Abstract
Two genes in the Escherichia coli genome, ypdE and ypdF, have been cloned and expressed, and their products have been purified. YpdF is shown to be a metalloenzyme with Xaa-Pro aminopeptidase activity and limited methionine aminopeptidase activity. Genes homologous to ypdF are widely distributed in bacterial species. The unique feature in the sequences of the products of these genes is a conserved C-terminal domain and a variable N-terminal domain. Full or partial deletion of the N terminus in YpdF leads to the loss of enzymatic activity. The conserved C-terminal domain is homologous to that of the methionyl aminopeptidase (encoded by map) in E. coli. However, YpdF and Map differ in their preference for the amino acid next to the initial methionine in the peptide substrates. The implication of this difference is discussed. ypdE is the immediate downstream gene of ypdF, and its start codon overlaps with the stop codon of ypdF by 1 base. YpdE is shown to be a metalloaminopeptidase and has a broad exoaminopeptidase activity.
Aminopeptidases form an abundant enzyme family in microorganisms (11), and multiple aminopeptidases are found in most sequenced microbial genomes. Aminopeptidases play key roles in protein degradation (5) and protein maturation (4), etc. The expanded families of aminopeptidases, with distinct sequence signatures and biochemical function, match the diversity in the chemical composition of the substrate peptides (18). For most aminopeptidases found by computer search, their substrate specificities have not been determined. Attempts to deduce substrate specificity on the basis of sequence similarity are hampered by the lack of clear sequence signatures that correlate with experimentally determined function.
Through computer analysis of microbial genes, we suggested the existence of segmentally variable genes (SVGs), whose products have a modular architecture composed of highly variable domains and well-conserved domains (24). The variable domains are typically over 70 amino acids (aa) in length and lack conserved sequence features that might suggest their function. Among many, we noticed one SVG family, members of which all encode proteins with a conserved C-terminal domain and a variable N-terminal domain (Fig. 1). Since it includes gene ypdF from Escherichia coli, we call it the YpdF family. In the products of these genes, the conserved C-terminal domains show strong global similarity to the 264-amino-acid-long methionine aminopeptidase (Map) in E. coli (4). The variable N-terminal domains, with average lengths of about 100 aa, do not show any detectable similarity to any known domains. It is known that the protein product of E. coli map is a metalloaminopeptidase and is activated in vitro by cobalt ions (19). The presence of an extra N-terminal domain plus a characteristic C-terminal domain similar to that of Map in the YpdF family resembles the domain structure observed in one of the two methionine aminopeptidases in Saccharomyces cerevisiae (6, 16). Most genes in the YpdF family are unknown, with no experimental evidence on detailed substrate specificities. It seemed possible that SVG family members encode similar aminopeptidase activity.
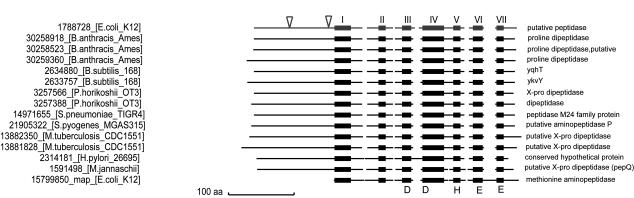
FIG. 1.
Schematic alignment of YpdF and its homologs in several selected completely sequenced microbial species. Black boxes represent conserved blocks reported by BLOCKS (13) and are numbered I to VII according to their sequential order. Conserved residues involved in metal ion binding are shown at the bottom of the blocks. Lines connecting the boxes are variable segments. Sequences are identified on the left, with the GenBank accession number followed by the species of origin. The sequence on the first line is YpdF of E. coli. The triangles in the N terminus show the start points of N-terminal deletions. The last sequence is the original methionyl aminopeptidase of E. coli. Notice the absence of the extra N-terminal domain in this sequence.
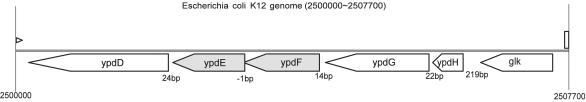
Examining the genomic context of ypdF in the E. coli genome shows that the start codon of its downstream neighboring gene, ypdE, overlaps with the stop codon of ypdF by 1 base (Fig. 2). Hence, expression of these two genes may be coupled and they may encode functionally related gene products. A similarity search reveals that ypdE homologs are present in over 60 microbial genomes and that YpdE has a subtle similarity (∼25% identity) to a previously reported archaeal-type deblocking aminopeptidase (17). Like ypdF, most genes from the YpdE family remain uncharacterized.
FIG. 2.
Genomic region in E. coli that includes the two genes studied in this paper (ypdE and ypdF, shaded). The numbers shown between the genes are intergenic distances in base pairs. The arrowheads of the boxes indicate the direction of transcription. ypdE and ypdF overlap by 1 base with TAATG, in which TAA is the stop codon of ypdF and ATG is the start codon of ypdE. Gene names are from the ASAP database (10). ypdF and ypdE correspond to b2385 and b2384, respectively, in GenBank genome record U00096. Annotations for other genes are as follows: ypdD, PTS system component IIA and enzyme I; ypdG, PTS system component IIC; ypdH, PTS system component IIB; glk, glucokinase.
In this study, we have expressed the active gene products of both ypdE and ypdF and shown that each encodes a metalloaminopeptidase. We have examined their substrate specificities for both.
MATERIALS AND METHODS
Cloning of ypdF, deleted versions of ypdF, and ypdE in E. coli.
For ypdF, the coding sequence was amplified by PCR from E. coli strain K-12 MG1655 genomic DNA using the forward primer b2385f and the reverse primer b2385r (Table 1). The purified PCR product was ligated into the pET28a (Novagen) plasmid between the EcoRI and HindIII sites. For ypdE, the coding sequence was amplified by PCR using the forward primer b2384f and the reverse primer b2384r (Table 1). The purified PCR product was ligated into the pET28a (Novagen) plasmid between the PstI and HindIII restriction sites.
TABLE 1.
Primers used in this study
| Name | Sequence |
|---|---|
| b2385f | CCCGAATTCATGACATTACTCGCTTCGCTGCGCGAC |
| b2385r | CCCAAGCTTATGCCTCTCCCGTGAGCAACACTG |
| b2384f | CCCGGATCCATGGATTTATCGCTATTAAAAGCGTTG |
| b2384r | CCCAAGCTTTCATCTGAAATCCGTCAGTTG |
| b2385_d1f | CCCGAATTCGAAACCGCGCACCGCTGGCAGTCTGAA |
| b2385_d2f | CCCGAATTCGTGGATTCGCGCTATTACGTTGAGGTGGAA |
| b2385_mf | GAAGTGCGCACGTGCGCAAGGCTACCAGCTG |
| b2385_mr | GCCTTGCGCACGTGCGCACTTTCACGGCTAATCAC |
Two N-terminal deletions of ypdF were made by amplifying the open reading frames (ORFs) using the forward primer b2385_d1f (starting from residue 103) or b2385_d2f (starting from residue 60) and the reverse primer b2385r. We first used pET28a to express the six-His-tagged recombinant protein; however, the yield was extremely low. We then cloned the purified PCR products into the plasmid pMALc2x (New England Biolabs) to produce MBP (maltose-binding protein)-fused protein. As a control, the intact ypdF gene was also cloned into the pMALc2x plasmid. To confirm the identity of all the cloned products, the inserts in the purified expression plasmids were analyzed by DNA sequencing (New England Biolabs).
Expression and purification of recombinant proteins.
pET28a carrying an ORF encoding a six-His-tagged protein was transformed into E. coli strain ER2566 (New England Biolabs). Transformed cells were cultured in LB medium supplemented with 100 μg/ml kanamycin at 37°C to mid-log phase. Protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and the cells were incubated at 30°C overnight. Cells from 10 ml of culture were harvested by centrifugation at 4,000 × g for 20 min and stored at −20°C for 30 min. Frozen cells were resuspended in 0.7 ml of lysis buffer (300 mM NaCl, 50 mM NaH2PO4, 10 mM imidazole, pH 8.0) and then briefly sonicated in ice. The cell lysates were centrifuged at 14,000 × g for 20 min at 4°C, and the supernatant was loaded onto an Ni-nitrilotriacetic acid column. The column was then washed twice with 10 ml of washing buffer (300 mM NaCl, 50 mM NaH2PO4, 20 mM imidazole, pH 8.0). The column bound protein was eluted in 0.7 ml of elution buffer (300 mM NaCl, 50 mM NaH2PO4, 250 mM imidazole, pH 8.0). The final protein concentrations as determined from Bradford assays are approximately 0.4 μg/μl for YpdF and 0.3 μg/μl for YpdE.
For pMALc2x constructs, transformed cells were cultured in LB medium supplemented with 100 μg/ml ampicillin at 37°C to mid-log phase. The remaining purification steps follow the same procedure as described in reference 12.
TLC assay for aminopeptidase activity.
Purified recombinant protein was assayed on a panel of peptides (BACHEM; New England Biolabs) to test their substrate specificity. All substrate peptides were dissolved in Tris buffer (10 mM, pH 7.6) to 1 mg/ml. Reactions were done in a total volume of 30 μl, with 25 μl of peptide substrate, 3 μl of purified enzyme, and 2 μl of metal ion (10 mM). The reaction mixture was incubated at 37°C for 30 min and then resolved by thin-layer chromatography (TLC) on a Silica Gel 60 plate (EMD Chemicals Inc.) using ethanol-isopropanol-water at a volume ratio of 1/2.1/0.9. The plate was then sprayed with ninhydrin (0.2% in ethanol) and heated. Images of the plate were taken under UV light at 366 nm.
Mutagenesis in the N-terminal domain of YpdF.
We used a two-step PCR procedure (12) to generate mutations in YpdF targeted at the conserved motif in the N-terminal domain. Briefly, the segment encompassing the sequence for this motif was first deleted from ypdF and then replaced with a synthetic double-stranded oligonucleotide with the desired mutations. In the first step, a pET28a plasmid with the inserted ypdF ORF was used as the template; oligonucleotides b2385f and b2385_mr were used as the forward and backward primers to obtain the 5′ region of ypdF; oligonucleotides b2385_mf and b2385r were used as primers to obtain the 3′ region of ypdF. In the second PCR step, a 1:1 mixture of purified 5′ and 3′ PCR products from the first step was used as the template; oligonucleotides b2385f and b2385r were used as the forward and backward primers. Compared with intact ypdF, the PCR product (ypdFΔ) from the second PCR step has a short internal segment deleted. Meanwhile, a new recognition sequence for PmlI (CACĜTG) was created at the deletion site. The purified PCR product was ligated into pET28a to generate pET28a-ypdFΔ. Next, we synthesized two complementary oligonucleotides with the desired amino acid changes and ligated it into the PmlI-digested plasmid pET28a-ypdFΔ as a replacement for the deleted segment. The resulting plasmids were then analyzed by DNA sequencing after purification.
RESULTS
Domain structure of YpdF and its similar sequences.
As shown in Fig. 1, members of the YpdF family have conserved C-terminal domains of about 260 aa and variable N-terminal domains of about 100 aa. The conserved C-terminal domain shows overall similarity to the 264 aa encoded by E. coli map, and all five key residues involved in metal ion binding are conserved (24) (Fig. 1).
Genes showing strong similarity to ypdF are widely distributed in other sequenced genomes. Part of the list is shown in Fig. 1. Interestingly, several genomes have multiple ypdF homologs (Fig. 1). The percent identity in the N-terminal-domain sequences of the multiple copies within the same genome is usually not high (<40%) and much less than the percent identity in the C-terminal-domain sequences, suggesting that these duplicated genes may have diverged.
YpdE is similar to an archaeal deblocking aminopeptidase, and the ypdEF structure is not conserved in other microbial genomes.
YpdE in E. coli shows moderate similarity (∼24% identity) to the previously reported metal ion-dependent deblocking aminopeptidase (PH0519) in the archaeon Pyrococcus horikoshii (1, 17) and to an aminopeptidase in Haloarcula marismortui (∼21% identity in an ∼150-aa region) (8). It is known that PH0519 shows broad aminopeptidase activity on nonblocked peptides and blocked peptides by acyl group (1). All three residues which are suggested to be involved in metal ion binding (17) are conserved (data not shown). However, computational analysis suggests that YpdE is not clearly orthologous to PH0519 because there exist multiple ypdE paralogs in both the P. horikoshii and E. coli genomes. Although the ypdE and ypdF ORFs overlap by 1 base in E. coli (including strains K-12, CFT073, and O157H7), this feature seems unique to E. coli and is not found in closely related species such as Salmonella enterica serovar Typhi or Shigella flexneri, etc. In the E. coli genome, the genes flanking ypdEF (Fig. 2) encode a sugar phosphotransferase (PTS) system that is responsible for carbohydrate transport (9).
Expression and purification of YpdF and YpdE in E. coli.
We expressed the active recombinant YpdE and YpdF proteins in E. coli. The six-His-tagged proteins were purified by one-step chromatographic procedures using an Ni-nitrilotriacetic acid column. YpdF has 361 aa with a calculated molecular mass of 39.6 kDa, and YpdE has 345 aa with a molecular mass of 37.4 kDa. The purified recombinant YpdF and YpdE proteins exhibit single bands on sodium dodecyl sulfate-polyacrylamide gels with estimated molecular masses of 41 kDa and 38 kDa (data not shown).
YpdF has limited Xaa-Pro aminopeptidase and methionyl aminopeptidase activities.
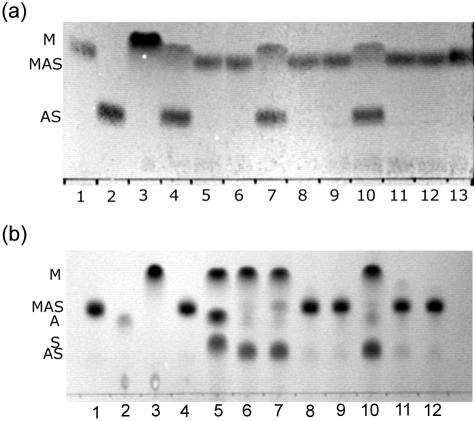
We found that YpdF has limited methionyl aminopeptidase activity when it was tested on a variety of peptides starting with l-methionine (Fig. 3a). A TLC assay shows that YpdF is capable of hydrolyzing the N-terminal methionine when the next amino acid is alanine, proline, or serine (Fig. 3a). It does not show detectable methionine-releasing activity when the second amino acid is glycine, lysine, or leucine, etc., among all tested peptides (Fig. 3a and Table 2) or when the first methionine residue is modified as formylmethionine or acetylmethionine (Table 2). Compared with the original methionyl aminopeptidase in E. coli (4), YpdF has a much narrower specificity in methionine cleavage. The map product in E. coli exhibits higher methionyl aminopeptidase activity when the second amino acid is glycine, alanine, proline, or serine, etc. (14). However, YpdF does not exhibit a similar property; for instance, it does not cleave methionine when the second amino acid is glycine (Fig. 3a).
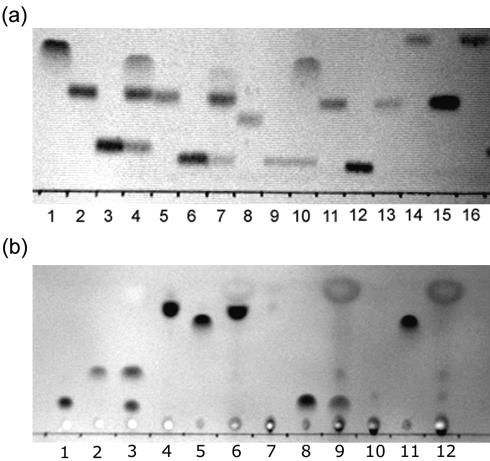
FIG. 3.
TLC plate image showing substrate specificity of YpdF. (a) The lanes are as follows (from left to right): 1, Met; 2, Met-Ala-Ser; 3, Ala-Ser; 4, Met-Ala-Ser+YpdF; 5, Met-Ser-Gly; 6, Ser-Gly; 7, Met-Ser-Gly+YpdF; 8, Met-Pro-Gly; 9, Pro-Gly; 10, Met-Pro-Gly+YpdF; 11, Met-Gly-Gly; 12, Gly-Gly; 13, Met-Gly-Gly+YpdF; 14, Met-Gly-Met; 15, Gly-Met; 16, Met-Gly-Met+YpdF. (b) The lanes are as follows (from left to right): 1, Ala-Pro-Ala; 2, Ala; 3, Ala-Pro-Ala+YpdF; 4, Tyr-Pro-Ile-Ser-Leu; 5, Tyr; 6, Tyr-Pro-Ile-Ser-Leu+YpdF; 7, Asn-Pro-Thr-Asn-Leu-His; 8, Asn; 9, Asn-Pro-Thr-Asn-Leu-His+YpdF; 10, Leu-Asp-Leu-Leu-Phe-Leu; 11, Leu; 12, Leu-Asp-Leu-Leu-Phe-Leu+YpdF.
TABLE 2.
Oligpeptides used in this study
| Enzymea and test result | Peptide(s)b |
|---|---|
| YpdF | |
| + | Met-Ala-Ser, Met-Pro-Gly, Met-Ser-Gly |
| + | Met-Pro-Gly-Ala-Arg-Ala-Asp-Ala-Ala-Leu-Ser-Met-Ala-Asp-Ala |
| + | Ala-Pro-Ala |
| + | Asn-Pro-Thr-Asn-Leu-His |
| − | Met-Arg-Phe, Met-Gly-Gly, Met-Gly-Met, Met-Met-Ala, Met-Leu-Gly, Met-Phe-Gly, For-Met-Ala-Ser, Ac-Met-Ala-Ser |
| − | Met-Cys-Gly-Lys |
| − | Met-Lys-Arg-Ser-Arg-Gly-Pro-Ser-Pro-Arg-Arg |
| − | Ala-Gly-Ser-Glu |
| − | Ala-Ala-Ala |
| − | Ser-Ser-Ser |
| − | Arg-Gly-Asp-Ser-Pro-Ala-Ser-Lys-Lys-Pro |
| − | Leu-Asp-Leu-Leu-Phe-Leu |
| − | Trp-Ala-Gly-Gly-Asp-Ala-Ser-Gly-Glu |
| − | Gly-Pro-Ile-Ser |
| − | Arg-Pro-Lys-Pro-Gln-Gln-Phe |
| − | Leu-Pro-Pro-Ser-Arg |
| − | Val-Pro-Leu |
| − | Ser-Pro-Val-Thr-Leu-Asp-Leu-Arg-Tyr |
| − | Tyr-Pro-Ile-Ser-Leu |
| − | Asp-Pro-Ala-Phe-Asn-Ser-Trp-Gly-NH2 |
| YpdE | |
| + | Met-▾Ala-▾Ser |
| + | Lys-▾Ala-Arg-Val-Nle-p-nitro-Phe-Glu-Ala-Nle-NH2 |
| + | Gln-▾Ala-▾Thr-▾Val-▾Gly-▾Asp-▾Val-▾Asn-▾Thr-▾Asp-▾Arg-Pro-Gly-Leu-Leu-Asp-Leu-Lys |
| + | Met-▾Ala-▾Gly-Pro-His-Pro-Val-Ile-Val-Ile-Thr-Gly-Pro-His-Glu-Glu |
| + | Asp-▾Ala-▾Glu-▾Phe-Arg-His-Asp-Ser-Gly-Tyr-Glu |
| + | Tyr-▾Leu-▾Leu-Pro-Ala-Glu-Val-Asn-Ile-Asp |
| − | Ac-Ala-Ala-Ala, Ac-Met-Ala-Ser, Ac-Pro-Leu-Gly |
| − | Ac-Lys-D-Ala-D-Ala, Ac-Asp-Arg-Gly-Asp-Ser, For-Met-Ala-Ser |
| − | His-Lys-Ala-Arg-Val-Leu-p-nitro-Phe-Glu-Ala-Ser-NH2 |
| − | Trp-Met-Asn-Ser-Thr-Gly-Phe-Thr-Lys-Val-Cys-Gly-Ala-Pro-Pro-Cys |
| − | Pro-Ala-Glu-Asp-Leu-Ala-Arg-Tyr-Tyr-Ser-Ala-Leu-Arg-His-Tyr-Ile-Asn-Leu-Ile-Thr-Arg-Gln-Arg-Tyr-NH2 |
| − | Ac-Ala-Ala-Asp-Ile-Ser-Gln-Trp-Ala-Gly-Pro-Leu |
| − | Ac-Ala-Thr-Leu-Asp-Ala-Leu-Leu-Ala-Leu-Arg-Arg-Ile-Gln-NH2 |
| − | Met-Ala-Pro-Arg-Gly-Phe-Ser-Cys-Leu-Leu-Leu-Leu-Thr-Ser-Glu-Ile-Asp-Leu-Pro-Val-Lys-Arg-Arg-Ala |
A plus sign means that the enzyme shows aminopeptidase activity on this substrate by TLC assay or MALDI-TOF experiments; a minus sign means that no cleavage was observed.
The symbol ▾ marks the site where the cleavage can be confirmed by MALDI-TOF spectrums. Ac stands for acetyl group, and For stands for formyl group.
Figure 3a reveals that the substrate preference of YpdF for methionyl aminopeptidase activity is Pro > Ala > Ser. We then tested if YpdF is able to cleave when residues other than methionine precede the second proline. As shown in Fig. 3b and Table 2, it is able to hydrolyze the Xaa-Pro peptide bond when the first amino acid is alanine (A), asparagine (N) (Fig. 3b), or methionine (M) (Fig. 3a) but not others (Table 2). Previous work has shown that aminopeptidase P (PepP) in E. coli is able to cleave essentially all Xaa-Pro peptides (23). Compared with E. coli PepP, YpdF has limited Xaa-Pro specificity. Other than the methionyl and Xaa-Pro aminopeptidase activity, YpdF does not show aminopeptidase activity on other peptides (Table 2), e.g., Ala-Ala-Ala or Ser-Ser-Ser, suggesting that it is not an aminopeptidase with broad specificity.
YpdE is an aminopeptidase with broad specificity but has no detectable deblocking aminopeptidase activity.
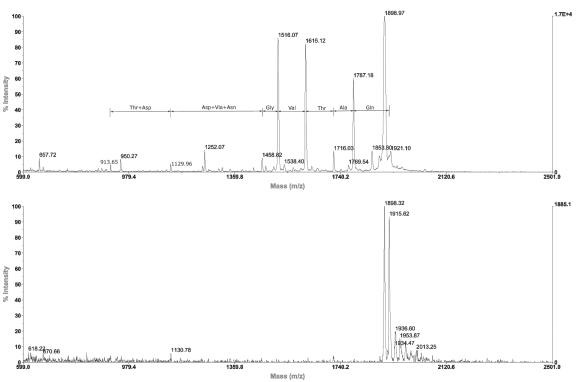
We tested if YpdE has deblocking activity against an array of acetyl or formyl group-blocked oligopeptides using TLC and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (data not shown). However, no detectable deblocking aminopeptidase activity was observed. Instead, we noticed that YpdE has a broad aminopeptidase activity on nonblocked peptides by progressively cleaving amino acids off the peptide substrate (Fig. 4). Its aminopeptidase activity stops at the residue before the first proline in the peptide, which was also observed for the related deblocking aminopeptidase homolog in archaea (1). This may be due to the unique imide bond in proline, which restricts the overall N-terminal conformation of the oligopeptides and disrupts the contacts between residues and active sites of the enzyme. From a panel of oligopeptides we examined by MALDI-TOF assay, we observed that YpdE can cleave most amino acids in the N terminus of the peptide (Table 2). Note that it does not cleave when proline is the first N-terminal residue (Table 2).
FIG. 4.
MALDI-TOF analysis of YpdE activity. The substrate used was the peptide Gln-Ala-Thr-Val-Gly-Asp-Val-Asn-Thr-Asp-Arg-Pro-Gly-Leu-Leu-Asp-Leu-Lys. The spectrum on the top is the reaction mixture of the substrate with enzyme supplemented with Co2+. The spectrum on the bottom is the control with just the substrate.
Divalent metal ion requirement of YpdE and YpdF.
The effects of Co2+, Mn2+, Mg2+, Ni2+, Ca2+, Fe2+, and Zn2+ on both YpdF and YpdE using the substrate Met-Ala-Ser were examined, as shown in Fig. 5. Mn2+ and Ni2+ can both substitute for Co2+ in assays for YpdF, while other divalent metal ions cannot. When metal ions are not present, YpdF loses its activity (Fig. 5a, lane 12).
FIG. 5.
Divalent metal ion dependence for YpdF and YpdE. (a) Divalent metal ion requirement for YpdF. The substrate used was Met-Ala-Ser. Metal ions included Co2+ (lane 4), Cu2+ (lane 5), Zn2+ (lane 6), Ni2+ (lane 7), Mg2+ (lane 8), Ca2+ (lane 9), Mn2+ (lane 10), and Fe2+ (lane 11). Other lanes are as follows: lane 1, MAS only; lane 2, AS only; lane 3, methionine only; lane 12, MAS with no metal ions but YpdF; lane 13, MAS with no enzyme but Co2+. (b) Divalent metal ion requirement for YpdE. The substrate used was Met-Ala-Ser. Metal ions included Co2+ (lane 5), Mn2+ (lane 6), Cu2+ (lane 7), Fe2+ (lane 8), Ca2+ (lane 9), Ni2+ (lane 10), Zn2+ (lane 11), and Mg2+ (lane 12). Other lanes are as follows: lane 1, MAS only; lane 2, alanine only; lane 3, methionine only; lane 4, MAS with YpdE but no metal ion.
In Fig. 5b, we show that YpdE can be activated by Co2+, Ni2+, Mn2+, and Cu2+. Notice that the substrate used in Fig. 5b is Met-Ala-Ser. YpdE is able to completely digest this tripeptide into individual amino acids in the presence of Co2+ and displays three spots on a TLC plate (Fig. 5b, lane 5).
Partial deletion and mutagenesis in the N-terminal domain of YpdF.
We asked whether the conserved C-terminal domain in YpdF is a stand-alone domain for the aminopeptidase activity. We then constructed two forms of YpdF with the N-terminal domain completely or partially deleted (Fig. 1a). We expressed and purified them as MBP (maltose-binding protein) fusion proteins (unpublished data). For control purposes, we also expressed the intact ypdF and purified the product as an MBP fusion protein. Purified MBP-YpdF shows the same activity as six-His-tagged YpdF (data not shown). However, YpdF with the N-terminal domain completely or partially deleted (Fig. 1a) loses its original aminopeptidase activity (data not shown). This suggests that the N-terminal domain is essential for the in vitro function of YpdF. However, we cannot rule out the possibility that the loss of the N terminus disrupts the overall folding of the protein.
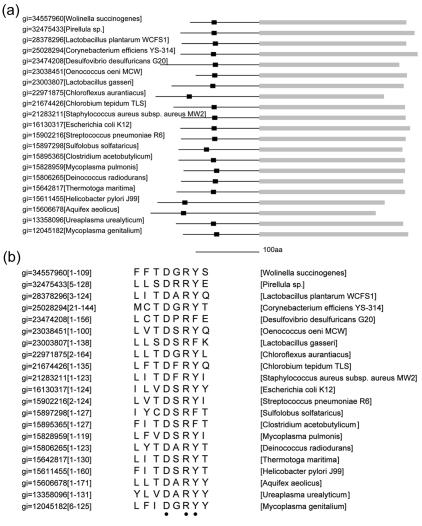
Using the motif finding program MEME (2) and the N-terminal sequences extracted from YpdF and its homologs, we found a short motif conserved within a subgroup of the YpdF homologs from a number of distantly related species (Fig. 6a). In the motif, two sites that are completely conserved are a negatively charged aspartate (D) and a positively charged arginine (R) (Fig. 6b). Another site appears to be dominated by aromatic residues: tyrosine (Y) or phenylalanine (F) (Fig. 6b). The fact that this motif is conserved across a diverse group of species in YpdF homologs suggests that it might be functionally important. We did not find any match to motifs of known function in motif databases. To investigate the possible role that this motif might play, we expressed a mutated YpdF protein in which the D, R, and Y sites were changed to A (DRY2A) and compared the mutated YpdF protein with the wild type. We found that the substrate specificity profile does not change between the wild type and this mutant. However, the mutated YpdF protein appears to have lost its methionyl aminopeptidase activity against the substrate Met-Ala-Ser while the activity against Met-Pro-Gly remains at the same level (data not shown). More work is needed to examine the functional role of this motif within the N terminus.
FIG. 6.
Conserved motif in the N-terminal domain of YpdF. (a) Location of the conserved motif in a number of YpdF homologs. The black box is the conserved short motif, and the line represents the variable N-terminal domain. Gray blocks represent the conserved C-terminal domains. (b) Amino acid alignment of the conserved motif. Two completely conserved sites, aspartate (D) and arginine (R), and one position dominated by aromatic residues are indicated by dots at the bottom. The numbers in brackets are the residue numbers corresponding to this N-terminal domain. Motif finding was performed using MEME (2).
DISCUSSION
We have studied two new aminopeptidases in E. coli. YpdF has limited aminopeptidase activity on peptide substrates starting with Met-Xaa or Xaa-Pro but not on other peptides. It has been suggested that the E. coli methionyl aminopeptidase and proline aminopeptidase PepP belong to the same family and adopt a similar fold (3). YpdF further reveals an inherent relationship between the two.
During previous screens for methionyl aminopeptidase activity, YpdF may be missed because of the limited nature of the peptides used (4, 22). Ben-Bassat et al. (4) used the peptide Met-Gly-Met-Met to screen an E. coli clone library, but this is not a substrate for YpdF. Similarly, during the screen for Xaa-Pro aminopeptidases, polyproline was used (22). Again, this is not a substrate for YpdF because YpdF cannot process peptides with an N-terminal proline.
The cellular roles of both YpdF and YpdE remain elusive. In a recent study (7), neither mRNA nor protein product of YpdF and YpdE were detected in the crude cell lysate of E. coli. However, the fact that both genes are evolutionarily conserved across distantly related species suggests they may be functionally important (15). Among all substrates tested, the maximum length of the peptides that YpdE can digest is 18 and the minimum is 2. It has a broad aminopeptidase activity and therefore could be involved in the ATP-independent downstream processing in cytosolic protein degradation pathways.
The segmentally variable feature of the YpdF family suggests that the attachment of an extra N-terminal domain may provide a convenient way of evolving new functions from existing protein scaffolds. If this is generally true, we may see it in other peptidase families in the database. The exact function of the N-terminal domains of YpdF and its homologs remains elusive. The N-terminal domains of methionyl aminopeptidases in S. cerevisiae harbor zinc finger motifs and short stretches of basic amino acids, which are suggested to be responsible for binding to the ribosome (20). However, in YpdF and its homologs, we did not observe a similar compositional bias in the N termini. E. coli PepP is similar to the type Ib methionyl aminopeptidase in domain structure but has a much longer N terminus than that in most type Ib methionyl aminopeptidases. Could the length of the N-terminal domains be the leading factor that distinguishes the two, or have all of the PepP proteins, which seem to have evolved from the methionyl aminopeptidase family, preserved some of the methionyl aminopeptidase activity? This should be tested experimentally. The crystal structure is available for aminopeptidase P, and it forms a tetramer (dimer of dimers) (21). Part of the N-terminal domain is responsible for contacts between subunits. Again, this may suggest another possible function of the N terminus of the YpdF family of proteins.
Acknowledgments
Y.Z. thanks Shelley Cushing and Jack Benner at NEB for help with MALDI-TOF experiments and David Landry at NEB for help with TLC analysis. We thank two anonymous reviewers for helping us interpret our experimental results.
This work was supported by New England Biolabs Inc.
REFERENCES
- 1.Ando, S., K. Ishikawa, H. Ishida, Y. Kawarabayasi, H. Kikuchi, and Y. Kosugi. 1999. Thermostable aminopeptidase from Pyrococcus horikoshii. FEBS Lett. 447:25-28. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28-36. [PubMed] [Google Scholar]
- 3.Bazan, J. F., L. H. Weaver, S. L. Roderick, R. Huber, and B. W. Matthews. 1994. Sequence and structure comparison suggest that methionine aminopeptidase, prolidase, aminopeptidase P, and creatinase share a common fold. Proc. Natl. Acad. Sci. USA 91:2473-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Bassat, A., K. Bauer, S. Y. Chang, K. Myambo, A. Boosman, and S. Chang. 1987. Processing of the initiation methionine from proteins: properties of the Escherichia coli methionine aminopeptidase and its gene structure. J. Bacteriol. 169:751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandu, D., and D. Nandi. 2003. PepN is the major aminopeptidase in Escherichia coli: insights on substrate specificity and role during sodium-salicylate-induced stress. Microbiology 149:3437-3447. [DOI] [PubMed] [Google Scholar]
- 6.Chang, Y. H., U. Teichert, and J. A. Smith. 1990. Purification and characterization of a methionine aminopeptidase from Saccharomyces cerevisiae. J. Biol. Chem. 265:19892-19897. [PubMed] [Google Scholar]
- 7.Corbin, R. W., O. Paliy, F. Yang, J. Shabanowitz, M. Platt, C. E. Lyons, Jr., K. Root, J. McAuliffe, M. I. Jordan, S. Kustu, E. Soupene, and D. F. Hunt. 2003. Toward a protein profile of Escherichia coli: comparison to its transcription profile. Proc. Natl. Acad. Sci. USA 100:9232-9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franzetti, B., G. Schoehn, J. F. Hernandez, M. Jaquinod, R. W. Ruigrok, and G. Zaccai. 2002. Tetrahedral aminopeptidase: a novel large protease complex from archaea. EMBO J. 21:2132-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ginsburg, A., and A. Peterkofsky. 2002. Enzyme I: the gateway to the bacterial phosphoenolpyruvate:sugar phosphotransferase system. Arch. Biochem. Biophys. 397:273-278. [DOI] [PubMed] [Google Scholar]
- 10.Glasner, J. D., P. Liss, G. Plunkett III, A. Darling, T. Prasad, M. Rusch, A. Byrnes, M. Gilson, B. Biehl, F. R. Blattner, and N. T. Perna. 2003. ASAP, a systematic annotation package for community analysis of genomes. Nucleic Acids Res. 31:147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzales, T., and J. Robert-Baudouy. 1996. Bacterial aminopeptidases: properties and functions. FEMS Microbiol. Rev. 18:319-344. [DOI] [PubMed] [Google Scholar]
- 12.Guan, C., S. Kumar, R. Kucera, and A. Ewel. 2004. Changing the enzymatic activity of T7 endonuclease by mutations at the beta-bridge site: alteration of substrate specificity profile and metal ion requirements by mutation distant from the catalytic domain. Biochemistry 43:4313-4322. [DOI] [PubMed] [Google Scholar]
- 13.Henikoff, S., J. G. Henikoff, and S. Pietrokovski. 1999. Blocks+: a non-redundant database of protein alignment blocks derived from multiple compilations. Bioinformatics 15:471-479. [DOI] [PubMed] [Google Scholar]
- 14.Hirel, P. H., M. J. Schmitter, P. Dessen, G. Fayat, and S. Blanquet. 1989. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl. Acad. Sci. USA 86:8247-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jordan, I. K., I. B. Rogozin, Y. I. Wolf, and E. V. Koonin. 2002. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 12:962-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, X., and Y. H. Chang. 1995. Amino-terminal protein processing in Saccharomyces cerevisiae is an essential function that requires two distinct methionine aminopeptidases. Proc. Natl. Acad. Sci. USA 92:12357-12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Onoe, S., S. Ando, M. Ataka, and K. Ishikawa. 2002. Active site of deblocking aminopeptidase from Pyrococcus horikoshii. Biochem. Biophys. Res. Commun.. 290:994-997. [DOI] [PubMed] [Google Scholar]
- 18.Rawlings, N. D., D. P. Tolle, and A. J. Barrett. 2004. MEROPS: the peptidase database. Nucleic Acids Res. 32:D160-D164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roderick, S. L., and B. W. Matthews. 1993. Structure of the cobalt-dependent methionine aminopeptidase from Escherichia coli: a new type of proteolytic enzyme. Biochemistry 32:3907-3912. [DOI] [PubMed] [Google Scholar]
- 20.Vetro, J. A., and Y. H. Chang. 2002. Yeast methionine aminopeptidase type 1 is ribosome-associated and requires its N-terminal zinc finger domain for normal function in vivo. J. Cell. Biochem. 85:678-688. [DOI] [PubMed] [Google Scholar]
- 21.Wilce, M. C., C. S. Bond, N. E. Dixon, H. C. Freeman, J. M. Guss, P. E. Lilley, and J. A. Wilce. 1998. Structure and mechanism of a proline-specific aminopeptidase from Escherichia coli. Proc. Natl. Acad. Sci. USA 95:3472-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yaron, A., and D. Mlynar. 1968. Aminopeptidase-P. Biochem. Biophys. Res. Commun. 32:658-663. [DOI] [PubMed] [Google Scholar]
- 23.Yoshimoto, T., A. T. Orawski, and W. H. Simmons. 1994. Substrate specificity of aminopeptidase P from Escherichia coli: comparison with membrane-bound forms from rat and bovine lung. Arch. Biochem. Biophys. 311:28-34. [DOI] [PubMed] [Google Scholar]
- 24.Zheng, Y., R. J. Roberts, and S. Kasif. 2004. Segmentally variable genes: a new perspective on adaptation. Public Library Sci. Biol. 2:E81. [DOI] [PMC free article] [PubMed] [Google Scholar]