Abstract
The recrudescence of severe invasive group A streptococcal (GAS) diseases has been associated with relatively few strains, including the M1T1 subclone that has shown an unprecedented global spread and prevalence and high virulence in susceptible hosts. To understand its unusual epidemiology, we aimed to identify unique genomic features that differentiate it from the fully sequenced M1 SF370 strain. We constructed DNA microarrays from an M1T1 shotgun library and, using differential hybridization, we found that both M1 strains are 95% identical and that the 5% unique M1T1 clone sequences more closely resemble sequences found in the M3 strain, which is also associated with severe disease. Careful analysis of these unique sequences revealed three unique prophages that we named M1T1.X, M1T1.Y, and M1T1.Z. While M1T1.Y is similar to phage 370.3 of the M1-SF370 strain, M1T1.X and M1T1.Z are novel and encode the toxins SpeA2 and Sda1, respectively. The genomes of these prophages are highly mosaic, with different segments being related to distinct streptococcal phages, suggesting that GAS phages continue to exchange genetic material. Bioinformatic and phylogenetic analyses revealed a highly conserved open reading frame (ORF) adjacent to the toxins in 18 of the 21 toxin-carrying GAS prophages. We named this ORF paratox, determined its allelic distribution among different phages, and found linkage disequilibrium between particular paratox alleles and specific toxin genes, suggesting that they may move as a single cassette. Based on the conservation of paratox and other genes flanking the toxins, we propose a recombination-based model for toxin dissemination among prophages. We also provide evidence that a minor population of the M1T1 clonal isolates have exchanged their virulence module on phage M1T1.Y, replacing it with a different module identical to that found on a related M3 phage. Taken together, the data demonstrate that mosaicism of the GAS prophages has contributed to the emergence and diversification of the M1T1 subclone.
Group A streptococci (GAS) are serious human pathogens that cause infections ranging from mild pharyngitis to chronic rheumatic heart disease and, in some cases, severe streptococcal toxic shock syndrome (STSS) and necrotizing fasciitis (16). The severe forms of streptococcal diseases reemerged in the late 1980s (33, 41), and many studies attempted to explain this phenomenon by identifying specific serotypes or changes within a serotype that may account for this drastic change in GAS epidemiology (29, 30, 40). Although several serotypes are capable of causing more severe infections in the susceptible host, one particular subclone of the M1 serotype, the globally disseminated clonal M1T1 strain (12), has persisted uninterruptedly as the most frequently isolated serotype from invasive and noninvasive GAS infections worldwide (13, 15, 35). This is uncommon for GAS serotypes, including several that are frequently isolated from severe invasive GAS infection, which exhibit cyclic prevalence patterns (2, 6, 28). It is intriguing, therefore, to determine what is so unique about this clonal M1T1 strain.
While in the past, the remarkable prevalence, persistence, and virulence of M1T1 isolates were attributed to a number of individual genetic factors (34, 36, 42), it is likely that the unique features of this clone result from complex traits encoded by a number of interacting genetic features and controlled by complex regulatory networks. With the advent of sophisticated genomic tools, we aimed to identify—at the genomic level—the unique bacterial genetic factors that distinguish this M1T1 clone from other M1 isolates and that might provide clues as to its prevalence, persistence, and virulence.
Unlike the M1T1 clonal strain, another closely related fully sequenced M1 strain, SF370, isolated from a wound infection (21, 43), has not been frequently isolated from severe invasive GAS infections (27) and has not shown the same pattern of prevalence and persistence seen in the M1T1 clonal strain. We took advantage of the high similarity, yet striking difference in epidemiology, between these two M1T1 strains and used differential microarray hybridization to identify unique genetic features of the clonal M1T1 strain without the need to sequence its entire genome. As expected, the majority of the differences were attributed to prophage sequences. The importance of prophage content in the diversification of various subclones of GAS M3 serotype has been recently demonstrated by Beres et al. (6). Here, we identified three distinct prophages integrated into the M1T1 genome, two of which are not found in the M1 SF370 strain; the third has two variants that distinguish two M1T1 lineages and that has likely emerged due to phage exchange between two distinct M serotypes. We also discovered that the genomes of these prophages are highly mosaic, with different regions being related to distinct GAS phages. Furthermore, we identified a highly conserved open reading frame (ORF) adjacent to the toxins (paratox; prx) in the majority of GAS prophages and found that allelic variants of paratox are in linkage disequilibrium with specific toxin genes. Based on these observations, we propose a model of recombination-induced toxin exchange among the GAS prophages.
MATERIALS AND METHODS
More detailed methods are provided in the supplemental material.
Bacterial strains and culture conditions.
Extensively characterized clonal M1T1 clinical isolates from invasive GAS infection cases were used in this study (12, 17, 32). One representative isolate, M1T1-6050, from the clonal M1T1 strain (12) was used in generating the genomic library and was compared, in the microarray experiments, to strain SF370 (ATCC 700294), isolated from an infected wound (21, 43). However, in certain studies, the microarray results (obtained from M1T1-6050 DNA) were confirmed by use of DNA from several isolates belonging to the same M1T1 clone (12). For simplification, M1T1-6050 is referred to as M1T1 throughout this article.
All GAS isolates were grown in Todd-Hewitt broth (Difco Laboratories) supplemented with 1.5% yeast extract (THY). Escherichia coli, in which the M1T1-6050 genomic library was generated, was grown in Luria-Bertani (LB) broth supplied with 50 μg/ml carbenicillin (Sigma).
Generation of the M1T1 GAS library and construction of microarrays.
In collaboration with Lucigen Corporation (Middleton, WI), we generated a genomic library for the M1T1-6050 isolate. M1T1 chromosomal DNA was extracted by a modified phenol-chloroform method (11), randomly sheared to 1 to 3 kb, and then cloned in pSMART-LC (Lucigen). DH10B electrocompetent cells were transformed with the ligated vectors at Lucigen Corporation to produce the M1T1 library. Colonies (n = 6144) were picked manually and subcultured in 96-well plates. With an average GAS genome length of 1.9 × 106 bp and an average insert size of 2,000 bp, 6,144 clones provide 99.84% genome coverage, as calculated from the Poisson distribution (22).
The glass microarrays were manufactured in the Molecular Resource Center and the Vision Core Facility at the University of Tennessee. We used a MicroGridII microarrayer (BioRobotics, Genomic Solutions) to spot the probes (library PCR products) onto superamine glass slides (Telechem International Inc.).
Labeling, hybridization, and image analysis.
Sheared chromosomal DNA from both M1 SF370 and M1T1-6050 strains was labeled by random priming with either Cy3 or Cy5 fluorescent nucleotides, as detailed in the supplemental Materials and Methods. Equal amounts of the labeled genomic DNA from both M1 SF370 and M1T1 strains were mixed and used to hybridize the unlabeled DNA probes on the microarray slides. The slides were dried then scanned by GenePix4000B scanner (Axon Instruments, Inc.). All steps were performed in the dark. The experiment was repeated twice, with three replicate microarrays each time. The scanned images were analyzed with the GenePixPro 4.0 software (Axon Instruments).
Probes that hybridized preferentially to labeled M1T1 DNA were chosen, and the corresponding clones were recovered from the master plates, amplified by the TempliPhi system (Amersham) and sequenced on ABI PRISM 3100 Genetic Analyzer (Applied Biosystems).
Sequence assembly, annotation, and bioinformatic analysis.
The sequence of each probe was compared to the nonredundant GenBank database by use of BLASTN and BLASTX software (1). BLASTN and BLASTX results were parsed in independent files by PERL bioinformatics scripts (R.A.E., unpublished scripts). We assembled the probe sequences into larger fragments then into prophages using Phred, Phrap, and Consed sequence analysis software package (23) and the Vector NTI (VNTI) Suite (Informax Inc.). We closed the gaps and corrected low-quality sequences in the assembled fragments by additional PCR amplifications followed by primer extension sequencing. Finally, we used VNTI to assemble all fragments and additional sequences into prophages and to identify and annotate all ORFs.
For sequence alignment and phylogenetic analysis, we used the AlignX feature of VNTI, ClustalW (44), PHYLIP (20), and njplot (39). To investigate phage mosaicism, we used BLASTN rather than BLASTX because nucleotide sequence similarity is a more relevant indicator of phage-phage relationships, especially in the case of closely related proteins, it shows the similarity of noncoding areas, and it is less affected by frameshift mutations or by accidental sequencing errors.
Confirmation of phage excision.
To confirm phage excision from GAS chromosome, we performed PCRs using primer pairs that flank the phage attachment sites (attP), the bacterial attachment sites (attB), and the attL sites. In an integrated prophage, attP primers are divergent and will be unable to yield a single PCR product, while the primers that flank attB would have to amplify the whole prophage (>30 kb). Only attL-flanking primers are expected to yield a product in an integrated prophage. Conversely, in a circularized phage, only those primers flanking attP and attB are expected to give a product. Therefore, a positive result from the attP primer pair was taken as evidence of the phage's ability to be excised from the genome; excision was further confirmed by amplification of an appropriate-sized product when attB primer pairs were used, indicating that the phage had been excised from the chromosome restoring the original boundaries of the bacterial attB sequence.
Phage nomenclature.
In this article, we follow current convention to designate GAS prophage by names consisting of the bacterial host's name followed by a serial number that reflects the prophage chromosomal location, in clockwise order (4, 10). Since some prophages of strains SF370 and MGAS8232 in the GenBank database are given discrepant names, we list the discrepancies in Table 1. As for the phages identified in this study, we called them—according to current convention—by the strain name (M1T1) followed by the letters X, Y, and Z in the clockwise order of their chromosomal locations. To make prophages M1T1.X, M1T1.Y, and M1T1.Z easier to discuss, we also designated them SPhinX, MemPhiS, and PhiRamid, respectively.
TABLE 1.
Discrepancies in prophage names
| Prophage namein this article | Prophage name in GenBank |
|---|---|
| Phi 370.2 | Phi370.3 |
| Phi 370.3 | Phi370.2 |
| Phi 8232.1 | 8232 Phi SpeA |
| Phi 8232.2 | 8232 Phi SpeC |
| Phi 8232.3 | 8232 Phi SpeLM |
| Phi 8232.4 | 8232 Phi370.2-like |
| Phi 8232.5 | 8232 Phi SDA |
Because prophages SPsP1, SPsP2, SPsP3 (Phi NIH1.1), and SPsP4 are almost identical to prophages 315.6, 315.5, 315.4, and 315.3, respectively, we did not include them in some homology analyses to avoid redundancy.
Nucleotide sequence accession numbers.
The sequence data from this study have been submitted to GenBank under accession numbers AY616023 and AY621076.
RESULTS
Differential hybridization reveals unique segments in the M1T1 genome.
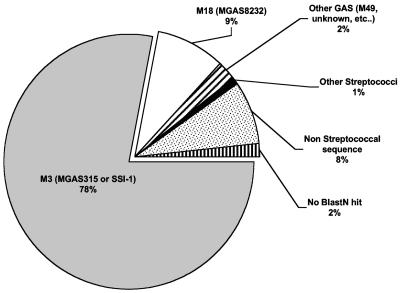
To compare the genomes of M1T1 and SF370, we constructed a genomic microarray from a shotgun DNA library of a representative M1T1 isolate. The number of probes spotted on the microarray, 6,144 probes (average size, 1 to 3 kb), is estimated to provide 99.8% coverage of the M1T1 genome. The design was such that only sequences found in the M1T1 clone and not in SF370 would be detected. When sheared chromosomal DNAs from M1T1 and M1 SF370 bacteria were labeled with Cy3 and Cy5, respectively, and used as targets in DNA hybridization experiments, 337 microarray probes preferentially hybridized with Cy3-labeled M1T1 DNA (Fig. S6A). However, when the dyes were flipped, only 315 probes hybridized with the Cy5-labeled M1T1 DNA (Fig. S6B). Because Cy3 tends to produce a stronger signal and may bind nonspecifically to certain sequences (19), only those probes (the 315 probes) that hybridized with both Cy3 in experiment 1 (Fig. S6A) and Cy5 in experiment 2 (Fig. S6B) were considered unique to M1T1 and designated positive. However, any spot that was suspected as positive on visual inspection of the scanned array images was also included in subsequent analyses to rule out that partial hybridizations were missed. Thus, 400 clones were sequenced and subjected to BLASTN homology analysis. Of those, 323 had their best BLASTN hits in strains or species other than GAS SF370 (Fig. 1); i.e., they were unique to the M1T1 clone. Interestingly, 78% of the non-SF370 matches were best matched to sequences found in MGAS315 and SSI-1 (Fig. 1), two M3 strains that have also been associated with STSS and necrotizing fasciitis cases.
FIG. 1.
Summary of differential hybridization results. Distribution of best BLASTN hits for the sequences that hybridized preferentially with M1T1 but not with SF370 DNA (raw data are provided in Table S1).
Most unique genetic features of M1T1 are phage-related sequences.
The 323 sequence fragments unique to M1T1 were assembled into 17 contigs (ranging from 1 to 13 kb). While 2 of these contigs belonged to an insertion sequence (see below), the remaining 15 were further assembled into three prophages that we named SPhinX (alias PhiM1T1.X), MemPhiS (alias PhiM1T1.Y), and PhiRamid (alias PhiM1T1.Z) (Fig. S7). Whereas MemPhiS is similar to Phi370.3 in SF370, SPhinX and PhiRamid are absent from the SF370 strain. SPhinX integrates into the tmRNA gene, PhiRamid integrates into a tRNA-Ser gene located between SPy1725 and SPy1726, and MemPhiS integrates in the same site as Phi370.3, between two protein-coding genes, cadA and hlpA (Fig. S8). Based on their attachment sites, we found that all three prophages—like all other known GAS prophages—are located in one replichore (one half of the chromosome) and are oriented so that their integrase genes are pointing (5′ to 3′) toward the bacterial origin of replication while the majority of their genes are transcribed in the direction of the chromosome replication (Fig. S7). It has been proposed that this orientation increases the efficiency of transcription by allowing the majority of genes to be transcribed in the same direction of the chromosome replication, possibly to avoid collision between polymerases (8, 9).
Mosaicism of the three M1T1 phages.
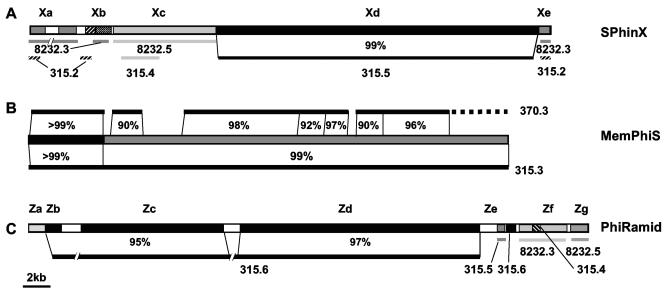
As mentioned above, SPhinX and PhiRamid bear very little sequence similarity to SF370 prophages, with the exception of very few areas that are highly conserved in most GAS phages. When the genomes of these two M1T1-specific phages were compared to the GenBank sequences, a striking genetic mosaicism was seen (Fig. 2). For example, SPhinX has an area with substantial similarity (99% identity along ∼23 kb) to Phi315.5 in the MGAS315 M3 strain (Fig. 2A, segment Xd). This area of high similarity between the two SpeA-encoding prophages includes most of the phage structural genes (head and tail morphogenesis), as well as the lysis cassette and the speA virulence gene. However, whereas SPhinX carries the speA2 allele, Phi315.5 carries speA3. The remainder of the SPhinX genome is rather different from that of Phi315.5: its replication module is mostly similar to Phi8232.5 (Fig. 2A, segment Xc), and its lysogeny module is mostly similar to SSA-carrying Phi315.2 and to Phi8232.3, which encodes SpeL and SpeM (Fig. 2A, segments Xa and Xe).
FIG. 2.
Mosaic nature of M1T1 prophages. The diagram shows the patterns and extent of similarity between different segments of SPhinX (A), MemPhiS (B), and PhiRamid (C) and their closest homologs among GAS prophages. Best BLASTN hits are shown below or above each phage segment, and—in some cases—the percentage of nucleotide identity is indicated. White boxes represent sequences with no BLASTN hits.
PhiRamid, like SPhinX, shares chimeric similarity with at least three prophages, Phi315.6, Phi8232.3, and Phi8232.5. The similarity of PhiRamid to Phi315.6, the Sdn-carrying phage in the M3 MGAS315 strain, ranges from 95 to 97% and is extended over more than 33 kb. Aside from major similarity to Phi315.6, PhiRamid's lysis module is similar to that of Phi8232.3 and Phi315.4, but its lysogenic conversion module is similar to Phi8232.5. Finally, the hylP module of PhiRamid shares little sequence similarity with any known GAS phage sequences (Fig. 2C, segment Ze).
As for MemPhiS, the first 5-kb segment of this prophage is virtually identical to MF3-carrying Phi370.3 (>99%) and thus was not picked by differential hybridization but rather by sequencing. Many regions in the remaining ∼30 kb of MemPhiS are highly similar to regions in Phi370.3 (90% to 98% identity at the nucleotide level); however, this phage is more similar (>99%) to MF4-carrying Phi315.3 (Fig. 2B). Interestingly, the majority of the M1T1 isolates tested are mf3+/mf4−, whereas much fewer M1T1 isolates are mf3−/mf4+; the gene products of mf3 and mf4 are only 20% identical at the amino acid level.
In all three prophages, there were areas with no or very poor BLASTN hits, e.g., parts of the lysogeny module of SPhinX and PhiRamid and parts of PhiRamid's lysogenic conversion module (white boxes in Fig. 2). Some of these unique islands are AT rich, similar to repeats found in the M protein and the SOF-encoding genes. It is tempting to speculate that these islands, which are flanked by highly conserved genes, result from interphage recombination, but this remains to be investigated.
Excision of SPhinX, PhiRamid, and MemPhiS from the GAS chromosome.
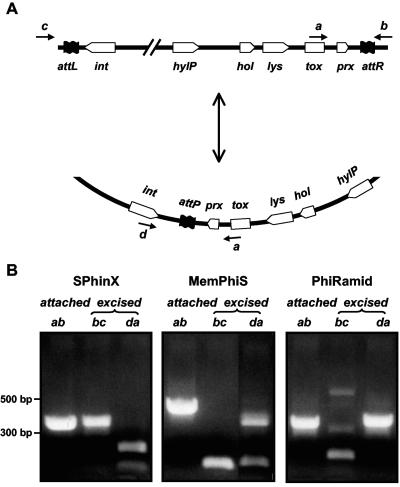
Based on the above sequence information, we designed PCR primers that would only generate products if the phages were circularized and we found that at least a proportion of each of the three M1T1 phage populations is present in circular form, i.e., is excised from the chromosome (Fig. 3). The sequence of the PCR products that encompass the attachment sites (attP) of all three circularized phages not only confirmed their excision but also provided direct evidence that the putative phage attachment sites and their core repeats were as predicted by sequence similarity of the redundant prophage ends.
FIG. 3.
PCRs showing phage excision and integration. (A, upper part) Map of the different genes in the lysis and the lysogeny modules of GAS prophages. (A, lower part) Map of the same genes' relative positions when the phage is circularized. Genes are not drawn to scale. Positions of PCR primers are shown by small black arrows (a, b, c, and d). (B) PCRs show the presence of each phage (SPhinX, MemPhiS, and PhiRamid) in both attached and excised forms. All PCR products were sequenced and their sequences validated.
Hot spots for recombination in the lysogenic conversion modules of GAS prophages.
Since we are primarily interested in the pathogenic potential of the globally disseminated M1T1 strain, we analyzed more extensively the sequences of the toxin-encoding lysogenic conversion modules to gain insights into how prophages acquire and exchange virulence factors. In all GAS prophages, the genes encoding virulence factors (i.e., toxins) are located between the phage lysis cassettes and the phage attachment sites (7, 10). Thus, whenever the phage is excised and circularized, these genes would be flanked by the lysis cassette from one side and the integrase gene from the other (Fig. 3A). Accordingly, we included both the lysis cassettes and the integrase sequences in bioinformatic analyses to investigate the mechanism of virulence factor mobilization to and from phages.
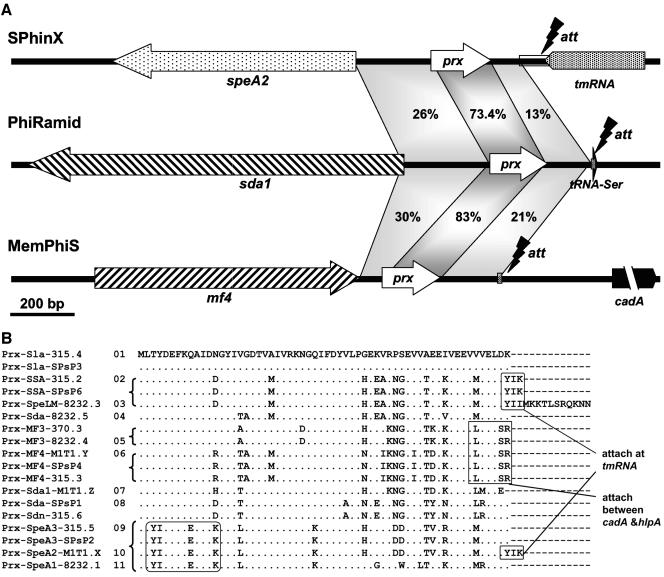
We compared the nucleotide sequences of the three lysogenic conversion modules of SPhinX, MemPhiS, and PhiRamid and identified a highly conserved ORF, lacking a signal peptide, located between the toxin gene and the phage attachment site (Fig. 4A), and—based on its location—we called it paratox. Paratox was found in 18 out of 24 GAS prophages in strains SF370, MGAS8232, MGAS315, and SSI-1, and in all cases, it was located adjacent to a toxin gene. Paratox homologs were also found in some phages in Streptococcus agalactiae and Streptococcus thermophilus. GAS prophages that do not have virulence genes (e.g., Phi315.1, PhiSPsP6, and Phi370.4) lack paratox-like genes. Curiously, no paratox homologs were identified in SpeC-carrying Phi370.1 and Phi8232.1 or in SpeH- and SpeI-carrying Phi370.2; none of these phages is in the M1T1 strain.
FIG. 4.
Paratox: a highly conserved ORF in M1T1 prophages. (A) A comparison between the lysogenic conversion modules and attachment sites of the three M1T1 prophages shows a highly conserved ORF that best matches a hypothetical phage protein located between each toxin and the phage attachment site. We named this hypothetical protein paratox (prx). Shaded areas indicate nucleotide similarity, and the percentage nucleotide identity is given. (B) Alignment of paratox protein alleles shows highly conserved amino acid sequence (represented by dots). Representative motifs linked to particular toxins or to phage attachment sites are boxed. All sequences are extracted from GenBank; in cases where the prx sequences were not annotated as ORFs, we picked them based on their similarity to the annotated ones. Each Prx will be referred to as (Prx_tox_Phi#), where tox is the name of the adjacent toxin and Phi# is the phage name and number (e.g., Prx_SpeA2_M1T1.X is the product of the paratox gene adjacent to SpeA2 in Phi M1T1.X, alias SPhinX). Serial numbers (1 to 11) were given to the distinct paratox alleles shown.
When their amino acid sequences were aligned, the 18 paratox proteins could be clustered into subgroups and were accordingly classified into 11 alleles (Fig. 4B). From the paratox phylogenetic tree (Fig. S9A), linkage disequilibrium between the paratox and toxin genes can be noticed, suggesting that prx and tox are inherited and/or mobilized as a single module. Whether other adjacent genes are also linked to the prx and tox cassette is a complex question. On the one hand, no sequence conservation was observed in the area between the paratox and the phage attachment site (att), and there was no linkage disequilibrium between att and tox; instead, most att sites in GAS prophages appear to cluster with their adjacent integrase proteins (Fig. S9B). On the other hand, at least three highly conserved genes are located on the other side of the toxin genes (distal to paratox); these genes encode lysin (Lys), holin (Hol), and hyaluronidase (HylP).
The conservation of the prx gene and its linkage disequilibrium to the tox gene also suggest that prx may be one of two hot spots of recombination (arms) flanking the virulence genes and promoting their dissemination between prophages by recombination. This hypothesis can only be valid if the sequence flanking the tox gene, opposite to prx (Fig. 5), is conserved and is at least in partial linkage disequilibrium with tox. To investigate this possibility and identify the putative second hot spot of recombination, we aligned the predicted amino acid sequences of the hol, lys, and hylP gene products in all known GAS prophages (Fig. S9C to E). The phylogenetic analyses confirmed their high conservation among different prophages and thus suggested that any of them could be the second recombination hot spot (Fig. 5). In addition, a unique feature of the three M1T1 prophages is that unlike all published sequenced GAS strains that possess two or more highly similar alleles of each of the Lys, Hol, and HylP proteins carried on different phages per strain, the M1T1 does not show this redundancy. Instead, each of the three M1T1 phages has its unique lysin, holin, or hyaluronidase. Altogether, this strain contains two nonhomologous holins, three weakly similar lysins, and two divergent hyaluronidases (Fig. S9).
FIG. 5.
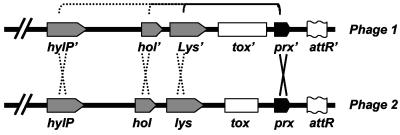
Putative model for toxin exchange between phages. Possible scenarios that may contribute to toxin exchange between different prophages by recombination are shown. Two recombination hot spots are shown on both sides of the toxin genes: one of them is the prx gene, and the other may be either lys, hol, or hylP.
Additional minor differences between M1T1 and SF370.
Besides the identification of mosaic prophages, the microarray data identified additional differences between the two subclones of the M1 serotype. For example, a copy of the insertion sequence IS1548 was detected in M1T1, adjacent to the gene encoding the ribosomal protein RpmB (SPy1888). Although the M1 SF370 strain has six copies of IS1548, none are inserted in the rpmB gene. Other differences between M1T1 and SF370 include a minor variation of only seven noncontiguous nucleotides in the slo gene contig, whose detection demonstrates the sensitivity of the differential hybridization technique.
DISCUSSION
We designed this study to focus on sequences present in the M1T1 strain and not in the M1 SF370 strain, whose genome sequence has been published (21). Whereas conventional differential genomic studies use glass slides on which the genome of a standard strain is arrayed and then hybridize it to different closely related strains (31), we used the reverse approach; i.e., in our study, the unknown strain (M1T1) is arrayed and it is being compared to the standard one (M1 SF370). This strategy allowed us to identify unique sequences that are not present in the SF370 strain and that may endow the M1T1 strain with its unusual epidemiology, without the need to sequence the whole M1T1 genome. This method is more economical and less time consuming than full genome sequencing; however, it is obviously not a method of choice for single-nucleotide-polymorphism analysis, which can certainly affect the regulation and function of virulence genes. However, this was not one of the goals of the present study, which aimed to identify global sequence differences between a highly virulent and a relatively less virulent M1 strain of GAS.
Overall, the majority of the differences between M1T1 and SF370 are phage-related sequences. Interestingly, 78% of the phage-related sequences unique to M1T1 are shared by the two sequenced M3 strains MGAS315 (5) and SSI-1 (37). Our findings are supported by an earlier report that an invasive M1 subclone differs from other members of the same M1 serotype by two prophages, T13 and T14 (14). Although no sequence was provided for T13 and T14, it is likely that they are closely related to SPhinX and PhiRamid, which distinguish M1T1 from SF370. These two prophages carry the speA2 and sda1 genes; homologs of these genes, which, respectively, encode a potent superantigen and a DNase, are also present in the M3 strains (speA3 and sdn). SpeA is a well-characterized superantigen that plays a pivotal role in STSS pathogenesis (38), and we recently demonstrated the DNase activity of Sda1 and showed that its unique carboxy terminus potentiates its nuclease activity (3). Inasmuch as M1T1 and M3 strains have been frequently isolated from severe invasive streptococcal infections, it is reasonable to suspect that these prophages and/or the toxins they encode may be conferring an added virulence on these strains. Despite the similarities between the M1T1 and M3 prophages, important differences were found, including differences in the attachment sites of phage pairs with similar structural genes (SphinX and Phi315.5, as well as PhiRamid and Phi315.6), unique integrase genes, and unique modules found only in the M1T1 phages (Fig. 2).
Genetic mosaicism in M1T1 prophages.
Analysis of M1T1 phage genomes suggests that they have diversified by exchanging information and shuffling genetic modules in a pattern that makes each M1T1 prophage a unique entity, sharing blocks of sequences with different prophages but also possessing unique sequences with no known homologs in the current databases. This phenomenon, also known as genetic mosaicism, is a hallmark of tailed phages (24), to which the streptococcal phages belong, and is likely to increase phage fitness and to enhance the dissemination of the genes located within the shuffled modules (25, 26). In addition to the expected similarities of M1T1 prophages to other GAS prophages, we identified sequences within the M1T1 phages that were best matched to phage-related sequences in other bacterial species. For example, the integrase gene of PhiRamid was mostly similar (53%) to phage λSa2 of S. agalactiae. These observations lead us to suggest that the newly identified M1T1 prophages and/or related phages may have taken habitat in other GAS strains, as well as in different bacterial species, where they may have plucked certain sequences or modules and left others behind.
Role of prophage in M1T1 GAS evolution and subclone emergence.
The recent completion of several GAS genomes demonstrated how bacteriophages account for major differences between the M serotypes (4, 5, 37), and—even within the same serotype—subclones emerge that have different prophage contents (6). This notion is illustrated in this study by the identification of SPhinX and PhiRamid that distinguish two subclones of the M1 serotype, M1T1 and SF370.
A third prophage, MF4-encoding MemPhiS identified in few M1T1 isolates, signalizes the presence of two lineages of the M1T1 subclone: a major mf3+/mf4− and a minor mf3−/mf4+ lineage. MemPhiS is more similar to mf4-carrying Phi315.3, found in the M3 strains, than to mf3-carrying Phi370.3, found in the SF370 strain. All three phages belong to a family of r1t-like phages (18) that are the most highly conserved prophages in GAS, as each GAS strain sequenced so far has an r1t-like prophage that is inserted between the hlpA and cadA genes. The fact that MemPhiS and Phi315.3 are virtually identical (99% nucleotide identity) suggests that they share a recent common ancestor or that one was derived from the other. To our knowledge, this is the first example of a virtually identical prophage present in two different M serotypes.
The mf4 gene was first detected in the genome of MGAS315, and—until this report—has only been found in M3 strains. However, Beres et al. showed recently that only 4/255 M3 isolates screened lacked the mf4 gene (6), and it would be interesting to test whether these 4 strains are carrying mf3 as our data suggest that a total or partial prophage exchange event occurred between the M1T1 and M3 strains. The facts that both M1T1 and M3 strains studied were isolated from invasive GAS infections and were from Canadian patients living in the Ontario area (6) make this exchange a likely scenario.
Role of prophage in harboring, disseminating, and remodeling GAS virulence factors.
Another question addressed in this study is how phages acquire and exchange virulence genes. In the case of streptococcal phage-encoded toxins, it is believed that these genes were acquired by inaccurate phage excision in a bacterial host with a lower G+C content (10). Data from recent environmental phage genomes show that some phage-encoded toxins are found in marine phages isolated from distant closed habitats (Forrest Rohwer, personal communication). While it is not possible to know exactly how an ancestral prophage acquired a bacterial toxin with a secretion signal, evidence from various phage and bacterial genomes suggests that the toxins are mostly spread by horizontal gene transfer between different prophages. We believe that the exchange of toxins not only helps their spread in nature but also promotes their diversification as in the case of streptodornases (3).
The finding of highly conserved sequences on both sides of the toxin genes in the phages reported here supports the homologous recombination model for toxin mobilization between various phages (7, 10). Among sequences flanking the toxins was a highly conserved ORF that we named paratox (prx). Despite their high similarity, paratox proteins could be classified into alleles, and we found linkage disequilibrium between particular paratox alleles and specific toxin genes, suggesting that the prx and toxin genes are inherited and disseminated as one cassette. Interestingly, whereas most of the paratox sequence is in linkage disequilibrium to the toxin gene, its C-terminal sequence appears to be in linkage with the phage attachment site (boxes in Fig. 4B), suggesting that recombination takes place within the paratox sequence. As more GAS genomes become available, and more paratox alleles are identified, this notion may be further validated.
Conclusion
Our goal was to identify—at the genomic level—unique features that distinguish the clonal M1T1 strain from the closely related SF370 strain. We identified three prophages in M1T1 that contribute largely to its uniqueness. The finding that prophage may play a role in subclone diversification, and the fact that the prophage cassettes can be shared between the M1T1 and M3 serotypes, brings into question the validity of the current GAS classification system, particularly when attempts are made to associate certain serotypes with specific clinical manifestations of GAS infections. In our opinion, the M serotype designation is no longer sufficient or clinically useful. The advent of genomic tools and the completion of several GAS genome sequences have unraveled the extent of the horizontal transfer of mobile genetic elements (IS and phages) among the various strains, and it is perhaps incumbent upon us to explore a new classification schema that better represents the basis for GAS virulence and their involvement in specific diseases.
Supplementary Material
Acknowledgments
We thank David Mead from Lucigen for help in generating the M1T1 library, William Orr for help with spotting the microarrays, Robert Belland for advice on labeling and hybridization, and Mary MacNealy for critical review of the manuscript.
This work was supported by grant AI40198-07 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (to M.K.), by the Research and Development Office, Medical Research Service, Department of Veterans Affairs (Merit Award to M.K.), and by a UTHSC Center of Excellence in Genomics and Bioinformatics grant (M.K., R.A.E., and R.K.A.). An abstract including parts of this study was awarded the ASM student travel grant (R.K.A.) in the ASM Functional Genomics and Bioinformatics Conference, 2004, Portland, OR.
Footnotes
Supplemental material for this paper may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony, B. F., E. L. Kaplan, L. W. Wannamaker, and S. S. Chapman. 1976. The dynamics of streptococcal infections in a defined population of children: serotypes associated with skin and respiratory infections. Am. J. Epidemiol. 104:652-666. [DOI] [PubMed] [Google Scholar]
- 3.Aziz, R. K., S. A. Ismail, H. W. Park, and M. Kotb. 2004. Post-proteomic identification of a novel phage-encoded streptodornase, Sda1, in invasive M1T1 Streptococcus pyogenes. Mol. Microbiol. 54:184-197. [DOI] [PubMed] [Google Scholar]
- 4.Banks, D. J., S. B. Beres, and J. M. Musser. 2002. The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence. Trends Microbiol. 10:515-521. [DOI] [PubMed] [Google Scholar]
- 5.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beres, S. B., G. L. Sylva, D. E. Sturdevant, C. N. Granville, M. Liu, S. M. Ricklefs, A. R. Whitney, L. D. Parkins, N. P. Hoe, G. J. Adams, D. E. Low, F. R. DeLeo, A. McGeer, and J. M. Musser. 2004. Genome-wide molecular dissection of serotype M3 group A Streptococcus strains causing two epidemics of invasive infections. Proc. Natl. Acad. Sci. USA 101:11833-11838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brüssow, H., C. Canchaya, and W. D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, A. M. 2002. Preferential orientation of natural lambdoid prophages and bacterial chromosome organization. Theor. Popul. Biol. 61:503-507. [DOI] [PubMed] [Google Scholar]
- 9.Canchaya, C., G. Fournous, and H. Brüssow. 2004. The impact of prophages on bacterial chromosomes. Mol. Microbiol. 53:9-18. [DOI] [PubMed] [Google Scholar]
- 10.Canchaya, C., C. Proux, G. Fournous, A. Bruttin, and H. Brussow. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67:238-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caparon, M. G., and J. R. Scott. 1991. Genetic manipulation of pathogenic streptococci. Methods Enzymol. 204:556-586. [DOI] [PubMed] [Google Scholar]
- 12.Chatellier, S., N. Ihendyane, R. G. Kansal, F. Khambaty, H. Basma, A. Norrby-Teglund, D. E. Low, A. McGeer, and M. Kotb. 2000. Genetic relatedness and superantigen expression in group A Streptococcus serotype M1 isolates from patients with severe and nonsevere invasive diseases. Infect. Immun. 68:3523-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cleary, P. P., E. L. Kaplan, J. P. Handley, A. Wlazlo, M. H. Kim, A. R. Hauser, and P. M. Schlievert. 1992. Clonal basis for resurgence of serious Streptococcus pyogenes disease in the 1980s. Lancet 339:518-521. [DOI] [PubMed] [Google Scholar]
- 14.Cleary, P. P., D. LaPenta, R. Vessela, H. Lam, and D. Cue. 1998. A globally disseminated M1 subclone of group A streptococci differs from other subclones by 70 kilobases of prophage DNA and capacity for high-frequency intracellular invasion. Infect. Immun. 66:5592-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cockerill, F. R., III, K. L. MacDonald, R. L. Thompson, F. Roberson, P. C. Kohner, J. Besser-Wiek, J. M. Manahan, J. M. Musser, P. M. Schlievert, J. Talbot, B. Frankfort, J. M. Steckelberg, W. R. Wilson, and M. T. Osterholm. 1997. An outbreak of invasive group A streptococcal disease associated with high carriage rates of the invasive clone among school-aged children. JAMA 277:38-43. [PubMed] [Google Scholar]
- 16.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies, H. D., A. McGeer, B. Schwartz, K. Green, D. Cann, A. E. Simor, D. E. Low, and the Ontario Group A Streptococcal Study Group Ontario Group. 1996. Invasive group A streptococcal infections in Ontario, Canada. N. Engl. J. Med. 335:547-554. [DOI] [PubMed] [Google Scholar]
- 18.Desiere, F., W. M. McShan, D. van Sinderen, J. J. Ferretti, and H. Brussow. 2001. Comparative genomics reveals close genetic relationships between phages from dairy bacteria and pathogenic streptococci: evolutionary implications for prophage-host interactions. Virology 288:325-341. [DOI] [PubMed] [Google Scholar]
- 19.Dobbin, K., J. H. Shih, and R. Simon. 2003. Questions and answers on design of dual-label microarrays for identifying differentially expressed genes. J. Natl. Cancer Inst. 95:1362-1369. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein, J. 1997. An alternating least squares approach to inferring phylogenies from pairwise distances. Syst. Biol. 46:101-111. [DOI] [PubMed] [Google Scholar]
- 21.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraser, C. M., and R. D. Fleischmann. 1997. Strategies for whole microbial genome sequencing and analysis. Electrophoresis 18:1207-1216. [DOI] [PubMed] [Google Scholar]
- 23.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 24.Hendrix, R. W. 2003. Bacteriophage genomics. Curr. Opin. Microbiol. 6:506-511. [DOI] [PubMed] [Google Scholar]
- 25.Hendrix, R. W., G. F. Hatfull, and M. C. Smith. 2003. Bacteriophages with tails: chasing their origins and evolution. Res. Microbiol. 154:253-257. [DOI] [PubMed] [Google Scholar]
- 26.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoe, N., K. Nakashima, D. Grigsby, X. Pan, S. J. Dou, S. Naidich, M. Garcia, E. Kahn, D. Bergmire-Sweat, and J. M. Musser. 1999. Rapid molecular genetic subtyping of serotype M1 group A streptococcus strains. Emerg. Infect. Dis. 5:254-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikebe, T., N. Murai, M. Endo, R. Okuno, S. Murayama, K. Saitoh, S. Yamai, R. Suzuki, J. Isobe, D. Tanaka, C. Katsukawa, A. Tamaru, A. Katayama, Y. Fujinaga, K. Hoashi, J. Ishikawa, and H. Watanabe. 2003. Changing prevalent T serotypes and emm genotypes of Streptococcus pyogenes isolates from streptococcal toxic shock-like syndrome (TSLS) patients in Japan. Epidemiol. Infect. 130:569-572. [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, D. R., D. L. Stevens, and E. L. Kaplan. 1992. Epidemiological analysis of group A streptococcal serotypes associated with severe systemic infections, rheumatic fever or uncomplicated pharyngitis. J. Infect. Dis. 166:374-382. [DOI] [PubMed] [Google Scholar]
- 30.Kaplan, E. L., J. T. Wotton, and D. R. Johnson. 2001. Dynamic epidemiology of group A streptococcal serotypes associated with pharyngitis. Lancet 358:1334-1337. [DOI] [PubMed] [Google Scholar]
- 31.Kim, C. C., E. A. Joyce, K. Chan, and S. Falkow. 2002. Improved analytical methods for microarray-based genome-composition analysis. Genome Biol. 3:RESEARCH0065. [DOI] [PMC free article] [PubMed]
- 32.Kotb, M., A. Norrby-Teglund, A. McGeer, H. El-Sherbini, M. T. Dorak, A. Khurshid, K. Green, J. Peeples, J. Wade, G. Thomson, B. Schwartz, and D. E. Low. 2002. An immunogenetic and molecular basis for differences in outcomes of invasive group A streptococcal infections. Nat. Med. 8:1398-1404. [DOI] [PubMed] [Google Scholar]
- 33.Low, D. E., B. Schwartz, and A. McGeer. 1997. The reemergence of severe group A streptococcal disease: an evolutionary perspective, p. 93-123. In W. M. Scheld, D. Armstrong, and J. M. Hughes (ed.), Emerging infections, vol. 1. ASM Press, Washington, D.C.
- 34.Lukomski, S., N. P. Hoe, I. Abdi, J. Rurangirwa, P. Kordari, M. Liu, S. J. Dou, G. G. Adams, and J. M. Musser. 2000. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect. Immun. 68:535-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muotiala, A., H. Seppala, P. Huovinen, and J. Vuopio-Varkila. 1997. Molecular comparison of group A streptococci of T1M1 serotype from invasive and noninvasive infections in Finland. J. Infect. Dis. 175:392-399. [DOI] [PubMed] [Google Scholar]
- 36.Musser, J. M., V. Kapur, S. Kanjilal, U. Shah, D. M. Musher, N. L. Barg, K. H. Johnston, P. M. Schlievert, J. Henrichsen, and D. Gerlach. 1993. Geographic and temporal distribution and molecular characterization of two highly pathogenic clones of Streptococcus pyogenes expressing allelic variants of pyrogenic exotoxin A (scarlet fever toxin). J. Infect. Dis. 167:337-346. [DOI] [PubMed] [Google Scholar]
- 37.Nakagawa, I., K. Kurokawa, A. Yamashita, M. Nakata, Y. Tomiyasu, N. Okahashi, S. Kawabata, K. Yamazaki, T. Shiba, T. Yasunaga, H. Hayashi, M. Hattori, and S. Hamada. 2003. Genome sequence of an M3 strain of Streptococcus pyogenes reveals a large-scale genomic rearrangement in invasive strains and new insights into phage evolution. Genome Res. 13:1042-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norrby-Teglund, A., and M. Kotb. 2000. Host-microbe interactions in the pathogenesis of invasive group A streptococcal infections. J. Med. Microbiol. 49:849-852. [DOI] [PubMed] [Google Scholar]
- 39.Perrière, G., and M. Gouy. 1996. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78:364-369. [DOI] [PubMed] [Google Scholar]
- 40.Schwartz, B., R. R. Facklam, and R. F. Breiman. 1990. Changing epidemiology of group A streptococcal infection in the USA. Lancet 336:167-171. [DOI] [PubMed] [Google Scholar]
- 41.Stevens, D. L., and E. L. Kaplan. 2000. Streptococcal infections: clinical aspects, microbiology, and molecular pathogenesis. Oxford University Press, New York, N.Y.
- 42.Stockbauer, K. E., L. Magoun, M. Liu, E. H. Burns, Jr., S. Gubba, S. Renish, X. Pan, S. C. Bodary, E. Baker, J. Coburn, J. M. Leong, and J. M. Musser. 1999. A natural variant of the cysteine protease virulence factor of group A streptococcus with an arginine-glycine-aspartic acid (RGD) motif preferentially binds human integrins αvβ3 and αIIbβ3. Proc. Natl. Acad. Sci. USA 96:242-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suvorov, A. N., and J. J. Ferretti. 1996. Physical and genetic chromosomal map of an M type 1 strain of Streptococcus pyogenes. J. Bacteriol. 178:5546-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.