Abstract
Iron-sulfur flavoproteins (ISF) constitute a widespread family of redox-active proteins in anaerobic prokaryotes. Based on sequence homologies, their overall structure is expected to be similar to that of flavodoxins, but in addition to a flavin mononucleotide cofactor they also contain a cubane-type [4Fe:4S] cluster. In order to gain further insight into the function and properties of ISF, the three-dimensional structures of two ISF homologs, one from the thermophilic methanogen Methanosarcina thermophila and one from the hyperthermophilic sulfate-reducing archaeon Archaeoglobus fulgidus, were determined. The structures indicate that ISF assembles to form a tetramer and that electron transfer between the two types of redox cofactors requires oligomerization to juxtapose the flavin mononucleotide and [4Fe:4S] cluster bound to different subunits. This is only possible between different monomers upon oligomerization. Fundamental differences in the surface properties of the two ISF homologs underscore the diversity encountered within this protein family.
The ISF (iron-sulfur flavoprotein) family is distinct from other flavin mononucleotide (FMN)-containing flavoproteins based on overall sequence identity and an unusually compact cysteine motif (24). The prototype of ISF was characterized from Methanosarcina thermophila (ISF-Mt), a strictly anaerobic methane-producing thermophile belonging to the domain Archaea (16). Since then, homologs of ISF have been identified in the genomes of numerous anaerobic prokaryotes belonging to the domains Bacteria and Archaea (24). Remarkably, many species contain multiple ISF homologs; indeed, the Methanosarcina acetivorans genome is annotated with 19 homologs (12). The only ISF homolog identified from a member of the domain Eukarya is that from the intestinal anaerobic pathogen Entamoeba histolytica (24). Thus, it appears that the ISF family occurs predominantly in anaerobic prokaryotes.
ISF from M. thermophila has been reported to be a functional homodimer, with each monomer containing one FMN and one [4Fe:4S] cluster (1, 16). Ferredoxin can act as an electron donor, and redox potential measurements support the hypothesis that electron flow proceeds from ferredoxin to the low-potential (−394 mV) [4Fe:4S] cluster and then to the flavin (1). Although a downstream electron acceptor is unknown, the inability to detect flavin semiquinone during redox titrations suggests that the protein environment of ISF-Mt stabilizes the hydroquinone form and that ISF-Mt functions as a one-electron/two-electron switch. A role for ISF in electron transport coupled to methane formation has been postulated (16); however, the presence of multiple ISF homologs in metabolically diverse anaerobic prokaryotes (24) suggests broader physiological functions.
The cysteine motif (Cys47, Cys50, Cys53, and Cys59) in ISF-Mt is highly conserved among the deduced sequences of all ISF homologs, consistent with a role in ligation of the [4Fe:4S] cluster. The compact nature of the motif is unusual compared to motifs known to coordinate low-potential redox-active [4Fe:4S] clusters, where one of the cysteines is located remote in the sequence from the other three (typically Cys-X2-Cys-X2-Cys and a distant Cys). Electron paramagnetic resonance spectroscopic analysis of site-specific replacement variants supported the postulated roles for Cys50 and Cys59, although the results for Cys47 and Cys53 were inconclusive (17); thus, ligation of the [4Fe:4S] cluster remained unknown.
To establish the overall molecular architecture and the arrangement of the iron-sulfur cluster and flavin cofactors, we determined the crystal structure of ISF from M. thermophila. In addition, we also crystallized and solved the structures of ISF-Mt and ISF homolog 3 from Archaeoglobus fulgidus (ISF3-Af) in the cluster-free “apo” form with bound FMN.
MATERIALS AND METHODS
Protein production and purification.
ISF-Mt was overproduced in Escherichia coli and purified as follows. Plasmid pFC100, containing an ISF-Mt—intein-chitin-binding domain construct, was generated from pTYB2 of an IMPACT T7 kit from New England Biolabs (Beverly, MA). The isf gene from M. thermophila was PCR amplified from the pML701 plasmid (15) using primers designed to maintain an NdeI site at the start codon and to introduce an EcoRI site downstream of the stop codon. The resulting gene fragment was digested with NdeI and EcoRI and subsequently cloned into pTYB2. The sequence of pFC100 was confirmed by the automated dideoxy method at the nucleic acid facility of the Pennsylvania State University Biotechnology Institute. ISF-Mt was overproduced in E. coli BL21(DE3) grown in Terrific Broth containing ampicillin (100 μg/ml). The cells transformed with pFC100 were grown at 37°C with shaking at 300 rpm until an optical density at 600 nm of 0.6 to 0.8 was reached, at which time the growth conditions were adjusted to 16°C and 200 rpm. After 30 min, the culture was induced by addition of 1% (wt/vol) (final concentration) of Bactolactose (Difco, Detroit, MI) and then harvested 16 h after induction. The entire purification procedure was performed anaerobically. Where appropriate, an anaerobic chamber (Coy Laboratory Products, Ann Arbor, MI) was employed. Approximately 10 g (wet weight) of cells was suspended in 30 ml of 50 mM HEPES-NaOH (pH 7.5) containing 500 mM NaCl, 1% Triton X-100, and 2 mM benzamidine. The cells were lysed by two passages through a French pressure cell at 138 MPa. The lysate was centrifuged at 74,400 × g for 20 min at 4°C. The supernatant solution containing the ISF-Mt—intein-chitin-binding domain protein was filtered (0.45 μm) and applied at a flow rate of 0.5 ml/min to a column containing 20 ml of chitin bead resin from the IMPACT T7 kit. The column was then washed with 200 ml of 50 mM HEPES-NaOH (pH 7.5) containing 500 mM NaCl and 1% Triton X-100 at a flow rate of 2 ml/min. ISF-Mt was cleaved from the chitin-binding domain by washing the column with 60 ml of 50 mM HEPES-NaOH (pH 7.5) containing 500 mM NaCl and 30 mM dithiothreitol, followed by incubation of the column for 16 h at 21°C. ISF-Mt was then eluted from the column with 70 ml of 50 mM HEPES-NaOH (pH 7.5) containing 500 mM NaCl. Reconstitution of ISF with the iron-sulfur cluster and FMN was performed as described previously (24), except that 2-mercaptoethanol was omitted from the buffers. ISF3-Af was overexpressed and purified as described previously (24).
Crystallization.
For all crystallization experiments, the protein solution was exchanged with 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, 2 mM dithiothreitol using a Sephadex G-25 desalting column (Amersham Biosciences). Crystals of the ISF-Mt holoprotein were prepared by a batch method under anaerobic conditions by adding 20 to 25 μl of 28% polyethylene glycol 400 and 0.2 M CaCl2 in 0.1 M Tris-HCl buffer (pH 7.5) to 10 to 15 μl of a 15- to 30-mg protein/ml solution. The cluster-free form of ISF-Mt was crystallized in sitting drops at 20°C under anaerobic conditions. A solution containing 14% ethylene glycol and 6 mg/ml of protein was equilibrated against a reservoir containing 28% ethylene glycol. Yellow crystals grew within 5 days to 1 week and could be used for diffraction experiments; the crystallization buffer was used as a cryoprotectant.
Crystals of ISF3-Af were also grown by the sitting drop method under anaerobic conditions at 20°C. In this case, a solution containing 5% polyethylene glycol 3350, 0.05 M sodium-potassium phosphate buffer (pH 6.2), and 10 mg/ml of protein was equilibrated against a reservoir containing 10% polyethylene glycol 3350 and a 0.1 M concentration of the same buffer. For diffraction experiments, the crystals were transferred anaerobically into the reservoir solution containing increasing amounts of 2,4-methylpentanediol up to a final concentration of 15%.
Crystal structure determination.
ISF-Mt was crystallized in a primitive tetragonal cell with 422-point group symmetry and the following unit cell dimensions: a = b = 93.1 Å and c = 85.9 Å. Assuming a typical packing density, two ISF subunits should be present in the asymmetric unit. Data sets for multiple-wavelength anomalous dispersion were collected at four wavelengths, 1.7367 Å (peak), 1.7423 Å (inflection), 1.6531 Å (high-energy remote), and 1.7711 Å (low-energy remote), with the data processing statistics shown in Table 1. At all wavelengths used, the data collected for ISF-Mt showed the systematic extinction of odd l reflections along the 00l axis, in accordance with a space group assignment of P4222. However, difference Patterson maps (both anomalous and dispersive) were interpretable only when the screw axis in c (i.e., in space group P422) was neglected. These maps unambiguously indicated the position of one [4Fe:4S] cluster per asymmetric unit, which was used to calculate phases to a resolution of ∼4 Å. At this resolution, tracing of the peptide chain was not possible, but a regular four-iron cluster could be manually placed in the electron density to obtain positions of the individual iron atoms. After refinement of these positions with SHARP (14), phases were calculated to a 2.5-Å resolution, yielding readily interpretable electron density maps after solvent flattening with DM (3). A model with one ISF monomer in the asymmetric unit was built using O (13) and was refined to a crystallographic residual (Rcryst) of 0.32 (Rfree = 0.35) at a 2.0-Å resolution using CNS (2) (Table 2).
TABLE 1.
Data collection statistics for ISF-Mt and ISF3-Af
| Data set | ISF-Mt
|
ISF3-Af native (apo) | ||||
|---|---|---|---|---|---|---|
| Peak | Inflection | High remote | Low remote | apo | ||
| Wavelength (Å) | 1.7367 | 1.7423 | 1.6531 | 1.7711 | 1.0331 | 1.0332 |
| Resolution limits (Å) | 50.0-2.2 | 50.0-2.2 | 50.0-2.0 | 50.0-2.2 | 50.0-3.5 | 50.0-2.5 |
| Unique reflections | 36,080 | 36,070 | 47,274 | 35,979 | 6,009 | 18,517 |
| Completeness (%) | 98.2 | 98.2 | 96.5 | 97.6 | 99.8 | 99.8 |
| Observation redundancy | 4.2 | 4.2 | 4.3 | 4.1 | 11.2 | 6.5 |
| Rsym (overall) | 0.138 | 0.137 | 0.139 | 0.135 | 0.087 | 0.085 |
| Rsym (outer shell) | 0.389 | 0.372 | 0.524 | 0.393 | 0.503 | 0.450 |
| I/σ (I) (overall) | 8.9 | 8.8 | 8.5 | 8.7 | 11.0 | 14.5 |
| I/σ (I) (outer shell) | 3.2 | 3.2 | 1.5 | 2.5 | 4.7 | 2.0 |
TABLE 2.
Model refinement statistics for ISF-Mt and ISF3-Af
| Parameter | ISF-Mt | ISF3-Af |
|---|---|---|
| No. of atoms in model | 1,469 | 2,874 |
| No. of solvent molecules | 37 | 90 |
| Final Rcrysta | 0.323 | 0.197 |
| Final Rfreea | 0.352 | 0.243 |
| r.m.s.d. in bond lengths (Å)b | 0.0219 | 0.009 |
| r.m.s.d. in bond angles (°) | 1.620 | 1.856 |
| Avg B factor overall (Å2) | 45.8 | 46.3 |
| Avg B factor for protein (Å2) | 46.2 | 46.6 |
| Avg B factor for FMN (Å2) | 36.8 | 39.0 |
| Avg B factor for [4Fe:4S] (Å2) | 47.3 |
In ISF-Mt, the model comprises only half the content of the asymmetric unit. We interpreted the crystal as being constructed from alternating ordered and disordered layers, and we included only the ordered layer in our model. This led to the observed high R factors. Due to the lower resolution of the apo-ISF-Mt structure relative to the wild-type protein (3.5 Å compared to 2.0 Å), the former structure was not fully refined.
br.m.s.d., root mean square deviation.
The crystallographic analysis of ISF-Mt was greatly complicated by the presence of a substantial region of the unit cell with weak and uninterpretable electron density that had to contain the second ISF subunit anticipated to be in the asymmetric unit of the crystals. Although the positions of the subunits are apparently disordered when they are averaged over all the unit cells, their presence is required to maintain the lattice contacts necessary to stabilize the crystal. As the uninterpretable region is separated from the ordered subunits by a spacing of half the c axis, scattering from disordered molecules in this volume apparently must generate the observed systematic absences. Despite considerable effort, including various twinning scenarios, it has not yet been possible to generate a model for this crystallographic disorder that satisfactorily accounts for the observed scattering. The omission of this scattering material presumably accounts for the high R factors calculated from the refined model. Nevertheless, the correctness of the structure of the ordered molecule was verified by the subsequent successful structure determination for two unrelated crystal forms, ISF3-Af and apo-ISF-Mt.
Crystals of ISF3-Af belonged to space group P43212 with unit cell dimensions of a = b = 89.0 Å and c = 128.4 Å, while apo-ISF-Mt crystallized in hexagonal space group P6122 with unit cell dimensions of a = b = 127.8 Å and c = 94.7 Å. Both crystals contained two monomers of the protein in the asymmetric unit. Data collection statistics for these structures are listed in Table 1. From the model of the M. thermophila holoprotein, the structures of the [4Fe:4S]-cluster-free proteins from both M. thermophila and A. fulgidus could be solved by molecular replacement using MOLREP (3). ISF3-Af was refined to final R values of 0.20 (Rcryst) and 0.24 (Rfree) using CNS (Table 2).
RESULTS AND DISCUSSION
Structure description.
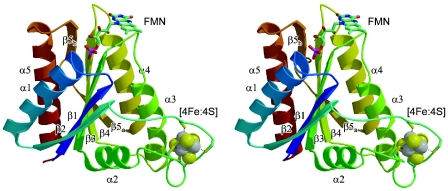
Both ISF-Mt and ISF3-Af adopt a typical flavodoxin-like β-α-β-fold with a parallel, five-strand beta-sheet with the topology 2-1-3-4-5. As in flavodoxins, the FMN is bound on the periphery of the protein at the distal end of strands β3 and β4 (Fig. 1). Consequently, a structural comparison of the ISF structures using the DALI server resulted in a number of high-scoring matches with flavodoxins, as well as with distant relatives, including P-loop nucleoside triphosphatases, lumazine synthase and the nitrogenase iron protein. The highest score (Z = 20.2), however, was obtained for the flavodoxin-like domain of rubredoxin:oxygen oxidoreductase (ROO) from Desulfovibrio gigas (10). In this structure, the FMN in the C-terminal domain is in close proximity to a di-iron center in an N-terminal, β-lactamase-like domain which is the active site of oxygen reduction. The FMN in ISF is located on the surface of the molecule with the si face exposed. The re face, packed against the protein, is involved in an aromatic stacking interaction with a tyrosine residue in both M. thermophila and A. fulgidus. In all ISF sequences analyzed to date, this residue has been found to be either tyrosine or histidine (1, 24).
FIG. 1.
Stereo view of the iron-sulfur flavoprotein monomer from M. thermophila, with colors ranging from blue at the N terminus to red at the C terminus. Secondary structure elements are labeled according to their order of appearance along the peptide chain. Within a single ISF monomer, the shortest distance between the FMN cofactor and the iron-sulfur cluster is ∼24 Å.
The loop connecting strand β2 with helix α2 contains an insertion in ISF that is absent in flavodoxins and ROO. Four cysteinyl residues in this loop coordinate a [4Fe:4S] cluster through a motif with the form Cys-X2-Cys-X2-Cys-X5-Cys. Bound to the protruding loop, this cluster is in close contact with helix α3, which is bent towards the cluster in a fashion that is not present in classical flavodoxins. This distortion is present in both the cluster-bound and cluster-free forms of ISF. Additionally, one of the most highly conserved residues among known ISF homologs is an arginine in this helix (Arg99 in ISF-Mt, Arg97 in ISF3-Af) (1, 24) whose side chain packs directly against the cluster. As evident from a comparison of the cluster-bound and apo forms of the ISF, the flavodoxin fold provides a stable core for these proteins, which retains its structure independent of the presence of the iron-sulfur cluster (Fig. 2).
FIG. 2.
Structural alignment of ISF-Mt (red) and the iron-free variant of ISF3-Af (black) with the C-terminal, flavodoxin-like domain of D. gigas ROO (blue). In the ISF-Mt structure, the four cysteine residues that coordinate the iron-sulfur cluster are labeled. Two protruding loops, labeled loop 1 and loop 2, are observed in the A. fulgidus structure. Their positions suggest involvement in stabilization of the tetrameric form of ISF. In the ROO domain, the cluster-binding loop is absent, but the overall fold is highly similar to that in ISF.
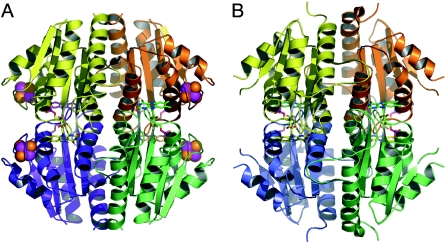
ISF-Mt and ISF3-Af were observed to assemble into a compact tetramer with D2 symmetry, with the flavin mononucleotides sandwiched between two adjacent protein monomers (Fig. 3). Since the same tetramer is observed in crystal forms belonging to three different space groups with different crystal contacts with neighboring molecules in each case, it seems unlikely that the observed tetramerization occurs only as a consequence of crystal formation. The stability of the tetrameric arrangement is underscored by the burial of 29% of the surface area of each monomer in subunit-subunit interfaces. The particular interface involved in packing of the flavins between adjacent subunits accounts for 12% of this area.
FIG. 3.
Tetramers of M. thermophila ISF (A) and A. fulgidus ISF3 (B). In panel B, the loop connecting the β2 sheet with helix α2 is disordered in the absence of the metal cluster.
Within the flavodoxin core fold, the most significant differences between the ISF structures and ROO are located in two loops that participate in subunit-subunit contacts. Loop 1 occurs in strand β5, where the ISF structures from M. thermophila and A. fulgidus show insertion of five and nine residues, respectively, forming a protruding loop that reaches out to another monomer, stabilizing the tetramer. Loop 2 is found only in the ISF3-Af structure and involves seven residues between strand β5 and terminal helix α5. These residues extend towards their counterparts in another monomer and further stabilize the quaternary structure (Fig. 2, 3).
Organization of the redox cofactors.
Within a single ISF monomer, the shortest distance between the flavin moiety and the iron-sulfur cluster is ∼24 Å. This separation is well outside the distance typically observed between physiological electron transfer partners (10 to 14 Å [21]), and the rate of electron transfer between these two cofactors is therefore likely too low to be mechanistically relevant (23). As a consequence of the tetrameric arrangement of ISF, however, the flavin group of one subunit closely approaches the iron-sulfur cluster of a neighboring monomer and vice versa. An analogous interaction between monomers of a dimer has been observed in the dimeric Ni-containing carbon monoxide dehydrogenase, in which an iron-sulfur cluster from one monomer is embedded between two clusters from the other monomer, forming a linear electron transport chain (4, 8, 9). In ISF-Mt, the shortest distance between the methyl group at C-7 of the isoalloxazine ring and the sulfur atom of Cys47, one of the protein ligands of the iron-sulfur cluster, is 3.8 Å, which would permit fast electron transfer. Similar positioning of the flavin moiety from one subunit with respect to a metal center on another is also observed in ROO, as well as in the Mo,Cu-containing carbon monoxide dehydrogenase (6, 7).
A further notable point concerns the accessibility of the flavin isoalloxazine ring. Electron transfer to and from flavins usually occurs through the methyl groups, and in flavodoxins the opposing edge of the flavin is shielded from the solvent by bulky side chains in the loop between sheet β4 and helix α4, among which is usually a conserved tryptophan or tyrosine (11). A Trp is also present in the flavodoxin-like domain of ROO, implying that this protein is indeed the product of a gene fusion between a nonheme iron protein and a flavodoxin. In ISF, however, not only is this residue missing, but the entire loop holding it is shifted away from the cofactor (Fig. 2). Shielding of the flavin in ISF is also not achieved through dimerization but only upon formation of the ISF tetramer (Fig. 4). On the other hand, access to the methyl groups of the flavin is efficiently blocked upon dimer formation, which brings these groups in close proximity to the [4Fe:4S] cluster of the other monomer. This arrangement of redox centers strongly supports the previous finding that electron transfer to the flavin occurs through the iron-sulfur cluster (1).
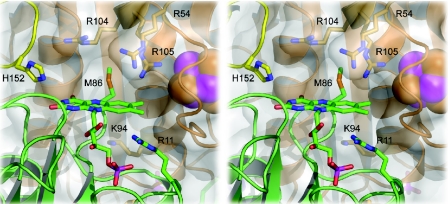
FIG. 4.
Detail of the flavin site of M. thermophila ISF. Although the FMN moiety is surrounded by a number of positively charged residues, the effective electrostatic surface potential in this region is negative (see Fig. 7).
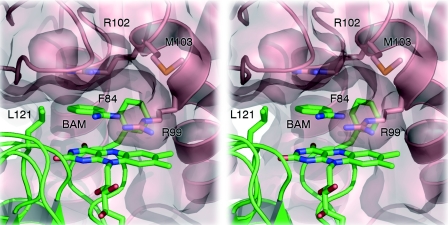
In ISF-Mt, the face of the flavin group is readily accessible from the protein surface and is exposed to solvent, while access to the methyl groups that form the presumed entry points for electrons is entirely blocked by the iron-sulfur cluster from the adjacent monomer (Fig. 4). In contrast, the positively charged vicinity of the flavin moiety in ISF3-Af is more shielded from the solvent, especially due to the presence of Arg99, whose side chain is located right on the flavin, presumably in a cation-π type of interaction (Fig. 5). In the structure of ISF3-Af, a hydrophobic cavity is formed with the flavin isoalloxazine as a floor, the guanidine group of Arg102 from the neighboring monomer as a ceiling, and Leu121 and Phe84 as walls. In the electron density, a small molecule could clearly be observed to bind in this pocket, forming a hydrogen bond to the side chain of Arg99 (Fig. 5). The electron density could be satisfactorily modeled with either benzoate or benzamidine; despite the unfavorable electrostatic interactions with Arg99, we modeled this density as benzamidine because this substance was used as a protease inhibitor during protein purification. In this binding mode, the ortho C-2 of the phenyl moiety of benzamidine comes to rest directly above the N-5 nitrogen of the flavin, which usually is the active position in flavoenzymes. However, there is currently no direct evidence for enzymatic activity of ISF3-Af.
FIG. 5.
Flavin site of A. fulgidus ISF3. In contrast to ISF-Mt, access to this site is limited due to the side chain of Arg99 from a neighboring monomer. Stacked between the flavin and a cation-π interaction with the side chain of Arg102, a benzamidine molecule (BAM) from the crystallization buffer was modeled hydrogen bonded to Arg99. The aromatic ring of the benzamidine molecule is situated in a hydrophobic pocket created by Leu121 and Phe84. The benzamidine sits at the entrance of a channel that crosses the entire ISF3 tetramer (see Fig. 6).
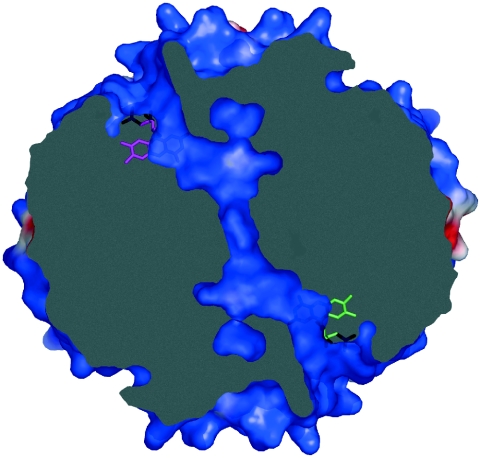
Intriguingly, in ISF3-Af the hydrophobic pocket at the flavin site lacks a back wall; rather, it forms the entrance to a pronounced, partially water-filled tunnel that crosses the entire tetramer along the interface of all subunits to reach the surface at a corresponding flavin on the opposite side of the molecules. The other two flavin sites of the tetramer are connected accordingly, and each of the tunnels has an additional branching point that reaches the surface at a different location (Fig. 6). ISF-Mt does not possess any of these channels.
FIG. 6.
A channel crosses the tetramer of A. fulgidus ISF3, connecting the FMN molecules of two opposing monomers. A second channel is found connecting the other two, symmetry-equivalent FMN molecules (not shown). The image shows a slice through the ISF3-Af tetramer, as shown in Fig. 7B, with identical surface coloring according to the electrostatic surface potential.
Surface properties.
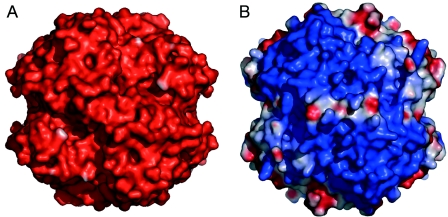
One striking difference between the otherwise highly similar ISF-Mt and ISF3-Af structures concerns their electrostatic surface potentials. For ISF-Mt, the electrostatic potential is strongly negative over the entire surface, including the immediate surroundings of the flavin si face, although the flavin itself is surrounded by several positively charged arginine and lysine residues (Fig. 4). In ISF3-Af, however, the overall surface potential is strongly positive, and little variation is observed around the edges of the tetramer (Fig. 7). In spite of the high structural similarity, ISF-Mt and ISF3-Af have drastically different surface properties that imply different functionalities. These properties likely represent important determinants for the interaction of each protein with redox partners or substrates, a proposal consistent with the finding that ISF homologs are ubiquitous among anaerobic prokaryotes with diverse metabolic capabilities (24). The widespread occurrence also suggests a universal function for the ISF family. A role in the oxidative stress response has been proposed based on gene clusters in several species containing an ISF homolog and genes encoding known oxidative stress proteins unique to anaerobes (5); for example, for the three ISF proteins in A. fulgidus, the ISF2 gene is clustered with genes encoding oxidative stress proteins. Recent results (unpublished) established that ISF-Mt reduces O2 to water. Assuming that this function is characteristic of the ISF family, it is likely that homologs evolved with specificity for the structurally diverse electron donors found in anaerobic prokaryotes belonging to the domains Archaea and Bacteria.
FIG. 7.
Electrostatic surface potentials for the tetramers of M. thermophila ISF (A) and A. fulgidus ISF3 (B), oriented as shown in Fig. 3. Potentials were calculated with the program DELPHI, using dielectric values (ɛ) of 4 for the protein interior and 80 for the surrounding solvent. To allow a more realistic comparison of surface properties, the cluster-binding loop of ISF-Mt was grafted onto the model of ISF3-Af, and the diverging amino acid side chains were replaced. Potentials are contoured from −75 kT (red) to 75 kT (blue).
Comparison of redox potentials of ISF and flavodoxins.
Flavodoxins are able to act as one-electron carriers by altering the redox potentials of the SQ and HQ forms of bound FMN compared to free FMN. The SQ form of bound FMN is stabilized and the HQ form is destabilized such that the OX/SQ and SQ/HQ potentials are reversed compared to those of free FMN. The flavodoxin potentials are −50 to −244 mV (OX/SQ) and −370 to −455 mV (SQ/HQ), in contrast to the measured values for free FMN (−238 and −314 mV for OX/SQ and −172 and −124 mV for SQ/HQ) (18). Flavodoxins destabilize the HQ state by preventing protonation at the N-1 nitrogen of the isoalloxazine ring, resulting in an unfavorable buried anionic HQ species (20). They stabilize the SQ state by preferentially binding FMN protonated at N-5 (the neutral SQ FMN) (19). Variation in redox potentials among flavodoxins can be explained by slight structural differences. The SQ/HQ potential is modulated by electrostatics, solvent accessibility, and aromatic interactions (24), whereas OX/SQ redox potentials are regulated by the stability of a loop that preferentially binds protonated and neutral SQ FMN (19).
The redox behavior of FMN in ISF-Mt deviates from that of flavodoxins by having no detectable SQ and a postulated OX/SQ midpoint potential below −380 mV. Also, the SQ/HQ pair lies above −175 mV (a difference of at least 205 mV) based on the overall OX/HQ midpoint potential of −277 mV (1). Thus, FMN in ISF behaves similar to free FMN in that the SQ is transient and the midpoint potential of the OX/SQ is lower than that of the SQ/HQ, but it is different from free FMN in having a larger difference between the OX/SQ and SQ/HQ midpoint potentials. This redox behavior in ISF suggests that the protonation states of the FMN may be different from that in flavodoxin. Although there are variations in electrostatics, solvent accessibility, and aromatic interactions between ISF and flavodoxin (Fig. 4), mutagenesis data for flavodoxins suggests that alterations in these properties do not result in a reversal of potentials (22, 25). If, however, ISF-bound FMN is anionic in the SQ state (not neutral as in flavodoxins) and neutral in the HQ state instead of anionic, the potentials observed for ISF could be explained. In this scenario, the SQ form of the FMN would be destabilized most by a negatively charged protein environment and would have a lower potential than the bound-HQ form and free SQ FMN. Although the electron paramagnetic resonance signal with a line width of 1.6 mT is indicative of an anionic or red semiquinone species (1), redox titrations versus pH will be important in confirming the protonation state of the SQ and HQ forms of FMN. If the protonation states of the SQ and HQ forms of FMN are different in ISF than in flavodoxin, the reversal in redox potentials could be readily explained.
Acknowledgments
We thank Hsiu-Ju Chiu for her assistance with data analysis.
This work was supported in part by a Marie Curie Intra-European Fellowship within FP6 (to S.L.A.A.), by NIH grants GM45162 (to D.C.R.) and F32-GM19044 (to C.L.D.), and by Department of Energy grant DE-FG02-95ER20198 (to J.G.F.). Stanford Synchrotron Radiation Laboratory operations are funded by the U.S. Department of Energy Office of Basic Energy Sciences and the NIH.
REFERENCES
- 1.Becker, D. F., U. Leartsakulpanich, K. K. Surerus, J. G. Ferry, and S. W. Ragsdale. 1998. Electrochemical and spectroscopic properties of the iron-sulfur flavoprotein from Methanosarcina thermophila. J. Biol. Chem. 273:26462-26469. [DOI] [PubMed] [Google Scholar]
- 2.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. Delano, P. Gros, R. W. Grosse Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 3.Collaborative Computational Project No. 4. 1994. The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. Ser. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 4.Darnault, C., A. Volbeda, E. J. Kim, P. Legrand, X. Vernede, P. A. Lindahl, and J. C. Fontecilla-Camps. 2003. Ni-Zn-[Fe4-S4] and Ni-Ni-[Fe4-S4] clusters in closed and open subunits of acetyl-CoA synthase/carbon monoxide dehydrogenase. Nat. Struct. Biol. 10:271-279. [DOI] [PubMed] [Google Scholar]
- 5.Ding, Y. H. R., and J. G. Ferry. 2004. Flavin mononucleotide-binding flavoprotein family in the domain Archaea. J. Bacteriol. 186:90-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobbek, H., L. Gremer, R. Kiefersauer, R. Huber, and O. Meyer. 2002. Catalysis at a dinuclear [CuSMo(==O)OH] cluster in a CO dehydrogenase resolved at 1.1 Å resolution. Proc. Natl. Acad. Sci. USA 99:15971-15976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobbek, H., L. Gremer, O. Meyer, and R. Huber. 1999. Crystal structure and mechanism of CO dehydrogenase, a molybdo iron-sulfur flavoprotein containing S-selanylcysteine. Proc. Natl. Acad. Sci. USA 96:8884-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobbek, H., V. Svetlitchnyi, L. Gremer, R. Huber, and O. Meyer. 2001. Crystal structure of a carbon monoxide dehydrogenase reveals a [Ni-4Fe-5S] cluster. Science 293:1281-1285. [DOI] [PubMed] [Google Scholar]
- 9.Doukov, T. I., T. M. Iverson, J. Seravalli, S. W. Ragsdale, and C. L. Drennan. 2002. A Ni-Fe-Cu center in a bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase. Science 298:567-572. [DOI] [PubMed] [Google Scholar]
- 10.Frazão, C., G. Silva, C. M. Gomes, P. Matias, R. Coelho, L. Sieker, S. Macedo, M. Y. Liu, S. Oliveira, M. Teixeira, A. V. Xavier, C. Rodrigues-Pousada, M. A. Carrondo, and J. Le Gall. 2000. Structure of a dioxygen reduction enzyme from Desulfovibrio gigas. Nat. Struct. Biol. 7:1041-1045. [DOI] [PubMed] [Google Scholar]
- 11.Freigang, J., K. Diederichs, K. P. Schäfer, W. Welte, and R. Paul. 2002. Crystal structure of oxidized flavodoxin, an essential protein in Helicobacter pylori. Protein Sci. 11:253-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galagan, J. E., C. Nusbaum, A. Roy, M. G. Endrizzi, P. Macdonald, W. FitzHugh, S. Calvo, R. Engels, S. Smirnov, D. Atnoor, A. Brown, N. Allen, J. Naylor, N. Stange-Thomann, K. DeArellano, R. Johnson, L. Linton, P. McEwan, K. McKernan, J. Talamas, A. Tirrell, W. J. Ye, A. Zimmer, R. D. Barber, I. Cann, D. E. Graham, D. A. Grahame, A. M. Guss, R. Hedderich, C. Ingram-Smith, H. C. Kuettner, J. A. Krzycki, J. A. Leigh, W. X. Li, J. F. Liu, B. Mukhopadhyay, J. N. Reeve, K. Smith, T. A. Springer, L. A. Umayam, O. White, R. H. White, E. C. de Macario, J. G. Ferry, K. F. Jarrell, H. Jing, A. J. L. Macario, I. Paulsen, M. Pritchett, K. R. Sowers, R. V. Swanson, S. H. Zinder, E. Lander, W. W. Metcalf, and B. Birren. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones, T. A., J.-Y. Zou, S. W. Cowan, and M. Kjelgaard. 1991. Improved methods for building proteins in electron density maps and location of errors in these models. Acta Crystallogr. Sect. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 14.La Fortelle, E. D., J. J. Irwin, and G. Bricogne. 1997. SHARP: a maximum-likelihood heavy-atom parameter refinement and phasing program for the MIR and MAD methods. Cryst. Comp. 7:1-9. [Google Scholar]
- 15.Latimer, M. T., and J. G. Ferry. 1993. Cloning, sequence analysis, and hyperexpression of the genes encoding phosphotransacetylase and acetate kinase from Methanosarcina thermophila. J. Bacteriol. 175:6822-6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Latimer, M. T., M. H. Painter, and J. G. Ferry. 1996. Characterization of an iron-sulfur flavoprotein from Methanosarcina thermophila. J. Biol. Chem. 271:24023-24028. [DOI] [PubMed] [Google Scholar]
- 17.Leartsakulpanich, U., M. L. Antonkine, and J. G. Ferry. 2000. Site-specific mutational analysis of a novel cysteine motif proposed to ligate the 4Fe-4S cluster in the iron-sulfur flavoprotein of the thermophilic methanoarchaeon Methanosarcina thermophila. J. Bacteriol. 182:5309-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludwig, M. L., and C. L. Luschinsky. 1992. Structure and redox properties of clostridial flavodoxin, p. 427-466. In F. Müller (ed.), Chemistry and biochemistry of flavoenzymes, vol. III. CRC Press, Boca Raton, Fla. [Google Scholar]
- 19.Ludwig, M. L., K. A. Pattridge, A. L. Metzger, M. M. Dixon, M. Eren, Y. C. Feng, and R. P. Swenson. 1997. Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes. Biochemistry 36:1259-1280. [DOI] [PubMed] [Google Scholar]
- 20.Ludwig, M. L., L. M. Schopfer, A. L. Metzger, K. A. Pattridge, and V. Massey. 1990. Structure and oxidation reduction behavior of 1-deaza-FMN flavodoxins: modulation of redox potentials in flavodoxins. Biochemistry 29:10364-10375. [DOI] [PubMed] [Google Scholar]
- 21.Page, C. C., C. C. Moser, X. X. Chen, and P. L. Dutton. 1999. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature 402:47-52. [DOI] [PubMed] [Google Scholar]
- 22.Swenson, R. P., and G. D. Krey. 1994. Site-directed mutagenesis of tyrosine-98 in the flavodoxin from Desulfovibrio vulgaris (Hildenborough)—regulation of oxidation-reduction properties of the bound FMN cofactor by aromatic, solvent, and electrostatic interactions. Biochemistry 33:8505-8514. [DOI] [PubMed] [Google Scholar]
- 23.Tezcan, F. A., B. R. Crane, J. R. Winkler, and H. B. Gray. 2001. Electron tunneling in protein crystals. Proc. Natl. Acad. Sci. USA 98:5002-5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao, T., F. Cruz, and J. G. Ferry. 2001. Iron-sulfur flavoprotein (Isf) from Methanosarcina thermophila is the prototype of a widely distributed family. J. Bacteriol. 183:6225-6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou, Z. M., and R. P. Swenson. 1995. Electrostatic effects of surface acidic amino acid residues on the oxidation-reduction potentials of the flavodoxin from Desulfovibrio vulgaris (Hildenborough). Biochemistry 34:3183-3192. [DOI] [PubMed] [Google Scholar]