Abstract
In Streptomyces coelicolor ParB is required for accurate chromosome partitioning during sporulation. Using a functional ParB-enhanced green fluorescent protein fusion, we observed bright tip-associated foci and other weaker, irregular foci in S. coelicolor vegetative hyphae. In contrast, in aerial hyphae regularly spaced bright foci accompanied sporulation-associated chromosome condensation and septation.
The normal growth and development of prokaryotes and eukaryotes requires proper replication and segregation of chromosomes. Progress in applying high-resolution fluorescence microscopy for the subcellular localization of proteins and specific chromosomal regions has revealed that chromosome segregation in rod-shaped bacteria is an active process (for reviews, see references 7, 21, 35, and 40). Among a number of proteins necessary for chromosome organization and localization during segregation, ParA and ParB were among the earliest identified and most extensively studied (reviewed in references 3 and 8). The ParAB homologues encoded by low-copy-number plasmids and required for their stable maintenance recruit plasmid molecules to specific positions (1/4 and 3/4) in the cell (29, 30). Chromosomal parAB genes have been identified and studied in many bacteria, including Pseudomonas putida (23), Caulobacter crescentus (28), and Bacillus subtilis (14, 31). Nevertheless, the exact function of the ParAB proteins in chromosome segregation is still not clear. Both genes are essential in C. crescentus (27), while soj (encoding the ParA homologue) and spo0J (encoding the ParB homologue) are not essential in B. subtilis but are required for proper chromosome partitioning during vegetative growth, particularly in rich medium; Spo0J is also required for initiation of sporulation (1, 14, 39).
ParB homologues are DNA-binding proteins interacting with partitioning sites (parS) (3, 8). Spo0J/ParB colocalizes with the oriC-proximal part of the B. subtilis and C. crescentus chromosomes (9, 22, 25, 28). In B. subtilis, Spo0J binds to eight parS sites in the 20% of the chromosome around the origin, clustering them into a large complex (24). The recently solved crystal structure of ParB from Thermus thermophilus revealed that the protein contains a helix-turn-helix (HTH) motif responsible for interaction with DNA (20). A mutant form of Spo0J defective in DNA binding is distributed evenly throughout the cytoplasm, as is a form of Spo0J containing a mutation in its C-terminal part, which is probably responsible for dimerization (1, 20). Thus, Spo0J/ParB forms a poorly characterized complex on DNA that presumably interacts with other segregation proteins or cellular structures. These activities of ParB depend on ParA (which belongs to the ATPase family), but the biochemical role of ParA remains obscure (3, 8).
Most studies of ParAB have focused on rod-shaped bacteria that divide by binary fission and have circular chromosomes whose replication cycle is followed by the cytologically obvious partitioning of sibling chromosomes. However, streptomycetes, gram-positive mycelial soil bacteria that produce many valuable antibiotics and other secondary metabolites, have large linear chromosomes with a centrally located oriC that does not show clear-cut partitioning during most of their morphologically complex life cycle (4, 12). Streptomyces species grow by tip extension to form a vegetative mycelium of branched hyphae in which septation occurs far from growing tips (5, 36). Compartments of vegetative hyphae contain several copies of uncondensed chromosomes. During further development of the Streptomyces coelicolor colony growing on an agar surface (but not in shaken submerged culture), new branches grow into the air to form a layer of white, fluffy aerial mycelium. After growth of an aerial hypha has stopped, a large number of FtsZ rings form a regular ladder in the long tip compartment, giving rise to sporulation septa that delimit unigenomic prespore compartments (38). Thus, sporulation requires a switch to a different mode of septation and demands DNA condensation and accurate chromosome partitioning into prespore compartments (26). During maturation of the spores, the compartments round up and the spore walls thicken and acquire color through synthesis of a polyketide pigment.
The large size and linearity of the Streptomyces chromosome (8.7 Mb for S. coelicolor, 9.0 Mb for S. avermitilis) (2, 33), the formation of multigenomic hyphae during growth (which, of course, do not have 1/4 and 3/4 positions), and the events that convert aerial hyphae into chains of prespore compartments all suggest that Streptomyces chromosome partitioning may be a complex and interesting issue. Partitioning genes parA and parB are present in the S. coelicolor A3(2) chromosome (17). As in other bacteria they are arranged in a two-gene operon. Disruption of the operon does not visibly affect colony growth, but chromosome partitioning aberrations are observed in about 13% of spores. Consistent with a role of ParB during sporulation, one of two parAB promoters is strongly expressed at the time immediately preceding sporulation (17). The S. coelicolor chromosome contains 24 16-bp parS sites, which is an unusually high number when compared with other analyzed bacterial chromosomes (15, 17). Most (21) of the parS sites are clustered within a 400-kb region around oriC. Our previous work describing ParB-DNA interactions, both in vitro and in vivo, has shown that ParB recognizes most of the parS sites. In fact, it binds to all the sites near to oriC; interaction with all but two oriC distal parS sites was detected in our in vivo assay. Additionally, probably because of oligomerization and/or some nonspecific DNA binding, the nucleoprotein complex seemed to spread along the DNA flanking parS sites, leading to the formation of large complexes encompassing a substantial segment of the chromosome around oriC (15). Remarkably, these complexes were detected in Streptomyces cells growing in liquid cultures, in vegetative hyphal compartments which usually have many copies of the chromosome, most of which are remote from sites of imminent cell division events, at which partitioning is most likely to be important.
To follow the assembly and localization of the ParB complexes during colony development, we have constructed a functional ParB-enhanced green fluorescent protein (EGFP) fusion. It enabled us to distinguish different behaviors of ParB in vegetative and aerial hyphae. Formation of the ParB complexes in aerial hyphae was correlated with particular stages of hyphal maturation and linked with efficient chromosome partitioning into spores.
A ParB-EGFP fusion protein is functional in S. coelicolor.
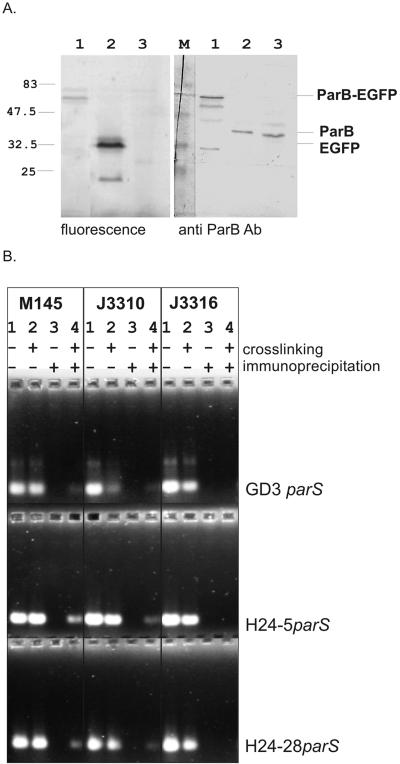
In order to localize oriC-linked ParB complexes in hyphae of S. coelicolor (15), we constructed strain J3310 (Table 1), which expressed chromosomally encoded EGFP-tagged ParB instead of the wild-type protein. To maximize the likelihood that the fusion protein would be functional, a 10-amino-acid flexible, proline- and glycine-rich linker was used as described earlier (13). For strain construction we used PCR targeting as described earlier (10, 11). Briefly, an egfp-aac3(IV)-oriT cassette (conferring apramycin resistance [Aprar]) was inserted downstream of parB in cosmid H24 (37) (Table 2). NdeI restriction sites flanking the aac3(IV)-oriT cassette allowed its excision by digestion of the engineered cosmid with NdeI and religation, providing a “clean” knock-in construct (H24parB-egfp). Subsequently, the kan gene in the SuperCos part of cosmid H24parB-egfp was exchanged for a vph-oriT cassette, yielding H24parB-egfp,kan::vio-oriT. This construct was used for conjugation into S. coelicolor J2538 [parAB::aac3(IV)]. Vior exconjugants were screened for the loss of both Vior and Aprar, indicating a double-crossover allelic exchange of the parAB locus of J2538 to give strain J3310 (Table 1). The presence of the EGFP-tagged ParB in S. coelicolor cell extracts was confirmed by scanning of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)-separated total proteins with a phosphorimager (FUJIFILM FLA5000) equipped with a 488-nm laser and by Western blotting as described earlier (15) (Fig. 1A).
TABLE 1.
Strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | supE44 ΔlacU169(φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Lab stock |
| BW25113/pIJ790 | K-12 derivative; ΔaraBAD ΔrhaBAD/λ-Red(gam bet exo) cat araC rep101(Ts) | 10 |
| ET12567/pUZ8002 | dam-13::Tn9 dcm cat tet hsdM hsdR zjj-201::Tn10/tra neo RP4 | 34 |
| S. coelicolor strains | ||
| M145 | SCP1− SCP2− | 2 |
| J2538 | M145 ΔparAB::apra | 17 |
| J3310 | M145 parB-egfp | This study |
| J3316 | M145 parB IGR(207-209)TYE-egfp | This study |
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea | Application |
|---|---|---|
| pparB-egfp-fw | CTTCTGGAGGGCGAGGACGAGGACGGGGACGCCGAGTCCCTGCCGGGCCCGGAGCTGCCGGGCCCGGAGGTGAGCAAGGGCGAGGAGCTG | Construction of egfp-aac3(IV)-oriT cassette |
| pegfp-apra-rv | GAAGCAGCTCCAGCCTACATATGATCTAGAGTCGCGGCCG NdeI | Construction of cassette |
| pegfp-apra-fw | CGGCCGCGACTCTAGATCATATGTAGGCTGGAGCTGCTTC NdeI | Construction of cassette |
| pparB-apra-rv | AAAGCAAAGGGCGGACCGTGTTCCGGTATGCACCGGACATATGATTCCGGGGATCCGTCGACC NdeI | Construction of cassette |
| pHTH-apra-fw | GAAGGACTTCAACTGCACGCATGACCAGCTGGCGGACCGTACGTAGGAACTTCATGAGCTCAGCC SnaBI | Introduction of mutation in parB HTH motif |
| pHTH-apra-rv | CTTCAGCAGACGCAGGGTGTTGGACACCTGCGGGCGGGACTCGTACGTAAGCTCCATCAGCAAAAGGGG SnaBI | Introduction of mutation in parB HTH motif |
Boldface indicates introduced changes, and italics indicate the restriction site.
FIG. 1.
Analysis of S. coelicolor strains expressing ParB-EGFP protein. A. SDS-PAGE and Western blotting of the cell extracts. Streptomyces cell extracts were subjected to SDS-PAGE. After fluorescence scanning of the gel (left panel), proteins were transferred to nitrocellulose and probed with anti-ParB antibody (right panel). Lane 1, J3310; lane 2, M145(pIJ8641) (J. Sun, unpublished data) expressing EGFP protein (32.5 kDa); lane 3, M145 wild type expressing wild-type ParB (40 kDa). B. Ethidium bromide-stained gel showing products of PCR performed on immunoprecipitated DNA fragments bound to ParB. Cross-linking followed by immunoprecipitation with anti-ParB antibodies was carried out on the wild-type control strain (M145), a strain expressing ParB-EGFP (J3310), and a strain expressing mutated ParB-EGFP (J3316). PCRs were carried out with primers flanking 6 of the 24 parS sites (examples for three of them, GD3, H24-5, and H24-28, are shown). Lanes: 1, input (not immunoprecipitated or cross-linked) DNA (positive control); 2, input (not immunoprecipitated) cross-linked DNA (positive control); 3, immunoprecipitated, not cross-linked DNA (negative control); 4, immunoprecipitated cross-linked DNA. The presence of a band in lane 4 indicates that the ParB protein of M145 and ParB-EGFP of J3310 were cross-linked to parS-containing DNA fragments.
A ParB-EGFP fusion protein of the expected size was produced at about wild-type levels, which implies correct autoregulation of ParB in the mutant strain (Fig. 1A). The ability of the fusion protein to bind DNA in vivo was tested by cross-linking of proteins to DNA in intact cells, followed by selective immunoprecipitation of protein-DNA complexes with antibodies raised against the ParB protein (performed as described earlier [15, 41]) (Fig. 1B). Complexes were indeed detected, recognition of parS sequences by ParB-EGFP seeming to be only slightly impaired in comparison to the wild-type ParB protein, as indicated by semiquantitative PCR. In addition, strain J3310 showed a close-to-wild-type frequency of anucleate spores (3%, compared to 1 to 2% in the wild type). This compares with the much higher frequencies observed with the parB deletion mutant J2537 (at least 13%) (17) and other parB mutants (D. Jakimowicz, unpublished data).
In vegetative mycelia, ParB-EGFP forms irregular foci that are often closely associated with hyphal tips.
Earlier studies indicated that although deletion of parB had an observable effect only in sporulating aerial hyphae, causing DNA partitioning defects, nevertheless ParB-DNA complexes were formed in submerged cultures in which no sporulation took place (15). This observation prompted us to examine the subcellular localization of ParB-EGFP in 1-day-old vegetative mycelia of strain J3310 growing on agar. Strains for microscopic observations were inoculated in the acute-angled junction of coverslips inserted at 45° in MM agar containing 1% mannitol (16). Confocal laser-scanning microscopy was carried out using a Leica SP2 microscope equipped with a 63× objective and 488- and 543-nm lasers.
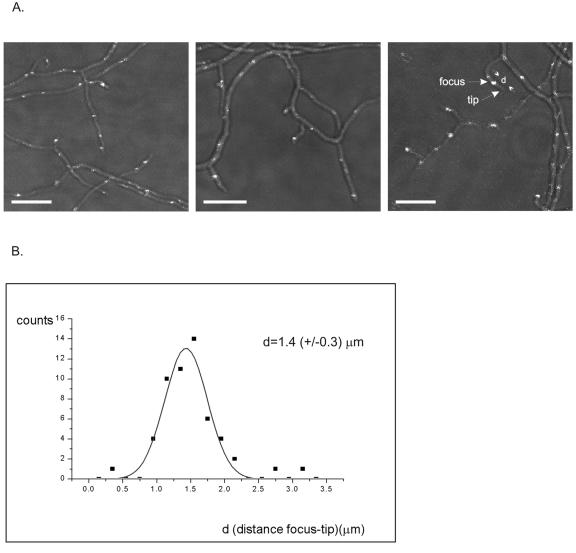
Green fluorescent foci of variable sizes, intensities, and distributions were seen in hyphal compartments (Fig. 2A). Cell wall staining (data not shown) did not indicate any spatial correlation between the foci and septa. The hyphal compartments contained several (usually four to eight) visible green foci. Distances between foci were variable, mostly ranging from 2 to 4 μm but quite often longer. Notably, 90% of hyphal branches had a strong focus near the tip and at a fairly constant distance from it (1.4 ± 0.3 μm; 52 measurements) (Fig. 2B).
FIG. 2.
Distribution of ParB-EGFP foci in vegetative mycelia. A. Strain J3310 was grown for 24 h, and the microscope samples were prepared without fixation and staining. Overlays of the fluorescence signals on phase-contrast images were generated to visualize the distribution of ParB-EGFP foci better. Bar, 5 μm. B. Distribution of distances measured from tips to the nearest ParB-EGFP foci (n = 52). The line is a Gaussian curve fitted to the plot.
ParB binding to the apical chromosomes may be a secondary effect of some difference in the organization or replication state of such chromosomes, or it may serve a special purpose, such as helping to anchor the apical chromosome to the elongating tip (or the nascent tip in emerging branches), thereby assuring correct chromosome position in relation to the sites of cell wall extension at hyphal tips.
The occurrence of the nonapical foci implies that ParB complexes form as part of the replication cycle of the chromosomes during vegetative growth. In S. coelicolor hyphae expressing ParB-EGFP, the distances between observed foci, although variable, were usually longer than the distances separating the oriC regions observed by Yang and Losick using fluorescence in situ hybridization (43) (to some extent these differences may be the result of different culture conditions). The generally low and irregular intensity of ParB foci indicates that some of them are not fully compacted, as if they form only transiently during chromosome replication and segregation. The formation of a ParB-DNA complex as one of the factors regulating DNA replication initiation has also been proposed for B. subtilis. Ogura et al. (32) found that high levels of ParB (Spo0J) interfered with the tight control of replication initiation, while Lee et al. (19) showed that spo0J deletion mutants had an increased chromosomal copy number, probably because of premature initiation. If such a model were true in Streptomyces, differential ParB complex aggregation could cause replication asynchrony within the compartment (a phenomenon that would also be consistent with the Yang and Losick [43] observation that double signals corresponding to the recently replicated oriC region were rare and not found in clusters). However, parB deletion mutants did not show any obvious abnormalities in hyphal growth and branching, and the functional significance, if any, of the complexes still needs to be determined.
ParB-EGFP assembles into arrays of bright complexes in sporulating aerial hyphae.
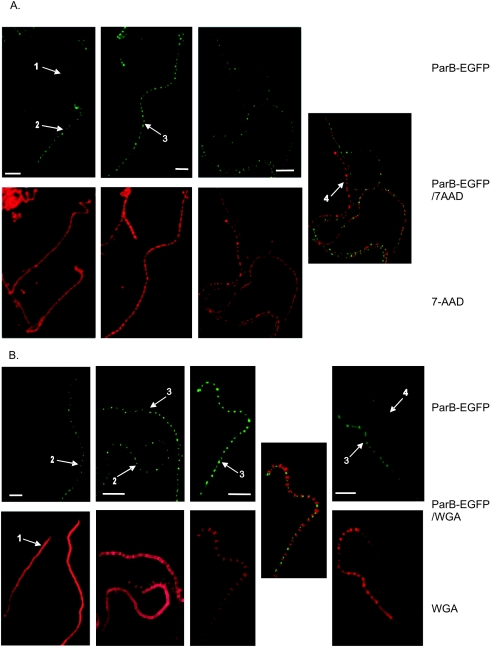
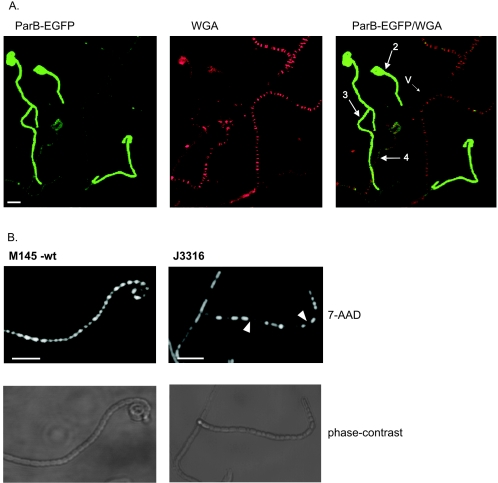
The generation of anucleate spores by a parB deletion mutant, and the strong up-regulation of one of two parAB promoters (p2) at a time correlating with the appearance of aerial mycelium growth in the wild type (17), indicated that ParB fulfills a particular role during aerial hyphae development and that its behavior in aerial hyphae might differ from that in vegetative hyphae. To demonstrate that the temporal correlation with the aerial growth is actually a spatial one, and to correlate the formation of ParB complexes with particular stages of aerial hyphal maturation, we prepared coverslip samples from cultures of strain J3310 (44-h growth). Staining procedures were as described previously (38). The samples were stained with 7-aminoactinomycin D (7-AAD) to visualize DNA or with wheat germ agglutinin-tetramethylrhodamine conjugate (WGA) to visualize the cell wall. WGA, used for cell wall staining, binds to small oligomers of N-acetylglucosamine in incompletely polymerized or lysozyme-treated peptidoglycan. We found that aerial hyphae were much more sensitive to WGA staining than were vegetative hyphae.
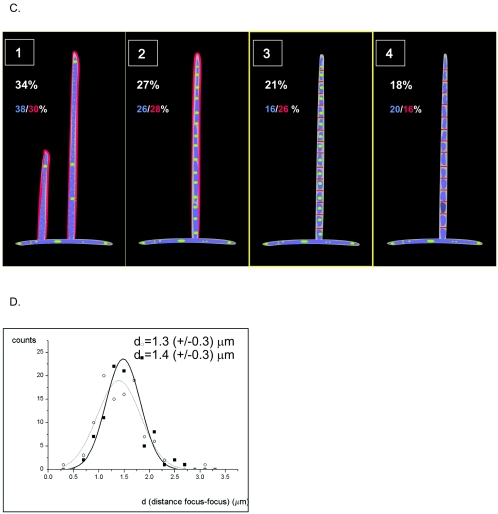
In young and short aerial hyphae recognized by strong cell wall staining, ParB foci of the vegetative hyphae were replaced by more diffuse fluorescence. In the longer and older branches of aerial hyphae, still showing the strong lateral wall staining but having no observable DNA condensation and sporulation septation, ParB assembled into bright and regularly spaced foci (Fig. 3). ParB complexes were also present in some hyphae containing visibly condensed and segregated chromosomes and septa. In such hyphae, foci overlapped with stained DNA and, as revealed by septal staining, single foci were usually situated in the middle of prespore compartments (although we found a few compartments containing two foci). The spacing measured between ParB foci in preseptation aerial hyphae was fairly uniform (1.3 ± 0.3 μm, measured for 100 foci) and similar to spacing in the hyphae containing fully developed prespore compartments (1.4 ± 0.3 μm; 104 foci measured) (Fig. 3D) and to distances measured between sporulation septa (1.3 ± 0.2 μm, measured for 94 septa) (38). Prespore chains lacking foci often showed faint diffuse fluorescence, which disappeared entirely in mature spore chains. Presumably, disaggregating of complexes was followed by eventual degradation.
FIG. 3.
Localization of ParB-EGFP foci in aerial hyphae of J3310. Numbered arrows refer to developmental stages shown in panel C. A. Images of ParB-EGFP fluorescence (top row), DNA stained with 7-AAD (bottom row), and an overlay of the two fluorescence signals. B. Images of ParB-EGFP fluorescence (top row), cell walls stained with WGA conjugate (bottom row), and an overlay of the two fluorescence signals. C. Diagram showing sequential stages of aerial hyphal development with respect to ParB-EGFP focus formation (see Results) (red, cell wall; green, ParB; blue, DNA). Percentages show the frequencies of occurrence of hyphae in the particular stage (white number, average; blue number, determined in DNA-stained images; red number, determined in cell wall-stained images). D. Gaussian distribution of distances measured between ParB-EGFP complexes in arrays of ParB-EGFP foci. Empty circles and thin line, measurement for hyphae in developmental stage 2 (n = 100); black squares and thick line, measurement obtained for hyphae containing septa (n = 104).
To define stages of aerial hypha development and the occurrence of ParB foci more precisely, we examined substantial numbers of presporulation aerial hyphae (211 in DNA-stained preparations and 316 in cell wall-stained preparations). They could be grouped into four developmental stages with respect to ParB-EGFP focus formation (examples are labeled 1 to 4 in Fig. 3C). A total of 61% of aerial hyphae showed continuous DNA staining (7-AAD staining) and no sign of sporulation septation but exhibited bright staining of apparently laterally growing cell walls (WGA staining). Of these, about half lacked EGFP foci (stage 1) and about half contained them (stage 2) (Fig. 3C). The third and fourth stages were defined by hyphae with condensed DNA or sporulation septa. Again, about half contained regular ParB-EGFP complexes. It seems that ParB complexes are relatively transient during aerial hyphal development and ParB aggregates on the oriC region, which reaches a location close to the prespore mid-point before chromosome condensation and cross-wall formation. We therefore believe that ParB complexes form around each replication origin after the final round of DNA replication in aerial hyphae has been initiated. Though sporulation-associated chromosome condensation in S. coelicolor does not depend on formation of the ParB complexes (it occurs normally in parB deletion mutants [17]), ParB-DNA complex assembly may be coupled to inhibition of further rounds of replication and may contribute to proper positioning of the oriC regions.
The helix-turn-helix region of ParB is crucial for complex formation and for proper DNA partitioning.
In order to prove that the observed ParB-EGFP foci reflected ParB complexes assembled on DNA rather than on some other anchor, and that they were not nonfunctional “inclusion bodies,” a mutation was introduced in the DNA binding domain of ParB. Based on sequence alignments and comparison to the crystal structure of the T. thermophilus protein, the middle of S. coelicolor ParB is predicted to contain an HTH motif (20). PCR targeting was used to insert an aac3(IV) cassette flanked by SnaBI restriction sites into the HTH motif of parB in cosmid H24parB-egfp,kan::vph-oriT. The SnaBI sites were flanked by a modified nucleotide sequence such that excision of the apra cassette with SnaBI, and religation, resulted in the substitution of three key amino acids, IGR (207 to 209), in the core of the HTH motif, by PYE (20) (Table 2). Chromosomal DNA of all strains constructed was checked by PCR, and cell extracts were checked by phosphorimager scanning of SDS-PAGE gels and by Western blotting using antibodies against the ParB protein. Cross-linking of J3316 and immunoprecipitation of mutated ParB protein was performed as described for strain J3310. Upon amplification of the immunoprecipitated material with primers flanking parS sequences, we obtained no PCR product, proving that binding of mutated ParB to DNA had been abolished (Fig. 1B). Microscopic examination revealed a complete absence of recognizable fluorescent foci in both vegetative and aerial mycelia. In vegetative hyphae only weak diffuse fluorescence could be observed, while growing aerial hyphae showed a very bright, but still diffuse signal (Fig. 4A). The strong fluorescence was observed both in hyphae containing unsegregated DNA and showing no septation and in hyphae with septa or condensed chromosomes. This strong and long-lived fluorescence was probably due to a lack of autoregulation by non-DNA-binding ParB protein (our transcription data clearly indicate autorepression of the parABp2 promoter [Jakimowicz, unpublished]). Our result is consistent with a requirement for DNA binding for ParB complex assembly, although we cannot rule out the possibility that the HTH mutation affects some other property in addition to DNA binding.
FIG. 4.
ParB-EGFP mutated in its DNA-binding motif is nonfunctional. A. Abolition of complex formation in strain J3316, as demonstrated in sample images of J3316 hyphae showing ParB-EGFP fluorescence, cell walls stained with WGA conjugate, and overlay of the two fluorescence signals. Numbered arrows indicate hyphae in various developmental stages (see Results and Fig. 3). v, vegetative hyphae. Bar, 5 μm. B. DNA segregation defect in strain J3316. Images of spores in the wild-type M145 and in J3316. Samples were stained for DNA visualization with 7-AAD. Arrows indicate some of the anucleate spores. Bar, 5 μm.
In order to test if the ParB complex assembly is needed for efficient partitioning, we screened strain J3316 for defective DNA segregation into spores. Indeed, spore nucleoid staining was much more heterogeneous than in the wild type, and about 14% percent of spores of strain J3316 were anucleate (Fig. 4B), confirming that ParB-DNA complex formation is essential for proper chromosome segregation at this stage. One way in which this could be achieved might be the directly ParB-dependent placement of the oriC region at regular intervals, with some aspect of the intervals between oriC regions favoring the formation of FtsZ rings. It is also possible that the ParB function in chromosome segregation is indirect and its only role is the proper spatial organization of the central, oriC proximal part of the chromosome, which is necessary for its correct segregation. In both models, parB mutants would have disordered positioning or organization of the oriC region. On one hand, this might permit enlargement of one oriC-oriC interval, leading to the formation of additional FtsZ rings and a DNA-free compartment or, on the other hand, it might prevent the formation of an FtsZ ring between closely associated and unsegregated sibling oriC regions, giving rise to a compartment with more than one chromosome. In fact, separation of nucleoids in Streptomyces is not seen until septa start forming, and ingrowth of septa can be observed over nonseparated chromosomes (18, 38).
In the case of B. subtilis, initial evidence that a key role of Spo0J was to anchor the oriC region to the cell pole (39) has not been supported by later work (6, 19, 42). It has recently been suggested that Spo0J contributes to compaction or spatial organization of the oriC region, which may influence origin separation (indeed, sister origins were observed to stay closer together in a spo0J mutant) and the initiation of replication (19). We may therefore speculate that Streptomyces ParB could also aid proper distribution of chromosomes along the aerial hyphae by a mechanism involving compaction of the DNA around oriC.
In summary, we have shown that ParB has unusually complex behavior in Streptomyces, in that it assembles into differently arranged complexes in vegetative and aerial hyphae. We have provided data indicating that direct interaction of ParB with DNA is necessary for efficient chromosome segregation in aerial hyphae. It is plausible that interaction of ParB with some cellular component present only in aerial hyphae is a prerequisite for proper complex assembly and/or localization. Further studies are necessary to find the other components of the segregation machinery and the signals that regulate ParB complex formation.
Acknowledgments
We are grateful to Mervyn Bibb, Klas Flardh, David Hopwood, and Tobias Kieser for valuable comments on the manuscript.
D.J. was supported by a Marie Curie Fellowship of the European Community Programme Human Potential under contract number HPMF-CT-2002-01676, and J.Z.-C. acknowledges support from the British-Polish Research Partnership Programme (Polish Committee of Scientific Studies, British Council). B.G. was supported by grants IGF12431 and EGH16080 from the Biotechnological and Biological Research Council of the United Kingdom.
REFERENCES
- 1.Autret, S., R. Nair, and J. Errington. 2001. Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis: evidence for separate domains involved in DNA binding and interactions with Soj protein. Mol. Microbiol. 41:743-755. [DOI] [PubMed] [Google Scholar]
- 2.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Bignell, C., and C. M. Thomas. 2001. The bacterial ParA-ParB partitioning proteins. J. Biotechnol. 91:34. [DOI] [PubMed] [Google Scholar]
- 4.Chater, K. F. 2001. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr. Opin. Microbiol. 4:667-673. [DOI] [PubMed] [Google Scholar]
- 5.Flardh, K. 2003. Growth polarity and cell division in Streptomyces. Curr. Opin. Microbiol. 6:564-571. [DOI] [PubMed] [Google Scholar]
- 6.Frandsen, N., I. Barak, C. Karmazyn-Campelli, and P. Stragier. 1999. Transient gene asymmetry during sporulation and establishment of cell specificity in Bacillus subtilis. Genes Dev. 13:394-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerdes, K., J. Moller-Jensen, G. Ebersbach, T. Kruse, and K. Nordstrom. 2004. Bacterial mitotic machineries. Cell 116:359-366. [DOI] [PubMed] [Google Scholar]
- 8.Gerdes, K., J. Moller-Jensen, and R. Bugge Jensen. 2000. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 37:455-466. [DOI] [PubMed] [Google Scholar]
- 9.Glaser, P., M. E. Sharpe, B. Raether, M. Perego, K. Ohlsen, and J. Errington. 1997. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev. 11:1160-1168. [DOI] [PubMed] [Google Scholar]
- 10.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gust, B., G. Chandra, D. Jakimowicz, T. Yuqing, C. J. Bruton, and K. F. Chater. 2004. Lambda red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv. Appl. Microbiol. 54:107-128. [DOI] [PubMed] [Google Scholar]
- 12.Hopwood, D. A., K. F. Chater, and M. J. Bibb. 1995. Genetics of antibiotic production in Streptomyces coelicolor A3(2), a model streptomycete. Biotechnology 28:65-102. [DOI] [PubMed] [Google Scholar]
- 13.Imai, Y., N. Ogasawara, D. Ishigo-Oka, R. Kadoya, T. Daito, and S. Moriya. 2000. Subcellular localization of DNA-initiation proteins of Bacillus subtilis: evidence that chromosome replication begins at either edge of the nucleoids. Mol. Microbiol. 36:1037-1048. [DOI] [PubMed] [Google Scholar]
- 14.Ireton, K., N. W. Gunther, and A. D. Grossman. 1994. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 176:5320-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jakimowicz, D., K. Chater, and J. Zakrzewska-Czerwinska. 2002. The ParB protein of Streptomyces coelicolor A3(2) recognizes a cluster of parS sequences within the origin-proximal region of the linear chromosome. Mol. Microbiol. 45:1365-1377. [DOI] [PubMed] [Google Scholar]
- 16.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 17.Kim, H. J., M. J. Calcutt, F. J. Schmidt, and K. F. Chater. 2000. Partitioning of the linear chromosome during sporulation of Streptomyces coelicolor A3(2) involves an oriC-linked parAB locus. J. Bacteriol. 182:1313-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwak, J., and K. E. Kendrick. 1996. Bald mutants of Streptomyces griseus that prematurely undergo key events of sporulation. J. Bacteriol. 178:4643-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, P. S., D. C. Lin, S. Moriya, and A. D. Grossman. 2003. Effects of the chromosome partitioning protein Spo0J (ParB) on oriC positioning and replication initiation in Bacillus subtilis. J. Bacteriol. 185:1326-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leonard, T. A., P. J. Butler, and J. Lowe. 2004. Structural analysis of the chromosome segregation protein Spo0J from Thermus thermophilus. Mol. Microbiol. 53:419-432. [DOI] [PubMed] [Google Scholar]
- 21.Lewis, P. J. 2001. Bacterial chromosome segregation. Microbiology 147:519-526. [DOI] [PubMed] [Google Scholar]
- 22.Lewis, P. J., and J. Errington. 1997. Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the SpoOJ partitioning protein. Mol. Microbiol. 25:945-954. [DOI] [PubMed] [Google Scholar]
- 23.Lewis, R. A., C. R. Bignell, W. Zeng, A. C. Jones, and C. M. Thomas. 2002. Chromosome loss from par mutants of Pseudomonas putida depends on growth medium and phase of growth. Microbiology 148:537-548. [DOI] [PubMed] [Google Scholar]
- 24.Lin, D. C., and A. D. Grossman. 1998. Identification and characterization of a bacterial chromosome partitioning site. Cell 92:675-685. [DOI] [PubMed] [Google Scholar]
- 25.Lin, D. C., P. A. Levin, and A. D. Grossman. 1997. Bipolar localization of a chromosome partition protein in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 94:4721-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCormick, J. R., E. P. Su, A. Driks, and R. Losick. 1994. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol. Microbiol. 14:243-254. [DOI] [PubMed] [Google Scholar]
- 27.Mohl, D. A., J. Easter, Jr., and J. W. Gober. 2001. The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus. Mol. Microbiol. 42:741-755. [DOI] [PubMed] [Google Scholar]
- 28.Mohl, D. A., and J. W. Gober. 1997. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell 88:675-684. [DOI] [PubMed] [Google Scholar]
- 29.Niki, H., and S. Hiraga. 1997. Subcellular distribution of actively partitioning F plasmid during the cell division cycle in E. coli. Cell 90:951-957. [DOI] [PubMed] [Google Scholar]
- 30.Niki, H., and S. Hiraga. 1999. Subcellular localization of plasmids containing the oriC region of the Escherichia coli chromosome, with or without the sopABC partitioning system. Mol. Microbiol. 34:498-503. [DOI] [PubMed] [Google Scholar]
- 31.Ogasawara, N., and H. Yoshikawa. 1992. Genes and their organization in the replication origin region of the bacterial chromosome. Mol. Microbiol. 6:629-634. [DOI] [PubMed] [Google Scholar]
- 32.Ogura, Y., N. Ogasawara, E. J. Harry, and S. Moriya. 2003. Increasing the ratio of Soj to Spo0J promotes replication initiation in Bacillus subtilis. J. Bacteriol. 185:6316-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Omura, S., H. Ikeda, J. Ishikawa, A. Hanamoto, C. Takahashi, M. Shinose, Y. Takahashi, H. Horikawa, H. Nakazawa, T. Osonoe, H. Kikuchi, T. Shiba, Y. Sakaki, and M. Hattori. 2001. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. USA 98:12215-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paget, M. S., L. Chamberlin, A. Atrih, S. J. Foster, and M. J. Buttner. 1999. Evidence that the extracytoplasmic function sigma factor sigma E is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pogliano, K., J. Pogliano, and E. Becker. 2003. Chromosome segregation in Eubacteria. Curr. Opin. Microbiol. 6:586-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prosser, J. I., and A. J. Tough. 1991. Growth mechanisms and growth kinetics of filamentous microorganisms. Crit. Rev. Biotechnol. 10:253-274. [DOI] [PubMed] [Google Scholar]
- 37.Redenbach, M., H. M. Kieser, D. Denapaite, A. Eichner, J. Cullum, H. Kinashi, and D. A. Hopwood. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21:77-96. [DOI] [PubMed] [Google Scholar]
- 38.Schwedock, J., J. R. McCormick, E. R. Angert, J. R. Nodwell, and R. Losick. 1997. Assembly of the cell division protein FtsZ into ladder-like structures in the aerial hyphae of Streptomyces coelicolor. Mol. Microbiol. 25:847-858. [DOI] [PubMed] [Google Scholar]
- 39.Sharpe, M. E., and J. Errington. 1996. The Bacillus subtilis soj-spo0J locus is required for a centromere-like function involved in prespore chromosome partitioning. Mol. Microbiol. 21:501-509. [DOI] [PubMed] [Google Scholar]
- 40.Sherratt, D. J. 2003. Bacterial chromosome dynamics. Science 301:780-785. [DOI] [PubMed] [Google Scholar]
- 41.Solomon, M. J., and A. Varshavsky. 1985. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc. Natl. Acad. Sci. USA 82:6470-6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, L. J., and J. Errington. 2002. A large dispersed chromosomal region required for chromosome segregation in sporulating cells of Bacillus subtilis. EMBO J. 21:4001-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang, M. C., and R. Losick. 2001. Cytological evidence for association of the ends of the linear chromosome in Streptomyces coelicolor. J. Bacteriol. 183:5180-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]