67: 397-404. [DOI] [PubMed] [Google Scholar]
- 38.Yu, C.-L., R. E. Parales, and D. T. Gibson. 2001. Multiple mutations at the active site of the naphthalene dioxygenase affect regioselectivity and enantioselectivity. J. Ind. Microbiol. Biotechnol. 27:94-103. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, N., B. G. Stewart, J. C. Moore, R. L. Greasham, D. K. Robinson, B. C. Buckland, and C. Lee. 2000. Directed evolution of toluene dioxygenase from Pseudomonas putida for improved selectivity toward cis-indandiol during indene bioconversion. Metab. Eng. 2:339-348. [DOI] [PubMed] [Google Scholar]
- 40.Zhou, N.-Y., J. Al-Dulayymi, M. S. Baird, and P. A. Williams. 2002. Salicylate 5-hydroxylase from Ralstonia sp. strain U2: a monooxygenase with close relationships to and shared electron transport proteins with naphthalene dioxygenase. J. Bacteriol. 184:1547-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zielinski, M., S. Kahl, H.-J. Hecht, and B. Hofer. 2003. Pinpointing biphenyl dioxygenase residues that are crucial for substrate interaction. J. Bacteriol. 185:6976-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zsolnai, T. 1961. New fungicides. II. Nitro-compounds. Biochem. Pharmacol. 5:287-304. [DOI] [PubMed] [Google Scholar]