Abstract
Peptide microcin J25 (MccJ25) inhibits bacterial RNA polymerase. We show that thermolysin-cleaved MccJ25 and MccJ25 lacking amino acids 13 to 17 also inhibit transcription. Our data and structural analysis of intact and thermolysin-digested MccJ25 suggest that distinct regions of MccJ25 are involved in transcription inhibition and cell entry.
Transcription of DNA into RNA, the first step and a major regulatory checkpoint of gene expression, is carried out by DNA-dependent RNA polymerases (RNAPs). The RNAP functional cycle consists of transcription initiation, processive elongation of nascent RNA chains, and transcription termination. In principle, each stage of the cycle can be regulated and/or targeted by inhibitors. The best-studied inhibitor of bacterial RNAP, rifampin, acts during transition from transcription initiation to elongation by binding to RNAP and preventing the synthesis of long RNAs. Rifampin and its derivatives are widely used to treat mycobacterial infections.
Cellular RNAPs are complex, multisubunit molecular machines that are related to each other through common ancestry. The RNAP molecule has a crab-claw-like shape, with a deep main channel separating the jaws of the claw (21). The DNA being transcribed binds in this channel. In addition to the main channel, a fully enclosed narrow secondary channel begins close to the catalytic center deep inside the main channel and opens on the enzyme's surface. The secondary channel is thought to supply RNA polymerization substrates, nucleoside triphosphates, to the catalytic center.
Microcin J25 (MccJ25) is a bactericidal peptide produced by some strains of the intestinal bacterium Escherichia coli (14). MccJ25-producing E. coli cells harbor a plasmid that is responsible for both MccJ25 production and resistance of MccJ25-producing cells to the drug (17, 18). MccJ25 production is activated when nutrients become scarce, which should give MccJ25-producing cells an advantage by inhibiting the growth of their brethren that are MccJ25 sensitive (6).
Most spontaneous MccJ25-resistant mutants carry mutations in genes coding for membrane transporters and are thus likely to be uptake mutants (7, 15, 16). A rare MccJ25-resistant mutant carrying a point mutation in the rpoC gene coding for the RNAP β′ subunit was isolated (8). Moreover, MccJ25 was shown to inhibit RNAP from gram-negative bacteria in a purified in vitro system (8, 20). In contrast, MccJ25 does not bind to mutant RNAP isolated from MccJ25-resistant cells (1, 20). The results firmly establish that RNAP is the cellular target of MccJ25. A large number of additional MccJ25-resistant mutations in RNAP genes were isolated (10, 20). The location of MccJ25 resistance mutations, around the circumference of the secondary channel, strongly suggests that transcription inhibition by MccJ25 takes place by occlusion of the secondary channel, resulting in complete cessation of substrate traffic to the enzyme's catalytic center. Recent biochemical and biophysical studies firmly place MccJ25 in the secondary channel, and structural modeling shows that bound MccJ25 should completely occlude the channel (1, 10). As expected, MccJ25 binding instantaneously stops RNAP in its tracks and prevents further elongation of transcription (1).
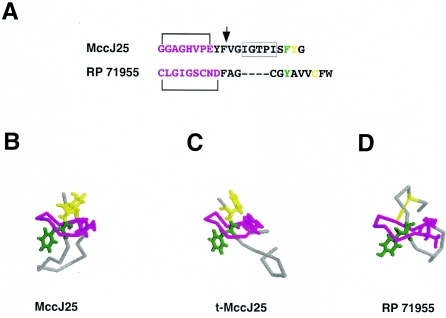
The structure of the 21-amino-acid MccJ25 has been determined (2, 12, 19). The MccJ25 molecule assumes an unusual “threaded lasso” fold (Fig. 1B). A lactam linkage between the first and the eighth MccJ25 amino acids creates a small ring; the C-terminal portion of the molecule forms a β-hairpin loop; the tail portion of the molecule passes through the ring and is topologically trapped (the bulky aromatic residues Phe19 and Tyr20 located at both sides of the ring do not allow the tail to be withdrawn from the ring) (Fig. 1B). The unusual structure is responsible for the extremely high stability of MccJ25: it withstands autoclaving without the loss of activity (5).
FIG. 1.
Comparison of intact MccJ25, t-MccJ25, and RP 71955 structures. A. Primary sequence of MccJ25 and RP71955. The lactam linkage resulting in ring formation is indicated. The site of thermolysin cleavage of MccJ25 is shown by a vertical arrow. MccJ25 amino acids removed in h16-MccJ25 are boxed. The color code is explained below. B, C, D. The structures of intact MccJ25 (B), t-MccJ25 (C), and RP71955 (D) were aligned using the program O. The ring portion of molecules is shown in magenta; the loop region is gray. The side chains participating in the formation of lactam bonds that close the rings are shown. The rest of the rings as well as the loop regions are shown in the Cα representation. In MccJ25 structures, the side chains of the two residues that trap the tail in the ring, Phe19 and Tyr20, are shown in green and yellow, respectively. In RP 71955, the counterpart of Tyr20 is absent; however, an intramolecular disulfide bridge (yellow) secures the tail in the ring. rms deviation between intact MccJ25 and RP 71955 is 1.182 Å for eight superimposed Cα atoms in the ring.
In general, MccJ25 is highly resistant to proteases (4). However, thermolysin quantitatively cleaves native MccJ25 after Phe10, in the loop region (Fig. 1A) (4). The resultant peptide, t-MccJ25, can be purified by high-performance liquid chromatography from residual undigested MccJ25 due to its distinct chromatographic behavior (4). Though t-MccJ25 consists of two polypeptide chains, topological trapping keeps the tail inside the ring and t-MccJ25 remains highly resistant to denaturation (5). The structure of t-MccJ25 was recently determined (Fig. 1C) (13). Comparisons of the intact and t-MccJ25 structures show substantial differences in the loop region: in intact MccJ25, the loop, which links the ring and the tail, forms a β hairpin; in t-MccJ25, the β hairpin is disrupted and MccJ25 residues 11 to 16 become disordered. In contrast, the two structures were identical in the ring and tail regions (with rms deviation of 0.43 Å for eight superimposed Cα atoms in the ring) (Fig. 1B and C). On the basis of dramatically decreased antibacterial action of t-MccJ25, it was suggested that residues 11 to 16 are involved in interaction with RNAP (13). Here, we tested this prediction by comparing the ability of intact MccJ25 and t-MccJ25 to affect cell growth and inhibit transcription by E. coli RNAP in vitro.
t-MccJ25 was prepared as described previously (13). Both intact MccJ25 and t-MccJ25 were chromatographically pure, and matrix-assisted laser desorption ionization mass spectrometry analysis revealed the presence of the expected mass peaks only (data not shown). The concentrations of the intact MccJ25 and t-MccJ25 used in the experiments described below were matched after quantitative amino acid analysis performed by the Protein Core of the University of Texas Medical School at Galveston, TX.
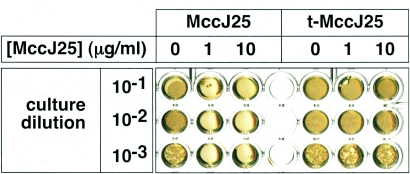
As can be seen from Fig. 2 and in agreement with previous data, t-MccJ25 had no biological activity. In this experiment, serial dilutions of MccJ25-sensitive E. coli MC4100 cells were seeded in microtiter plate wells containing Luria-Bertani agar with 0, 1, or 10 μg/ml of intact MccJ25 or t-MccJ25 and were allowed to grow overnight. As can be seen, the presence of 10 μg/ml t-MccJ25 in the medium had no effect on the growth of E. coli MC4100. In contrast, no colonies formed in the presence of as little as 1 μg/ml wild-type MccJ25. We conclude that the t-MccJ25 preparation that we used (i) is biologically inactive and (ii) contains less than 10% contaminating intact MccJ25, which, if present, would have inhibited cell growth in a well containing 10 μg/ml t-MccJ25.
FIG. 2.
t-MccJ25 is biologically inactive. Five microliters of the indicated dilutions of E. coli MC4100 overnight culture was spotted in microtiter plate wells containing Luria-Bertani agar with the indicated concentrations of intact MccJ25 or t-MccJ25. The results of overnight growth at 37°C are shown.
The process of intact MccJ25 entry into E. coli cells was shown to be dependent on the outer membrane protein channel FhuA (15); also involved are TonB, ExbB, and ExbD (these three proteins form a complex in the inner membrane that uses energy from proton motive force to open the FhuA channel in a ligand-dependent fashion) and SbmA (an inner membrane protein of unknown function) (16). Mutations in genes coding for these proteins lead to MccJ25 resistance (in fact, the high frequency of intake mutations made it difficult to identify the MccJ25 cellular target, RNAP). Therefore, the result of the experiment in Fig. 2 could be due to either inefficient (or unproductive) interaction between t-MccJ25 and RNAP or rapid degradation of t-MccJ25 in the cytoplasm, or it could be due to the inability of t-MccJ25 to enter the cell.
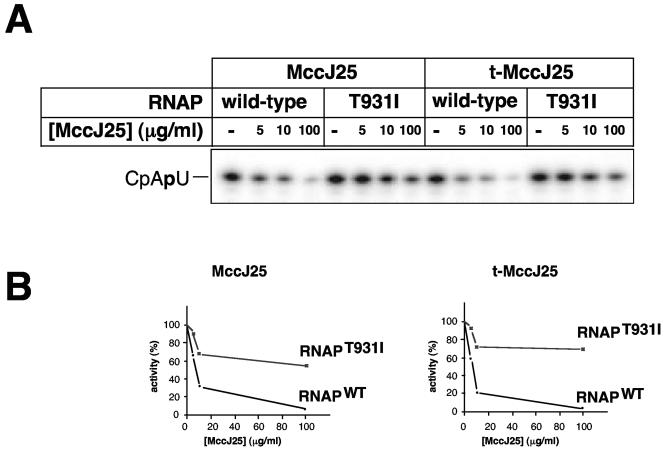
We performed an in vitro abortive initiation reaction from a T7 A1 promoter-containing DNA template in the presence of increasing concentrations of either intact MccJ25 or t-MccJ25. Transcription reactions were performed using wild-type σ70 RNAP holoenzyme and a mutant holoenzyme that carries a T931I substitution in the β′ subunit that leads to resistance to intact MccJ25 (8) by abolishing the drug interaction with RNAP (1, 20). The results are presented in Fig. 3. As can be seen, t-MccJ25 inhibited the production of the abortive CpApU transcript by the wild-type RNAP even better than intact MccJ25. Both microcins had a mild effect on transcription by the mutant enzyme. We therefore conclude that (i) since the ability of MccJ25 to bind to and inhibit RNAP is unaffected by thermolysin cleavage, the lack of bactericidal effect of t-MccJ25 is entirely due to defects in its uptake by sensitive cells and (ii) since the T931I substitution affects the interaction of both intact MccJ25 and t-MccJ25 to the same extent, t-MccJ25 interacts with RNAP in the same way as does the intact MccJ25.
FIG. 3.
t-MccJ25 inhibits transcription by E. coli RNAP. A. The results of in vitro abortive transcription from the T7 A1 promoter by wild-type RNAP and MccJ25-resistant RNAP harboring the β′ T931I in the absence or in the presence of increasing concentrations of intact MccJ25 or t-MccJ25 are shown. B. Radioactivity in CpApU bands in the gel presented in panel A was quantified using a PhosporImager and plotted as a function of MccJ25 concentration. The activity in the absence of MccJ25 was assumed to be 100%. WT, wild type.
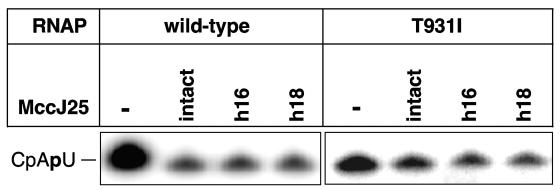
Previous work described two MccJ25 derivatives obtained by chemical degradation, h16-MccJ25 and h18-MccJ25, lacking β-hairpin region amino acids 13 to 17 and 15 to 17, respectively (Fig. 1A) (13). Limited amounts of h16-MccJ25 and h18-MccJ25 were prepared, their identity and purity were established by mass spectrometry, and their ability to inhibit in vitro transcription was tested. The results are presented in Fig. 4. As can be seen, both MccJ25 derivatives efficiently inhibited transcription by wild-type RNAP (∼15% residual transcription in the presence of 50 μg/ml MccJ25 or the corresponding concentration of its derivatives). In contrast, RNAP carrying the T931I substitution in β′ exhibited ∼60% residual activity in the presence of 50 μg/ml MccJ25 or its derivatives. We conclude that MccJ25 amino acids 13 to 17 are dispensable for transcription inhibition.
FIG. 4.
h18-MccJ25 and h16-MccJ25 inhibit transcription by E. coli RNAP. A. The results of in vitro abortive transcription from the T7 A1 promoter by wild-type RNAP and MccJ25-resistant RNAP harboring the β′ T931I in the absence or in the presence of intact MccJ25 or indicated MccJ25 derivatives are shown.
Our results clearly show that MccJ25 derivatives with lesions in the β-hairpin region are at least as active in transcription inhibition as intact MccJ25. The result therefore excludes the role of the β-hairpin region in RNAP binding and suggests that it is the unusual ring-tail part of the MccJ25 molecule that interacts with RNAP and blocks the secondary channel. The lack of bactericidal activity of t-MccJ25 suggests that the loop region of MccJ25 is important for the uptake, or alternatively, the loss of structure in this region may make MccJ25 proteolytically labile inside the cell. The latter alternative is unlikely, since the recent report of Bellomio et al. indicates that cells whose permeability was increased by mutation become sensitive to t-MccJ25 (3). The loop region shall thus be an attractive target for genetic and/or chemical modifications to obtain more permeable and hence more potent MccJ25 derivatives. Interestingly, the ring-tail part of MccJ25 is very similar to the corresponding part of RP 71955 (Fig. 1A and C) (9; see also reference 11) (also known as aborycin), a circular peptide produced by Streptomyces. Aborycin inhibits the growth of gram-positive bacteria but has no effect on gram-negative bacteria (11) and is thus complementary to MccJ25, which inhibits gram-negative but not gram-positive bacteria. Experiments aimed at determining the ability of aborycin to inhibit bacterial RNAP are currently in progress.
Acknowledgments
We are grateful to Seth Darst for help with structural alignments.
This work was supported by NIH RO1 grant GM64530, NIH FIRCA grant RO3 TW006828, and a Burroughs Wellcome Fund Career Award (to K.S.). E.S. was partially supported by the Charles and Johanna Busch postdoctoral fellowship.
REFERENCES
- 1.Adelman, K., J. Yuzenkova, A. La Porta, N. Zenkin, J. Lee, J. T. Lis, S. Borukhov, M. D. Wang, and K. Severinov. 2004. Molecular mechanism of transcription inhibition by peptide antibiotic microcin J25. Mol. Cell 16:753-762. [DOI] [PubMed] [Google Scholar]
- 2.Bayro, M. J., J. Mukhopadhyay, G. V. Swapna, J. Y. Huang, L. C. Ma, E. Sineva, P. E. Dawson, G. T. Montelione, and R. H. Ebright. 2003. Structure of antibacterial peptide microcin J25: a 21-residue lariat protoknot. J. Am. Chem. Soc. 125:12382-12383. [DOI] [PubMed] [Google Scholar]
- 3.Bellomio, A., P. A. Vincent, B. F. de Arcuri, R. A. Salomon, R. D. Morero, and R. N. Farias. 2004. The microcin J25 β-hairpin region is important for antibiotic uptake but not for RNA polymerase and respiration inhibition. Biochem. Biophys. Res. Commun. 325:1454-1458. [DOI] [PubMed] [Google Scholar]
- 4.Blond, A., J. Peduzzi, C. Goulard, M. J. Chiuchiolo, M. Barthelemy, Y. Prigent, R. A. Salomon, R. N. Farias, F. Moreno, and S. Rebuffat. 1999. The cyclic structure of microcin J25, a 21-residue peptide antibiotic from Escherichia coli. Eur. J. Biochem. 259:747-755. [DOI] [PubMed] [Google Scholar]
- 5.Blond, A., M. Cheminant, D. Destoumieux-Garzon, I. Segalas-Milazzo, J. Peduzzi, C. Goulard, and S. Rebuffat. 2002. Thermolysin-linearized microcin J25 retains the structured core of the native macrocyclic peptide and displays antimicrobial activity. Eur. J. Biochem. 269:6212-6222. [DOI] [PubMed] [Google Scholar]
- 6.Chiuchiolo, M. J., M. A. Delgado, R. N. Farias, and R. A. Salomon. 2001. Growth-phase-dependent expression of the cyclopeptide antibiotic microcin J25. J. Bacteriol. 183:1755-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delgado, M. A., J. O. Solbiati, M. J. Chiuchiolo, R. N. Farias, and R. A. Salomon. 1999. Escherichia coli outer membrane protein TolC is involved in production of the peptide antibiotic microcin J25. J. Bacteriol. 181:1968-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delgado, M. A., M. R. Rintoul, R. N. Farias, and R. A. Salomon. 2001. Escherichia coli RNA polymerase is the target of the cyclopeptide antibiotic microcin J25. J. Bacteriol. 183:4543-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frechet, D., J. D. Guitton, F. Herman, D. Faucher, G. Helynck, B. Monegier du Sorbier, J. P. Ridoux, E. James-Surcouf, and M. Vuilhorgne. 1994. Solution structure of RP 71955, a new 21 amino acid tricyclic peptide active against HIV-1 virus. Biochemistry 33:42-50. [DOI] [PubMed] [Google Scholar]
- 10.Mukhopadhyay, J., E. Sineva, J. Knight, R. M. Levy, and R. H. Ebright. 2004. Antibacterial peptide microcin J25 inhibits transcription by binding within and obstructing the RNA polymerase secondary channel. Mol. Cell 14:739-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Potterat, O., H. Stephan, J. W. Metzger, V. Gnau, H. Zahner, and G. Jung. 1994. Aborycin—a tricyclic 21-peptide antibiotic isolated from Streptomyces griseoflavus. Liebigs Ann. Chem. 1994:741-743. [Google Scholar]
- 12.Rosengren, K. J., R. J. Clark, N. L. Daly, U. Goransson, A. Jones, and D. J. Craik. 2003. Microcin J25 has a threaded sidechain-to-backbone ring structure and not a head-to-tail cyclized backbone. J. Am. Chem. Soc. 125:12464-12474. [DOI] [PubMed] [Google Scholar]
- 13.Rosengren, K. J., A. Blond, C. Afonso, J. C. Tabet, S. Rebuffat, and D. J. Craik. 2004. Structure of thermolysin cleaved microcin J25: extreme stability of a two-chain antimicrobial peptide devoid of covalent links. Biochemistry 43:4696-4702. [DOI] [PubMed] [Google Scholar]
- 14.Salomon, R. A., and R. N. Farias. 1992. Microcin 25, a novel antimicrobial peptide produced by Escherichia coli. J. Bacteriol. 174:7428-7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salomon, R. A., and R. N. Farias. 1993. The FhuA protein is involved in microcin 25 uptake. J. Bacteriol. 175:7741-7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salomon, R. A., and R. N. Farias. 1995. The peptide antibiotic microcin 25 is imported through the TonB pathway and the SbmA protein. J. Bacteriol. 177:3323-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solbiati, J. O., M. Ciaccio, R. N. Farias, and R. A. Salomon. 1996. Genetic analysis of plasmid determinants for microcin J25 production and immunity. J. Bacteriol. 178:3661-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solbiati, J. O., M. Ciaccio, R. N. Farias, J. E. Gonzalez-Pastor, F. Moreno, and R. A. Salomon. 1999. Sequence analysis of the four plasmid genes required to produce the circular peptide antibiotic microcin J25. J. Bacteriol. 181:2659-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson, K.-A., M. Kalkum, J. Ottesen, J. Yuzenkova, B. T. Chait, R. Landick, T. W. Muir, K. Severinov, and S. A. Darst. 2003. The structure of microcin J25, a peptide inhibitor of bacterial RNA polymerase. J. Am. Chem. Soc. 125:12475-12483. [DOI] [PubMed] [Google Scholar]
- 20.Yuzenkova, J., M. Delgado, S. Nechaev, D. Savalia, V. Epshtein, I. Artsimovitch, R. A. Mooney, R. Landick, R. N. Farias, R. Salomon, and K. Severinov. 2002. Mutations of bacterial RNA polymerase leading to resistance to microcin J25. J. Biol. Chem. 277:50867-50875. [DOI] [PubMed] [Google Scholar]
- 21.Zhang, G., E. A. Campbell, L. Minakhin, C. Richter, K. Severinov, and S. A. Darst. 1999. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell 98:811-824. [DOI] [PubMed] [Google Scholar]