Abstract
Rhodococcus equi is a facultative intracellular pathogen which proliferates rapidly in both manure-enriched soil and alveolar macrophages. Although both environments are characterized by extremely low concentrations of free iron, very little is known regarding the strategies employed by R. equi to thrive under these conditions. This paper reports the characterization of an R. equi transposome mutant that fails to grow at low iron concentrations. The transposome was shown to be inserted into iupA, the first gene of the iupABC operon encoding an ABC transport system highly similar to siderophore uptake systems. Disruption of the iupA gene also resulted in a failure of R. equi to utilize heme and hemoglobin as a source of iron. Introduction of the iupABC operon in trans restored the wild-type phenotype of the mutant strain. iupABC transcripts were 180-fold more abundant in R. equi grown in iron-depleted medium than in organisms grown in iron-replete medium. Proliferation of the iupABC mutant strain in macrophages was comparable to that of the wild-type strain. Furthermore, the iupABC mutant was not attenuated in mice, showing that the iupABC operon is not required for virulence.
Rhodococcus equi is a major cause of pyogranulomatous pneumonia in foals of up to 5 months of age. Although it is mainly an equine pathogen, R. equi sporadically infects other animals, such as pigs, goats, and cattle. In addition, R. equi is increasingly responsible for AIDS-associated pneumonia (33). The virulence of R. equi is dependent on an 80- to 85-kb plasmid harboring a 27-kb pathogenicity island allowing the bacterium to proliferate in macrophages (19, 49, 50). In addition to this pathogenic lifestyle, R. equi is also a saprophytic bacterium that rapidly multiplies in manure-enriched soils (25). The most likely route of infection is by inhalation of contaminated dust particles by susceptible hosts (33).
Both the saprophytic and pathogenic lifestyles of R. equi are characterized by a very low concentration of free iron due to the extremely low solubility of Fe3+ in aerobic environments at neutral pH. In mammalian tissues, the iron concentration is further reduced, since iron is bound to proteins such as transferrin, lactoferrin, ferritin, and hemoglobin (55). Bacteria therefore have evolved sophisticated strategies to obtain iron from the iron-limited environment. Many produce siderophores, low-molecular-weight compounds of large structural diversity that have a very high affinity for iron. Bacteria release siderophores into the extracellular environment, and following binding to iron, the siderophore-iron complex is taken up by the cell. Based on their structures, siderophores are classified as catecholates, hydroxamates, or compounds containing ligands of both categories (55). The mycolata, a phylogenetically distinct group of actinomycetes which includes species belonging to the genera Rhodococcus, Mycobacterium, and Corynebacterium, produce siderophores of all three categories. For example, a catecholate siderophore structurally related to enterobactin is produced by Corynebacterium glutamicum (7). Mycobacterium species produce mycobactins and carboxymycobactins, which are of the mixed type, and exochelin, a hydroxamate siderophore (reviewed in reference 37). The only rhodococcal siderophore to be characterized to date is heterobactin produced by Rhodococcus erythropolis, which has both catecholate and hydroxamate ligands (8).
An alternative strategy to overcome iron limitation is to use iron-containing host compounds and proteins, such as heme, hemoglobin, and transferrin (55). Heme and/or hemoglobin utilization has been demonstrated in several members of the mycolata, including Arcanobacterium pyogenes, Arcanobacterium haemolyticum, Mycobacterium haemophilum, Corynebacterium diphtheriae, and Corynebacterium ulcerans (12, 38, 42, 43). Transferrin is used as a source of iron by Mycobacterium tuberculosis and C. diphtheriae. In both species, this process is siderophore dependent (20, 43).
A common element in siderophore and heme utilization is the use of ATP binding cassette (ABC) transport systems to facilitate transport across the cell membrane. These systems consist of a permease made up of either a homo- or heterodimeric protein or a single two-domain protein and a cell membrane-associated ATPase to energize uptake. In addition, some systems use a binding protein that binds heme or siderophores and delivers these to the ABC transport systems (29, 47, 52).
To date, little is known regarding iron utilization by R. equi. Although it was reported that R. equi does not produce mycobactins (23), analysis of the partial genome sequence of R. equi ATCC 33701 revealed the presence of mycobactin as well as exochelin biosynthesis genes (36). However, whether these are functional is as yet unknown. In addition to these potential siderophore-mediated iron uptake systems, it was recently shown that R. equi is able to utilize transferrin and lactoferrin as sources of iron (27), although it is unknown what mechanisms are involved. This paper describes the identification of an iron-regulated operon encoding an ABC transport system that is required for growth under iron-limiting conditions. Although the operon is similar to hydroxamate siderophore transport systems, mutation of this operon also impairs the utilization of heme and hemoglobin by R. equi. Since mutation of this operon does not affect virulence, it is likely that this ABC transport system is required during saprophytic growth of R. equi.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Genotype or characteristics | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Bethesda Research Laboratories |
| R. equi | ||
| ATCC 33701 | Virulent strain, 81-kb virulence plasmid p33701 | American Type Culture Collection |
| ATCC 33701 (P−) | Avirulent strain, virulence plasmid cured | 50 |
| α5G12 | iupA::(EZ::TN Kan2) | This study |
| Plasmids | ||
| pBluescript II KS+ | Apr, αlacZ′ | Stratagene |
| pREV9 | Aprr, oriV(pMF1), oriV(ColE1), αlacZ′ of pBLUESCRIPT | 31 |
| pα5G12 | 4.4-kb KpnI fragment containing iupA::(EZ::TN), iupB, and iupC′ in pBluescript II KS+ | This study |
| pIUP3′C | 546-bp KpnI-PstI fragment containing the 3′ end of iupC in pBluescript II KS+ | This study |
| pREIUPABC | 4,187-bp fragment containing iupABC in pREV9 | This study |
| pREIUPAB5′C | 3,695-bp fragment containing iupABC′ in pREV9 | This study |
| Oligonucleotides | ||
| IUPRm3F | 5′-CCCTGCTGGTGTGGAATCG-3′ | This study |
| IUP4R | 5′-GAAAGCGAACCACAGGTACTGG-3′ | This study |
| IUPAex | 5′-GAACAACACGAGCACCAGCAG-3′ | This study |
| 16SrRNA200F | 5′-ACGAAGCGAGAGTGACGGTA-3′ | This study |
| 16SrRNA200R | 5′-ACTCAAGTCTGCCCGTATCG-3′ | This study |
Media and growth conditions.
Escherichia coli and R. equi strains were grown on Luria-Bertani (LB) medium (41) or on minimal medium supplemented with 20 mM lactate (LMM) at 37°C as previously described (28). LMM without FeSO4 (LMM-Fe) and containing 2,2-dipyridyl was used as low-iron medium. To examine the effect of iron availability on iupABC transcription, a 2-liter Erlenmeyer flask containing 400 ml of LMM was inoculated with R. equi pregrown on LMM. When the culture reached an optical density at 600 nm of 0.5, 50-ml samples were washed with phosphate-buffered saline (PBS) and resuspended in 50 ml of LMM for iron-replete conditions or in LMM-Fe medium containing 200 μM 2,2-dipyridyl. When appropriate, the following supplements were added: ampicillin, 50 μg ml−1; apramycin, 30 μg ml−1 (E. coli) or 80 μg ml−1 (R. equi); kanamycin, 50 μg ml−1 (E. coli) or 200 μg ml−1 (R. equi); X-Gal (5-chromo-4-chloro-3-indolyl-β-D-galactopyranoside), 20 μg ml−1; and isopropyl-β-D-thiogalactoside (IPTG), 0.1 mM. Hemin (Sigma) and equine hemoglobin (Sigma) were added to solid media containing 250 μM 2,2-dipyridyl to final concentrations of 400 and 80 μM, respectively. Agar was added for solid media (1.5% [wt/vol]).
DNA manipulations.
Plasmid DNA was isolated with the alkaline lysis method (4) or by using the Wizard Plus SV miniprep system (Promega). Chromosomal DNA was isolated as described previously (34). DNA-modifying enzymes were used according to the manufacturer's recommendations (Roche). Dideoxy sequencing reactions were done with the CEQ DTCS-Quickstart kit as described by the manufacturer (Beckman). The nucleotide sequence was determined using a Beckman CEQ 2000 automatic sequencer; nucleotide sequence data were compiled using the Staden package (48). Protein sequences were compared to entries in GenBank using BlastP (2). The prediction of membrane-spanning helices was done using TopPred2 (http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html) (11).
In order to complement the R. equi α5G12 strain, the entire iupABC operon was cloned into pREV9 shuttle vector (Table 1). Briefly, the transposome was removed by digesting pα5G12 with NcoI and BsmI and replaced with a 411-bp wild-type sequence to obtain pIUPAB5′C. Then, a 546-bp KpnI-PstI fragment containing the 3′ end of iupC was cloned into pBluescript II KS, yielding pIUP3′C. The entire operon was subsequently constructed by ligating the 3,212-bp KpnI fragment of pIUPAB5′C into the pIUP3′C KpnI site. This plasmid was called pIUPABC. Finally, a 4,187-bp PciI-NotI fragment from pIUPABC was filled in with the Klenow fragment of DNA polymerase and blunt-end ligated into the EcoRV site of pREV9. The resultant pREIUPABC was electroporated into R. equi. The PciI-NotI fragment of pIUPAB5′C was subcloned into pREV9, as above, to produce pREIUPAB5′C.
Transposome mutagenesis.
Random insertion mutagenesis using transposomes (EZ::TN <Kan-2> insertion kit; Epicentre) was carried out as described previously (32). Briefly, electrocompetent R. equi cells were electroporated with 20 ng of transposomes. The electroporation mixture was plated onto LB agar plates containing 200 μg/ml kanamycin to select for R. equi harboring the EZ::TN transposon.
RNA isolation.
Bacteria were harvested by centrifugation at 4°C at 3,220 × g for 5 min. Pellets were resuspended in 1 ml of Tri reagent (Sigma), and the suspension was transferred to a 2-ml screw-cap tube containing 0.5 ml of diethyl pyrocarbonate-treated 0.1-mm zirconia-silica beads (BioSpec). The cells were broken with two 30-s pulses in a Ribolyser (Hybaid) at the 6.5 speed setting and stored at −80°C until further processing. After thawing on ice, the supernatant was recovered and extracted with 200 μl of chloroform. Nucleic acids were precipitated with 0.5 ml of isopropanol, washed with 1.5 ml of 75% ethanol, dissolved in 90 μl of diethyl pyrocarbonate-treated water at 65°C for 5 min, and digested with 4 U of RNase-free Turbo DNase (Ambion) at 37°C for 30 min. RNA was purified with the RNA easy kit (QIAGEN) according to the manufacturer's instructions, except that a 15-min on-column digestion with 30 U of RNase-free DNase I (QIAGEN) was performed. RNA purity and concentration were determined by UV spectrophotometry and visualized in borax agarose gel electrophoresis to assess the quality of every preparation. Samples were stored in small aliquots at −80°C.
RT-PCR.
Reverse transcription (RT) reactions were performed with 1 U of Improm II reverse transcriptase following the manufacturer's recommendations using 500 ng of total RNA, 300 nM IUPAex reverse primer, and 16SrRNA200R (Table 1) in a final volume of 20 μl. cDNA (2 μl) was amplified by 30 cycles of PCR with Taq DNA polymerase using the oligonucleotides IUPAex and IUPRm3F or 16SrRNA200R and 16SrRNA200F (Table 1). Possible DNA contamination of RNA samples was ruled out by including controls with no addition of reverse transcriptase. Amplification products were analyzed by 1% agarose gel electrophoresis.
Real-time RT-PCR.
Total RNA (100 ng) was used in the reverse transcription step, after which amplification was performed using the QuantiTect SYBR Green real-time kit following the manufacturer's instructions (QIAGEN). Reaction mixtures were subjected to 40 cycles of 94°C for 15 s, 60°C for 15 s, and 72°C for 30 s in a LightCycler (Roche) with temperature transition rates of 20°C/s using the oligonucleotides IUPAex and IUPRm3F or 16SrRNA200R and 16SrRNA200F. To ensure that fluorescence signal was specific for the expected DNA products, melting curve analysis from 50 to 99°C (temperature transition, 0.2°C/s) was performed after amplification. Cycle threshold values were obtained and used to calculate the number of RNA copies per microgram of total RNA using a standard curve of known amounts of DNA target with r2 coefficients larger than 0.997 in the range of 5 × 103 to 5 × 108 molecules per reaction. 16S rRNA was used as a housekeeping gene to compare the amount of RNA in each reaction. Three independent experiments in duplicate were performed for each RNA sample.
Macrophage infection.
J774A.1 murine macrophages were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (vol/vol) fetal calf serum (FCS), 2 mM glutamine, and 10 μg/ml gentamicin. Eight-well chamber slides were seeded at a concentration of 1.5 × 105 macrophages per well and left for 24 h. R. equi 33701, 33701 (P−), and α5G12 cultures were grown in minimal medium to a density of 108 CFU per ml. R. equi was washed twice with divalent cation-free PBS and resuspended in phagocytosis buffer. Macrophage monolayers were washed with warm DMEM, and phagocytosis buffer (24) and normal mouse serum (5% [vol/vol]) were added. Macrophages were subsequently infected with wild-type and mutant strains at a multiplicity of infection of 10 bacteria per macrophage. Chamber slides were incubated for 45 min at 37°C in 5% CO2. Monolayers were then washed with phagocytosis buffer to remove unbound bacteria and incubated for a further 15 min to allow internalization of the bacteria. Phagocytosis buffer was then replaced with DMEM supplemented with 10% (vol/vol) FCS, 2 mM glutamine, and 10 μg/ml gentamicin. After 90 min and 48 h, infected monolayers were fixed with methanol (300 μl/well) for 20 min at 4°C. Monolayers were washed four times with PBS containing 5% FCS, and polyclonal rabbit anti-R. equi antibodies (Irish Equine Centre, Ireland) were added to each well. Slides were incubated at 37°C for 45 min and washed four times, and a secondary fluorescein isothiocyanate-conjugated goat anti-rabbit antibody was added. Slides were incubated for 45 min at 37°C in the dark. Monolayers were washed as before, and the removable chamber was removed and examined under a fluorescence microscope.
Virulence assessment in mice.
Two groups of seven 6- to 8-week-old female CD1 mice were injected intravenously with 100 μl of 5 × 106/ml R. equi ATCC 33701+ or 100 μl of 5 × 106 R. equi mutant α5G12. The mice were euthanized 4 days after infection, and their spleens and livers were aseptically removed and ground in PBS, pH 7.2. The suspended ground tissue was diluted in a 10-fold series, and 50-μl aliquots were plated on trypticase soy agar (Difco). Bacterial colonies were counted after 48 h of incubation at 37°C. In addition, two groups of seven CD1 female mice were infected intraperitoneally with 0.1 ml of 5 × 108 bacteria/ml of either R. equi ATCC 33701+ or R. equi mutant α5G12 and examined for 2 weeks for evidence of illness.
Nucleotide sequence accession number.
The GenBank accession number of the sequence reported in this paper is AY426738.
RESULTS
Isolation of an R. equi mutant unable to grow at low iron concentrations.
To date, very little is known regarding the mechanisms used by R. equi to adapt to growth at low iron concentrations. To obtain insight into this, a transposome library of R. equi was constructed by electroporation of R. equi with transposomes, a protein-DNA complex consisting of a hyperactive Tn5 transposase and a DNA fragment containing a kanamycin resistance gene flanked by 19-bp transposase recognition sequences (21). Kanamycin-resistant colonies were screened for mutants that were unable to grow on LB agar containing an 80 μM concentration of the iron chelator 2,2-dipyridyl. Growth of the wild-type strain is inhibited by 225 μM 2,2-dipyridyl. Out of 1,500 individual transposome mutants, one kanamycin-resistant colony that was unable to grow in the presence of 80 μM dipyridyl was selected. This mutant (R. equi α5G12) displayed the same growth rate on lactate minimal medium (0.27 ± 0.01 h−1) as the wild-type strain, indicating that the inability to grow in the presence of 2,2-dipyridyl was not due to a mutation in the central metabolic or biosynthetic pathways. Addition of FeCl3 to LB medium containing 80 μM 2,2-dipyridyl restored growth of R. equi α5G12, showing that failure to grow was due to iron limitation and not to an increased sensitivity of the mutant to 2,2-dipyridyl.
R. equi uses hemin and hemoglobin as sources of iron.
To determine whether R. equi is able to use hemin (iron protoporphyrin IX) and/or hemoglobin as sources of iron, it was grown on LB agar plates containing a 250 μM concentration of the iron chelator 2,2-dipyridyl. In the absence of alternative sources of iron, growth at this concentration of the chelator was completely inhibited. Addition of either hemin or hemoglobin restored growth, showing that R. equi can use these compounds as sources of iron. A virulence plasmid-free strain and the wild-type strain grew equally well, indicating that hemin and hemoglobin utilization is not plasmid encoded. Interestingly, R. equi α5G12 was unable to utilize either hemin or hemoglobin as a source of iron (Fig. 1B).
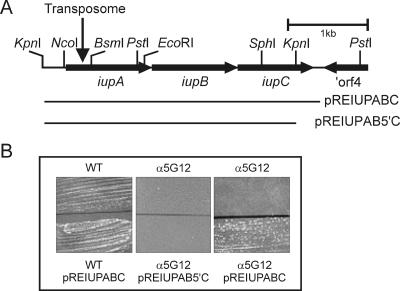
FIG. 1.
Physical map of the iupABC locus and complementation of R. equi α5G12. (A) The positions of the iupABC genes and ORF4 are indicated by arrows. The bars below the restriction map indicate the sizes of the inserts of pREIUPABC, containing the entire iupABC operon, and pREIUPAB5′C, which lacks the 3′ end of iupC. The downward-pointing arrow shows the position of the transposome insertion. (B) Complementation of R. equi α5G12. R. equi ATCC 33701 (WT) and R. equi α5G12 were grown on LB plates containing 250 μM 2,2-dipyridyl and 400 μM hemin. R. equi fails to grow at this concentration of 2,2-dipyridyl in the absence of an exogenous source of iron, such as hemin. The top half of the plate harbors the wild-type and mutant strains without plasmids, whereas the bottom half shows the growth of strains containing either pREIUPABC or pREIUPAB5′C.
R. equi α5G12 has a transposome insertion in an operon encoding an ABC transport system.
To determine where the transposome had inserted, chromosomal DNA was isolated from the mutant, digested with KpnI, and ligated into the KpnI restriction site of pBluescript II KS. E. coli was subsequently transformed with the ligation mixture, and transformants were screened for kanamycin resistance. Since the KpnI restriction site does not occur within the transposome, plasmids conferring kanamycin resistance to E. coli harbor the transposome and flanking DNA sequences. A colony was isolated harboring pBluescript containing a 4.4-kb KpnI insert (pα5G12). Restriction analysis revealed the presence of a transposome within the KpnI insert (Fig. 1).
In order to characterize the gene harboring the transposome insertion, the nucleotide sequence of the 4.4-kb KpnI fragment of pα5G12 was determined. Sequence analysis showed that the transposome interrupted an open reading frame (ORF1) at nucleotide position 502 (Fig. 1). Insertion of a transposome into the genome mediated by hyperactive Tn5 transposase results in the duplication of the 9-bp target sequence (39). Analysis of the DNA sequences flanking the transposome revealed a 9-bp repeat, showing that the mutation was due to a bona fide Tn5 transposition event. In order to obtain the complete sequence of ORF1, a DNA fragment was amplified from the genome of the wild-type strain using oligonucleotides complementary to sequences up- and downstream from the transposome insertion site. The sequence of the PCR product was subsequently determined to obtain the sequence of the wild-type gene. Two additional open reading frames downstream from ORF1 which were transcribed in the same direction were identified. The three open reading frames are translationally coupled, with overlapping start and stop codons, strongly indicating that these open reading frames form an operon. Downstream of ORF3 and transcribed in the opposite direction is the 3′ end of an open reading frame that may encode a protein that is 45 and 43% identical to the carboxy termini of the Nocardia farcinica and Mycobacterium avium proteins Nfa 41350 and Map0262, respectively (Fig. 1). These proteins are classified as belonging to a family of alkylsulfatase and related hydrolases (COG2015).
ORF1 (iupA) and ORF2 (iupB) share 25% identity and 37% similarity with each other and encode proteins with molecular weights of 36,580 and 37,176, respectively. Both are predicted to be integral membrane proteins with eight membrane-spanning helices. ORF3 (iupC) encodes a protein with a molecular weight of 30,362. Comparison of the three proteins to those in GenBank using BlastP showed a high degree of similarity to proteins encoding ABC transport systems, in particular to transport systems required for siderophore uptake. IupA and IupB belong to the FecCD binding protein-dependent transport systems (pfam01032), which are permease components of ABC transport systems. IupC contains an ATPase domain (pfam0005). The N. farcinica (26) and Streptomyces coelicolor (3) genomes contain gene clusters similar to iupABC. The former encodes three proteins (nfa25210, nfa25200, and nfa25190) that are 50, 55, and 74% identical to IupA, IupB, and IupC, respectively. As is the case for the iup gene cluster, the genes encoding the N. farcinica Iup homologues are also translationally coupled. The S. coelicolor genes cchC, cchD, and cchE are part of a gene cluster that is predicted to produce the tripeptide siderophore coelichelin (10). The cchC and cchD genes are 45 and 54% identical to IupA and IupB, respectively; cchE is 73% identical to IupC.
To rule out the possibility that the inability of R. equi α5G12 to grow in the presence of 80 μM 2,2-dipyridyl was due to a mutation of a gene other than iupA, plasmid pREIUPABC containing the complete iupABC gene cluster was introduced into the mutant. Growth of R. equi α5G12 (pIUPABC) was comparable to that of the wild-type strain in the presence of 2,2-dipyridyl, showing that the phenotype of the mutant was due to insertion of a transposome into iupA. In addition, the complemented mutant was able to utilize hemin and hemoglobin as sources of iron. Interestingly, plasmid pREIUPAB5′C, which contains iupAB and the 5′ end of iupC, failed to complement R. equi α5G12. The insertion of a transposome in iupA therefore had a polar effect on expression of the downstream genes iupBC, showing that iupABC is transcribed as an operon (Fig. 1).
Transcription of the iupABC operon is regulated by the concentration of iron.
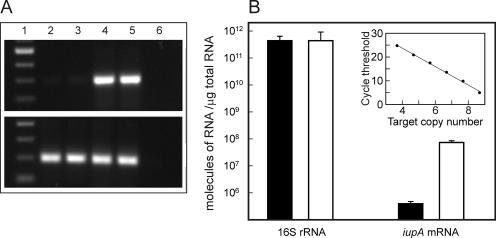
The transcriptional regulation of the iupABC operon was assessed by growing R. equi in lactate minimal medium until the optical density at 600 nm was 0.5. At this point, the culture was washed and aliquots were resuspended in either LMM or LMM-Fe containing 200 μM 2,2-dipyridyl. Two hours later, total RNA was extracted from both cultures, followed by RT-PCR to detect iupA mRNA. Whereas iupA mRNA was barely detectable in RNA extracted from cells growing in lactate minimal medium, it was clearly present in RNA obtained from cells grown in iron-depleted medium. In contrast, the levels of 16S rRNA appeared to be the same in both (Fig. 2A). Quantification of iupA mRNA by real-time PCR showed that iupA transcripts are 180-fold more abundant in R. equi grown under iron-depleted conditions than in organisms grown in iron-replete conditions (Fig. 2B).
FIG. 2.
Detection and quantification of iupA mRNA. (A) RT-PCR of iupA mRNA (top panel) and 16S rRNA (bottom panel) from two independent experiments. Lane 1, 100-bp size marker; lanes 2 and 3, RNA isolated from R. equi grown under iron-replete conditions; lanes 4 and 5, RNA isolated from R. equi grown under iron-depleted conditions; lane 6, RT-PCR carried out without reverse transcriptase. (B) Absolute quantification of iupA mRNA and 16S rRNA using RNA isolated from R. equi grown under iron-replete (black bar) or -depleted (white bar) conditions. Insert: relationship between the cycle threshold and number of template molecules used to calibrate the real-time PCR. Target copy number is shown as log10. Shown are the averages of results from three independent experiments in which each sample was analyzed in duplicate.
The iupABC operon is not required for virulence.
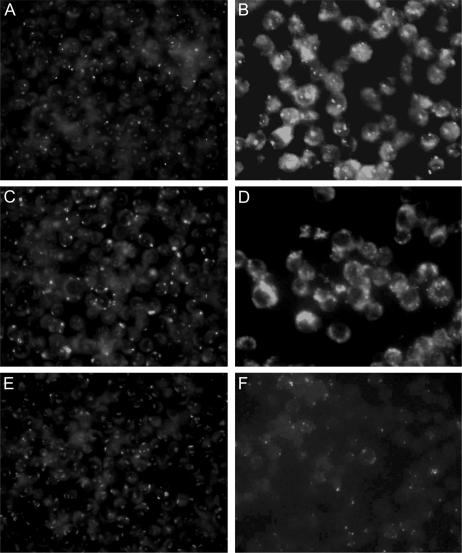
Many pathogens require siderophores for virulence. For example, an M. tuberculosis strain unable to produce mycobactins was not able to grow in macrophages (13). We therefore compared the intracellular growth of R. equi α5G12 with the virulent wild type and the avirulent plasmid-cured strains. Macrophages were incubated with R. equi and allowed to internalize bacteria, after which gentamicin was added to kill extracellular bacteria. Growth was subsequently assessed by immunofluorescent labeling of internalized R. equi. In contrast to the avirulent R. equi strain, R. equi α5G12 displayed the same degree of proliferation in macrophages 48 h following infection as the wild-type strain, showing that the iupABC operon is not required for growth in macrophages (Fig. 3).
FIG. 3.
Intracellular growth of R. equi in murine macrophages. Monolayers were infected with isogenic R. equi wild-type, avirulent, and α5G12 strains. Following a 45-min incubation to allow phagocytosis, unbound bacteria were removed by washing. Monolayers were left for 15 min to allow internalization of bound bacteria, washed, and maintained in medium containing gentamicin to kill any remaining extracellular bacteria. Intracellular bacteria were visualized by immunofluorescence. Shown are murine macrophages 90 min (A, C, and E) and 48 h (B, D, and F) after phagocytosis of wild-type R. equi (A and B), R. equi α5G12 (C and D), and avirulent R. equi (E and F).
To further substantiate these findings, mice were intravenously and peritoneally infected with R. equi. Although immunocompetent mice will eventually clear R. equi, the increase in R. equi numbers in liver and spleen 2 to 4 days after injection provides a good indication of R. equi virulence in vivo (19). There was no significant difference between the numbers of wild-type R. equi ATCC 33701 and the α5G12 mutant in the liver (mean ± standard deviation, 4.43 ± 0.83 versus 5.23 ± 0.97 log10/g) or spleen (3.66 ± 0.61 versus 3.99 ± 0.63 log10/g) of mice 4 days after intravenous injection of a similar number of bacteria. After intraperitoneal infection, however, three mice in the group infected with R. equi ATCC 33701+ became ill; two of these mice were euthanized for humane reasons 7 days postinjection, and the third died 5 days later. Two mice in the α5G12 group showed similar but less severe symptoms and subsequently recovered.
DISCUSSION
Although it has been reported that R. equi requires iron for growth (27), nothing is known regarding mechanisms employed by this facultative pathogen to grow at low iron concentrations. This paper describes a mutant that, unlike the wild-type strain, was unable to grow at relatively low concentrations of 2,2-dipyridyl. The mutant contained a transposome insertion in first gene of a three-cistron operon, iupABC, encoding proteins highly similar to ABC transport systems required for uptake of siderophores. The inability of R. equi α5G12 to grow in the presence of 80 μM 2,2-dipyridyl is therefore probably due to a failure to translocate a siderophore-iron complex into the cell. To date, siderophores have not been identified in R. equi, although analysis of a partial genome sequence revealed sequences that are similar to mycobactin and exochelin biosynthesis genes (36). The iupABC genes are highly similar to the cchCDE genes of S. coelicolor, which are part of gene cluster that is predicted to produce a novel hydroxamate siderophore, coelichelin (10). Recent structural analysis of coelichelin confirmed that coelichelin is indeed a hydroxamate siderophore (G. Challis, personal communication). It therefore appears likely that the ABC transport system encoded by the iupABC operon is a hydroxamate siderophore transport system. The iupABC operon encodes an ABC transport system belonging to the FecCD family of solute binding protein-dependent transport systems. The coelichelin biosynthesis gene cluster contains a gene encoding a putative solute binding lipoprotein (cchF), which is located downstream from cchE (10). However, a similar binding protein-encoding gene is not located downstream from iupC, suggesting that it may be encoded elsewhere on the R. equi genome.
This study showed that both heme and hemoglobin were used as a source of iron by R. equi. The utilization of heme and hemoglobin is not uncommon in bacteria belonging to the mycolic acid-containing actinomycetes, although it has been studied in detail only in C. diphtheriae and C. ulcerans (12, 38, 42, 43). In these pathogens, heme utilization is dependent on hmuTUV and hmuO. The hmuT gene encodes a heme-binding lipoprotein, and hmuUV specify a heme-specific ABC transport system. Heme is subsequently oxidized by HmuO, a mammalian type heme oxidase (14, 56). Gram-negative bacteria employ outer membrane receptors to bind hemoglobin and heme. Heme is often released from hemoglobin by secreted or outer membrane-bound proteases; alternatively, hemoglobin binds directly to specific outer membrane receptors. Following binding to outer membrane receptors, heme is subsequently transported across the outer membrane into the periplasm via an energy-dependent process facilitated by the TonB system, followed by uptake into the cell using heme-specific ABC transport systems (6, 18, 55). In contrast to other bacteria, heme and hemoglobin utilization by R. equi is dependent on a siderophore transport system (55). Furthermore, in comparison to other heme- and hemoglobin-utilizing bacteria, very high concentrations were required to provide the cell with iron. It seems therefore likely that iron is abstracted from heme by R. equi outside the cell in an aspecific manner that is not physiologically relevant.
Transcription of the iupABC operon was 180-fold higher during growth at low iron conditions than at high iron conditions, which is typical for siderophore biosynthesis and uptake genes (55). Iron-dependent transcriptional regulation of genes required for siderophore biosynthesis and uptake is mediated by DtxR in Corynebacterium (30, 35, 44-46) and the homologous proteins IdeR and DmdR in, respectively, Mycobacterium (1, 15, 16, 40) and Streptomyces (17, 22). DtxR complexed with Fe2+ binds to the operator sequences of iron-regulated genes to repress transcription (53). Since R. equi encodes a DtxR homologue that represses transcription in an iron-dependent manner, it appears likely that this protein is responsible for the iron-dependent transcriptional regulation of the iupABC operon (5).
Many pathogens depend on the production of siderophores to thrive in the host environment. For example, the mbtB gene specifying an enzyme participating in an early step of mycobactin biosynthesis is required for growth of M. tuberculosis in macrophages (13). However, there was no difference between R. equi α5G12 and the wild-type strain in the ability to proliferate in macrophages. In addition, there was no significant effect of disruption of the iupABC operon on virulence as assessed by clearance of R. equi α5G12 from the liver and spleen of mice. This strongly suggests that R. equi employs a separate iron uptake system for growth in the host. A similar situation exists for Burkholderia cenocepacia, Bacillus anthracis, and Pseudomonas aeruginosa. These pathogens produce two types of siderophores, yet only one is required for virulence (9, 51, 54). We therefore propose that the putative siderophore translocated by the iupABC-encoded ABC transport system is required for saprophytic growth, whereas a second siderophore is required for growth of R. equi in macrophages. Analysis of the partial genome sequence of R. equi identified sequences similar to mycobactin biosynthesis genes (36). The relevance of these for virulence is currently being analyzed.
Acknowledgments
This study was supported by a grant from Enterprise Ireland (SC/2003/0560).
We thank Gavin Byrne for helpful technical assistance and Thomas Buckley of the Irish Equine Centre for a gift of anti-R. equi antibodies.
REFERENCES
- 1.Adilakshmi, T., P. D. Ayling, and C. Ratledge. 2000. Mutational analysis of a role for salicylic acid in iron metabolism of Mycobacterium smegmatis. J. Bacteriol. 182:264-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 4.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening of recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boland, C. A., and W. G. Meijer. 2000. The iron regulatory protein IdeR (DtxR) of Rhodococcus equi. FEMS Microbiol. Lett. 191:1-5. [DOI] [PubMed] [Google Scholar]
- 6.Braun, V., and H. Killmann. 1999. Bacterial solutions to the iron-supply problem. Trends Biochem. Sci. 24:104-109. [DOI] [PubMed] [Google Scholar]
- 7.Budzikiewicz, H., A. Boessenkamp, K. Taraz, A. Pandey, and J. M. Meyer. 1997. Corynebactin, a cyclic catecholate siderophore from Corynebacterium glutamicum ATCC 14067 (Brevibacterium sp. DSM 20411). Z. Naturforsch. Sect. C 52:551-554. [Google Scholar]
- 8.Carrano, C. J., M. Jordan, H. Drechsel, D. G. Schmid, and G. Winkelmann. 2001. Heterobactins: a new class of siderophores from Rhodococcus erythropolis IGTS8 containing both hydroxamate and catecholate donor groups. Biometals 14:119-125. [DOI] [PubMed] [Google Scholar]
- 9.Cendrowski, S., W. MacArthur, and P. Hanna. 2004. Bacillus anthracis requires siderophore biosynthesis for growth in macrophages and mouse virulence. Mol. Microbiol. 51:407-417. [DOI] [PubMed] [Google Scholar]
- 10.Challis, G. L., and J. Ravel. 2000. Coelichelin, a new peptide siderophore encoded by the Streptomyces coelicolor genome: structure prediction from the sequence of its non-ribosomal peptide synthetase. FEMS Microbiol. Lett. 187:111-114. [DOI] [PubMed] [Google Scholar]
- 11.Claros, M. G., and G. von Heijne. 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10:685-686. [DOI] [PubMed] [Google Scholar]
- 12.Collins, M. D., D. Jones, and G. M. Schofield. 1982. Reclassification of ‘Corynebacterium haemolyticum ’ (MacLean, Liebow & Rosenberg) in the genus Arcanobacterium gen.nov. as Arcanobacterium haemolyticum nom.rev., comb.nov. J. Gen. Microbiol. 128:1279-1281. [DOI] [PubMed] [Google Scholar]
- 13.De Voss, J. J., K. Rutter, B. G. Schroeder, H. Su, Y. Zhu, and C. E. I. Barry. 2000. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc. Natl. Acad. Sci. USA 97:1252-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drazek, E. S., C. A. Hammack, and M. P. Schmitt. 2000. Corynebacterium diphtheriae genes required for acquisition of iron from haemin and haemoglobin are homologous to ABC haemin transporters. Mol. Microbiol. 36:68-84. [DOI] [PubMed] [Google Scholar]
- 15.Dussurget, O., M. Rodriguez, and I. Smith. 1996. An ideR mutant of Mycobacterium smegmatis has derepressed siderophore production and an altered oxidative-stress response. Mol. Microbiol. 22:535-544. [DOI] [PubMed] [Google Scholar]
- 16.Dussurget, O., J. Timm, M. Gomez, B. Gold, S. Yu, S. Z. Sabol, R. K. Holmes, W. R. Jacobs, Jr., and I. Smith. 1999. Transcriptional control of the iron-responsive fxbA gene by the mycobacterial regulator IdeR. J. Bacteriol. 181:3402-3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flores, F. J., and J. F. Martin. 2004. Iron-regulatory proteins DmdR1 and DmdR2 of Streptomyces coelicolor form two different DNA-protein complexes with iron boxes. Biochem. J. 380:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genco, C. A., and D. W. Dixon. 2001. Emerging strategies in microbial haem capture. Mol. Microbiol. 39:1-11. [DOI] [PubMed] [Google Scholar]
- 19.Giguère, S., M. K. Hondalus, J. A. Yager, P. Darrah, D. M. Mosser, and J. F. Prescott. 1999. Role of the 85-kilobase plasmid and plasmid-encoded virulence-associated protein A in intracellular survival and virulence of Rhodococcus equi. Infect. Immun. 67:3548-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gobin, J., and M. A. Horwitz. 1996. Exochelins of Mycobacterium tuberculosis remove iron from human iron-binding proteins and donate iron to mycobactins in the M. tuberculosis cell wall. J. Exp. Med. 183:1527-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goryshin, I. Y., J. Jendrisak, L. M. Hoffman, R. Meis, and W. S. Reznikoff. 2000. Insertional transposon mutagenesis by electroporation of released Tn5 transposition complexes. Nat. Biotechnol. 18:97-100. [DOI] [PubMed] [Google Scholar]
- 22.Gunter, K., C. Toupet, and T. Schupp. 1993. Characterization of an iron-regulated promoter involved in desferrioxamine B synthesis in Streptomyces pilosus: repressor-binding site and homology to the diphtheria toxin gene promoter. J. Bacteriol. 175:3295-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall, R. M., and C. Ratledge. 1986. Distribution and application of mycobactins for the characterization of species within the genus Rhodococcus. J. Gen. Microbiol. 132:853-856. [DOI] [PubMed] [Google Scholar]
- 24.Hondalus, M. K., M. S. Diamond, L. A. Rosenthal, T. A. Springer, and D. M. Mosser. 1993. The intracellular bacterium Rhodococcus equi requires Mac-1 to bind to mammalian cells. Infect. Immun. 61:2919-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes, K. L., and I. Sulaiman. 1987. The ecology of Rhodococcus equi and physicochemical influences on growth. Vet. Microbiol. 14:241-250. [DOI] [PubMed] [Google Scholar]
- 26.Ishikawa, J., A. Yamashita, Y. Mikami, Y. Hoshino, H. Kurita, K. Hotta, T. Shiba, and M. Hattori. 2004. The complete genomic sequence of Nocardia farcinica IFM 10152. Proc. Natl. Acad. Sci. USA 101:14925-14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jordan, M. C., J. R. Harrington, N. D. Cohen, R. M. Tsolis, L. J. Dangott, E. D. Weinberg, and R. J. Martens. 2003. Effects of iron modulation on growth and viability of Rhodococcus equi and expression of virulence-associated protein A. Am. J. Vet. Res. 64:1337-1346. [DOI] [PubMed] [Google Scholar]
- 28.Kelly, B. G., D. W. Wall, C. A. Boland, and W. G. Meijer. 2002. Isocitrate lyase of the facultative intracellular pathogen Rhodococcus equi. Microbiology 148:793-798. [DOI] [PubMed] [Google Scholar]
- 29.Köster, W. 2001. ABC transporter-mediated uptake of iron, siderophores, heme and vitamin B12. Res. Microbiol. 152:291-301. [DOI] [PubMed] [Google Scholar]
- 30.Kunkle, C. A., and M. P. Schmitt. 2003. Analysis of the Corynebacterium diphtheriae DtxR regulon: identification of a putative siderophore synthesis and transport system that is similar to the Yersinia high-pathogenicity island-encoded yersiniabactin synthesis and uptake system. J. Bacteriol. 185:6826-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mangan, M. W., G. A. Byrne, and W. G. Meijer. 2005. Versatile Rhodococcus equi-Escherichia coli shuttle vectors. Antonie Leeuwenhoek 87:161-167. [DOI] [PubMed] [Google Scholar]
- 32.Mangan, M. W., and W. G. Meijer. 2001. Random insertion mutagenesis of the intracellular pathogen Rhodococcus equi using transposomes. FEMS Microbiol. Lett. 205:243-246. [DOI] [PubMed] [Google Scholar]
- 33.Meijer, W. G., and J. F. Prescott. 2004. Rhodococcus equi. Vet. Res. 35:383-396. [DOI] [PubMed] [Google Scholar]
- 34.Nagy, I., G. Schoofs, F. Compernolle, P. Proost, J. Vanderleyden, and R. de Mot. 1995. Degradation of the thiocarbamate herbicide EPTC (S-ethyl dipropylcarbamothioate) and biosafening by Rhodococcus sp. strain NI86/21 involve an inducible cytochrome P-450 system and aldehyde dehydrogenase. J. Bacteriol. 177:676-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian, Y., J. H. Lee, and R. K. Holmes. 2002. Identification of a DtxR-regulated operon that is essential for siderophore-dependent iron uptake in Corynebacterium diphtheriae. J. Bacteriol. 184:4846-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahman, M. T., L. L. Herron, V. Kapur, W. G. Meijer, B. A. Byrne, J. Ren, V. M. Nicholson, and J. F. Prescott. 2003. Overview of a partial genome sequence of Rhodococcus equi ATCC 33701. Vet. Microbiol. 94:143-158. [DOI] [PubMed] [Google Scholar]
- 37.Ratledge, C. 2004. Iron, mycobacteria and tuberculosis. Tuberculosis (Edinburgh) 84:110-130. [DOI] [PubMed] [Google Scholar]
- 38.Reddy, C. A., C. P. Cornell, and M. Kao. 1977. Hemin-dependent growth stimulation and cytochrome synthesis in Corynebacterium pyogenes. J. Bacteriol. 130:965-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reznikoff, W. S., A. Bhasin, D. R. Davies, I. Y. Goryshin, L. A. Mahnke, T. Naumann, I. Rayment, M. Steiniger-White, and S. S. Twining. 1999. Tn5: a molecular window on transposition. Biochem. Biophys. Res. Commun. 266:729-734. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez, G. M., M. I. Voskuil, B. Gold, G. K. Schoolnik, and I. Smith. 2002. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70:3371-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Saubolle, M. A., T. E. Kiehn, M. H. White, M. F. Rudinsky, and D. Armstrong. 1996. Mycobacterium haemophilum: microbiology and expanding clinical and geographic spectra of disease in humans. Clin. Microbiol. Rev. 9:435-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmitt, M. P. 1997. Utilization of host iron sources by Corynebacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenases and is required for acquisition of iron from heme and hemoglobin. J. Bacteriol. 179:838-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitt, M. P., and R. K. Holmes. 1991. Iron-dependent regulation of diphtheria toxin and siderophore expression by the cloned Corynebacterium diphtheriae repressor gene dtxR in C. diphtheriae C7 strains. Infect. Immun. 59:1899-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitt, M. P., and R. K. Holmes. 1994. Cloning, sequence, and footprint analysis of two promoter/operators from Corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor (DtxR) and iron. J. Bacteriol. 176:1141-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmitt, M. P., B. G. Talley, and R. K. Holmes. 1997. Characterization of lipoprotein IRP1 from Corynebacterium diphtheriae, which is regulated by the diphtheria toxin repressor (DtxR) and iron. Infect. Immun. 65:5364-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider, E., and S. Hunke. 1998. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol. Rev. 22:1-20. [DOI] [PubMed] [Google Scholar]
- 48.Staden, R., K. F. Beal, and J. K. Bonfield. 2000. The Staden package, 1998. Methods Mol. Biol. 132:115-130. [DOI] [PubMed] [Google Scholar]
- 49.Takai, S., S. A. Hines, T. Sekizaki, V. M. Nicholson, D. A. Alperin, M. Osaki, D. Osaki, M. Nakamura, K. Suzuki, N. Ogino, T. Kakuka, H. Dan, and J. F. Prescott. 2000. DNA sequence and comparison of virulence plasmids from Rhodococcus equi ATCC 33701 and 103. Infect. Immun. 68:6840-6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takai, S., T. Sekizaki, T. Ozawa, T. Sugawara, Y. Watanabe, and S. Tsubaki. 1991. Association between a large plasmid and 15- to 17-kilodalton antigens in virulent Rhodococcus equi. Infect. Immun. 59:4056-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takase, H., H. Nitanai, K. Hoshino, and T. Otani. 2000. Impact of siderophore production on Pseudomonas aeruginosa infections in immunosuppressed mice. Infect. Immun. 68:1834-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tam, R., and M. H. Saier, Jr. 1993. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 57:320-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tao, X., and J. R. Murphy. 1992. Binding of the metalloregulatory protein DtxR to the diphtheria tox operator requires a divalent heavy metal ion and protects the palindromic sequence from DNase I digestion. J. Biol. Chem. 267:21761-21764. [PubMed] [Google Scholar]
- 54.Visser, M. B., S. Majumdar, E. Hani, and P. A. Sokol. 2004. Importance of the ornibactin and pyochelin siderophore transport systems in Burkholderia cenocepacia lung infections. Infect. Immun. 72:2850-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wandersman, C., and P. Delepelaire. 2004. Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58:611-647. [DOI] [PubMed] [Google Scholar]
- 56.Wilks, A., and M. P. Schmitt. 1998. Expression and characterization of a heme oxygenase (Hmu O) from Corynebacterium diphtheriae. Iron acquisition requires oxidative cleavage of the heme macrocycle. J. Biol. Chem. 273:837-841. [DOI] [PubMed] [Google Scholar]