Abstract
Several mycoplasma species are known to glide on solid surfaces such as glass in the direction of the membrane protrusion, but the mechanism underlying this movement is unknown. To identify a novel protein involved in gliding, we raised monoclonal antibodies against a detergent-insoluble protein fraction of Mycoplasma mobile, the fastest glider, and screened the antibodies for inhibitory effects on gliding. Five monoclonal antibodies stopped the movement of gliding mycoplasmas, keeping them on the glass surface, and all of them recognized a large protein in immunoblotting. This protein, named Gli521, is composed of 4,738 amino acids, has a predicted molecular mass of 520,559 Da, and is coded downstream of a gene for another gliding protein, Gli349, which is known to be responsible for glass binding during gliding. Edman degradation analysis indicated that the N-terminal region is processed at the peptide bond between the amino acid residues at positions 43 and 44. Analysis of gliding mutants isolated previously revealed that the Gli521 protein is missing in a nonbinding mutant, m9, where the gli521 gene is truncated by a nonsense mutation at the codon for the amino acid at position 1170. Immunofluorescence and immunoelectron microscopy indicated that Gli521 localizes all around the base of the membrane protrusion, at the “neck,” as previously observed for Gli349. Analysis of the inhibitory effects of the anti-Gli521 antibody on gliding motility revealed that this protein is responsible for force generation or force transmission, a role distinct from that of Gli349, and also suggested conformational changes of Gli349 and Gli521 during gliding.
Mycoplasmas are parasitic or commensal bacteria with small genome sizes that lack a peptidoglycan layer (26). Several mycoplasma species have a membrane protrusion at a cell pole, such as the “attachment organelle” of Mycoplasma pneumoniae or the “head-like structure” of Mycoplasma mobile, and exhibit gliding motility, defined as a smooth translocation of cells on solid surfaces in the direction of the membrane protrusion (2, 12, 17, 19). Mycoplasmas have no flagella or pili on their cell surfaces and no homologs of genes that encode pili, flagella, or other genes related to bacterial motility. Nor have homologs of motor proteins that are common in eukaryotic motility been detected (3, 5, 7, 10, 25, 29). These observations suggest that mycoplasmas glide by an entirely unknown mechanism, although the gliding motility of M. mobile is thought to depend on the cellular concentration of ATP (9).
M. mobile was isolated from the gill organ of a fish (13). It provides an opportunity to study mycoplasma gliding motility, because this species is the fastest gliding and, unlike other species, glides without interruption (9, 21, 24, 27, 34). At all stages of growth, M. mobile glides smoothly and continuously on glass at an average speed of 2.0 to 4.5 μm/s or three to seven times the length of the cell per second, exerting a force of up to 27 pN (21).
Subcellular localization of surface proteins detected by monoclonal antibodies suggested that the cell surface is differentiated into three parts, “head,” “neck,” and “body,” starting from the pole of protrusion (16). Recently, we identified a novel protein, Gli349 with a molecular mass of 349 kDa, responsible for adhesion to animal cells (34). Gli349 clusters at the neck, which is believed to be specialized for binding and gliding (16, 34). Analysis of inhibitory effects of an anti-Gli349 antibody on gliding revealed that Gli349 is responsible for glass binding during gliding (34). Rapid-freeze-and-freeze-fracture rotary-shadow electron microscopy revealed many spike-like structures 50 nm in length sticking out around the neck and bound to the glass surface at their distal ends, while the spike structure cannot be found in a nonbinding and consequently nongliding mutant (20). These observations suggest that the spike includes the Gli349 molecule and functions as a “leg” in the gliding mechanism (19, 20, 34). However, it is likely that other proteins also are involved in the gliding mechanism, because biological motility systems generally comprise two or more proteins, and the gli349 gene is encoded in an operon composed of four open reading frames (ORFs) (10, 34).
In this study, we isolated monoclonal antibodies that blocked movement but not binding and identified their target, a novel protein of molecular mass 521 kDa.
MATERIALS AND METHODS
Strains and culture conditions.
M. mobile strain 163K (ATCC 43663) and its mutants were grown at 25°C in Aluotto medium (1, 24), consisting of 2.1% heart infusion broth, 0.56% yeast extract, 10% horse serum, 0.025% thallium acetate, and 0.005% ampicillin. Cells were cultured to reach an optimal density at 600 nm of 0.07 (corresponding to 7 × 108 CFU/ml). Mycoplasma pulmonis (ATCC 19612) was grown at 37°C in the same medium.
Making and screening of monoclonal antibody.
A triton-insoluble fraction was prepared as described previously (34), suspended in phosphate-buffered saline (75 mM Na-phosphate, pH 7.4, 68.4 mM NaCl), and emulsified in complete Freund's adjuvant. Antibodies were raised by two separate methods, using different rodents. In the first method, an emulsion containing 1 mg of protein per animal was injected into footpads of female Wistar rats (8 weeks old), as previously described (14). Two weeks later, the animals were sacrificed, and the iliac lymph nodes were excised. In the second method, female BALB/c mice (6 weeks old) were injected intraperitoneally with an emulsion containing 300 μg of protein, as previously described (6, 16). A second injection was done 3 weeks later with 150 μg of protein emulsified with Freund's incomplete adjuvant. One week after the second injection, the mice were injected with 150 μg of protein without adjuvant. The mice were sacrificed 1 week later, and the spleen cells were excised and dissociated. In both cases, the isolated immunized cells were fused with mouse myeloma cells and screened for antibodies (6, 14, 16). The first screening of hybridoma cells was done for both antibody production ability and the subcellular localization of the target structure visualized by immunofluorescence microscopy, using the hybridoma supernatant (30, 32). The second screening was done by a tunnel test as described previously, which is based on the inhibitory effect to gliding (16, 34). Briefly, mycoplasma cells in Aluotto medium were allowed to adhere to a glass coverslip, which served as the top of a rudimentary flow chamber constructed with a microscope slide and double-stick tape. Then, hybridoma supernatant diluted with 10 volumes of Aluotto medium was allowed to flow into the chamber. Hybridomas whose supernatant had inhibitory effects on gliding mycoplasmas were selected.
Identification of Gli521 protein.
The target proteins of the antibodies that inhibit movement were detected by immunoblotting analysis. To identify the target protein of antibodies that can stop gliding, the whole-cell lysate containing 30 μg of protein per lane was fractionated by electrophoresis in sodium dodecyl sulfate (SDS) on 5.5% acrylamide gels. The protein band reactive to the antibody was subjected to the Cleveland method for partial digestion, as described previously (4, 34), transferred to an Immobilon-PSQ membrane (Millipore, Inc., Billerica, MA), stained by amido black, and subjected to Edman degradation. Edman degradation was also carried out with the protein band directly blotted to the membrane. To confirm the identity between Gli521 and the target of the inhibitory antibodies, partial fragments of the gli521 gene were amplified by PCR, ligated into pGEM-T Easy (Promega), excised by using NcoI and XhoI restriction enzymes, and inserted into an expression vector pGEX-KG using the restriction sites. The recombinant proteins were expressed in Escherichia coli cells (BL21), and whole-cell lysates were examined by immunoblotting for reactivity to the inhibitory antibodies.
DNA sequencing.
To identify the mutation in the gli521 gene of the m9 mutant of M. mobile, genomic DNA was prepared by the Genomic-tip system (QIAGEN, Hilden, Germany) and amplified by PCR as 1-kb fragments and then sequenced for the 30-kb region previously sequenced (34). The genomic DNA of M. pulmonis was prepared by phenol extraction (28) from a culture of the type culture strain. The gap regions of M. pulmonis ORFs between MYPU_2120 and MYPU_2130 and between MYPU_2140 and MYPU_2130 were amplified by PCR, using the primers CCACAATCAATTAAAGACTTGAG and TGTTCTTTAACAGCATCACC and primers CACCTAACTTTTGCAAGTGC and GTAAGATCAGAAGAGACAAC, respectively. The PCR products were purified by agarose gel electrophoresis and subjected to sequencing analyses for both strands, using the primers listed above.
Fluorescence, electron, and video microscopy.
Antibodies were purified from the hybridoma supernatant by using a HiTrap protein G column (Amersham). Immunofluorescence microscopy was done with permeabilized cells, as described previously (30, 32, 34). Anti-Gli521 antibody (0.3 μg/ml) was used as the primary antibody, and anti-rat antibody conjugated by Alexa594 (Molecular Probes) was used as the secondary antibody. Double staining was done by simultaneous additional staining using 0.1 μg/ml anti-Gli349 antibody (34) and anti-mouse antibody conjugated by Alexa488. Immunoelectron microscopy was done as previously described with additional treatment by 0.1% Triton X-100 in phosphate-buffered saline for 15 min at room temperature prior to the first antibody treatment (34). The effects of antibodies on gliding were studied in a tunnel slide and recorded by video microscopy, as reported previously (21, 23, 34).
RESULTS
Making inhibitory antibody.
To identify a novel protein involved in the gliding mechanism, we made and screened antibodies that can stop mycoplasma gliding without removing cells from the glass. We previously obtained the anti-Gli349 antibody by immunizing mice with whole mycoplasma cells (16). Here, we sought to raise antibodies that were different from those found earlier by immunizing both rats and mice with a triton-insoluble fraction. Four and two hybridoma cell lines established from rat and mouse, respectively, were found to recognize structures localized at the neck, a region believed to be responsible for glass binding and gliding (16, 34). Antibodies were added to cells in a tunnel slide to examine their effects on gliding (16, 34). One of the antibodies rapidly displaced cells from glass. Immunoblot analysis revealed that this antibody recognized Gli349, which was previously shown to be involved in gliding (34). The remaining five antibodies inhibited gliding, but cells remained bound to the glass for more than several minutes (Fig. 1). Incubation with low concentrations of these antibodies eventually resulted in displacement of cells from glass, as presented later.
FIG. 1.
Inhibitory effects of an antibody on gliding. Phase-contrast images of cells before (left) and 15 s after (right) addition of 300 μg/ml of the inhibitory antibody, obtained by superimposing video frames over a 2-s time span. The last frame was displayed in a different color. Bar, 5 μm.
Identification of Gli521.
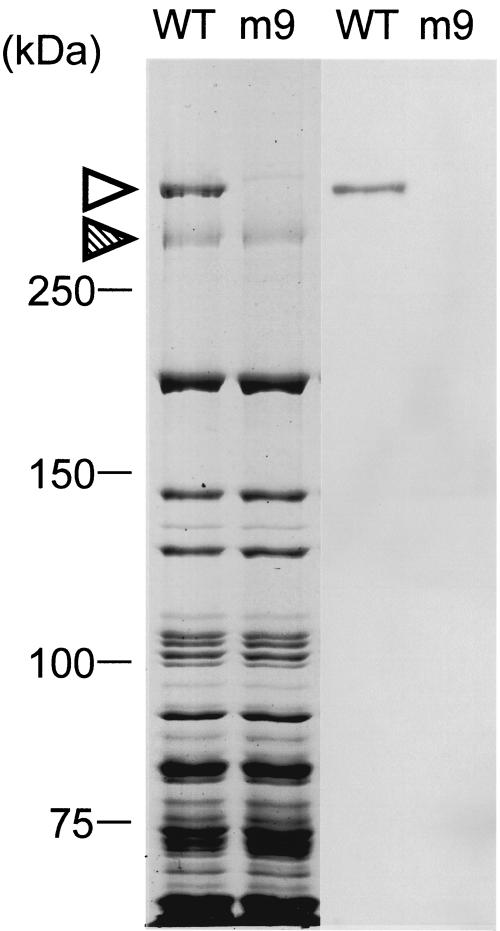
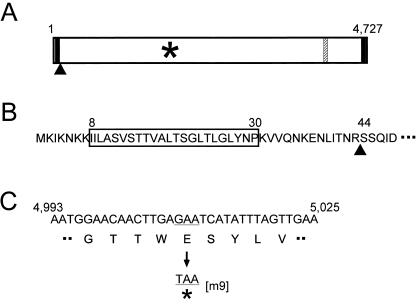
We probed the whole mycoplasma cell lysate by immunoblotting using the five antibodies that inhibited gliding without rapidly releasing cells from glass. Each of these antibodies recognized a protein band with a smaller electrophoretic mobility than that of Gli349 (Fig. 2). The amount of this protein was comparable to that of Gli349 in the Coomassie brilliant blue (CBB)-stained gel image. Edman degradation of a peptide produced by partial digestion of this protein band by V8 protease showed the sequence SLVVA. This sequence corresponded to an internal sequence spanning from amino acid residue 859 to 863 of an ORF immediately downstream of the gli349 gene (Fig. 3). We named this novel protein Gli521, because the product from this ORF is predicted to have 4,727 amino acid residues and a molecular mass of 520,568 Da. Edman degradation of the N terminus of this protein showed the sequence SSQID, corresponding to amino acids 44 to 48 of the gli521 ORF. This result indicated that the Gli521 protein is cleaved between amino acid residues 43 and 44 in the cell (Fig. 4). Therefore, the actual molecular mass should be 516 kDa.
FIG. 2.
Immunoblot analysis of the wild-type (WT) strain and mutant m9 by inhibitory antibody. Whole-cell lysates were analyzed by SDS-5.5% polyacrylamide gel electrophoresis (left two lanes) staining with CBB and immunoblotting (right two lanes) using the anti-Gli521 monoclonal antibody (mAb-R19). The protein bands of Gli521 and Gli349 are indicated by open and hatched triangles, respectively. Molecular mass is indicated on the left.
FIG. 3.
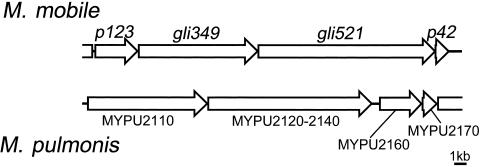
Gene arrangement around gliding genes in M. mobile and M. pulmonis. ORFs in the flanking regions of gli349 and gli521 were named from the predicted molecular weight. MYPU_2120, MYPU_2130, and MYPU_2140 form a single large ORF in M. pulmonis (ATCC 19612).
FIG. 4.
Sequence of the gli521 gene. (A) Schematic of the gli521 gene. Predicted transmembrane segments (solid lines), nonsense mutation (asterisk), N terminus of mature protein (filled triangle), and epitope of antibody (hatched line) are shown. (B) Amino acid sequence near the N terminus. The predicted transmembrane segment, indicated by the box, is preceded by a cluster of lysine residues. The processed site is indicated by a triangle. (C) Nucleotide and amino acid sequences around the mutation point of m9. A codon, GAA, encoding glutamic acid, is changed to TAA, a stop codon.
To confirm that the inhibitory antibodies recognize Gli521, recombinant protein fragments comprising the amino acid residues 17 to 193, 678 to 944, 1678 to 1896, 3736 to 4020, 4197 to 4460, and 4526 to 4696 of Gli521 were produced in E. coli cells and probed by the antibodies in immunoblotting. The fragment from amino acids 3736 to 4020 reacted with three of five inhibitory antibodies. This result confirms the identity between the target of inhibitory antibodies and Gli521 and also that the gli521 gene is translated as a single peptide at least to amino acid residue 3736. The other two antibodies did not react with any of the recombinant proteins, showing that these two antibodies originated from different spleen cells from those reactive to the fragment from amino acid 3736 to 4020. Considering that one of these antibodies was derived from a different animal, the nonreactive antibodies should be derived from independent cells. We used an antibody, named mAb-R19, reactive to the recombinant protein for the following experiments unless otherwise mentioned.
The predicted sequence for the Gli521 protein showed a high asparagine content (13.5%), lack of cysteine residues, and a low pI value (5.16). Using SOSUI, an algorithm that predicts membrane spanning segments, a transmembrane segment preceded by a positively charged region was predicted from amino acid residue 8 to 30 (8), suggesting the existence of a protein sorting and peptidase target signal. Another membrane-spanning segment was predicted near the C terminus extending from amino acid residue 4696 to 4718.
Ortholog in M. pulmonis.
The sequence of the gli521 gene is similar to three ORFs, MYPU_2120, MYPU_2130, and MYPU_2140, coded in tandem in the genome of another gliding mycoplasma, M. pulmonis, which is a pathogen of mouse (3). MYPU_2120, MYPU_2130, and MYPU_2140 have 21%, 19%, and 23% identities with regions spanning the codons for amino acid residues 191 to 1390, 1413 to 3041, and 3347 to 4725 of the 4,727 of Gli521, respectively. Since these observations suggested that the three ORFs of M. pulmonis might form one large ORF, we sequenced the gap regions of the genome from M. pulmonis (ATCC 19612). Our sequence analysis showed that the stop codon, TAA of MYPU_2130, starting from the nucleotide at position 240173 in the genome sequence is a glutamic acid codon, GAA, resulting in the fusion of ORFs MYPU_2130 and MYPU_2140. This correction has been accepted by the group that sequenced the genome (http://genolist.pasteur.fr/MypuList/). Our sequencing analysis of another gap between MYPU_2120 and MYPU_2130 showed that a sequence, CCCAAGACCCAAGA starting from nucleotide position 235433 in the genome sequence, is CCCAAGA, resulting in a fused ORF of MYPU_2120 and MYPU_2130. Therefore, M. pulmonis (ATCC 19612) should have an ortholog of Gli521, with molecular mass of 495 kDa and 23% identity with Gli521 of M. mobile (Fig. 3). The difference between the sequencing results might be caused by the difference in strains, because the genome project was carried out on M. pulmonis strain UAB CTIP (3). Considering that the preceding ORF of M. pulmonis, MYPU_2110, is the ortholog of the gli349 gene, this locus is well conserved in these two species. Indeed, the predicted amino acid sequences at positions 857 to 1126 and 11 to 321 of the p123 and p42 genes showed 24% and 24% identities with MYPU_2160 and MYPU_2170, respectively.
Absence of other orthologs.
No significant similarity to other protein sequences was detected in the genomes of other gliding mycoplasmas (5, 7, 25, 29), and no motifs, such as a P-loop involved in motor proteins, were found in gli521.
Mutant lacking Gli521 protein.
Previously, we isolated 10 mutants characterized by nonbinding, reduced binding, or faster gliding speed (24). The presence of the Gli521 protein was examined in mutants m6, m9, m12, m13, m14, m23, m26, m27, m29, and m34 by SDS-polyacrylamide gel electrophoresis followed by immunoblotting. The Gli521 band was detected by the antibody in nine mutants but not in mutant m9 (Fig. 2). The m9 mutant has been typed as one with no binding activity and irregular cell morphology (24). Sequencing of the 30-kb region containing the gli521 gene of the m9 mutant revealed that the codon at nucleotide position 1670, GAA, coding glutamic acid in the wild-type strain, is replaced by a stop codon, TAA, resulting in a truncated product (Fig. 4); no other mutations were identified. The truncated protein fragment was estimated to have a molecular mass of 179 kDa, but such a fragment was not detected by immunoblotting of the mutant. This was expected, since the epitope for the antibody extends from amino acid residue 3736 to 3863, which is on the C-terminal side of the nonsense mutation. The other antibodies also failed to detect the truncated band. The absence of the Gli521 protein in m9 is evident in the CBB-stained gel (Fig. 2, left). Although another band of less density can be seen at a position similar to Gli521 in the lane of m9, this band did not react with any anti-Gli521 antibodies, suggesting that it is derived from another ORF.
Subcellular localization of Gli521 protein.
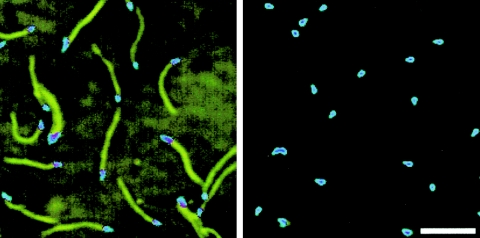
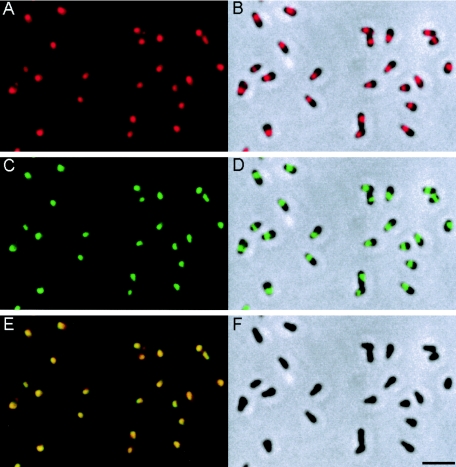
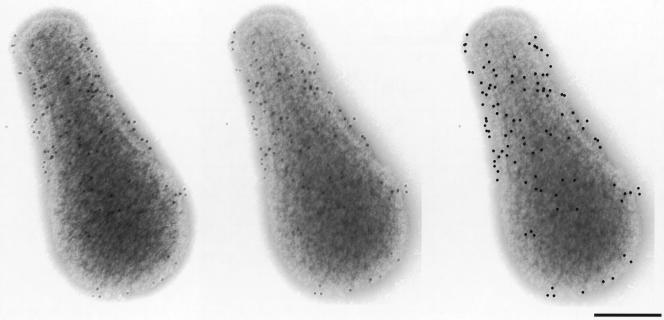
Immunofluorescence microscopy using the anti-Gli521 antibody revealed that the Gli521 protein is localized around the cell neck (Fig. 5A and B), the base of the membrane protrusion, as observed for Gli349 (Fig. 5C and D) (34). To examine the positional relationship between these two proteins, double staining for Gli521 and Gli349 proteins was performed (Fig. 5). The merged image (Fig. 5E) shows that the fluorescence label for Gli521 (Fig. 5A and B) overlays that for Gli349 (Fig. 5C and D) in all cells, showing colocalization of these two proteins. To examine localization of Gli521 at higher resolution, we performed immunogold electron microscopy (Fig. 6). Gli521 molecules were detected primarily around the neck, consistent with the results from immunofluorescence microscopy (Fig. 5). Three-dimensional observation using a tilted image set showed that the gold particles are localized all around the neck, rather than only at one side. A similar result was reported for Gli349 (34).
FIG. 5.
Subcellular localization of Gli521 and Gli349 proteins revealed by immunofluorescence microscopy. The cells were simultaneously stained for both proteins. (A) Fluorescence image detected for Gli521. (B) Merger of image shown in panel A with the phase-contrast image of panel F. (C) Fluorescence image detected for Gli349. (D) Merger of images shown in panels C and F. (E) Merger of images shown in panels A and C. (F) Phase-contrast image of cells. Bar, 2 μm.
FIG. 6.
Subcellular localization of Gli521 by immunoelectron microscopy. Gli521 was labeled with 10-nm gold particles. The left two images are a stereo pair. Since some of the gold particles are difficult to see, they are marked by black dots in the right image at the positions at which they appear in the middle image. Most cells found on an electron microscopy grid showed a similar distribution of gold particles. A stereoscopic view was achieved by tilting the specimen 10 degrees. Bar, 0.2 μm.
Analysis of inhibitory effect of anti-Gli521 antibody on binding and gliding.
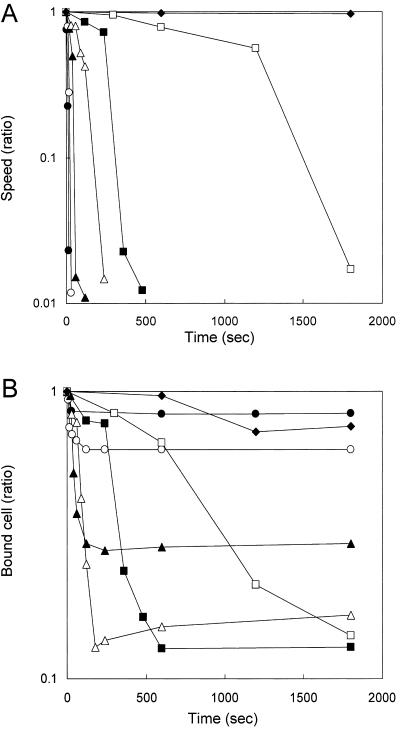
The effect of antibody on gliding motility was analyzed in a tunnel slide, with the results shown in Fig. 7. Various concentrations of anti-Gli521 antibody were added to gliding mycoplasmas at time zero, and the motility was monitored. The antibody reduced the gliding speed with time in a concentration-dependent manner (Fig. 7A): the higher the concentration, the more rapidly cells stopped moving. Cell displacement followed a similar time course: at higher concentrations, fewer cells were displaced, but they were displaced more rapidly (Fig. 7B). We analyzed the other four antibodies against Gli521 and concluded that they have similar effects on gliding, although the concentration dependency was not uniform. Monoclonal antibodies recognizing other surface proteins (mock antibodies) did not show any effects on gliding (data not shown). Thus, these effects are specific for the antibodies against Gli521.
FIG. 7.
Inhibitory effects of anti-Gli521 antibody on gliding speed and glass binding. (A) Gliding speed after the addition of different amounts of anti-Gli521 antibody, shown as a function of time. The speed shown is the fraction of the initial speed, averaged over the cell population. (B) The number of cells bound to glass after the addition of different amounts of antibody, shown as a function of time. The number shown is the fraction of the initial number. The number of cells bound to a glass surface with an area of 3,200 μm2 was counted. The initial number of cells was more than 100. Cells were treated with 0 (⧫), 1 (□), 3 (▪), 10 (▵), 30 (▴), 100 (○), and 300 (•) μg/ml antibody.
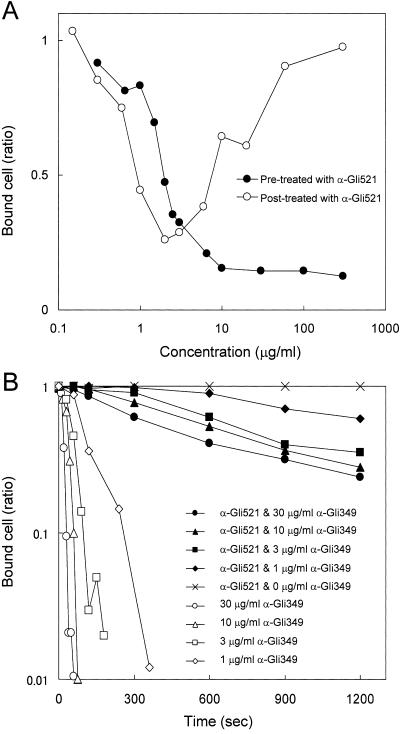
Next, we mixed antibodies with cells in suspension and examined their effects on the binding of mycoplasmas to glass (Fig. 8). Mycoplasma cell suspensions were incubated with various concentrations of antibody for 5 min at room temperature and inserted into a tunnel slide, and the numbers of cells bound after 10 min were counted (Fig. 8A). Binding was inhibited in a concentration-dependent manner. As a control, mycoplasma suspensions were inserted into a tunnel slide without antibodies and left for 10 min, various concentrations of antibody were added, and the numbers of cells bound were counted after 5 min. When the antibody was applied at concentrations higher than 2 μg/ml, it stopped gliding while keeping the cells on the glass, as shown in Fig. 7A and B.
FIG. 8.
Inhibitory effects of anti-Gli521 antibody on the adsorption of cells to glass. The number of cells bound to a glass surface with an area of 3,200 μm2 was counted. (A) Inhibitory effects on glass binding of antibody added before or after glass binding of mycoplasma cells. Cells in suspension were mixed with various concentrations of antibody, inserted into a tunnel slide, and then the numbers of cells bound to glass were counted (filled circles), or cells bound to glass in a tunnel slide were exposed to various concentrations of antibody, and the numbers of cells bound to the glass were counted (open circles). The fraction of cells remaining on the glass is shown for each concentration. (B) Inhibitory effect of anti-Gli521 antibody on the subsequent removal of cells from glass by anti-Gli349 antibody. The fraction of cells remaining on the glass is shown for each time point.
We found previously that a monoclonal antibody against Gli349 finally removed the gliding cells from glass (34). So, next we examined the effects of anti-Gli521 antibody on the action of the anti-Gli349 antibody (Fig. 8B). Gliding mycoplasmas were treated with 300 μg/ml of anti-Gli521 antibody for 10 min and then with various concentrations of anti-Gli349 antibody mixed with 300 μg/ml of anti-Gli521 antibody. The numbers of cells bound to glass were counted for each time point. Displacement of cells by the anti-Gli349 antibody was drastically reduced by pretreatment by the anti-Gli521 antibody. This observation suggests that the binding of anti-Gli521 antibody to Gli521 inhibited either the binding of anti-Gli349 antibody to Gli349 or the release from glass normally caused by anti-Gli349 antibody.
DISCUSSION
The novel protein, Gli521, is assumed to be involved in the gliding mechanism for the following reasons. (i) Monoclonal antibodies raised against this protein inhibited gliding (Fig. 1). (ii) A mutant whose Gli521 protein is truncated cannot bind to substrates and consequently cannot glide (24). (iii) Gli521 protein is coded in tandem with and downstream of a protein responsible for glass binding during gliding, Gli349 (Fig. 3). (iv) Gli521 localizes at the subcellular position, the neck, specialized for binding and gliding (Fig. 5 and 6) (16, 34).
However, the role of Gli521 in the gliding mechanism is obviously different from that of Gli349, because at higher concentrations all five of the antibodies against Gli521 inhibited movement but not glass binding, unlike the anti-Gli349 antibody. This conclusion is also supported by the observation that the anti-Gli349 antibody inhibits the adsorption of red blood cells to M. mobile colonies, so-called hemadsorption (34), but the anti-Gli521 antibody does not (data not shown). These observations suggest that the role of Gli521 in the gliding mechanism is force generation or force transmission rather than the binding to glass.
Then, why do the mutants m9 and m13, lacking Gli521 and Gli349, respectively, show similar phenotypes, namely, a deficiency in binding (24)? These observations can be explained by assuming that the function of Gli349 depends upon Gli521, which has the role of supporting Gli349; i.e., the lack of functional Gli521 depletes the function of Gli349. This assumption is supported by the observations that the amount of Gli521 in a cell is comparable to that of Gli349 (Fig. 2) and that the subcellular localization of Gli349 is lost in the m9 mutant, which is truncated for the Gli521 (our unpublished data). It is possible that Gli349, the major component of the spike, i.e., of the “leg” in the gliding machinery, is driven by the force generated by Gli521 itself or by that generated by another protein and transmitted through Gli521.
Possibly, the genes coding P123 and P42 in the flanking regions of Gli349 and Gli521 are also involved in the gliding mechanism, because they seem to be in the same operon (Fig. 3). P123 shares the features of amino acid sequence with Gli349, namely, an N-terminal transmembrane segment preceded by a positively charged region and high asparagine content. Moreover, a BLAST search showed 37% similarity between the region from amino acid 533 to 1128 of P123 and the region from amino acid 723 to 1300 of Gli349. These proteins may interact with the same target via these conservative domains. Experimental studies should be done to address these possibilities.
We previously quantified the number of Gli349 molecules on the M. mobile cell surface and concluded that around 450 molecules exist on a cell (34). Assuming that Gli521 forms a one-to-one complex with Gli349, Gli521 would occupy the whole neck region 200 nm in diameter and 300 nm in length, because a globular 516-kDa protein can be calculated to have a diameter about 20 nm. Occupation of the neck region by the gliding units is suggested by the observation that the proteins possibly involved in the mechanism of antigenic variation do not distribute into this region (16). Such a “collar” structure might function as a scaffold of gliding units for exerting force. This possibility is different from the observation of M. pneumoniae, where cytoskeletal structures, the so-called electron-dense core, can be found in the center of the membrane protrusion (11, 15, 18, 19, 22, 32, 33).
The addition of anti-Gli521 antibody decreased the gliding speed (Fig. 7), and eventually stopped the gliding cells on the glass, in a concentration-dependent manner. These observations can be explained by the assumption that the binding of antibody to a Gli521 molecule fixes the putative conformational change of Gli521 at a stage in the mechanical cycle where the leg binds to the glass surface and generates drag force. If the antibody generates drag force through the binding, how does the addition of antibody at a low concentration cause the removal of gliding cells from the glass? Possibly, a low concentration of antibody causes slow occupation of gliding units by the antibody, and the competition between the drag generated by the disabled legs and the thrust generated by the working legs distorts the linkage between the cell and the glass, inducing release of the cell from the surface. When higher concentrations of antibody are added, the quick occupation by the antibody does not cause this type of competition or finishes it before the dissociation of a cell from glass can occur.
When mycoplasma cells in suspension were treated by the anti-Gli521 antibody before they were exposed to glass, the binding of cells to glass was inhibited, even though the antibody kept cells on the glass when it was added after they began gliding (Fig. 8A). On the other hand, removal of cells by the anti-Gli349 antibody was inhibited by the anti-Gli521 antibody if the anti-Gli521 antibody was added to the gliding mycoplasma cells prior to the addition of anti-Gli349 antibody (Fig. 8B). These observations support the hypothesis that Gli521 executes conformational changes during gliding and that the antibody blocks those changes, fixing the states of the leg.
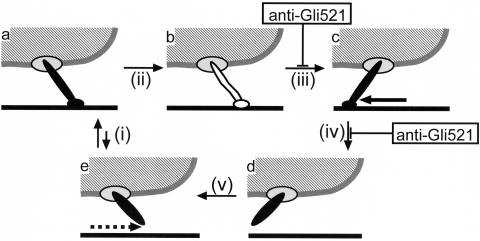
The glass binding during gliding presumably depends upon the Gli349 molecule, because the antibody against this protein displaces all of the gliding cells from the glass (34). Therefore, it is likely that the movement of Gli349 along the glass is involved in the gliding mechanism. Considering the images from the rapid-freeze-and-freeze-fracture rotary-shadow electron microscopy (20), we have proposed a simple model for the mechanical cycle (19, 34), as shown in Fig. 9. Here, gliding can be hypothesized to be composed of a series of states (a to e) and transition steps (i, initial binding; ii, conversion from initial to tight binding; iii, stroke; iv, release; and v, return). Anti-Gli521 antibody could inhibit the stroke (iii) or release (iv) step and fix the cells in state b or c.
FIG. 9.
One possible mechanical cycle to explain the effects of antibody. A mycoplasma cell is represented by a hatched area bounded by a gray line. A leg mainly composed of a Gli349 molecule is represented as a rod sticking out from the cell, driven by or through Gli521 represented by an ellipse on the cell. The glass surface is represented by a solid line. The cyclic movement of the leg, including binding, stroke, and release, propels the cell body. The leg is shown differently for free, bound, and tightly bound forms. The binding of anti-Gli521 antibody inhibits step iii or iv and stops the leg, fixing the mycoplasma on the glass.
Although several mycoplasma species have been reported to have the ability of gliding (12, 19), an ortholog of Gli521 or Gli349 cannot be found in the genomes of gliding mycoplasmas except M. pulmonis (3, 5, 7, 25, 29). These facts show that the primary structures of gliding proteins have diverged among mycoplasma species. However, the gliding motility of mycoplasmas can be based on a common mechanism, because the features of gliding are similar among gliding mycoplasma species, and recently we found that an anti-P1 adhesin monoclonal antibody has effects on the gliding of M. pneumoniae that are very similar to those of anti-Gli349 antibody on the gliding of M. mobile (31). The identification of the force-generating or -transmitting protein, Gli521, should help to elucidate the mechanism of mycoplasma gliding.
Acknowledgments
We thank Howard C. Berg of Harvard University for critical reading of manuscript and Tsuguo Sasaki of the National Institute of Infectious Diseases for providing the NS-1 strain of mouse myeloma cells.
This work was supported in part by grants-in-aid for JSPS fellows from the Japan Society for the Promotion of Science (to S.S.), for scientific research (C) (to M.M.), and science research on priority areas (motor proteins, genome science, and infection and host response) from the Ministry of Education, Science, Sports, Culture, and Technology (to M.M.).
REFERENCES
- 1.Aluotto, B. B., R. G. Wittler, C. O. Williams, and J. E. Faber. 1970. Standardized bacteriologic techniques for the characterization of mycoplasma species. Int. J. Syst. Bacteriol. 20:35-58. [Google Scholar]
- 2.Bredt, W. 1979. Motility, p. 141-145. In M. F. Barile, S. Razin, J. G. Tully, and R. F. Whitcomb (ed.), The mycoplasmas, vol. 1. Academic Press, New York. N.Y.
- 3.Chambaud, I., R. Heilig, S. Ferris, V. Barbe, D. Samson, F. Galisson, I. Moszer, K. Dybvig, H. Wroblewski, A. Viari, E. P. Rocha, and A. Blanchard. 2001. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic. Acids Res. 29:2145-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cleveland, D. W., S. G. Fischer, M. W. Kirschner, and U. K. Laemmli. 1977. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J. Biol. Chem. 252:1102-1106. [PubMed] [Google Scholar]
- 5.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, R. D. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J.-F. Tomb, B. A. Dougherty, K. F. Bott, P.-C. Hu, T. S. Lucier, S. N. Peterson, H. O. Smith, C. A. Hutchison III, and J. C. Venter. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 6.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- 7.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B.-C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirokawa, T., B. C. Seah, and S. Mitaku. 1998. SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 9.Jaffe, J. D., M. Miyata, and H. C. Berg. 2004. Energetics of gliding motility in Mycoplasma mobile. J. Bacteriol. 186:4254-4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaffe, J. D., N. Stange-Thomann, C. Smith, D. DeCaprio, S. Fisher, J. Butler, S. Calvo, T. Elkins, M. G. FitzGerald, N. Hafez, C. D. Kodira, J. Major, S. Wang, J. Wilkinson, R. Nicol, C. Nusbaum, B. Birren, H. C. Berg, and G. M. Church. 2004. The complete genome and proteome of Mycoplasma mobile. Genome Res. 14:1447-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenri, T., S. Seto, A. Horino, Y. Sasaki, T. Sasaki, and M. Miyata. 2004. Use of fluorescent-protein tagging to determine the subcellular localization of Mycoplasma pneumoniae proteins encoded by the cytadherence regulatory locus. J. Bacteriol. 186:6944-6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirchhoff, H. 1992. Motility, p. 289-306. In J. Maniloff, R. N. McElhaney, L. R. Finch, and J. B. Baseman (ed.), Mycoplasmas: molecular biology and pathogenesis. American Society for Microbiology, Washington, D.C.
- 13.Kirchhoff, H., R. Rosengarten, W. Lotz, M. Fischer, and D. Lopatta. 1984. Flask-shaped mycoplasmas: properties and pathogenicity for man and animals. Israel J. Med. Sci. 20:848-853. [PubMed] [Google Scholar]
- 14.Kishiro, Y., M. Kagawa, I. Naito, and Y. Sado. 1995. A novel method of preparing rat-monoclonal antibody-producing hybridomas by using rat medial iliac lymph node cells. Cell Struct. Funct. 20:151-156. [DOI] [PubMed] [Google Scholar]
- 15.Krause, D. C., and M. F. Balish. 2004. Cellular engineering in a minimal microbe: structure and assembly of the terminal organelle of Mycoplasma pneumoniae. Mol. Microbiol. 51:917-924. [DOI] [PubMed] [Google Scholar]
- 16.Kusumoto, A., S. Seto, J. D. Jaffe, and M. Miyata. 2004. Cell surface differentiation of Mycoplasma mobile visualized by surface protein localization. Microbiology 150:4001-4008. [DOI] [PubMed] [Google Scholar]
- 17.McBride, M. J. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 55:49-75. [DOI] [PubMed] [Google Scholar]
- 18.Miyata, M. 2002. Cell division, p. 117-130. In R. Herrmann and S. Razin (ed.), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum Publishers, London, United Kingdom.
- 19.Miyata, M. Gliding motility of mycoplasmas: the mechanism cannot be explained by current biology. In A. Blanchard and G. Browning (ed.), Mycoplasmas: pathogenesis, molecular biology, and emerging strategies for control, in press. Horizon Scientific Press, Norwich, United Kingdom.
- 20.Miyata, M., and J. Petersen. 2004. Spike structure at interface between gliding Mycoplasma mobile cell and glass surface visualized by rapid-freeze-and-fracture electron microscopy. J. Bacteriol. 186:4382-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyata, M., W. S. Ryu, and H. C. Berg. 2002. Force and velocity of Mycoplasma mobile gliding. J. Bacteriol. 184:1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyata, M., and S. Seto. 1999. Cell reproduction cycle of mycoplasma. Biochimie 81:873-878. [DOI] [PubMed] [Google Scholar]
- 23.Miyata, M., and A. Uenoyama. 2002. Movement on the cell surface of gliding bacterium, Mycoplasma mobile, is limited to its head-like structure. FEMS Microbiol. Lett. 215:285-289. [DOI] [PubMed] [Google Scholar]
- 24.Miyata, M., H. Yamamoto, T. Shimizu, A. Uenoyama, C. Citti, and R. Rosengarten. 2000. Gliding mutants of Mycoplasma mobile: relationships between motility and cell morphology, cell adhesion and microcolony formation. Microbiology 146:1311-1320. [DOI] [PubMed] [Google Scholar]
- 25.Papazisi, L., T. S. Gorton, G. Kutish, P. F. Markham, G. F. Browning, D. K. Nguyen, S. Swartzell, A. Madan, G. Mahairas, and S. J. Geary. 2003. The complete genome sequence of the avian pathogen Mycoplasma gallisepticum strain R(low). Microbiology 149:2307-2316. [DOI] [PubMed] [Google Scholar]
- 26.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosengarten, R., and H. Kirchhoff. 1987. Gliding motility of Mycoplasma sp.nov. strain 163K. J. Bacteriol. 169:1891-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Sasaki, Y., J. Ishikawa, A. Yamashita, K. Oshima, T. Kenri, K. Furuya, C. Yoshino, A. Horino, T. Shiba, T. Sasaki, and M. Hattori. 2002. The complete genomic sequence of Mycoplasma penetrans, an intracellular bacterial pathogen in humans. Nucleic Acids Res. 30:5293-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seto, S., T. Kenri, G. Layh-Schmitt, and M. Miyata. 2001. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J. Bacteriol. 183:1621-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seto, S., T. Kenri, T. Tomiyama, and M. Miyata. 2005. Involvement of P1 adhesin in gliding motility of Mycoplasma pneumoniae as revealed by the inhibitory effects of antibody under optimized gliding conditions. J. Bacteriol. 187:1875-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seto, S., and M. Miyata. 2003. Attachment organelle formation represented by localization of cytadherence protein and formation of electron-dense core in the wild-type and mutant strains of Mycoplasma pneumoniae. J. Bacteriol 185:1082-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimizu, T., and M. Miyata. 2002. Electron microscopic studies of three gliding mycoplasmas, Mycoplasma mobile, M. pneumoniae, and M. gallisepticum, by using the freeze-substitution technique. Curr. Microbiol. 44:431-434. [DOI] [PubMed] [Google Scholar]
- 34.Uenoyama, A., A. Kusumoto, and M. Miyata. 2004. Identification of a 349-kilodalton protein (Gli349) responsible for cytadherence and glass binding during gliding of Mycoplasma mobile. J. Bacteriol. 186:1537-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]