Abstract
The first elongation step of fatty acid biosynthesis by a type II dissociated fatty acid synthases is catalyzed by 3-ketoacyl-acyl carrier protein (ACP) synthase III (KASIII, FabH). This enzyme, encoded by the fabH gene, catalyzes a decarboxylative condensation between an acyl coenzyme A (CoA) primer and malonyl-ACP. In organisms such as Escherichia coli, which generate only straight-chain fatty acids (SCFAs), FabH has a substrate preference for acetyl-CoA. In streptomycetes and other organisms which produce a mixture of both SCFAs and branched-chain fatty acids (BCFAs), FabH has been shown to utilize straight- and branched-chain acyl-CoA substrates. We report herein the generation of a Streptomyces coelicolor mutant (YL/ecFabH) in which the chromosomal copy of the fabH gene has been replaced and the essential process of fatty acid biosynthesis is initiated by plasmid-based expression of the E. coli FabH (bearing only 35% amino acid identity to the Streptomyces enzyme). The YL/ecFabH mutant produces predominantly SCFAs (86%). In contrast, BCFAs predominate (∼70%) in both the S. coelicolor parental strain and S. coelicolor YL/sgFabH (a ΔfabH mutant carrying a plasmid expressing the Streptomyces glaucescens FabH). These results provide the first unequivocal evidence that the substrate specificity of FabH observed in vitro is a determinant of the fatty acid made in an organism. The YL/ecFabH strain grows significantly slower on both solid and liquid media. The levels of FabH activity in cell extracts of YL/ecFabH were also significantly lower than those in cell extracts of YL/sgFabH, suggesting that a decreased rate of fatty acid synthesis may account for the observed decreased growth rate. The production of low levels of BCFAs in YL/ecFabH suggests either that the E. coli FabH is more tolerant of different acyl-CoAs substrates than previously thought or that there is an additional pathway for initiation of BCFA biosynthesis in Streptomyces coelicolor.
The type II dissociated fatty acid synthase (FAS) found in most bacteria and plant plastids comprises a set of discrete enzymes responsible for catalyzing each of the individual steps of fatty acid biosynthesis (13). The elongation steps are carried out by ketoacyl acyl carrier protein (ACP) synthases, which typically catalyze a decarboxylative condensation of malonyl-ACP, and an acyl-ACP. FabH (KASIII) is an exception in that it uses a short-chain acyl coenzyme A (CoA) substrate and is thought to initiate the biosynthetic process by catalyzing the first decarboxylative condensation. It has been suggested that the substrate specificity of FabH is a major determinant in the types of fatty acids made by an organism (1, 6). The Escherichia coli FabH (ecFabH) has been shown in vitro to have a strong preference for acetyl-CoA and propionyl-CoA, which correlates with the observation that only straight-chain fatty acids (SCFAs) are produced in this organism (20). In contrast a number of microorganisms, including Streptomyces glaucescens and Bacillus subtilis, produce a mixture of both SCFAs and branched-chain fatty acids (BCFAs) (1, 6). Isobutyryl-CoA and methylbutyryl-CoA, formed to a significant degree from branched-chain amino acid degradation, are used as primers for BCFA biosynthesis (2, 21). Consistent with this observation, the Streptomyces glaucescens FabH (sgFabH) was shown to have less stringent substrate specificity than the ecFabH (6), utilizing both straight- and branched-chain acyl-CoAs. Subsequent analyses revealed that FabH enzymes from other organisms which produce both BCFAs and SCFAs had similar substrate flexibilities (1, 8).
These data suggest that a functional exchange of FabH isozymes with different acyl-CoA specificities could lead to a dramatic change in fatty acid profiles of a microorganism. However, this critical experiment has not been yet successfully carried out. Attempts to significantly alter fatty acid profiles by expression of a heterologous FabH in the presence of the natural FabH have been marginally successful. Expression of either the sgFabH or a Bacillus subtilis FabH in E. coli does not result in production of significant levels of BCFAs (1, 18). In the first case BCFA production was dependent upon addition of appropriate biosynthetic precursors to the fermentation media and was not increased by the presence of the sgFabH (18). In the second case a claim for low-level production of a C17 BCFA was made but remains to be verified (1). Attempts have also been made to increase the ratio of SCFAs relative to BCFAs in Streptomyces glaucescens by expression of the ecFabH, but these did not result in any significant alteration of the fatty acid profile (N. Smirnova and K. A. Reynolds, unpublished results). Contributing reasons for the lack of significant change in these experiments include the lack of appropriate precursors (particularly for BCFA biosynthesis in E. coli) and competition with the endogenous FabH. A more significant change in fatty acid profiles was accomplished by plasmid-based expression of an sgFabH mutant bearing a Cys122Gln mutation in Streptomyces glaucescens, where a fivefold increase in SCFAs was observed (19). This change was attributed to the sgFabH mutant being unable to catalyze the normal decarboxylative condensation reaction yet retaining the ability to decarboxylate malonyl-ACP to acetyl-ACP (a presumed precursor for SCFA biosynthesis). Even in this case BCFAs predominated, presumably due to superior catalytic capabilities of the nonmutant sgFabH. Subsequent attempts to introduce the mutation into the chromosomal fabH gene were unsuccessful (N. Smirnova and K. A. Reynolds, unpublished results).
The proposed role of FabH in catalyzing the initial step of fatty acid biosynthesis by a type II FAS has made it an attractive target for generating novel antibacterial agents for a number of years (14, 15). Recently, clear evidence has been obtained that FabH is an essential enzyme for fatty acid biosynthesis in E. coli, Streptomyces coelicolor, and Lactococcus lactis (11, 16). In the cases of E. coli and S. coelicolor it was shown that the chromosomal fabH gene could be removed only if an additional copy of fabH was present; for S. coelicolor it was a second copy of the natural fabH gene (16), and in the case of E. coli it was the Salmonella enterica fabH (11). S. enterica is a close relative of E. coli, and it is known that the genes are fully interchangeable between the two species (11). We sought to extend this work with a more ambitious exchange of the S. coelicolor FabH with the E. coli FabH. These two enzymes have limited amino acid sequence homology (35% amino acid identity) and different acyl-CoA and ACP specificities. Despite these differences, this exchange was possible and resulted in the generation of an S. coelicolor mutant (YL/ecFabH) which generates predominantly SCFAs (∼87%). As such, this experiment provides the first unequivocal evidence that the substrate specificity of FabH observed in vitro is a determinant of the fatty acid made in an organism. Low-level production of BCFAs in the S. coelicolor YL/ecFabH mutant is a surprising finding given the strict substrate specificity of the E. coli FabH and indicates the important role precursor availability plays in determining fatty acid profiles of a microorganism. Finally, the ability to change fatty acid profiles in Streptomyces suggests a way to engineer enhanced production of specific commercial natural products which are made using the same precursors used to initiate BCFA biosynthesis (2, 23).
MATERIALS AND METHODS
Materials.
The E. coli and S. glaucescens ACPs were obtained as described previously (12). Malonyl-CoA and CoASH were obtained from Sigma Chemical Co. (St. Louis, MO). Radiolabeled [14C]acetyl-CoA (specific activity, 50mCi/mmol) was obtained from Moravek Biochemicals. All other chemicals were of reagent grade or better and were obtained from Fisher Scientific (Pittsburgh, PA).
Strains, plasmids, and culture conditions.
All E. coli strains and S. coelicolor used in this study were grown according to standard protocols (10, 17). The template plasmids and strains used for PCR-targeted disruption (including E. coli BW25113, pIJ790, and pIJ773) were provided by John Innes Center Plant Biosciences Limited, Norwich, England. The S. coelicolor cosmid 4A7 was provided by Helen Kieser at the John Innes Center, Norwich, England. Streptomyces coelicolor M511 (3) was provided by Greg Challis (University of Warwick, England). The plasmid pSW7, expressing the Streptomyces glaucescens fabH from PermE*, has been described previously and was generated by inserting the corresponding fabH gene as a BglII fragment into the Streptomyces expression plasmid pSE34 (19). The plasmid pSE4, expressing the E. coli fabH under PermE* control, was similarly constructed. This fabH gene was obtained as a BamHI PCR fragment by using E. coli chromosomal DNA and was inserted into pSE34 to create pSE4. The primer set for this experiment was AGAGAGGATCCACCCATGTATACGAAGATTATTG (forward) and CTCTCTGGATCCTAGAAACGAACCAG (reverse).
Construction of Streptomyces coelicolor fabH deletion mutants (YL/sgFabH and YL/ecFabH).
A linear fragment of DNA carrying the aac(3)IV resistance gene and oriT flanked by FRT sites were amplified by PCR from the pIJ773 disruption cassette (5), using ScFabHtargetF (5′-CCGAGCACACCCAGGCCTGACAAGGAGCGCGAGAGCATGATTCCGGGGATCCGTCGACC-3′) and ScFabHtargetR (5′-CTCCGTTCCGGACCGGGGTGATCCGGCACGGAGTGCCTATGTAGGCTGGAGCTGCTTC-3′) (underlining indicates S. coelicolor homologous sequence). The resulting 1.3-kb PCR product was gel purified and used for a fabH gene replacement in S. coelicolor cosmid 4A7 according to standard methodologies (5). Briefly, the 1.3-kb PCR product was used to replace fabH, first in S. coelicolor cosmid 4A7 and then in both S. coelicolor M511/pSE4 and M511/pSW7 (5). The resulting mutants, YL/sgFabH [ΔfabH::oriT aac(3)IV/pSW7] and YL/ecFabH [ΔfabH::oriT aac(3)IV/pSE4], were shown to be Kans Aprar, indicating a double-crossover allelic exchange in S. coelicolor. Allelic replacement of the fabH gene in YL/sgFabH and YL/ecFabH was confirmed by PCR amplification of chromosomal DNA with oligonucleotides ScFabHF (5′-AAGGAGCGCGAGAGCATGTCG-3′) and ScFabHR (5′-ACCGTGATCCGGCACGGAGTG-3′), which primed approximately 100 bp outside the region of recombination.
Growth of S. coelicolor M511, YL/sgFabH, and YL/ecFabH.
Determination of the growth rates of the S. coelicolor M511/pSE34, YL/sgFabH, YL/ecFabH strains was carried out by inoculating a 100-μl spore suspension into 100 ml of yeast extract-malt extract (YEME) medium containing thiostrepton (12 μg/ml) and incubating on an orbital shaker (220 rpm) for 7 days at 30°C. To measure cell growth, duplicate samples (1 ml) of fermentation broth were collected every 24 h, and optical absorbance was measured at 600 nm.
FabH enzyme assays were conducted on cell extracts generated from M511/pSE34, YL/sgFabH, and YL/ecFabH after 5 days of fermentation. Cells were collected by centrifugation at 5,000 × g for 20 min at 4°C, washed with Na+-phosphate buffer pH 7.5, resuspended in 5 ml buffer (50 mM Na+-phosphate pH 7.5, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 5% glycerol), and disrupted by sonication in an ice bath. Centrifugation at 20,000 × g for 20 min provided the cell extract, which was adjusted to a final protein concentration of 5 mg/ml and stored at −80°C.
FabH enzyme assays.
FabH assays were carried out on cell extracts of S. coelicolor M511/pSE34, YL/sgFabH, and YL/ecFabH by using a standard coupled trichloroacetic acid precipitation assay which determines the rate of formation of radiolabeled 3-ketoacyl-ACP from malonyl-ACP and radiolabeled acetyl-CoA (6). In this coupled assay Streptomyces glaucescens FabD is used to generate the malonyl-ACP substrate from malonyl-CoA and either the E. coli or S. glaucescens ACP (7).
Fatty acid analysis.
Fatty acid profiles were obtained by standard protocols (21).
RESULTS AND DISCUSSION
Deletion of fabH in S. coelicolor M511/pSW7 and M511/pSE4.
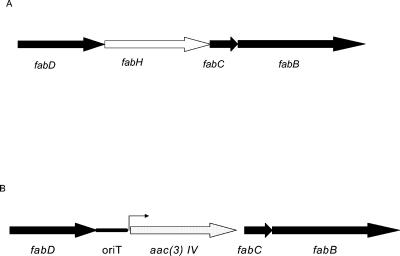
Previous data (16) have suggested that the S. coelicolor fabH gene, which lies in a cluster of fatty acid biosynthetic genes (Fig. 1), is an essential gene and is required for initiation of fatty acid biosynthesis. The fabH gene could be deleted only if a second copy of it was introduced into the cell. In this work and comparable experiments with E. coli, the additional copy of fabH was delivered as a chromosomal copy distant from its loci (11, 16).
FIG. 1.
Organization of four fatty acid biosynthetic genes in S. coelicolor M511 (A) and YL/sgFabH and YL/ecFabH(B).
In the current work we first used a pSW7 expression plasmid to provide the Streptomyces glaucescens FabH (which has 100% amino acid homology to the S. coelicolor FabH) for initiation of fatty acid biosynthesis in S. coelicolor and predicted that this would permit allelic replacement of fabH from the fatty acid biosynthetic loci. Thus, S. coelicolor YL/sgFabH was generated from S. coelicolor M511/pSW7 by replacing the chromosomal copy of fabH with oriT and aacIV (Fig. 1) through homologous recombination as described in Material and Methods. The fabH gene in S. coelicolor is 5′ of fabC, which encodes ACP, and fabB (encoding a ketoacyl-ACP synthase I). These genes are also presumably required for growth of S. coelicolor and thus are expressed in YL/sgFabH, as this strain had growth rates (Fig. 2) and fatty acid profiles (see Table 1 and Fig. 3A) comparable to those of the M511/pSE34 strain.
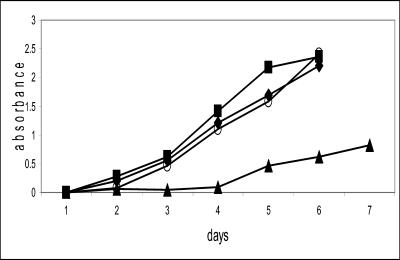
FIG. 2.
Growth of S. coelicolor M511/pSW7 (▪), M511/pSE4 (♦), YL/sgFabH (○), and YL/ecFabH (▴) in liquid YEME medium.
TABLE 1.
Fatty acid compositions of M511/pSE34, YL/sgFabH, and YL/ecFabH mutants of S. coelicolor in YEME medium
| Fatty acid(s) | % of total fatty acid poola in:
|
||
|---|---|---|---|
| M511/pSE34 | YL/sgFabH | YL/ecFabH | |
| Total BCFA | 74.4 | 77.8 | 12.5 |
| Isomyristate (C14) | 3.5 | 3.6 | 4.2 |
| Isopentadecanaoate (C15) | 10.0 | 15.4 | |
| Anteisopentadecanaoate (C15) | 17.3 | 17.9 | |
| Isopalmitate (C16) | 20.3 | 17.0 | 8.3 |
| Iso/anteisoheptadecanaoate (C17) | 23.3 | 23.9 | |
| Total SCFAs | 25.6 | 22.2 | 87.5 |
| Myristate (C14) | 1.6 | 1.9 | 7.8 |
| Pentadecanaoate (C15) | 1.0 | 2.5 | 6.8 |
| Palmitoleate (C16:1) | 4.7 | 2.1 | 19.7 |
| Palmitate (C16) | 18.3 | 15.7 | 46.3 |
| Otherb | <1 | <1 | 6.9 |
Values are representative of those from analyses of triplicate fermentations of each strain.
Minor components including C17:1, C18:0, C18:1.
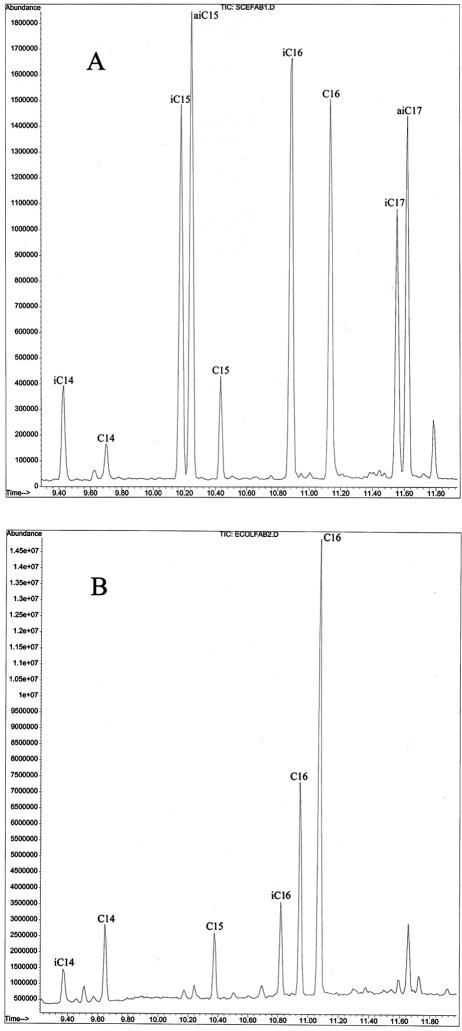
FIG. 3.
Fatty acid profiles of YL/sgFabH (A) and YL/ecFabH (B). Fatty acid abbreviations: iC14, isomyristate; C14, myristate; iC15, isopentadecanoate; aiC15, anteisopentadecanoate; iC16, isopalmitate; C16:1, palmitoleate; C16:0, palmitate; iC17 isopentadecanoate; aiC17, anteisopentadecanoate; C17, pentadecanoate.
The success of this initial experiment served as a basis for a more ambitious experiment involving an analogous allelic replacement in S. coelicolor M511 carrying pSE4 expressing the E. coli FabH. Although this enzyme has different substrate specificities from and low homology (35% amino acid identity) to the S. coelicolor FabH, the YL/ecFabH strain was readily obtained from M511/pSE4.
Growth rates of the YL/sgFabH and YL/ecFabH strains.
The growth rate of YL/sgFabH (carrying multiple copies of the Streptomyces fabH gene) on either solid agar or liquid R2YE medium was not distinguishable from that of M511/pSW7 or M511/pSE4 (plasmid controls with no fabH replacement) As noted above, this observation indicates that replacement of the chromosomal copy of fabH does not have a significant polar effect on the expression of fabC and fabB. In stark contrast, YL/ecFabH (carrying multiple copies of the E. coli FabH) grew substantially slower on both solid agar and liquid medium (Fig. 2).
To investigate possible reasons for this observation, we analyzed the levels of FabH activities in all strains used in the study, as described in Materials and Methods. In this analysis the cultures were fermented for 5 days, allowing comparable amounts of cells to be used for generating cell extracts, despite the lower growth rate of YL/ecFabH relative to other strains. We have previously observed residual FabH activity in cell extracts generated from S. glaucescens after 4 days of growth (the maximal specific activity was observed after 24 h of growth) (4). Using acetyl-CoA and malonyl-ACP generated from FabC (the Streptomyces ACP), we also saw residual FabH activity in cell extracts of S. coelicolor M511/pSE34. In contrast, cell extracts of YL/sgFabH consistently exhibited 15-fold-higher FabH levels, consistent with efficient expression of the S. glaucescens FabH from a constitutive promoter on a multicopy plasmid. In contrast, the levels of FabH activity in cell extracts of YL/ecFabH were almost indistinguishable from those in extracts of M511/pSE4. The low-level activity in this strain may arise as result of both poorer expression of the E. coli FabH and a lower catalytic activity with FabC than with the E coli ACP (AcpP). However, when the assays were repeated with AcpP, a comparable result of significantly higher FabH activity for cell extracts of YL/sgFabH relative to those of M511/pSE4 and YL/ecFabH was obtained. Thus, poor expression of the E. coli FabH in YL/ecFabH, possibly due to different codon utilization, may give a decreased rate of fatty acid biosynthesis and be a contributing factor to the lower growth rate of this strain. Studies involving codon modification of the E. coli fabH sequence and use of other fabH homologs (encoding enzymes with acetyl-CoA substrate preference) are ongoing and will ultimately address this issue.
Fatty acid profiles of YL/sgFabH and YL/ecFabH.
We have previously demonstrated that Streptomyces does not have an obligatory growth requirement for BCFAs and that a mutant producing only SCFAs is obtained by blocking degradation of branched-chain amino acids (providing the primers for BCFA biosynthesis) (2). Introduction of the E. coli fabH expression plasmid (pSE4) into S. coelicolor M511 and other Streptomyces spp., however, does not lead to significant changes in the fatty acid profile (data not shown). The possible reasons for this observation likely include relatively low levels of catalytic activity of expressed E. coli FabH relative to endogenous S. coelicolor FabH. In contrast, expression of the E. coli FabH in YL/ecFabH (in which the corresponding S. coelicolor fabH gene has been replaced by an apramycin resistance gene) leads to a dramatic change in the fatty acid profile (Table 1; Fig. 3).
As predicted, the fatty acid profiles for YL/sgFabH and M511/pSE34 were identical and consisted primarily of BCFA (∼76%) (Table 1) There have been several reported observations that increased levels of FabH activity in bacteria lead to increased proportions of shorter-chain fatty acid products. Despite the large increase in FabH activity in YL/sgFabH relative to M511/pSE4, this phenomenon was not observed. We have previously noted that this phenomenon was also not observed with expression of this S. coelicolor FabH or S. glaucescens FabH in E. coli (6, 19).
The fatty acid profile of YL/ecFabH (Table 1) was different and consisted of more than 87.5% SCFA, which presumably reflects the substrate preference of E. coli FabH towards acetyl-CoA and propionyl-CoA (fatty acids containing even and odd numbers of carbons are observed). This observation thus provides the first in vivo evidence that the substrate specificity of an initiation enzyme contributes significantly to determining the kind of fatty acids synthesized in organism. In the YL/ecFabH strain, palmitoleate is the second most predominant fatty acid (palmitate being the first). The levels of this unsaturated fatty acid (UFA) increase from 4% of the total fatty acid pool in M511/pSE34 to 20% in YL/ecFabH. The BCFAs generated by organisms such as streptomycetes are important for regulation of membrane fluidity. In bacilli BCFA production is an obligate growth requirement (22). We have previously shown that a Streptomyces avermitilis bkd mutant which produces only SCFAs compensates for the loss of BCFAs by increased production of UFAs (2). The large increase in levels of UFAs in YL/ecFabH demonstrates that this compensation mechanism occurs even when the BCFAs are at significantly lower levels but are still present.
The production of BCFAs (representing approximately 12.5% of the total fatty acid pool) by YL/ecFabH is an intriguing observation. In addition, there is a much larger decrease in BCFAs generated from C5 methylbutyryl-CoA precursors (iC15, aiC15, iC17, and aiC17) than from isobutyryl-CoA precursors (iC14 and iC16) (Table 1; Fig. 3). E. coli generates no detectable levels of BCFAs, and the E. coli FabH has been shown to have a significant preference for the acetyl-CoA and propionyl-CoA substrates (1, 9). In fact, activity with isobutyryl-CoA (45 μM) is reported to be less than 0.2% of that with acetyl-CoA (45 μM) (1). Nonetheless, it has been shown that E. coli can make trace levels of BCFAs when grown in the presence of isobutyrate and that cell extracts can convert perdeuterated isobutyryl-CoA to isopalmitate (18). It has also been shown for both E. coli and streptomycetes that substrate availability plays a contributing role in the type of fatty acids generated (2, 18, 21). Thus, in S. coelicolor YL/ecFabH (and also possibly in E. coli) the E. coli FabH may initiate some level of BCFA biosynthesis. Alternatively, BCFA biosynthesis may be initiated by (i) a protein encoded from other genes identified from the S. coelicolor genome as having homology to fabH or (ii) a FabH-independent pathway such as a FabB-catalyzed condensation of a branched-chain acyl-CoA with malonyl-ACP. Such alternative initiation pathways would presumably be inefficient, as neither we nor others (16) have been able to successfully delete the S. coelicolor FabH without first providing an additional FabH catalytic activity. Nonetheless, it is clear that in S. coelicolor YL/ecFabH SCFAs predominate, consistent with the demonstrated substrate specificity of the E. coli FabH
Conclusion.
We have demonstrated that deletion of the fabH gene of S. coelicolor is possible in the presence of a plasmid expressing the fabH gene of S. glaucescens. In the absence of this plasmid the gene deletion could not be accomplished, thus confirming that in Streptomyces coelicolor, as well as in E. coli and Lactococcus lactis (11, 16), this is an essential gene. More important, we have shown for the first time a replacement of FabH with a heterologous enzyme with only 35% of amino acid identity and dramatically different acyl-CoA substrate specificity. The resulting dramatic shift in the fatty acid profile provides the first unequivocal in vivo evidence that the observed substrate specificity of FabH determines the type of fatty acid product made by a type II FAS. However, the presence of BCFAs in YL/ecFABH indicates that other factors, such as the availability of different precursors for fatty acid synthesis, also play a role in determining the fatty acid profile of a microorganism. The ability to change fatty acid profiles by heterologous FabH exchange has numerous potential applications. In Streptomyces it may provide an approach to engineer enhanced production of specific commercial natural products which are made using the same precursors used to initiate BCFA biosynthesis (2, 23).
REFERENCES
- 1.Choi, K. H., R. J. Heath, and C. O. Rock. 2000. β-Ketoacyl-acyl carrier protein synthase III (FabH) is a determining factor in branched-chain fatty acid biosynthesis. J. Bacteriol. 182:365-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cropp, T. A., A. Smogowicz, E. Hafner, C. Denoya, H. McArthur, and K. Reynolds. 2000. Fatty acid biosynthesis in a branched chain α-keto acid dehydrogenase mutant of Streptomyces avermitilis. Can. J. Microbiol. 46:506-514. [PubMed] [Google Scholar]
- 3.Floriano, B., and M. Bibb. 1996. afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 21:385-396. [DOI] [PubMed] [Google Scholar]
- 4.Florova, G., G. Kazanina, and K. A. Reynolds. 2002. Enzymes involved in fatty acid and polyketide biosynthesis in Streptomyces glaucescens: role of FabH and FabD and their acyl carrier protein specificity. Biochemistry 41:10462-10471. [DOI] [PubMed] [Google Scholar]
- 5.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han, L., S. Lobo, and K. A. Reynolds. 1998. Characterization of 3-ketoacyl acyl carrier protein synthase III from Streptomyces glaucescens: Its role in the initiation of fatty acid biosynthesis. J. Bacteriol. 180:4481-4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He, X., A. M. Reeve, U. R. Desai, G. E. Kellogg, and K. A. Reynolds. 2004. 1,2-Dithiole-3-ones as potent inhibitors of the bacterial 3-ketoacyl acyl carrier protein synthase III (FabH). Antimicrob. Agents Chemother. 48:3093-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He, X., and K. A. Reynolds. 2002. Purification, characterization, and identification of novel inhibitors of the beta-ketoacyl-acyl carrier protein synthase III (FabH) from Staphylococcus aureus. Antimicrob. Agents Chemother. 46:1310-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heath, R. J., and C. O. Rock. 1996. Inhibition of β-ketoacyl-acyl carrier protein synthase-III (FabH) by acyl-acyl carrier protein in Escherichia coli. J. Biol. Chem. 271:10996-11000. [DOI] [PubMed] [Google Scholar]
- 10.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 11.Lai, C. Y., and J. E. Cronan. 2003. Beta-ketoacyl-acyl carrier protein synthase III (FabH) is essential for bacterial fatty acid synthesis. J. Biol. Chem. 278:51494-51503. [DOI] [PubMed] [Google Scholar]
- 12.Lobo, S., G. Florova, and K. A. Reynolds. 2001. A Streptomyces collinus thiolase with novel acetyl-CoA:acyl carrier protein transacylase activity. Biochemistry 40:11955-11964. [DOI] [PubMed] [Google Scholar]
- 13.Magnusson, K., S. Jackowski, C. O. Rock, and J. E. Cronan. 1993. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol. Rev. 57:522-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Payne, D. J., P. V. Warren, D. J. Holmes, Y. Ji, and J. T. Lonsdale. 2001. Bacterial fatty-acid biosynthesis: a genomics-driven target for antibacterial drug discovery. Drug Discov. Today 6:537-544. [DOI] [PubMed] [Google Scholar]
- 15.Qiu, X., C. A. Janson, A. K. Konstantinidis, S. Nwagwu, C. Silverman, W. W. Smith, S. Khandekar, J. Lonsdale, and S. S. Abdel-Meguid. 1999. Crystal structure of beta-ketoacyl-acyl carrier protein synthase III. A key condensing enzyme in bacterial fatty acid biosynthesis. J. Biol. Chem. 274:36465-36471. [DOI] [PubMed] [Google Scholar]
- 16.Revill, W. P., M. J. Bibb, A. K. Scheu, H. J. Kieser, and D. A. Hopwood. 2001. β-Ketoacyl acyl carrier protein synthase III (FabH) is essential for fatty acid biosynthesis in Streptomyces coelicolor A3(2). J. Bacteriol. 183:3526-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook, J., and D. W Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Smirnova, N., and K. A. Reynolds. 2001. Branched-chain fatty acid biosynthesis in Escherichia coli. J. Ind. Microbiol. Biotechnol. 26:2335-2342. [DOI] [PubMed] [Google Scholar]
- 19.Smirnova, N., and K. A. Reynolds. 2001. Engineered fatty acid biosynthesis in Streptomyces by altered catalytic function of beta-ketoacyl-acyl carrier protein synthase III. J. Bacteriol. 183:2335-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsay, J. T., W. Oh, T. J. Larson, S. Jackowski, and C. O. Rock. 1992. Isolation and characterization of the β-keto-acyl carrier protein synthase III gene (fabH) from Escherichia coli K-12. J. Biol. Chem. 267:6807-6814. [PubMed] [Google Scholar]
- 21.Wallace, K. K., B. Zhao, H. A. I. McArthur, and K. A. Reynolds. 1995. In vivo analysis of straight-chain and branched-chain fatty acid biosynthesis in three actinomycetes. FEMS Microbiol. Lett. 131:227-234. [DOI] [PubMed] [Google Scholar]
- 22.Willecke, K., and A. B. Pardee. 1971. Fatty acid-requiring mutant of Bacillus subtilis defective in branched chain α-keto acid dehydrogenase. J. Biol. Chem. 246:5264-5272. [PubMed] [Google Scholar]
- 23.Zhang, Y. X., C. D. Denoya, D. D. Skinner, R. W. Fedechko, H. A. McArthur, M. R. Morgenstern, R. A. Davies, S. Lobo, K. A. Reynolds, and C. R. Hutchinson. 1999. Genes encoding acyl-CoA dehydrogenase (AcdH) homologues from Streptomyces coelicolor and Streptomyces avermitilis provide insights into the metabolism of small branched-chain fatty acids and macrolide antibiotic production. Microbiology 145:2323-2334. [DOI] [PubMed] [Google Scholar]