Abstract
The RcsCDB His-Asp phosphorelay is shown to positively regulate the bdm (biofilm-dependent modulation) and sra (stationary-phase-induced ribosome-associated protein) genes in Escherichia coli. The regulation is direct and requires an RcsB box next to the bdm −35 element. In addition, bdm is shown to be activated by osmotic shock in an Rcs-dependent way.
The RcsCDB His-Asp phosphorelay system, initially identified as a positive regulator of the capsular exopolysaccharide (EPS) biosynthesis gene cluster (wza-wca) in Escherichia coli (8), is conserved in several γ-proteobacteria, including animal and plant pathogens. In E. coli, the Rcs system is required for recovery from chlorpromazine-induced stress (3). It is also involved in multidrug resistance (11) and participates in biofilm development (6). In Salmonella enterica serovar Typhimurium, it is involved in later stages of infection in mice and in cationic peptide resistance (5). The regulator RcsB is activated upon the transfer of a phosphoryl group from its cognate sensor, RcsC (19), via a histidine-containing phosphotransmitter (Hpt) domain protein, RcsD (K. E. Rudd, http://bmb.med.miami.edu/EcoGene/EcoWeb/; formerly called YojN [21]). Activation of the RcsCDB pathway, usually observed by monitoring the expression of the wza-wca gene cluster and the development of a mucoid phenotype, occurs under some environmental conditions, such as dehydration and osmotic shock (15, 18). Curiously, among the well-characterized targets, only the EPS synthesis genes have so far been found to be regulated following osmotic shock through the Rcs system; in addition, this regulation seems to be strain dependent and can be rather modest (4, 21), suggesting that the outcome of the Rcs response to osmolarity is modulated by unidentified factors. Recently, Ferrières and Clarke (6) reported that interaction with a solid surface activates the Rcs pathway, an observation consistent with the involvement of the Rcs system in the development of biofilms.
Targets regulated by the Rcs system are of two types, depending on responsiveness to the RcsB-cofactor RcsA. For the RcsA-independent class of RcsB targets, the site required for RcsB activity (the RcsB box) is located next to the −35 sequence, centered at −41/−42 (1, 4). For the RcsA-dependent class, this site (the RcsAB box) is located either further upstream (23) or downstream from the promoter (7).
Transcriptome analyses indicated that up to 2.5% of the E. coli genome might be regulated by the Rcs system (6, 10; our unpublished results). Approximately half of the putative targets have no defined function, suggesting that the Rcs system might be involved in adaptation to environmental conditions not usually encountered in the laboratory. The vast majority of the remaining targets are involved in envelope composition or trafficking.
In the present study, we characterize the regulation by the RcsCDB His-Asp phosphorelay of the biofilm-dependent modulation gene bdm as well as the stationary phase-inducible ribosome-associated protein gene sra. We also show that bdm is activated by osmotic shock and that this osmoregulation requires the Rcs system.
RcsB positively regulates the bdm gene independently of RcsA.
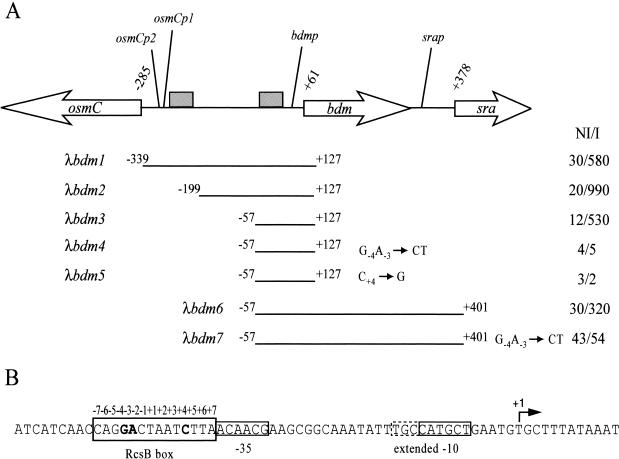
In order to define the Rcs regulon, the transcriptome of an MG1655 derived-strain was analyzed following overproduction of RcsB (data not shown). Among the targets identified in this analysis, the gene bdm (yddX), encoding a putative 71-amino-acid protein, attracted our attention for several reasons. First, bdm is located at 33.5 min in the E. coli genetic map, next to the osmoregulated gene osmC but oriented opposite to it (Fig. 1). We have previously shown that the osmC gene is directly activated by RcsB, and it was therefore of interest to investigate the mechanism of activation of two divergent transcripts by RcsB (4, 20). Second, bdm was also identified as a putative target of the Rcs system in the transcriptome analysis of Hagiwara et al. (10) following activation of the phosphorelay at a low temperature in the presence of glucose and Zn2+, but not in that of Ferrières and Clarke (6), in which activation of the Rcs system was followed by overexpression of the membrane protein gene djlA (13). Third, bdm was shown to be down-regulated in biofilms (16), and the Rcs system was shown to be required for efficient biofilm development (6). In order to characterize the role of the Rcs system in the regulation of bdm, several transcriptional fusions between the putative bdm promoter region and lacZ were constructed and installed in monocopy on the chromosome (17). The activities of these fusions were monitored following the overexpression of RcsB from the pHRcsB plasmid (1). As shown in Fig. 1, a fusion with a region extending from −339 to +127 relative to the transcription start point (determined in this study [Fig. 2 ]) was activated 19.3-fold upon overexpression of RcsB (λbdm1). Therefore, bdm is positively regulated by RcsB. When the fusion λbdm1 was tested after overexpression of RcsA from the pHRcsA plasmid (4), no effect was observed, indicating that bdm is not regulated by the RcsB auxiliary factor RcsA (data not shown).
FIG. 1.
Identification of the bdm RcsB box. (A) The genetic organization of the osmC-sra region is presented at the top of the figure. Grey boxes represent the osmC and the putative bdm RcsB boxes. Coordinates of osmC, bdm, and sra translational start sites are given and refer to the bdm transcriptional start site (+1). Below are indicated the coordinates of the fragments used in the different transcriptional fusions with lacZ (λbdm1 to -7). (B) The relevant sequences are shown. The bdm −35 and extended −10 sequences are boxed, the transcriptional start point is indicated (+1), and the three most conserved nucleotides of the RcsB box are in boldface. On the right-hand side of the figure are indicated the β-galactosidase-specific activities of the fusions expressed in Miller units 90 min after the induction (I) or noninduction (NI) of the expression of rcsB from the pHRcsB plasmid (1) in MG1655 ΔlacIZ(MluI).
FIG. 2.
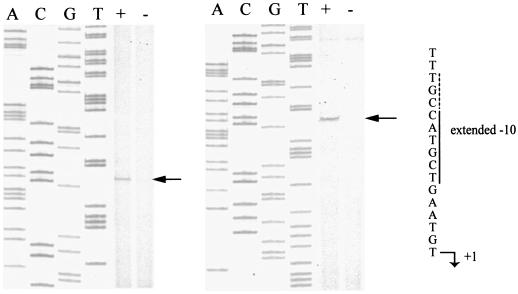
Determination of the bdm transcriptional start point. Reverse transcription was performed with total RNA purified from the MG1555 ΔlacIZ(MluI) strain containing the plasmid version of the λbdm1 fusion and overproducing (+) or not overproducing (−) RcsB from the pHRcsB plasmid (1). Two different primers were used. The relevant sequences are highlighted.
Identification of the bdm transcription start site.
To confirm the transcriptional regulation of bdm by RcsB and to localize the concerned promoter, mapping of the bdm transcription start site was performed by reverse transcription. RcsB production from pHRcsB was induced or left uninduced in cultures of a strain containing the plasmid version of the λbdm1 fusion. Total RNA isolated from these cells was used as templates in extension reactions with two different primers. As shown in Fig. 2, both primers identified the same RNA species, which started with a T residue. The transcript was barely visible except in samples in which rcsB was overexpressed, in agreement with the positive transcription regulation of the promoter by RcsB. A putative −10 sequence, 5′-CATGCT-3′ with three conserved nucleotides, including the two most conserved at positions 2 (A) and 6 (T), is found 6 nucleotides upstream of the T residue. The likelihood that this is a promoter is strengthened by the presence of a 5′-TGN-3′ motif characteristic of extended −10 promoters (14) at the 5′ end of the −10 hexamer (5′-TGCCATGCT-3′). A poorly conserved −35 element is found 17 nucleotides upstream of the −10 sequence (5′-ACAACG-3′), with only the fourth and fifth nucleotides matching the canonical sequence 5′-TTGACA-3′. However, strong deviations from the consensus of the −35 sequence are not unusual with extended −10 promoters (14) and with activated promoters.
Sequences required for regulation of bdm and osmC do not overlap.
In order to determine the sequence involved in the regulation of bdm by RcsB, a 5′-end deletion analysis was performed. As shown in Fig. 1, no sequence beyond −57 is required for the regulation of bdm by RcsB, as a deletion removing regions beyond that coordinate left a sequence still sensitive to the overexpression of RcsB (λbdm3). As expected, a deletion removing regions beyond −199 was also sensitive to RcsB (λbdm2). Both deletions in λbdm2 and λbdm3 fusions remove the RcsB box, which is required for the regulation of osmC by RcsB (4), indicating that sequences required for the regulation of bdm can function independently of those required for the regulation of osmC.
Inspection of the sequence in the vicinity of the promoter revealed a putative RcsB box next to the bdm −35 element, notably with the conserved motif GA-5N-C (1, 4, 7), suggesting that bdm might be directly regulated by RcsB (Fig. 1 and 3). The relevance of this sequence to RcsB regulation was tested by changing either the conserved dinucleotide GA to CT or the conserved C to a G in the λbdm3 fusion. As shown in Fig. 1, regulation by RcsB was lost in both fusions (λbdm4 and λbdm5, respectively). Thus, bdm regulation by RcsB requires a region containing a characteristic RcsB box upstream of the promoter −35 element.
FIG. 3.
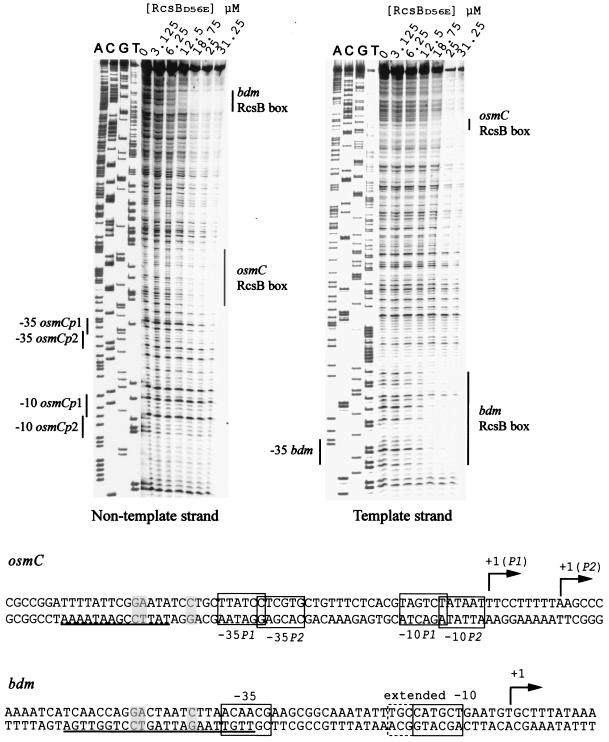
DNase I protection assay by RcsB at the bdm region. Reactions were performed as described before (7). Final concentrations of the constitutive form of RcsB (RcsBD56E) are indicated. Vertical lines show the protected regions. The sequences of the osmC and bdm promoter regions are shown. The three most conserved nucleotides of the RcsB boxes are shaded. The protected regions are underlined.
Identification of the RcsB box in vitro.
The identification of the bdm RcsB box was confirmed by DNase I protection assays. The experiment was performed with a purified mutant form of RcsB, RcsBD56E, in which the conserved Asp residue was replaced by a Glu residue. This mutation makes the protein more active, probably by mimicking the phosphorylated state of the protein (4, 9). A protected region was observed next to the bdmp −35 element in both the template and the nontemplate strand (Fig. 3). In agreement with the genetic data, the protected region includes the RcsB box. The probe used in the assay also contains the osmC RcsB box, and as expected, the profile of the protected region matches that reported in the work of Sturny et al. (20) at the osmC nontemplate strand. We note that the protected region for bdm was visible at lower concentrations of RcsBD56E than for osmC, suggesting that RcsBD56E has a higher affinity for the bdm RcsB box than for that of osmC (Fig. 3).
The bdm and sra genes form an operon which is regulated by RcsB.
bdm is located upstream of sra, a gene encoding a ribosome-associated protein whose expression is induced in stationary phase (Fig. 1) (12). A promoter responsible for sra induction in the stationary phase was identified in the intergenic region between bdm and sra. However, deletion analysis indicated that a second region located between positions −455 and −325 from the sra ATG initiation codon also contributes to sra expression (12). This region corresponds to −77 to +13 relative to the bdm transcription start site and therefore encompasses the bdm promoter and the bdm RcsB box. These observations suggest that bdm and sra constitute an operon in which bdm and sra are coregulated by RcsB. In agreement with this hypothesis, a fusion extending from the bdm regulatory region to the sra 5′ end is activated 12.5-fold when rcsB is overexpressed, whereas the same fusion with a mutated RcsB box was completely insensitive to RcsB (Fig. 1, λbdm6 and λbdm7, respectively). Both fusions were still activated at the stationary phase, in agreement with the presence in the constructions of the previously reported specific sra stationary-phase-inducible promoter (12). This activation by stationary phase was not observed with fusions containing only the bdm promoter (data not shown). The conservation of the genetic organization of bdm/sra in several members of the Enterobacteriaceae might suggest that both genes are involved in the same biological function. Unfortunately, no phenotypes that might have indicated the functions of those genes have yet been associated with bdm or sra mutants (12; our unpublished results).
The osmotic induction of bdm requires RcsB.
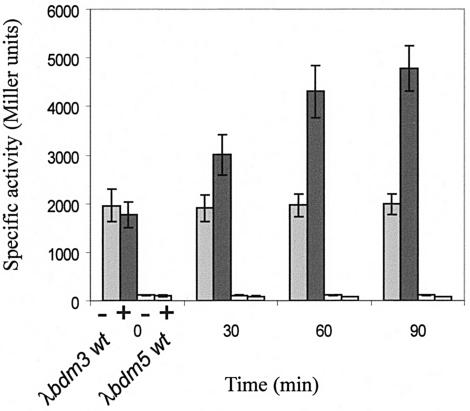
Contradictory results on the effect of changes in osmolarity on the expression of bdm have been reported. Using a bdm::lacZ fusion, Prigent-Combaret et al. (16) showed that bdm was repressed by high osmolarity in either biofilms or liquid medium. In contrast, Weber and Jung (22) reported a Northern analysis showing that bdm was activated following an osmotic upshift. A similar result was also obtained in the microarray-based study of Cheung et al. (2). We found that the expression of the λbdm3 fusion in minimal medium following an osmotic shock increased 90 min after the addition of NaCl to 0.5 M (Fig. 4). Similar results were obtained with λbdm2 and λbdm1 fusions (data not shown). Therefore, in minimal medium bdm is activated by osmotic shock, in agreement with studies of Weber and Jung (22) and Cheung et al. (2). In addition, the osmotic induction was not observed either with the λbdm5 fusion in which the RcsB box was mutated (Fig. 4) or in the rcsB mutant strain (data not shown). Therefore, the osmotic activation of bdm is RcsB dependent. In contrast, with the λbdm1 fusion, osmotic induction was still observed in an rpoS background, indicating that RcsB induction is not mediated by the stationary-phase sigma factor RpoS (data not shown). No effect of the osmotic shock on bdm expression was observed when a Luria-Bertani broth-derived rich medium was used, either at 30°C or 37°C (data not shown). This is the second example, the first being the EPS biosynthesis wza-wzc gene cluster, of RcsB targets being up-regulated by osmolarity through the Rcs system (18). However, whereas induced expression of EPS genes was reported to be transient, with a maximum between 30 and 75 min, expression of bdm continued to increase beyond 280 min, suggesting that different mechanisms are involved in the RcsB-dependent osmoregulation of the bdm and the wza-wzc genes. Further studies will be required in order to understand these differences as well as to explain why the other characterized targets of the Rcs system, including osmC, are not osmoregulated through the Rcs pathway.
FIG. 4.
Osmotic regulation of bdm expression by RcsB. Cultures were performed at 37°C in M63 supplemented with glucose and shocked at time zero with 0.5 M NaCl (+) or not shocked (−). Strains are MG1655 ΔlacIZ(MluI) containing either λbdm3 or λbdm5 fusions (Fig. 1).
Finally, the Rcs system was shown to be required for efficient biofilm development and to be activated by contact with solid surfaces (6), suggesting that the system is activated in biofilms. This proposal seems to be in contradiction with the observation that bdm is down-regulated in biofilms (16). This implies that, in biofilms, bdm might be subject to repression by a second regulator.
Acknowledgments
We are grateful to D. Lane for helpful discussions on the manuscript and to M. Cashel for strain CF6343 [MG1655 ΔlacIZ(MluI)].
This work was supported in part by the Université Paul Sabatier and the French Ministère de l'Enseignement Supérieur et de la Recherche (Programme de Recherche Fondamentale en Microbiologie, Maladies Infectieuses et Parasitaires) and by grants from the Institut Universitaire de France and from the Fondation pour la Recherche Médicale to C.G. and A.F.-C., respectively.
REFERENCES
- 1.Carballès, F., C. Bertrand, J. P. Bouché, and K. Cam. 1999. Regulation of Escherichia coli cell division genes ftsA and ftsZ by the two-component system rcsC-rcsB. Mol. Microbiol. 34:442-450. [DOI] [PubMed] [Google Scholar]
- 2.Cheung, K. J., V. Badarinarayana, D. W. Selinger, D. Janse, and G. M. Church. 2003. A microarray-based antibiotic screen identifies a regulatory role for supercoiling in the osmotic stress response of Escherichia coli. Genome Res. 13:206-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conter, A., R. Sturny, C. Gutierrez, and K. Cam. 2002. The RcsCB His-Asp phosphorelay system is essential to overcome chlorpromazine-induced stress in Escherichia coli. J. Bacteriol. 184:2850-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davalos-Garcia, M., A. Conter, I. Toesca, C. Gutierrez, and K. Cam. 2001. Regulation of osmC gene expression by the two-component system rcsB-rcsC in Escherichia coli. J. Bacteriol. 183:5870-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Detweiler, C. S., D. M. Monack, I. E. Brodsky, M. Hanza, and S. Falkow. 2003. virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol. Microbiol. 48:385-400. [DOI] [PubMed] [Google Scholar]
- 6.Ferrières, L., and D. J. Clarke. 2003. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol. Microbiol. 50:1665-1682. [DOI] [PubMed] [Google Scholar]
- 7.Francez-Charlot, A., B. Laugel, A. Van Gemert, N. Dubarry, F. Wiorowski, M. P. Castanie-Cornet, C. Gutierrez, and K. Cam. 2003. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 49:823-832. [DOI] [PubMed] [Google Scholar]
- 8.Gottesman, S., P. Trisler, and A. Torres-Cabassa. 1985. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes. J. Bacteriol. 162:1111-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupte, G., C. Woodward, and V. Stout. 1997. Isolation and characterization of rcsB mutations that affect colanic acid capsule synthesis in Escherichia coli K-12. J. Bacteriol. 179:4328-4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagiwara, D., M. Sugiura, T. Oshima, H. Mori, H. Aiba, T. Yamashino, and T. Mizuno. 2003. Genome-wide analyses revealing a signaling network of the RcsC-YojN-RcsB phosphorelay system in Escherichia coli. J. Bacteriol. 185:5735-5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirakawa, H., K. Nishino, J. Yamada, T. Hirata, and A. Yamaguchi. 2003. Beta-lactam resistance modulated by the overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J. Antimicrob. Chemother. 52:576-582. [DOI] [PubMed] [Google Scholar]
- 12.Izutsu, K., C. Wada, Y. Komine, T. Sako, C. Ueguchi, S. Nakura, and A. Wada. 2001. Escherichia coli ribosome-associated protein SRA, whose copy number increases during stationary phase. J. Bacteriol. 183:2765-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelley, W. L., and C. Georgopoulos. 1997. Positive control of the two-component RcsC/B signal transduction network by DjlA: a member of the DnaJ family of molecular chaperones in Escherichia coli. Mol. Microbiol. 25:913-931. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell, J. E., D. Zheng, S. J. Busby, and S. D. Minchin. 2003. Identification and analysis of ‘extended −10’ promoters in Escherichia coli. Nucleic Acids Res. 31:4689-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ophir, T., and D. L. Gutnick. 1994. A role for exopolysaccharides in the protection of microorganisms from desiccation. Appl. Environ. Microbiol. 60:740-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prigent-Combaret, C., O. Vidal, C. Dorel, and P. Lejeune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181:5993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and gene fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 18.Sledjeski, D. D., and S. Gottesman. 1996. Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 178:1204-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stout, V., and S. Gottesman. 1990. RcsB and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J. Bacteriol. 172:659-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sturny, R., K. Cam, C. Gutierrez, and A. Conter. 2003. NhaR and RcsB independently regulate the osmCp1 promoter of Escherichia coli at overlapping regulatory sites. J. Bacteriol. 185:4298-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeda, S. I., Y. Fujisawa, M. Matsubara, H. Aiba, and T. Mizuno. 2001. A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC→YojN→RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol. Microbiol. 40:440-450. [DOI] [PubMed] [Google Scholar]
- 22.Weber, A., and K. Jung. 2002. Profiling early osmostress-dependent gene expression in Escherichia coli using DNA macroarrays. J. Bacteriol. 184:5502-5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wehland, M., and F. Bernhard. 2000. The RcsAB box. Characterization of a new operator essential for the regulation of exopolysaccharide biosynthesis in enteric bacteria. J. Biol. Chem. 275:7013-7020. [DOI] [PubMed] [Google Scholar]