Abstract
In addition to its role as carbon and energy source, fructose metabolism was reported to affect other cellular processes, such as biofilm formation by streptococci and bacterial pathogenicity in plants. Fructose genes encoding a 1-phosphofructokinase and a phosphotransferase system (PTS) fructose-specific enzyme IIABC component reside commonly in a gene cluster with a DeoR family regulator in various gram-positive bacteria. We present a comprehensive study of fructose metabolism in Lactococcus lactis, including a systematic study of fru mutants, global messenger analysis, and a molecular characterization of its regulation. The fru operon is regulated at the transcriptional level by both FruR and CcpA and at the metabolic level by inducer exclusion. The FruR effector is fructose-1-phosphate (F1P), as shown by combined analysis of transcription and measurements of the intracellular F1P pools in mutants either unable to produce this metabolite or accumulating it. The regulation of the fru operon by FruR requires four adjacent 10-bp direct repeats. The well-conserved organization of the fru promoter region in various low-GC gram-positive bacteria, including CRE boxes as well as the newly defined FruR motif, suggests that the regulation scheme defined in L. lactis could be applied to these bacteria. Transcriptome profiling of fruR and fruC mutants revealed that the effect of F1P and FruR regulation is limited to the fru operon in L. lactis. This result is enforced by the fact that no other targets for FruR were found in the available low-GC gram-positive bacteria genomes, suggesting that additional phenotypical effects due to fructose metabolism do not rely directly on FruR control, but rather on metabolism.
Carbohydrate utilization systems are of particular importance to provide carbon and energy to bacteria. Among them, glucose and lactose systems have been widely studied, while other sugar utilization systems have been studied mostly with regard to their implication in targeted processes. Considering fructose availability in most ecosystems associated with plants, fructose metabolism and its regulation received little attention to date. The utilization of fructose is best documented in Escherichia coli, with the existence of three routes. In the main route, fructose is taken up via the membrane-spanning protein FruA and concomitantly phosphorylated to fructose-1-phosphate. Phosphorylation takes place by transfer of the phosphate group from phosphoenolpyruvate to fructose, which involves concerted action of two cytoplasmic proteins, EI of the phosphotransferase system (PTS) and a membrane-associated diphosphoryl transfer protein (FruB). The fructose-1-phosphate thus formed is further phosphorylated by ATP and 1-phosphofructokinase (FruK) to fructose-1,6-bisphosphate (17). The fruBKA operon of enteric bacteria is regulated at the transcriptional level primarily by the catabolite repressor-activator Cra (previously designated FruR) and by the cyclic AMP-CRP complex, which plays a secondary role (9, 25).
Although non-PTS uptake systems for fructose utilization have been described (5), sugar-specific PTS appear to be the most frequent system of fructose utilization. Several fructose (fru) operons encoding EIIFru enzyme and 1-phosphofructokinase have been described in different bacterial groups, such as Spiroplasma citri (a mollicute), Streptococcus mutans, and Streptococcus gordonii (firmicutes). In the first bacterium, the fru operon has been shown to be involved in phytopathogenicity of S. citri, the causal agent of the citrus “stubborn” disease (11). In the two latter bacteria, high-affinity sugar utilization systems such as the PTSFru enhance survival of oral streptococci during periods between meals, while acid production from sugar contributes directly to human tooth decay. Moreover, it was shown that the fru operon is involved in biofilm formation by S. gordonii (19), a process allowing bacterial accumulation, proliferation, and persistence on oral surfaces. Interestingly, in these distantly related bacteria, the two fructose utilization genes are preceded by a regulator of the DeoR repressor family. The three genes were shown to be cotranscribed from an upstream promoter. In silico analysis shows that the genetic organization of most fructose utilization operons is the same in different genera, such as Bacillus, Staphylococcus, Lactococcus, Enterococcus, Lactobacillus, Streptococcus, Streptomyces, Corynebacterium, Clostridium, and Fusobacterium (our personal analysis, but most information can be retrieved at http://theseed.uchicago.edu or http://string.embl.de).
The regulation of the fru operon and the role of the FruR regulator have been investigated in S. citri and S. gordonii (12, 19). The transcription of these operons is enhanced by the presence of fructose in the culture medium. Surprisingly, FruR was found to be an activator in S. citri, whereas it was shown to be a repressor in S. gordonii (12, 19). This difference could not be explained from the sequence analysis of the two proteins, which share more than 35% identity (55% similarity) over their entire length. Moreover, putative motifs of regulation were suggested in both cases, but there are no experimental data available to confirm their involvement in fru operon transcription. Finally, the signal sensed by FruR has not been identified.
In this paper, we present a study of the fru operon from Lactococcus lactis. The genes were initially annotated lacR, lacC, and fruA (2), as the products encoded by the first two genes share high identity to the repressor-of-lactose-utilization operon and tagatose-6-phosphate kinase from L. lactis, respectively (28). We show that fruR (lacR), fruC (lacC), and fruA are involved in the main fructose utilization pathway in L. lactis. Furthermore, we present evidence that FruR is the repressor of the fructose operon and that its activity is modulated by fructose-1-phosphate. In this regulatory process, FruA, encoding a potential EIIABCFru PTS unit, is necessary to produce fructose-1-phosphate, while FruC, encoding a putative 1-phosphofructokinase, plays an indirect role in fructose operon regulation even in the absence of fructose in the culture medium. The specificity of FruR regulation in L. lactis was shown by DNA microarrays, and the DNA-binding site for FruR was identified by genetic experiments. Finally, comparative genomics analysis indicates that the L. lactis fructose regulation model may exist in many low-GC gram-positive bacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this work are listed in Table 1. E. coli was grown in Luria-Bertani broth at 37°C (21). L. lactis cells were grown in M17 broth (Difco Laboratories, Detroit, Mich.) supplemented with 0.5% glucose or in a chemically defined medium (CDM) (26) at 30°C, unless stated otherwise. To test the effects of various carbon sources, sugars (Merck) were added to CDM at final concentrations of 0.5%. When appropriate, the medium contained erythromycin (75 μg · ml−1 for E. coli or 3.5 μg · ml−1 for L. lactis), ampicillin (100 μg · ml−1 for E. coli), chloramphenicol (10 μg · ml−1 for L. lactis), or tetracycline (5 μg · ml−1 for E. coli and L. lactis).
TABLE 1.
Bacteria strains and plasmids used in this study
| Strain or plasmid | Relevant markers, phenotypes, and characteristics | Reference or origin |
|---|---|---|
| E. coli strain | ||
| TG1 | supE Δthi(lac-proAB) hsdD5 (F′+traD36 proAB lacIqZDM15) | 13 |
| L. lactis strains | ||
| IL1403 | L. lactis ssp. lactis | 6 |
| JIM8233 | Emr, IL1403 containing pJIM5500 integrated at the Pfru locus | This work |
| JIM8234 | Emr, IL1403 after homologous recombination of pJIM5501 | This work |
| JIM8235 | Emr, IL1403 after homologous recombination of pJIM5502 | This work |
| JIM8236 | Emr, IL1403 after homologous recombination of pJIM5503 | This work |
| JIM8239 | Emr, IL1403 after homologous recombination of pJIM5506 | This work |
| JIM8240 | Emr, IL1403 after homologous recombination of pJIM5507 | This work |
| JIM8245 | Emr Tetr, JIM8239 containing the replicative pJIM5517 | This work |
| JIM8246 | Emr Tetr, JIM8240 containing the replicative pJIM5517 | This work |
| JIM7794 | IL1403 ccpA | J. Bardowskia |
| JIM8253 | Emr, JIM7794 containing pJIM5500 integrated at the Pfru locus | This work |
| JIM8254 | Emr, JIM7794 after homologous recombination of pJIM5506 | This work |
| JIM8641 | Emr, IL1403 after homologous recombination of pJIM5548 | This work |
| JIM8642 | Emr, IL1403 after homologous recombination of pJIM5549 | This work |
| JIM8643 | Emr, IL1403 after homologous recombination of pJIM5550 | This work |
| JIM8644 | Emr, IL1403 after homologous recombination of pJIM5551 | This work |
| Replicative plasmids | ||
| pGEM-T | Apr, M13ori pBR322ori, linear T-overhang vector | Promega |
| pJIM1276 | Cmr, thermosensitive replicative plasmid in L. lactis | 20 |
| pJIM2374 | Emr, integrative promoter probe vector containing the luxAB genes | 8 |
| pJIM6001 | Tetr, replicative plasmid in L. lactis | M.-C. Chopina |
| pJIM5517 | Tetr, SalI fusion of pJIM6001 and pGEM-T containing the fruR gene and its promoter on a fragment obtained by amplification of IL1403 chromosomal DNA using lacR1/lacC4 primer pair | This work |
| Integrative plasmids | ||
| pJIM5500 | Emr, SalI fusion of pJIM2374 and pGEM-T containing Pfru, obtained by amplification of IL1403 chromosomal DNA using lacR1/lacR3 primers | This work |
| pJIM5501 | Emr, SalI fusion of pJIM2374 and pGEM-T containing a fruR internal fragment, obtained by amplification of IL1403 chromosomal DNA using lacR2/lacR3 primer pair | This work |
| pJIM5502 | Emr, SalI fusion of pJIM2374 and pGEM-T containing a fruC internal fragment, obtained by amplification of IL1403 chromosomal DNA using lacC1/lacC2 primer pair | This work |
| pJIM5503 | Emr, SalI fusion of pJIM2374 and pGEM-T containing a fruA internal fragment, obtained by amplification of IL1403 chromosomal DNA using fruA1/fruA2 primer pair | This work |
| pJIM5506 | Emr, NcoI digestion of pJIM5501 to remove pGEM-T | This work |
| pJIM5507 | Emr, NcoI digestion of pJIM5502 to remove pGEM-T | This work |
| pJIM5548 | Emr, SalI fusion of pJIM2374 and pGEM-T containing a part of the intergenic region upstream of fruR, obtained by amplification of pJIM5500 using M1/lacR4 primer pair | This work |
| pJIM5549 | Emr, SalI fusion of pJIM2374 and pGEM-T containing a part of the intergenic region upstream of fruR, obtained by amplification of pJIM5500 using M2/lacR4 primer pair | This work |
| pJIM5550 | Emr, SalI fusion of pJIM2374 and pGEM-T containing a part of the intergenic region upstream of fruR, obtained by amplification of pJIM5500 using M3/lacR4 primer pair | This work |
| pJIM5551 | Emr, SalI fusion of pJIM2374 and pGEM-T containing a part of the intergenic region upstream of fruR, obtained by amplification of pJIM5500 using M4/lacR4 primer pair | This work |
Personal communication.
Sugar metabolism profiles were determined using API 50 CH as recommended by the manufacturer (BioMérieux, Marcy l'Etoile, France). The resulting fermentation patterns were inspected following incubation at 30°C for 4 and 24 h. Fermentation of carbohydrates was detected by acid production and a change in color of the pH indicator.
DNA manipulation procedures.
Procedures for DNA manipulations, transformation of E. coli cells, and cloning were done essentially as described elsewhere (21). Electrotransformation of L. lactis was carried out as described previously (15). Southern hybridization and detection were performed according to the Amersham ECL protocol (Amersham, Freiburg, Germany). DNA was sequenced on both strands using the 370A DNA sequencer according to the manufacturer's instructions.
Construction of lux transcriptional fusions and negative mutants in L. lactis.
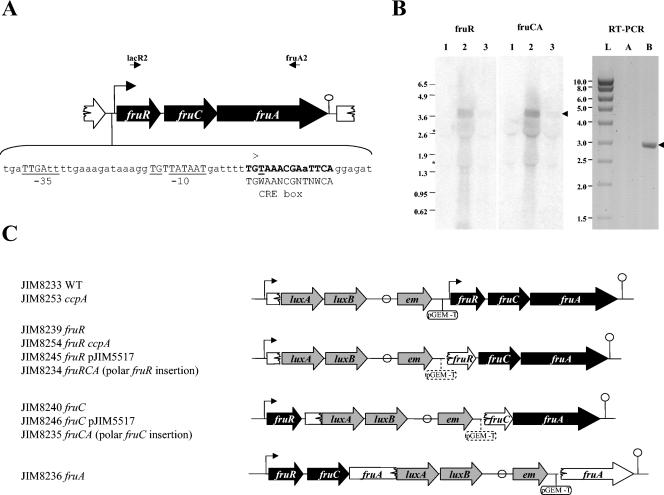
The integrative plasmids used to obtain strains carrying lux transcriptional fusions and the inactivated gene(s) are described in Table 1. Briefly, PCR products containing the PfruR promoter or an internal fragment of the gene to be inactivated were cloned in the pGEM-T vector in E. coli and their sequences were verified. The specific primers used in this work are listed in Table 2. The integrative plasmids were obtained by fusing the derived pGEM-T vectors with the L. lactis integrative vector pJIM2374 at the SalI restriction sites. Out of the two possible plasmids obtained by this method, we chose those where the luciferase gene of pJIM2374 and the fru gene insert have the same orientation. In some constructions, the pGEM-T vector was removed to allow transcription of the downstream genes from the erythromycin gene promoter. These plasmids were integrated in the chromosome of L. lactis by a single crossover event with pJIM1276 as a helper plasmid (14). The resulting strains were verified by PCR amplification and Southern blotting. The strains JIM8233 and JIM8253 contained the luxAB genes downstream of the cloned promoter region followed by a copy of the intact gene (Fig. 1). The strains JIM8234 to -8240 and JIM8245, JIM8246, and JIM8254 contained the duplicated target gene deleted either at its 3′ end or at its 5′ end (Fig. 1). The strains JIM8641 to -8644 contained luxAB downstream of the intact promoter region followed by the fru operon transcribed from a modified promoter (see Fig. 4, below).
TABLE 2.
Sequences of the primers used in this study
| Primer | Sequence (5′→3′) | Location |
|---|---|---|
| lacR1 | GACTACTATTTGGAAAGCGC | yjiD |
| lacR2 | TGATCTATATCGACGCTTAGG | fruR |
| lacR3 | TTAATTGTCAAAGCCGTTAGC | fruR |
| lacR4 | GTGTAAGTTCTGCACCACC | fruR |
| lacC1 | CACTAGGCTTTCTAGGTGG | fruC |
| lacC2 | GGTCGAATCACCAGCACC | fruC |
| lacC3 | AAGTTTTGGTCCAGCCGC | fruC |
| lacC4 | GAAGTAGTCAAGTGCTGGG | fruC |
| fruA1 | AAGCCTTGGAAATGGGCG | fruA |
| fruA2 | GTATTGATTGCTGCCATTGGG | fruA |
| M1 | TGAGTTTTTTTTGGAAGAAAAT GATAGTTAATG | fru promoter region |
| M2 | TGGAAGAAAATGATAGTTAATG | fru promoter region |
| M3 | TGATTGATTTTGAAAGATAAAG | fru promoter region |
| M4 | AAGAAGAAAAAAATAGTTAAAAA TTGATTTAAAAAGATAAAGGa | fru promoter region |
| tuf1 | CGCGAACGTGGTATCACA | tuf |
| tuf2 | GTCCATTTGGGCAGCACC | tuf |
Inserted mutations are underlined.
FIG. 1.
(A) Genetic organization of the L. lactis IL1403 fructose operon. fruR encodes a protein homologous to a transcriptional regulator, fruC, a 1-phosphofructokinase, and fruA, a fructose-specific enzyme II (EIIABC components). The transcriptional start point (+1) determined by 5′-RACE and the corresponding −10 and −35 boxes are indicated and underlined. A putative rho-independent terminator is indicated by a circle. A putative CRE box is shown in bold. The primers lacR1 and fruA2 are shown by arrows. (B) Northern blot with fruR (lacR2-lacR3) and fruCA (lacC1-fruA2) probes and, right panel, PCRs on the fructose gene transcripts using primers complementary to fruR (lacR2) and fruA (fruA2). RNAs were prepared from IL1403 cells grown in CDM-glucose (lane 1), CDM-fructose (lanes 2, A, and B), and CDM-glucose-fructose (lane 3). Lane L, Smart ladder (Promega), sizes of which are indicated in the left margin; lane A, PCR on 500 ng of RNA; lane B, RT-PCR on 500 ng of RNA diluted 40-fold. The asterisks on the left of the Northern blot indicate the positions of 16S and 23S RNAs that produced slightly artifactual bands. Arrows to the right of the panel indicate the position of the expected band corresponding to the fruRCA transcript (3.7 kb) and RT-PCR product (3 kb). (C) Schematic representation of inserted constructions in the fru operon and the corresponding strains used in this study. The promoter of the fru operon (Pfru) is indicated by an arrow. A putative rho-independent terminator is indicated by a circle. Genes of the fru operon are shown either in black when they are intact or in white when they are inactivated. The inserted genes are represented in gray. The pGEM-T plasmid is indicated by a line and a circle in either full features, when it is present in all the constructions, or in stippled features when it is not present in all the constructions.
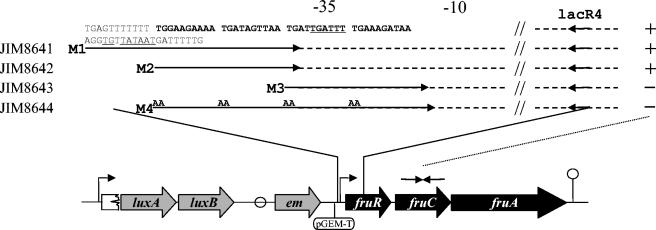
FIG. 4.
Schematic representation of the modified fru operon promoter sequences used to characterize the DNA-binding motif recognized by FruR. The promoter region inserted upstream of the fru operon is shown in the upper panel. The putative binding site of FruR is in bold. Strains were obtained by integration in the IL1403 chromosome of the plasmid pJIM2374 carrying pGEM-T, and the promoter regions amplified by the primers are indicated by arrows. Conserved sequences are shown by dotted lines, and TG→AA mutations in the M4 primer are indicated. The −10 and −35 regions are underlined. + or - at the right of the fragments indicate that the ratios of expression, as measured by QRT-PCR with lacC1 and lacC2 primers (complementary to the fruC gene) in cells grown in CDM with trehalose and fructose versus those grown in CDM-trehalose, are similar to that of the wild-type strain or close to 1, respectively.
Determination of luciferase activity in L. lactis.
Luciferase assays were carried out on a Lumat LB9501 apparatus (Berthold Technologies, Bad Wildbad, Germany). One milliliter of L. lactis culture was mixed with 5 μl of nonaldehyde, and the light emission was immediately measured. The value of the peak obtained was standardized to the optical density at 600 nm (OD600) of the culture. Values were measured at an OD600 of 0.3 to 0.4.
Fructose-1-phosphate assays on perchloric acid extracts.
Cells were grown in CDM containing appropriate sugars at a final concentration of 0.5%. For each condition, two independent cultures were carried out. When cultures reached an OD600 of 0.3 to 0.4, they were rapidly chilled to 4°C in an ethanol-dry ice bath and subsequently harvested by centrifugation. Pellets were resuspended in 2 volumes of ice-cold 20% perchloric acid solution. The perchloric acid extracts were neutralized by the addition of 2.5 volumes of tri-n-octylamine-CH3Cl (1:3.6 [vol/vol]) mixture according to the methods of Khym (16). After centrifugation, supernatants were extracted with the same volume of CH3Cl and centrifuged. This neutralization step was repeated twice. The final supernatants were kept at −80°C until further analysis. Assays of fructose-1-phosphate were carried out using recombinant purified E. coli 1-phosphofructokinase according to the methods described by Veiga-da-Cunha et al. (30).
RNA isolation.
Cells were grown in CDM containing appropriate sugars until the OD600 reached 0.3 to 0.4. They were quickly centrifuged for 2 min at 6,000 × g, frozen in liquid nitrogen, and broken using 500 mg of glass beads, 500 μl of phenol-chloroform, 30 μl of 3 M sodium acetate, and 15 μl of 20% sodium dodecyl sulfate. RNAs were isolated using the High Pure RNA isolation kit (Roche, Mannheim, Germany) according to the manufacturer's instructions.
RT-PCR and 5′-RACE.
Reverse transcriptase PCR (RT-PCR) was carried out on 500 ng of total RNA with the OneStep RT-PCR kit (QIAGEN, Courtaboeuf, France) as recommended by the manufacturer, using primers fruA2 and lacR2. Reaction conditions were a reverse transcription of 30 min at 45°C; an initial PCR activation step of 15 min at 95°C; 25 cycles of 10 s at 94°C, 50 s at 50°C, and 3 min at 68°C; and a final extension step of 10 min at 68°C. The 5′/3′ RACE kit (Roche) was used according to the supplier's instructions. A 1.5-μg aliquot of total RNA was used to obtain the cDNA by primer extension with primer lacC2. Following the 3′ tailing reaction with a dATP string, the cDNA was amplified by PCR using the reverse primer lacR3 and the forward primer [oligo(dT)-anchor primer] supplied with the kit. The 5′ end of the transcript was then determined by sequencing the PCR product.
QRT-PCR.
Differential expression of genes was checked by real-time quantitative RT-PCR (QRT-PCR). De novo cDNAs were prepared as described previously. One microliter of 40-fold-diluted cDNAs was used for each 25-μl PCR mixture containing 1× Absolute QPCR SYBR Green ROX (ABgene, Epsom, Surrey, United Kingdom) and a 200 nM concentration of each primer. All reactions were carried out in duplicate using an ABI Prism 7700 sequence detector (Applied Biosystems, Foster City, Calif.) with the following cycle parameters: one cycle of 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and annealing and extension at 60°C for 60 s. Prior to comparative analysis, each primer pair was tested to determine its efficiency using a genomic DNA scale. The efficiency of the primer pairs was in the range of 80% to 100%. Results were calculated from at least two independent RNA extractions for which measures by QRT-PCR were carried out in duplicate. The tuf gene was used as an internal control with the tuf1 and tuf2 primers.
Microarray experiments and analysis.
DNA microarrays contained 2,126 L. lactis IL1403 gene PCR products spotted in duplicate to glass slides as previously described (18). Single-strand reverse transcription and labeling of 20 μg of total RNA were done using the Superscript 3 reverse transcriptase (Invitrogen, Cergy Pontoise, France) and the Amersham CyScribe labeling kit according to the manufacturers' instructions. Slides were prehybridized for 1 h in 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate, and 0.1 mg ml−1 bovine serum albumin. After removal of the prehybridization buffer, slides were hybridized for 16 h at 42°C in SlideHyb buffer I (Ambion, Huntingdon, Cambridgeshire, United Kingdom) containing Cy3/Cy5-labeled cDNA mix. All comparisons were performed at least twice (once each with Cy3 and Cy5) to check for possible differences in labeling efficiency between fluorophores. Slides were scanned using a confocal laser scanner (Virtek Chipreader, Virtek, Waterloo, Ontario, Canada). Fluorescent signal intensity data were quantified using Imagene 5.5 software (Biodiscovery, Los Angeles, Calif.). Each expression ratio was represented by at least four separate measurements (duplicate spots on each of two slides). The data sets were normalized, and a statistical analysis (z-test) was done using the PreP software (10). Genes having a threefold ratio and a P value of <0.001 were considered to be differentially expressed. DNA microarray data are available on the website http://genome.jouy.inra.fr/efp/base/www.
RESULTS
Genetic organization of the fructose operon.
In L. lactis IL1403, the fruR, fruC, and fruA genes are coexpressed as a single transcript as deduced from Northern blot analysis (Fig. 1B) and RT-PCR using the reverse primer fruA2 and the forward primer lacR2, which are complementary to fruA and fruR, respectively (Fig. 1). To determine the transcriptional start site of the operon, 5′ rapid amplification of cDNA end (RACE) was carried out using primer lacC2, which is complementary to fruC. The transcriptional start point is located 22 bp or possibly 21 bp (due to a possible artifact from RACE when the start is a T nucleotide) upstream of the fruR start codon (Fig. 1A). A classical vegetative promoter is present immediately upstream of the transcriptional start, with a perfectly consensual −10 extended box and a −35 box (TTGATT) matching four (in boldface) of the six bases compared to the consensus (Fig. 1A). A rho-independent terminator structure is present downstream of fruA at a location compatible with the observed size of the transcript (Fig. 1A).
Role of FruC and FruA in fructose catabolism.
In order to determine the role of fruC and fruA, the two genes were inactivated by integration into the chromosome of plasmids derived from pJIM2374 (Fig. 1C). The growth patterns of the resulting strains were followed in CDM containing different sugars: glucose, mannose, trehalose, maltose, or fructose. The strains JIM8240, JIM8236, and JIM8235 inactivated for fruC, fruA, and fruCA, respectively, did not grow in CDM containing 0.5% fructose, whereas their growth on other sugars was unaffected (data not shown). Moreover, API 50 CH profiles, allowing the determination of the fermentation for 22 sugars, showed that the mutants were only affected in fructose metabolism (data not shown). We conclude that FruC and FruA are specifically involved in fructose catabolism.
To test if another lower-affinity pathway for fructose catabolism is present in L. lactis, the fruC or fruA mutants were grown in a medium containing a higher fructose concentration (2%). The high fructose amount allowed weak growth of both mutants (doubling time sixfold higher than the wild-type strain), suggesting that an alternative route for fructose catabolism exits.
Regulation of the expression of the fructose utilization operon. (i) Regulation by FruR.
In order to determine the role of the potential regulator FruR in the expression of the fru operon, fruR was inactivated by integration of plasmids derived from pJIM2374 carrying the luxAB genes (strains JIM8234 and JIM8239) into the bacterial chromosome (Fig. 1). In the wild-type strain (JIM8233) and the mutant strains, the expression of the fru operon can be quantified by measuring luciferase activity encoded by the luxAB genes. In CDM containing trehalose or glucose, the luciferase activity was, respectively, 160-fold and 100-fold higher in JIM8239 than in the wild-type strain, JIM8233 (Table 3). Similar results were obtained with strain JIM8234, although it did not grow in medium containing fructose as the sole carbon source, which is likely to be due to a polar effect of pGEMt (Table 3). The strong expression of the promoter fusion in the two fruR mutants compared to the wild type suggests that FruR acts as a repressor.
TABLE 3.
Expression of the fructose operon in the presence of different sugars and in different mutants
| Strain | Genetic makeup | Luciferase activitya (lux/OD unit [103]) measured in CDM containing:
|
||||
|---|---|---|---|---|---|---|
| Trehalose | Trehalose and fructose | Glucose | Glucose and fructose | Fructose | ||
| JIM8233 | Wild type | 42 | 1,910 | 14 | 185 | 2,349 |
| JIM8239 | fruR | 6,684 | 4,202 | 1,427 | 1,616 | 3,481 |
| JIM8245 | fruR; pJIM5517 | 16 | 1,248 | ND | ND | ND |
| JIM8253 | ccpA | 41 | ND | 41 | 74 | 2,243 |
| JIM8254 | ccpA, fruR | 8,844 | 9,822 | 9,190 | 9,871 | 11,157 |
| JIM8240 | fruC | 5,087 | 8,271 | 590 | 686 | — |
| JIM8235 | fruCA (polar fruC insertion) | 7,260 | 7,161 | 1,026 | 1,475 | — |
| JIM8234 | fruRCA (polar fruR insertion) | 7,426 | 6,595 | 1,768 | 1,928 | — |
| JIM8246 | fruC; pJIM5517 | 1,181 | 3,401 | ND | ND | — |
| JIM8236 | fruA | 52 | 82 | 10 | 14 | — |
Values are means of at least three independent experiments showing less than 20% variation. ND, not determined; —, no growth.
(ii) Role of fructose-1-phosphate as the inducer.
The addition of fructose in CDM containing glucose or trehalose resulted, respectively, in a 13-fold and a 45-fold increase of luciferase activity in strain JIM8233 (Table 3). To determine if fructose itself or an intracellular derivative of fructose was the inducer, luciferase activity transcribed from the fru operon promoter was measured in strain JIM8236, which is inactivated for fruA and thus lacks the fructose-specific enzyme IIABC component of the PTS. Under this condition, despite the presence of fructose, the expression of the fru operon remained low and similar to that measured under the repressing condition for the wild-type strain (Table 3). Similar results were obtained with a higher fructose concentration in the medium (2%), indicating that fructose-1-phosphate is the inducer (data not shown). These results indicate that the inducer is not fructose itself but an intracellular derivative of fructose formed in the presence of PTSFru. Intracellular assays showed that in the wild-type strain the level of fructose-1-phosphate rose from 35 nmol/g to 793 nmol/g when the wild-type strain was shifted from CDM-glucose to CDM-fructose. Therefore, fructose-1-phosphate is the most probable inducer in this regulation, since it is then transformed by FruC into fructose-1,6-biphosphate, a glycolysis compound.
Interestingly, the disruption of fruC, encoding the 1-phosphofructokinase (strain JIM8240), resulted in the induction of the expression of the operon independently of the sugar present in the growth medium (Table 3). Since this pattern of expression is similar to that of a fruR mutant, we have verified that the inactivation of fruC does not affect the expression of fruR in cis. For this purpose the plasmid pJIM5517, expressing fruR, was introduced in different genetic backgrounds. This plasmid was able to restore a wild-type repressing activity when introduced in the fruR background of JIM8245 (Table 3). Clearly, the introduction of pJIM5517 in the fruC background of JIM8246 did not suppress the constitutive high expression from the fru operon promoter. This result shows that the induction observed in JIM8240 is not due to fruR mis-expression but to the lack of activity of its product, FruR.
Furthermore, the inactivation of fruC could lead to an accumulation of FruR effector in the cell and thus to the inactivation of its repressor activity. To test this hypothesis, the intracellular concentration of fructose-1-phosphate was measured. The fructose-1-phosphate content of cells grown in glucose was 156 and 35 nmol of fructose-1-phoshate/g (dry weight) in JIM8240 (fruC) and in JIM8233 (wild type), respectively. A similar result was obtained in trehalose-grown cells, where the level of fructose-1-phospate was of 156 nmol/g in the fruC mutant, against 73 nmol/g in the wild-type strain. Thus, it appears that a basal expression of FruC is required to degrade fructose-1-phosphate, which is likely generated by spontaneous dephosphorylation of the fructose-1,6-bisphosphate coming from the glycolysis pathway or a side activity of a dephosphorylating enzyme with a phosphatase-like activity that remains to be identified. The induction of the fru operon in the fruC background leading to an increase of fructose-1-phosphate in the cell is a supplementary argument supporting that fructose-1-phosphate is the FruR effector.
(iii) Catabolite repression by CcpA.
Expression of sugar catabolic genes is usually repressed by glucose in the medium. Part of this repression is mediated by the CcpA repressor in low-GC gram-positive bacteria. In the case of the fru promoter, the presence of glucose in CDM supplemented with fructose resulted in a 13-fold decrease of luciferase activity (Table 3), suggesting that expression of the fru operon was down-regulated by catabolite repression. To test whether the glucose repression is CcpA dependent, the transcriptional fusion between the promoter of the fru operon and luxAB genes was compared in a ccpA background and in a wild-type background. In the presence of an intact fruR gene, the luciferase activity in strains JIM8233 (wild type) and JIM8253 (ccpA) remained low and almost identical for all the sugars tested except fructose (Table 3). However, in a fruR background, the luciferase activity increased sixfold when ccpA was inactivated (JIM8239 versus JIM8254) in glucose-supplemented CDM. These results indicate that CcpA effectively represses the expression of the fru operon in the presence of glucose.
Specificity of FruR regulation at the genome scale.
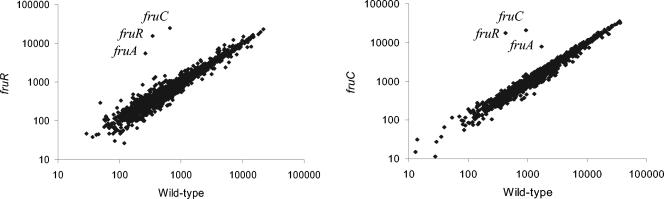
To analyze whether genes other than those of the fru operon are regulated by FruR, we determined genome-wide expression profiles, using total RNAs extracted from the fruR and fruC mutants (JIM8239 and JIM8240, respectively). Since the presence of glucose in the medium could interfere with the expression of sugar utilization genes, this experiment was performed in CDM with trehalose. The only genes that appeared to be differentially expressed relative to the wild-type IL1403 strain were those from the fru operon (Fig. 2). A similar result was obtained by comparing the fructose-1-phosphate-accumulating fruC (JIM8240) mutant to the wild-type strain (Fig. 2). Therefore, it appears that the FruR regulatory target is unique in L. lactis and that accumulation of fructose-1-phosphate has no effect other than modulating the regulatory activity of FruR.
FIG. 2.
Differential gene expression plots in the fruR (A) and the fruC (B) mutants compared to the wild-type strain. Relative signal intensities (mean normalized quanta) are plotted for genes as measured for the wild-type (x axis) and mutant (y axis) strains. The names of the genes that differ significantly (based on a threefold and a z-test [see Materials and Methods]) in the strains are indicated. Of these, only fruR, fruC, and fruA were induced more than threefold. In the fruR mutant their corresponding ratio and P values are 45 and 4 × 10−29 for fruR, 38 and 3 × 10−20 for fruC, and 21 and 8 × 10−23 for fruA. In the fruC mutant their corresponding ratio and P values are 23 and 1 × 10−49 for fruR, 41 and 8 × 10−69 for fruC, and 5 and 4 × 10−14 for fruA.
Determination of FruR DNA-binding site.
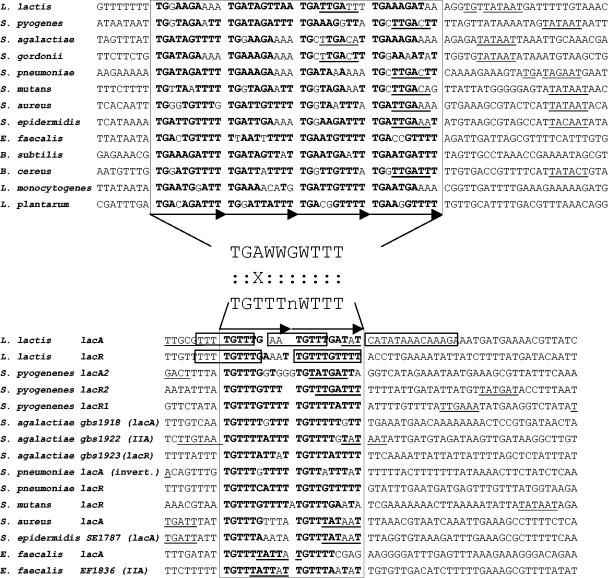
In order to further characterize FruR targets, we undertook determination of the DNA-binding site for this regulator. First, we searched for conserved motifs present upstream of the fru operon of several low-GC gram-positive bacteria that have an orthologous regulator, with the aid of the MEME tool (1). Two sets of motifs were found by searching motifs of different sizes. One, with the consensus TGWAAACGWWTWCA, corresponds to the CRE element (TGWAANCGNTNWCA) (31) and was present at a distance ranging from +23 to −59 from the fruR start codons in the various operons. The other appeared to be composed of four adjacent repeats of 10 bp with a consensus TGAWWGWTTT and was present at positions −113 to −20 from the fruR start codon. Importantly, this motif overlapped the −35 box sequence in cases where a promoter could be deduced from the −10 and −35 consensus sequences (8 out of the 12 promoters studied) (Fig. 3).
FIG. 3.
Alignment of the promoter regions of L. lactis fruR (A) and lacR and lacA (B) with the corresponding homologues in Streptococcus agalactiae NEM316 (accession no. AL732656), Streptococcus pneumoniae R6 (accession no. AE007317), S. mutans UA159 (accession no. AE014133), Streptococcus pyogenes MGAS315 (accession no. AE014074), S. gordonii Challis NCTC 7868 (http://tigrblast.tigr.org), Staphylococcus aureus Mu50 (accession no. BA000017), Staphylococcus epidermidis ATCC 12228 (accession no. AE015929), Bacillus cereus ATCC 14579 (accession no. AE016877), B. subtilis 168 (accession no. AL009126), Listeria monocytogenes EGD-e (accession no. NC_003210), Enterococcus faecalis VE583 (accession no. AE016830), and Lactobacillus plantarum WCFS1 (accession no. AL935263). IIA indicates that the corresponding genes encode a PTS enzyme IIA component. The putative −10 and −35 regions are underlined. The FruR and LacR operators are boxed, and direct repeats are represented by horizontal arrows. Regions protected against DNase I cleavage by the LacR repressor in lacR and lacA genes of L. lactis are framed (29). Bases in bold are those conserved in the determined consensus sequence. Between panels A and B are indicated the potential consensus sequences recognized by FruR and LacR as determined by the alignments of the 48 and 30 presented repeats, respectively. The similarities and the significant differences are indicated by a colon and an X, respectively.
To test whether the motif identified by in silico analysis was necessary for the repression by FruR, we measured the effect of several modifications of this motif on the strength of FruR repression. The strains JIM8641 and JIM8642 were controls containing the wild-type motif, JIM8643 contained a motif missing the two first repeats, and JIM8644 carried a mutated motif in which the conserved TG from each repeat was replaced by AA (Fig. 4A). In these strains an intact copy of the promoter drives the transcription of luxAB genes, used to follow FruR expression, while the expression of the fru operon is mediated from the fragment containing the control or modified promoters (Fig. 4). In the two control strains (JIM8641 and JIM8642), the QRT-PCR measures using the primers lacC1 and lacC3 complementary to the fruC gene showed that fru operon transcription was repressed in the absence of fructose (Fig. 4). In contrast, in the two mutant strains (JIM8643 and JIM8644), FruR repression was no longer exerted (Fig. 4). These results indicate that the motif found in silico is very likely the FruR-binding site.
To examine whether other genes might be regulated by FruR, we searched for a motif, TGDNWRDWWDTKRWWDNWWDTGVNDRDNWWTGVNNGWNWD, obtained by the alignment of the potential FruR motifs of 12 low-GC gram-positive bacteria (Fig. 3), in all potential promoter regions of the corresponding genomes. No motifs other than those upstream of the fructose operon were found, suggesting a high specificity of FruR regulation in these bacteria.
DISCUSSION
We characterized the operon fruRCA of L. lactis, which is involved in fructose utilization. The fruR gene encodes a regulator belonging to the DeoR family, the fruC gene, a 1-phosphofructokinase, and the fruA gene, the enzyme II for fructose transport.
The involvement of FruC and FruA in fructose catabolism was confirmed by the growth defect of the corresponding mutants in a fructose-containing medium. However, at a high fructose concentration, a weak growth of the mutants was observed, possibly due to the lack of specificity of other sugar transport systems present in L. lactis. A similar observation was reported for E. coli (17), where a second route for fructose utilization involves the PTS and membrane-spanning proteins that recognize a variety of sugars possessing the 3,4,5-d-arabino-hexose configuration. Fructose metabolism via this route is only observed when fructose is supplied in large amounts and, in this case, fructose-6-phosphate was formed instead of fructose-1-phosphate. In a number of streptococci, the mannose PTS was reported to catalyze the transport and the phosphorylation of glucose, mannose, 2-deoxyglucose and, to a lesser extent, fructose (7, 27). In L. lactis, a mannose PTS is present in the genome (2) and could be an alternative pathway for the transport of fructose in L. lactis.
The expression of the fru operon is repressed in a medium containing glucose. Such regulation, generally found for most sugar utilization operons, is designated catabolite repression and leads to the preferred use of highly metabolizable sugars. In low-GC gram-positive bacteria, it is exerted at the transcriptional level by the regulator CcpA and at the cellular level by inducer expulsion-exclusion (4). Our expression assays indicate that the fru operon of L. lactis is regulated directly at the transcriptional level by CcpA. This result is supported by the presence of a CRE box overlapping the site +1 of transcription in the promoter region of the operon. However, the level of expression in CDM-glucose is increased only 4-fold by a ccpA mutation, versus 100-fold for a fruR mutation, confirming the predominant role of FruR over CcpA in the regulation of the fru operon. Moreover, a very strong level of control is still exerted by glucose in the ccpA mutant, leading to a 30-fold decrease of luciferase activity when glucose is added to CDM-fructose. The most efficient level of catabolite control is thus mediated at the metabolic level, possibly by inducer exclusion, since the only way to degrade the fructose-1-phosphate appears to be its metabolism toward glycolysis (see the discussion below).
In L. lactis, FruR is the main repressor of the fructose operon. This level of regulation is directly dependent on the presence of fructose in the medium, as shown by the total lack of modulation of luciferase activity in the fruR mutant. In S. gordonii, FruR was also shown to act as a repressor of fructose metabolism (19). It is thus somewhat surprising that FruR from the mollicute S. citri might be an activator (12), as the two proteins have similar structure as shown by multiple alignments with all FruR proteins completed by secondary structure prediction searches (ClustalW and Predator). FruR of S. citri was proposed to bind to two repeats overlapping the −35 box of the fructose operon promoter, a location that usually leads to steric hindrance, making the promoter inaccessible to RNA polymerase. This apparent contradiction could be resolved if the mutant form of the S. citri FruR were trans-dominant over the wild type, as has been described for certain AgaR repressor mutants (24).
Interestingly, not only the fruR mutant but also fruA and fruC mutants are affected in the transcriptional regulation of the fructose operon promoter. Indeed, a nonpolar fruA mutation impairs growth on fructose and also leads to a constitutive low expression of the fru operon, even in the presence of fructose in the medium. This result suggests that uptake and phosphorylation of fructose to fructose-1-phosphate by FruA is necessary to relieve repression by FruR. The analysis of the fruC mutant, impaired in 1-phosphofructokinase activity, confirms this hypothesis. This mutant, which is also unable to grow on fructose as the sole carbon and energy source, displays a constitutive high expression of the fru operon, independent of the fructose content of the medium. Fructose-1-phosphate accumulates in the fruC mutant, even in a medium depleted of fructose. This constitutive induction also occurs in the double fruCA mutant (polar fruC mutation), a result that rules out the possibility that the induction is due to contamination of the medium by fructose. The lack of induction in the fruA mutant and its constitutive induction in the presence of fructose-1-phosphate indicate that fructose-1-phosphate modulates the DNA-binding activity of FruR. This agrees well with the observation that the DNA-binding activity of regulators of the DeoR family proteins is usually regulated by sugar-phosphates produced by the regulated pathway. For example, LacR from the lac operon in L. lactis is induced by tagatose-6-phosphate (29). In the case of fructose metabolism, fructose-1-phosphate is the only specific intermediate that could be used by the cell for this purpose, since this pathway branches to glycolysis in the next step. Lastly, the analysis of the fruC mutant shows that fruC plays a crucial role in the regulation of the fructose operon of L. lactis. Indeed, no other enzyme appears to be able to dephosphorylate fructose-1-phosphate, either formed by FruA or present as an offshoot of glycolysis. It follows that a basal level of FruC is necessary in the cell to degrade the inducer and mediate catabolic control of the fructose operon by inducer exclusion and transcriptional control by CcpA.
The repression by FruR appears to depend on the presence of four repeats of 10 bp. This structure, well conserved in various low-GC gram-positive bacteria, has a consensus motif (TGAWWGWTTT)4. The deletion of two repeats completely prevents repression. Mutations in the first two bases of the consensus (TG→AA) also abolish the repression by FruR. These two bases are almost perfectly conserved, since they are present in 48/48 and 47/48 of the analyzed sequences, respectively (Fig. 3A). Another member of the DeoR family of regulators, LacR from L. lactis, binds to two operators localized in lacR and lacABCDFEGX promoters (framed in Fig. 3B) (29). Two repeats of a 10-bp consensus (TGTTTNWTTT) are present in the lacR and lacA promoters of a set of various gram-positive bacteria (Fig. 3B). This consensus shares similarities with the FruR consensus, suggesting that the two members of the DeoR family recognize similar DNA sequences. This hypothesis is enforced by the similarities of the second helix of the FruR and LacR helix-turn-helix, which is known to be directly involved in the recognition and contact of the DNA target region in the DeoR family regulators (data not shown) (3).
In S. gordonii, fructose metabolism through the PTSFru pathway appears to be essential for biofilm formation, as inactivation of either 1-phosphofructokinase or fructose-specific enzyme II leads to a biofilm-defective phenotype (19). It was suggested that the fructose operon could be involved in a sensory mechanism enabling the switch from a sessile to a planktonic phenotype or that 1-phosphofructokinase plays a role in cross-regulation of a two-component system by phosphorylation of a response regulator. A wider set of genes is thus suspected to be regulated by the fructose system in this bacterium. Our analysis has failed to identify other genes that could be regulated by FruR in completely sequenced genomes of low-GC gram-positive bacteria.
It is interesting that in enteric bacteria, transcription of the fru operon is regulated by a pleitropic regulator, Cra (formerly named FruR), and to a lesser extent by the cyclic AMP-CRP complex (9, 25). Cra represses the expression of genes encoding glycolytic enzymes (i.e., key enzymes in the Embden-Meyerhof and Entner-Doudoroff pathways) and activates expression of genes encoding biosynthetic and oxidative enzymes (i.e., key enzymes in the Krebs cycle, the glyoxalate shunt, the gluconeogenic pathway, and electron transfer) (23, 25). Furthermore, the effect of Cra on transcription is counteracted by a micromolar concentration of fructose-1-phosphate (22). In order to detect possible side effects due to fructose metabolism or its regulation in L. lactis, DNA microarray experiments were carried out. The inactivation of fruC leads to an increase in expression of the fru operon comparable to that due to fruR inactivation, which is most probably a consequence of an elevated content of fructose-1-phosphate in the cells. However, the effect of fruC or fruR inactivation appeared to be restricted to the fru operon under the conditions tested. Moreover, addition of fructose to CDM containing trehalose specifically induces the fru operon and marginally represses the expression of trehalose and some other sugar utilization genes in the wild-type IL1403 strain (data not shown). Thus, metabolism of fructose does not appear to trigger other signaling pathways acting on other genes in L. lactis. The involvement of fructose metabolism as a sensory mechanism in biofilm formation may thus be either specific to S. gordonii or, more likely, the result of a metabolic effect rather than a sensory role.
Acknowledgments
We thank Jorge Jorge García de la Nava Ruiz, Oswaldo Trelles, Valentin Loux, and Philippe Bessières for their help on the bioinformatics for microarray analysis, Alain and Marie-Christine Chopin for the gift of plasmid pJIM6001, and Jacek Bardowski for the gift of strain JIM7794.
This work was supported by grant QLK3-2001-010473 under the EU sub-program area “Quality of Life and Management of Living Resources—The Cell Factory.”
REFERENCES
- 1.Bailey, T. L., and C. Elkan. 1995. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 3:21-29. [PubMed] [Google Scholar]
- 2.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennan, R. G., and B. W. Matthews. 1989. Structural basis of DNA-protein recognition. Trends Biochem. Sci. 14:286-290. [DOI] [PubMed] [Google Scholar]
- 4.Bruckner, R., and F. Titgemeyer. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209:141-148. [DOI] [PubMed] [Google Scholar]
- 5.Chiou, C. Y., H. H. Wang, and G. C. Shaw. 2002. Identification and characterization of the non-PTS fru locus of Bacillus megaterium ATCC 14581. Mol. Genet. Genomics 268:240-248. [DOI] [PubMed] [Google Scholar]
- 6.Chopin, A., M. C. Chopin, A. Moillo-Batt, and P. Langella. 1984. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 11:260-263. [DOI] [PubMed] [Google Scholar]
- 7.Cochu, A., C. Vadeboncoeur, S. Moineau, and M. Frenette. 2003. Genetic and biochemical characterization of the phosphoenolpyruvate:glucose/mannose phosphotransferase system of Streptococcus thermophilus. Appl. Environ. Microbiol. 69:5423-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delorme, C., S. D. Ehrlich, and P. Renault. 1999. Regulation of expression of the Lactococcus lactis histidine operon. J. Bacteriol. 181:2026-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feldheim, D. A., A. M. Chin, C. T. Nierva, B. U. Feucht, Y. W. Cao, Y. F. Xu, S. L. Sutrina, and M. H. Saier, Jr. 1990. Physiological consequences of the complete loss of phosphoryl-transfer proteins HPr and FPr of the phosphoenolpyruvate:sugar phosphotransferase system and analysis of fructose (fru) operon expression in Salmonella typhimurium. J. Bacteriol. 172:5459-5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia de la Nava, J., S. van Hijum, and O. Trelles. 2003. PreP: gene expression data pre-processing. Bioinformatics 19:2328-2329. [DOI] [PubMed] [Google Scholar]
- 11.Gaurivaud, P., J. L. Danet, F. Laigret, M. Garnier, and J. M. Bove. 2000. Fructose utilization and phytopathogenicity of Spiroplasma citri. Mol. Plant Microbe Interact. 13:1145-1155. [DOI] [PubMed] [Google Scholar]
- 12.Gaurivaud, P., F. Laigret, M. Garnier, and J. M. Bove. 2001. Characterization of FruR as a putative activator of the fructose operon of Spiroplasma citri. FEMS Microbiol. Lett. 198:73-78. [DOI] [PubMed] [Google Scholar]
- 13.Gilson, T. J. 1984. Studies on the Epstein-Barr virus genome. Ph.D. thesis. University of Cambridge, Cambridge, United Kingdom.
- 14.Godon, J. J., C. J. Pillidge, K. Jury, C. A. Shearman, and M. J. Gasson. 1995. Molecular analysis of the Lactococcus lactis sex factor. Dev. Biol. Stand. 85:423-430. [PubMed] [Google Scholar]
- 15.Holo, H., and I. F. Nes. 1995. Transformation of Lactococcus by electroporation. Methods Mol. Biol. 47:195-199. [DOI] [PubMed] [Google Scholar]
- 16.Khym, J. X. 1975. An analytical system for rapid separation of tissue nucleotides at low pressures on conventional anion exchangers. Clin. Chem. 21:1245-1252. [PubMed] [Google Scholar]
- 17.Kornberg, H. L. 2001. Routes for fructose utilization by Escherichia coli. J. Mol. Microbiol. Biotechnol. 3:355-359. [PubMed] [Google Scholar]
- 18.Kuipers, O. P., A. de Jong, R. J. Baerends, S. A. van Hijum, A. L. Zomer, H. A. Karsens, C. D. den Hengst, N. E. Kramer, G. Buist, and J. Kok. 2002. Transcriptome analysis and related databases of Lactococcus lactis. Antonie Leeuwenhoek 82:113-122. [PubMed] [Google Scholar]
- 19.Loo, C. Y., K. Mitrakul, I. B. Voss, C. V. Hughes, and N. Ganeshkumar. 2003. Involvement of an inducible fructose phosphotransferase operon in Streptococcus gordonii biofilm formation. J. Bacteriol. 185:6241-6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1991. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Ramseier, T. M., D. Negre, J. C. Cortay, M. Scarabel, A. J. Cozzone, and M. H. Saier, Jr. 1993. In vitro binding of the pleiotropic transcriptional regulatory protein, FruR, to the fru, pps, ace, pts and icd operons of Escherichia coli and Salmonella typhimurium. J. Mol. Biol. 234:28-44. [DOI] [PubMed] [Google Scholar]
- 23.Ramseier, T. M., S. Bledig, V. Michotey, R. Feghali, and M. H. Saier, Jr. 1995. The global regulatory protein FruR modulates the direction of carbon flow in Escherichia coli. Mol. Microbiol. 16:1157-1169. [DOI] [PubMed] [Google Scholar]
- 24.Ray, W. K., and T. J. Larson. 2004. Application of AgaR repressor and dominant repressor variants for verification of a gene cluster involved in N-acetylgalactosamine metabolism in Escherichia coli K-12. Mol. Microbiol. 51:813-826. [DOI] [PubMed] [Google Scholar]
- 25.Saier, M. H., Jr., and T. M. Ramseier. 1996. The catabolite repressor/activator (Cra) protein of enteric bacteria. J. Bacteriol. 178:3411-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sissler, M., C. Delorme, J. Bond, S. D. Ehrlich, P. Renault, and C. Francklyn. 1999. An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis. Proc. Natl. Acad. Sci. USA 96:8985-8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vadeboncoeur, C., and M. Pelletier. 1997. The phosphoenolpyruvate:sugar phosphotransferase system of oral streptococci and its role in the control of sugar metabolism. FEMS Microbiol. Rev. 19:187-207. [DOI] [PubMed] [Google Scholar]
- 28.van Rooijen, R. J., S. van Schalkwijk, and W. M. de Vos. 1991. Molecular cloning, characterization, and nucleotide sequence of the tagatose 6-phosphate pathway gene cluster of the lactose operon of Lactococcus lactis. J. Biol. Chem. 266:7176-7181. [PubMed] [Google Scholar]
- 29.van Rooijen, R. J., K. J. Dechering, C. Niek, J. Wilmink, and W. M. de Vos. 1993. Lysines 72, 80 and 213 and aspartic acid 210 of the Lactococcus lactis LacR repressor are involved in the response to the inducer tagatose-6-phosphate leading to induction of lac operon expression. Protein Eng. 6:201-206. [DOI] [PubMed] [Google Scholar]
- 30.Veiga-da-Cunha, M., A. Hoyoux, and E. Van Schaftingen. 2000. Overexpression and purification of fructose-1-phosphate kinase from Escherichia coli: application to the assay of fructose 1-phosphate. Protein Expr. Purif. 19:48-52. [DOI] [PubMed] [Google Scholar]
- 31.Weickert, M. J., and G. H. Chambliss. 1990. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 87:6238-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]