Abstract
Representatives of the genus Beijerinckia are known as heterotrophic, dinitrogen-fixing bacteria which utilize a wide range of multicarbon compounds. Here we show that at least one of the currently known species of this genus, i.e., Beijerinckia mobilis, is also capable of methylotrophic metabolism coupled with the ribulose bisphosphate (RuBP) pathway of C1 assimilation. A complete suite of dehydrogenases commonly involved in the sequential oxidation of methanol via formaldehyde and formate to CO2 was detected in cell extracts of B. mobilis grown on CH3OH. Carbon dioxide produced by oxidation of methanol was further assimilated via the RuBP pathway as evidenced by reasonably high activities of phosphoribulokinase and ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO). Detection and partial sequence analysis of genes encoding the large subunits of methanol dehydrogenase (mxaF) and form I RubisCO (cbbL) provided genotypic evidence for methylotrophic autotrophy in B. mobilis.
The current definition of the genus Beijerinckia characterizes its members as nonsymbiotic, aerobic, chemo-heterotrophic bacteria with the ability to fix atmospheric dinitrogen (6). Beijerinckia species utilize a wide range of multicarbon compounds, but sugars are the preferred growth substrates. Members of this genus are typical rod-shaped cells with round ends containing polar lipoid bodies. Another distinctive feature of these bacteria is their acid tolerance, which allows them to grow and to fix dinitrogen at pH 3.0 to 4.0. The first isolates of this genus were obtained from acidic soils of tropical regions (1, 24). Later studies revealed that these bacteria are widely distributed in both acidic and neutral soils of different tropical and nontropical regions (5). The four recognized species of this genus, i.e., Beijerinckia indica, Beijerinckia mobilis, Beijerinckia derxii, and Beijerinckia fluminensis, were taxonomically described and intensively studied half a century ago. Since that time no further studies into the physiology and metabolism of this group of bacteria have been undertaken.
Recently, a novel subgroup of acidophilic serine pathway methanotrophic bacteria that are phylogenetically closely related to the genus Beijerinckia was discovered in acidic Sphagnum peat bogs and forest soils (10, 11, 13). The 16S rRNA sequence similarity values between Beijerinckia spp. and the acidophilic methanotrophs Methylocella and Methylocapsa range from 96.0 to 97.3%, and the representatives of these three genera form a monophyletic cluster within the alphaproteobacteria. Interestingly, methanotrophic and heterotrophic members of this monophyletic cluster possess some morphological similarities and share several physiological characteristics, including acid tolerance and ability to fix dinitrogen. Comparison of 16S rRNA phylogeny with phylogenies based on two different structural genes of nitrogenase (nifH and nifD) has led to the conclusion that these two metabolically different groups of bacteria might have originated from a common acidophilic dinitrogen-fixing ancestor (12). However, none of the currently known Beijerinckia spp. are capable of growth on methane, and no evidence for any kind of metabolic similarity between Beijerinckia and acidophilic methanotrophs has been obtained so far. The only indication for the possible occurrence of C1 metabolism in Beijerinckia was reported for B. mobilis which, in contrast to all other species of this genus, is capable of weak growth on formate (6). This report and the close phylogenetic relationship between Beijerinckia spp. and acidophilic methanotrophs prompted us to examine the capability of all currently available type strains of Beijerinckia to grow on methanol. The data presented here show that at least one of the currently known species of the genus Beijerinckia, B. mobilis, is capable of methylotrophic metabolism coupled with the ribulose bisphosphate (RuBP) pathway of C1 assimilation. This is the first report of methylotrophy and autotrophy in Beijerinckia.
Growth of Beijerinckia species on methanol.
We tested five type strains of different Beijerinckia species (B. mobilis DSM 2326T, B. indica subsp. indica ATCC 9039T, B. indica subsp. lacticogenes DSM 1719T, B. derxii subsp. derxii DSM 2328T, and B. derxii subsp. venezuelae DSM 2329T) for their ability to grow on methanol (0.1%, vol/vol) as the sole carbon and energy source in either nitrogen-free half-strength (1:2) liquid mineral medium M1 (9) or in the same medium supplemented with 0.05% (wt/vol) KNO3. For comparison, these strains were grown on the same mineral medium, but with glucose (0.1%, wt/vol) added as the carbon source. Flasks were incubated at 24°C on a rotary shaker at 120 rpm. Growth was monitored by measuring optical density at 600 nm.
All strains of Beijerinckia showed exponential growth on medium with glucose within 2 to 4 days of incubation, while most of these strains failed to grow on the same medium with methanol within 6 weeks. The only Beijerinckia species capable of growth on methanol was B. mobilis. Growth occurred under a wide range of methanol concentrations ranging from 0.01 to 3% (vol/vol). Optimum growth was observed between 0.05 and 0.5% (vol/vol) CH3OH.
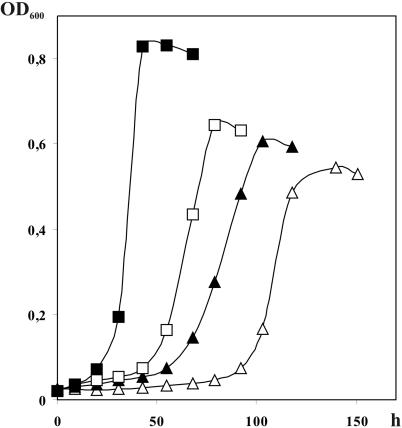
B. mobilis grew on methanol under both nitrogen-fixing and nitrogen-sufficient conditions (Fig. 1). In contrast to growth dynamics of this culture on glucose, the exponential growth on methanol was always preceded by a lag phase of 2 to 4 days. This lag phase was longer for cultures grown on nitrogen-free medium than on nitrate-containing medium. However, the maximum specific growth rates attained on methanol and on glucose did not differ significantly and were 0.054 to 0.072 and 0.071 to 0.087 h−1, respectively. B. mobilis could be maintained continuously on methanol as the sole carbon and energy source without loss of viability.
FIG. 1.
Growth dynamics of B. mobilis in batch cultures on mineral media supplemented with glucose (squares) or with methanol (triangles). Closed squares and triangles indicate growth on nitrogen-sufficient medium (KNO3, 500 mg/liter), while open squares and triangles show growth curves obtained on nitrogen-free medium.
Cell morphology and ultrastructure.
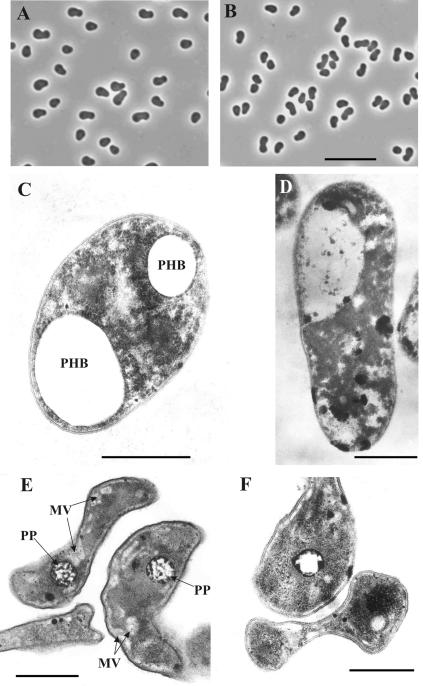
Both glucose- and methanol-grown cells of B. mobilis had the same distinctive bipolar appearance typical for the representatives of this genus, except that the cells grown on methanol were slightly smaller in size (Fig. 2A and B). Thin sections of these cells were prepared and examined using the procedure described by Khmelenina et al. (18). Major differences were observed between cells grown in nitrogen-sufficient and nitrogen-free media (Fig. 2C and D, in comparison to E and F). The cells grown under nitrogen-fixing conditions possessed a highly irregular sinusoidal form, while the cultures grown in nitrogen-sufficient medium were more regular rod-shaped cells with two terminal lipoid bodies (poly-β-hydroxybutyrate spherical inclusions) (Fig. 2C). In general, the cell ultrastructure of B. mobilis grown under nitrogen-fixing conditions was highly similar to the cell ultrastructure described before for acidophilic methanotrophs of the genus Methylocella (10, 13).
FIG. 2.
Phase-contrast micrographs of B. mobilis grown on glucose (A) and on methanol (B) in nitrogen-free medium for 6 days. Bar, 10 μm. Electron micrographs of ultrathin sections of cells of B. mobilis grown on glucose (C and E) and on methanol (D and F) in nitrogen-sufficient (C and D) or nitrogen-free (E and F) medium. MV, membrane vesicles; PHB, poly-β-hydroxybutyrate; PP, granules of polyphosphates. Bars, 0.5 μm.
PCR-mediated screening for mxaF genes in representatives of Beijerinckia and comparative sequence analysis of MxaF from B. mobilis.
The mxaF gene encodes the large subunit of the pyrroloquinoline quinone (PQQ)-linked enzyme methanol dehydrogenase, which catalyzes the oxidation of methanol to formaldehyde in all gram-negative methylotrophic bacteria that have been studied (2, 3). Using primers f1003 and r1561 and the protocol developed by McDonald and Murrell (20), we were able to amplify an approximately 550-bp mxaF gene fragment from DNA of B. mobilis, as well as from DNA of most reference methylotrophic bacteria tested in this study, i.e., Methylobacterium extorquens AM1 (NCIMB 9133), Methylobacterium dichloromethanicum DM4 (DSM 6343T), Methylorhabdus multivorans VKM B-2030T (ATCC 51890), Methylopila capsulata ATCC 700716T, Paracoccus kondratievae NCIMB 13773T, and Methylobacillus glycogenes ATCC 29475T. However, no products were obtained in PCR with DNA from other species of Beijerinckia or with DNA of Albibacter methylovorans DSM 13819T, and two amplicons of incorrect sizes were obtained in PCR with DNA from Methylophaga marina ATCC 35842T. A recent report noted that the same primer set yielded mxaF fragments from only two of four different methylotrophic Afipia species (21), so primers f1003 and r1561 appear to be less universal than previously thought. Thus, to provide further evidence for the absence of mxaF in other species of Beijerinckia, a few novel primers were designed that corresponded to highly conserved regions within an alignment generated for complete mxaF gene sequences of the alphaproteobacterial methylotrophs M. extorquens AM1 (M31108), Methylobacterium organophilum XX (M22629), Methylobacterium nodulans (AF220764), and Paracoccus denitrificans PD1207 (M17339) (numbers in parentheses are EMBL, GenBank, and DDBJ database entries). One newly designed forward primer, mxaF-f769 (5′-TGGGAGGGCGAYGCCTGGAA-3′), and three newly designed reverse primers, mxaF-r1392, mxaF-r1585, and mxaF-r1690 (5′-CTTSGGGCCCGGATACATG-3′, 5′-CTTCCASAGNAGKTCRCCNGTGTC-3′, and 5′-CCCGGCCARCCGCCGAC-3′, respectively) were applied along with the primers f1003 and r1561 in all possible combinations in PCR with template DNA from different species of Beijerinckia and alphaproteobacterial methylotrophic reference strains. Amplicons of the expected size were obtained from DNA of B. mobilis and two strains of Methylobacterium, i.e., M. extorquens AM1 and M. dichloromethanicum DM4, using all primer combinations tested. Amplification of a mxaF fragment from DNA of A. methylovorans was possible only with the primer combination mxaF-f769 and mxaF-r1561. With respect to other alphaproteobacterial methylotrophs tested, the most consistent results were obtained using primers mxaF-f769 and mxaF-r1690. However, neither of these primer combinations yielded a product in PCR with DNA from other Beijerinckia species, i.e., B. indica subsp. indica, B. indica subsp. lacticogenes, B. derxii subsp. derxii, and B. derxii subsp. venezuelae. The mxaF gene amplicons were purified using QIAquick spin columns (QIAGEN) and sequenced on an ABI Prism 377 DNA sequencer (PE Applied Biosystems). Processing of sequence data was performed using the ARB program package (http://www.arb-home.de). Phylogenetic analyses were carried out using the PHYLIP package for phylogenetic inference (14).
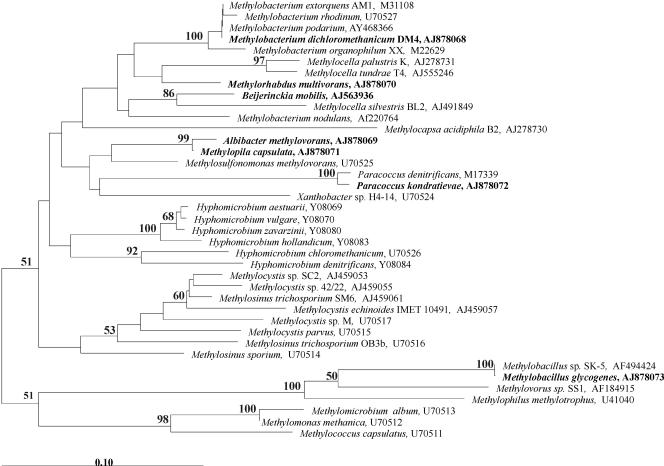
Comparative sequence analysis grouped the inferred peptide sequence of mxaF from B. mobilis within a cluster characterized by representatives of the alphaproteobacterial genera Methylobacterium, Methylocella, and Methylorhabdus (Fig. 3). However, the bootstrap value for the common branch point of this cluster was relatively low (50 to 54%), independently of the method used for the calculation of distance (PAM or Kimura). The identity values between the MxaF sequence of B. mobilis and the MxaF sequences from other members of this cluster ranged from 85.7 to 91.8%, while the highest sequence identity values were observed between MxaF of B. mobilis and those of M. nodulans (91.8%) and Methylocella silvestris (91.3%). The MxaF identity values between B. mobilis and gammaproteobacterial methylotrophs ranged from 74.5 to 79%, while the corresponding identity values to the betaproteobacterial methylotrophs were 73 to 74%.
FIG. 3.
Neighbor-joining tree constructed based on 168 deduced amino acid sites of partial MxaF sequences, showing the position of B. mobilis in relation to other representative methylotrophic organisms of the alpha-, beta-, and gammaproteobacteria. Microorganisms for which sequence data were obtained in this study are indicated in bold. Bootstrap values (1,000 data resamplings) of >50% are shown. Bar, 0.1 substitutions per amino acid position.
Enzymatic profile.
Cell extracts were prepared from cells of B. mobilis grown at 28°C in nitrogen-free medium with 0.2% (vol/vol) methanol or with 1% (vol/vol) glucose using an earlier-described procedure (13). The following enzymes were assayed spectrophotometrically at 30°C in cell extracts using standard methods: methanol dehydrogenase EC 1.1.99.8 (4); formaldehyde dehydrogenase EC 1.2.1.1 and formate dehydrogenase EC 1.2.1.2 (17); transketolase EC 2.2.1.1 and transaldolase EC 2.2.1.2 (8); hydroxypyruvate reductase EC 1.1.1.29 and serine-glyoxylate aminotransferase EC 2.6.1.45 (7); malyl-coenzyme A (CoA) synthetase EC 6.2.1.9 and malyl-CoA lyase EC 4.3.24 (22); and glycerate kinase EC 2.7.1.33 and 3-hexulose phosphate synthase EC 4.1.2.X (15). Specific activities of ribulose bisphosphate carboxylase EC 4.1.1.49, phosphoribulokinase EC 2.7.1.19, and phosphoenolpyruvate carboxylase EC 4.1.1.31 were assayed using a radioisotopic method (19).
The activities of the enzymes potentially involved in primary C1 oxidation and assimilation in B. mobilis are shown in Table 1. This bacterium possessed a PQQ-containing methanol dehydrogenase, which required alkaline pH and NH4+ ions for in vitro activity with phenazine methosulfate (PMS) as an artificial electron acceptor. Two formaldehyde-oxidizing enzymes were present, i.e., NAD(P)-, glutathione (GSH)-dependent formaldehyde dehydrogenase, and the PMS-linked form. Also, the PMS-linked formate dehydrogenase was detected. As expected, these enzyme activities were absent in glucose-grown cells. Thus, B. mobilis appears to possess the complete suite of enzymes required to conduct methanol oxidation to CO2 through formaldehyde and formate and to provide metabolic energy for methylotrophic growth. The presence of the two enzymes unique to the RuBP cycle, i.e., phoshoribulokinase and ribulose-1,5-bisphosphate carboxylase (RubisCO), suggested that the carbon derived from the oxidation of methanol is assimilated at the level of CO2 via the RuBP pathway. This was further confirmed by the finding that the RubisCO activity in methanol-grown cells was 1 order of magnitude higher than that in glucose-grown cells (Table 1). It appears that the RuBP cycle operates with transketolase-transaldolase rearrangements converting glyceraldehyde-3-phosphate to xylulose-5-phosphate to regenerate the primary acceptor of CO2, i.e., ribulose-1,5-bisphosphate. We also observed rather low activities of the serine pathway-specific enzymes, i.e., serine-glyoxylate aminotransferase, NADH-dependent hydroxypyruvate reductase, malyl-CoA synthetase, malyl-CoA lyase, and glycerate kinase. In addition, the activity of phosphoenolpyruvate carboxylase, which is involved in the serine pathway of C1 assimilation and is responsible for heterotrophic CO2 fixation, was enhanced in methanol-grown cells. Thus, at present, we cannot exclude the possibility that the serine pathway plays a minor, supplementary role. In contrast, a key enzyme of the ribulose monophosphate (RuMP) cycle, 3-hexulose phosphate synthase, was not detected in methanol-grown cells, suggesting that the RuMP cycle is not involved in primary C1 assimilation in B. mobilis.
TABLE 1.
Activities of enzymes of primary and intermediate metabolism in cell extracts of B. mobilis grown on glucose or on methanol
| Enzyme | Cofactor(s) | Activity (nmol min−1 mg of protein−1) on:
|
|
|---|---|---|---|
| Methanol | Glucose | ||
| Methanol dehydrogenase | PMS | 27 | 0 |
| Formaldehyde dehydrogenase | PMS | 17 | 0 |
| NAD+, GSH | 210 | 0 | |
| NADP+, GSH | 108 | 0 | |
| Formate dehydrogenase | PMS | 32 | 0 |
| NAD+ | 0 | 0 | |
| Phoshoribulokinase | ATP | 103 | 3 |
| Ribulose-1,5-bisphosphate carboxylase | 160 | 30 | |
| Transketolase | 107 | 111 | |
| Transaldolase | 86 | 94 | |
| 3-Hexulose phosphate synthase | 0 | 0 | |
| Hydroxypyruvate reductase | NADH | 18 | 10 |
| NADPH | 0 | 0 | |
| Serine-glyoxylate aminotransferase | 63 | 40 | |
| Malyl-CoA synthetase/malyl-CoA lyase | ATP, CoA | 49 | 56 |
| Glycerate kinase | ATP | 64 | 40 |
| Phosphoenolpyruvate carboxylase | 103 | 47 | |
Comparative sequence analysis of the cbbL gene from B. mobilis.
Since high RubisCO activity was found in methanol-grown cells of B. mobilis, we made an attempt to detect genes coding for this enzyme in a given organism using three different primer sets developed by Spiridonova et al. (23). Only the primer set proposed for the specific detection of the red-like group of cbbL genes coding for the large subunit of form I RubisCO, i.e., the forward primer RubIrF (5′-GCVACCTGGACSGTSGTVTGG-3′) and the reverse primer RubIrR (5′-TCGCCYTCSAGCTTGCCSAC-3′), yielded an amplicon of the expected size (approximately 820 bp) from DNA of B. mobilis. Comparative sequence analysis grouped the inferred peptide sequence of cbbL from B. mobilis within a red-like cluster of CbbL represented by the alphaproteobacterial organisms of the genera Bradyrhizobium, Methylocapsa, Xanthobacter, and Oligotropha. The identity values between the CbbL sequence of B. mobilis and the CbbL sequences from other members of this cluster ranged from 78.1 to 94.9%. An intriguing fact is that the highest sequence identity (94.9%) was observed between CbbL fragments from B. mobilis and the chemolithoautotrophic bacterium Oligotropha carboxidovorans, which carries cbb genes on a 133,058-bp self-transmissible megaplasmid, pHCG3 (16). No product was obtained in PCR with cbb-specific primers and template DNA from other Beijerinckia species, i.e., B. indica subsp. indica, B. indica subsp. lacticogenes, B. derxii subsp. derxii, and B. derxii subsp. venezuelae.
Our results demonstrate that, in addition to chemoheterotrophy, B. mobilis is able to employ chemoautotrophy as an alternative type of nutrition. Autotrophic growth is driven by energy from the oxidation of methanol via a PQQ-containing methanol dehydrogenase, an NAD+/GSH-linked formaldehyde dehydrogenase, and a PMS-linked formate dehydrogenase, while CO2 derived from oxidation of CH3OH is further assimilated via the RuBP pathway. Therefore, B. mobilis is a facultatively chemoautotrophic methylotroph that in addition to growth on C1 reduced compounds (methanol and formate) has the ability to use a wide range of multicarbon substrates.
Two explanations for the origin of the methylotrophic autotrophy in B. mobilis can be considered. One explanation would involve facilitated, plasmid-mediated genetic exchange. This would explain why the ability to oxidize methanol and the ability to assimilate CO2 via the RuBP pathway were found in only one species of the genus Beijerinckia. Another explanation would be the occurrence of a common acidophilic, dinitrogen-fixing, autotrophic ancestor of Methylocapsa and Beijerinckia, which has been suggested previously due to the high identity of nif genes in these bacteria (12). It should be noted that the initial examination of C1 assimilation pathways in Methylocapsa acidiphila B2 did not reveal the presence of the RuBP pathway (11). However, Spiridonova et al. (23) recently reported the detection of a red-like cbbL gene in strain B2. Thus, the occurrence of a RuBP pathway in Methylocapsa needs to be verified under different growth conditions.
In summary, we have demonstrated that there is more metabolic versatility within representatives of the genus Beijerinckia than was previously thought. At present, methylotrophic autotrophy can be attributed to only one species of this genus. However, future studies on unexplored diversity within this genus may significantly extend the number of Beijerinckia species capable of C1 metabolism.
Nucleotide sequence accession number.
The nucleotide sequence of the cbbL gene fragment from B. mobilis has been determined and deposited in the EMBL, GenBank, and DDBJ sequence databases under the accession number AJ878074.
Acknowledgments
This research was supported in part by the Russian Fund of Basic Research (grant no. 04-04-04000), the Program “Molecular and Cell Biology” of the Russian Academy of Sciences, and the Deutsche Forschungsgemeinschaft (436 RUS 113/543/0-3).
We are grateful to N. V. Doronina, who kindly provided a set of reference methylotrophic strains for this study.
REFERENCES
- 1.Alston, R. A. 1936. Studies on Azotobacter in Malayan soils. J. Agric. Sci. 26:268-280. [Google Scholar]
- 2.Anthony, C. 1982. The biochemistry of methylotrophs. Academic Press, Inc. Ltd., London, United Kingdom.
- 3.Anthony, C., and P. Williams. 2003. The structure and mechanism of methanol dehydrogenase. Biochim. Biophys. Acta 1647:18-23. [DOI] [PubMed] [Google Scholar]
- 4.Anthony, C., and L. J. Zatman. 1964. The microbial oxidation of methanol. The methanol-oxidizing enzyme of Pseudomonas sp. M27. Biochem. J. 92:614-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becking, J. H. 1961. Studies on nitrogen-fixing bacteria of the genus Beijerinckia. I. Geographical and ecological distribution in soils. Plant Soil 14:49-81. [Google Scholar]
- 6.Becking, J. H. 1999. The genus Beijerinckia. In M. Dworkin et al. (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.0, 21 May 1999. Springer-Verlag, New York, N.Y. [Online.] http://141.150.157.117:8080/prokPUB/index.htm.
- 7.Blackmore, M. A., and J. R. Quayle. 1970. Microbial growth on oxalate by a route not involving glyoxylate carboligase. Biochem. J. 118:53-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colby, J., and L. J. Zatman. 1975. Enzymological aspects of the pathways for trimethylamine oxidation and C1-assimilation in obligate methylotrophs. Biochem. J. 148:513-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dedysh, S. N., N. S. Panikov, and J. M. Tiedje. 1998. Acidophilic methanotrophic communities from Sphagnum peat bogs. Appl. Environ. Microbiol. 64:922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dedysh, S. N., W. Liesack, V. N. Khmelenina, N. E. Suzina, Y. A. Trotsenko, J. D. Semrau, A. M. Bares, N. S. Panikov, and J. M. Tiedje. 2000. Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bogs, representing a novel subtype of serine-pathway methanotrophs. Int. J. Syst. Evol. Microbiol. 50:955-969. [DOI] [PubMed] [Google Scholar]
- 11.Dedysh, S. N., V. N. Khmelenina, N. E. Suzina, Y. A. Trotsenko, J. D. Semrau, W. Liesack, and J. M. Tiedje. 2002. Methylocapsa acidiphila gen. nov., sp. nov., a novel methane-oxidizing and dinitrogen-fixing acidophilic bacterium from Sphagnum bog. Int. J. Syst. Evol. Microbiol. 52:251-261. [DOI] [PubMed] [Google Scholar]
- 12.Dedysh, S. N., P. Ricke, and W. Liesack. 2004. NifH and NifD phylogenies: an evolutionary basis for understanding nitrogen fixation capabilities of methanotrophic bacteria. Microbiology 150:1301-1313. [DOI] [PubMed] [Google Scholar]
- 13.Dunfield, P. F., V. N. Khmelenina, N. E. Suzina, Y. A. Trotsenko, and S. N. Dedysh. 2003. Methylocella silvestris sp. nov., a novel methanotroph isolated from an acidic forest cambisol. Int. J. Syst. Evol. Microbiol. 53:1231-1239. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 15.Ferenci, T., T. Strom, and J. R. Quayle. 1974. Purification and properties of 3-hexulose phosphate synthase phospho-3-hexuloisomerase from Methylococcus capsulatus. Biochem. J. 144:477-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuhrmann, S., M. Ferner, T. Jeffke, A. Henne, G. Gottschalk, and O. Meyer. 2003. Complete nucleotide sequence of the circular megaplasmid pHCG3 of Oligotropha carboxidovorans: function in the chemolithoautotrophic utilization of CO, H2 and CO2. Gene 322:67-75. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, P. A., and J. R. Quayle. 1964. Microbial growth on C1-compounds. Oxidation of methanol, formaldehyde and formate of methanol-grown Pseudomonas AM1. Biochem. J. 93:281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khmelenina, V. N., M. Kalyuzhnaya, N. E. Suzina, Y. A. Trotsenko, and G. Gottschalk. 1999. Osmoadaptation in halophilic and alkaliphilic methanotrophs. Arch. Microbiol. 172:321-329. [DOI] [PubMed] [Google Scholar]
- 19.Loginova, N. V., B. B. Namsaraev, and Y. A. Trotsenko. 1978. Autotrophic metabolism in Microcyclus aquaticus. Microbiology 47:168-170. [PubMed] [Google Scholar]
- 20.McDonald, I. R., and J. C. Murrell. 1997. The methanol dehydrogenase structural gene mxaF and its use as a functional gene probe for methanotrophs and methylotrophs. Appl. Environ. Microbiol. 63:3218-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moosvi, S. A., C. C. Pacheco, I. R. McDonald, P. De Marco, D. A. Pearce, D. P. Kelly, and A. P. Wood. 2005. Isolation and properties of methanesulfonate-degrading Afipia felis from Antarctica and comparison with other strains of A. felis. Environ. Microbiol. 7:22-33. [DOI] [PubMed] [Google Scholar]
- 22.Salem, A. R., A. J. Hacking, and J. R. Quayle. 1973. Cleavage of malyl-coenzyme A into acetyl-coenzyme A and glyoxylate by Pseudomonas AM1 and other C1-unit-utilizing bacteria. Biochem. J. 136:89-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spiridonova, E. M., I. A. Berg, T. V. Kolganova, R. N. Ivanovsky, B. B. Kuznetsov, and T. P. Tourova. 2004. An oligonucleotide primer system for amplification of the ribulose-1,5-bisphosphate carboxylase/oxygenase genes of bacteria of various taxonomic groups. Microbiology 73:316-325. [English translation of Mikrobiologiya 73: 377-387.] [PubMed] [Google Scholar]
- 24.Starkey, R. L., and P. K. De. 1939. A new species of Azotobacter. Soil Sci. 47:329-343. [Google Scholar]