Abstract
Analysis of unidirectional genomic deletion events and single nucleotide variations suggested that the four subspecies of Francisella tularensis have evolved by vertical descent. The analysis indicated an evolutionary scenario where the highly virulent F. tularensis subsp. tularensis (type A) appeared before the less virulent F. tularensis subsp. holarctica (type B). Compared to their virulent progenitors, attenuated strains of F. tularensis exhibited specific unidirectional gene losses.
Francisella tularensis, the causative agent of tularemia, is considered a potential bioterrorist agent. Important premises are an extremely low infectious dose and a potential for airborne transmission (6). The two most clinically important entities of tularemia, type A and type B, correspond to the highly virulent subspecies F. tularensis subsp. tularensis and the moderately virulent F. tularensis subsp. holarctica, respectively (18, 24). Taxonomic work has identified two additional subspecies, F. tularensis subsp. mediasiatica, exhibiting a moderate virulence, and “F. tularensis subsp. novicida,” with a low virulence in animals and humans (24). The latter subspecies has less-fastidious extracellular growth requirements than the other subspecies and a distinct lipopolysaccharide O-antigen (10, 29).
Based on small subunit RNA sequences, F. tularensis is classified as a member of the γ-subgroup of proteobacteria (24). The two species F. tularensis and F. philomiragia and in addition a number of more recently identified tick endosymbionts are the only members of the genus Francisella, which diverges deeply among the γ-proteobacteria (22, 27).
Assuming that F. tularensis subspecies have evolved from a common ancestor, identification of genetic differences among subspecies might provide insights into species phylogeny and provides a basis for studies of more functional issues. Although exhibiting differences in virulence, geographical distribution, and a few biochemical tests, F. tularensis subspecies are highly similar in gene content (1). Mapping of genetic differences will allow future exploration of their relationships to functional correlates. In a recent DNA-based study using highly mutable variable-number tandem repeat (VNTR) sequences dispersed over the genome, F. tularensis subsp. tularensis (type A) showed more diversity than F. tularensis subsp. holarctica (type B), suggesting the former subspecies to be evolutionarily older (11). Significant linkage disequilibrium was detected among VNTR loci of F. tularensis, consistent with a predominantly clonal population structure.
The present work was based on findings from a whole-genome microarray study of multiple F. tularensis strains (1). The microarray study identified large size regions of difference (RDs) among F. tularensis strains. Notably, a pair of direct repeated sequences flanked seven of eight identified RDs. We hypothesized that the presence of direct repeats in a genomic region would represent a propensity for interstrain variation of that region. Therefore, direct repeated sequences were searched for in the genome sequence of strain SCHU S4, and when identified, the corresponding genomic regions were assayed for variability among various strains of F. tularensis. In parallel, single nucleotide variations (SNVs) were analyzed by sequencing of internal fragments of genes of F. tularensis and related bacteria of the γ-subgroup of proteobacteria.
Bacteria and identification of genomic regions with direct repeats.
Criteria for including F. tularensis strains in this study were to ensure maximum spatial and temporal diversity. Forty-five isolates (Table 1) representing the four F. tularensis subspecies were selected from the Francisella Strain Collection (FSC) in Umeå, Sweden. Bacteria were grown on modified Thayer-Martin agar. Cells were suspended into phosphate-buffered saline and heat killed. DNA was prepared using silica and guanidine isothiocyanate buffer as described previously (23).
TABLE 1.
Presence or absence of 17 regions of difference in 45 F. tularensis strains
| Species and isolate information (no. of isolates) | FSC no. | Alternative strain designation | RDa
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1a | 1b | 1c | 2 | 3 | 4 | 5 | 6 | 7 | 11 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |||
| Francisella tularensis subsp. tularensis (12) | |||||||||||||||||||
| Human ulcer, 1941, Ohio | 237 | SCHU S4 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Human ulcer, 1941, Ohio | 043 | SCHU, avirulent | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + |
| Unknown, 1960, Eigelsbach | 013 | FAM standard | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Squirrel, Georgia | 033 | SnMF | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Tick, 1935, B. C., Canada | 041 | Vavenby | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Hare, Canada | 042 | Utter | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Human pleural fluid, 1940, Ohio | 046 | Fox Downs | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Rabbit, 1953, Nevada | 054 | Nevada 14 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Lab acquired when handling Nevada 14 | 053 | F. tul AC | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Mite, 1988, Slovakia | 198 | SE-219/38 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Mite, 1988, Slovakia | 199 | SE-221/38 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Human lymph node, 1920, Utah | 230 | ATCC6223 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Francisella tularensis subsp. mediasiatica (4) | |||||||||||||||||||
| Experimental isolate, cap- | 122 | (TTC-R)6-4-1 | + | + | − | + | + | + | + | + | + | + | + | − | + | + | − | + | + |
| Miday gerbil, 1965, Kazakhstan | 147 | GIEM 543 | + | + | − | + | + | + | + | + | + | + | + | − | + | + | − | + | + |
| Ticks, 1982, Central Asia | 148 | 240 | + | + | − | + | + | + | + | + | + | + | + | − | + | + | − | + | + |
| Hare, 1965, Central Asia | 149 | 120 | + | + | − | + | + | + | + | + | + | + | + | − | + | + | − | + | + |
| Francisella tularensis subsp. holarctica (from Eurasia and North America) (24) | |||||||||||||||||||
| Tick, 1941, Montana | 012 | 425F4G | + | − | + | − | − | − | − | − | − | − | − | − | + | + | + | + | + |
| Beaver, 1976, Hamilton, Montana | 035 | B423A | + | − | + | − | − | − | − | − | − | − | − | − | + | + | + | + | + |
| Human ulcer, 1999, Karlstad, Sweden | 236 | + | − | + | − | − | − | − | − | − | − | − | − | + | + | + | + | + | |
| Human, Vosges, France | 247 | T 20 | + | − | + | − | − | − | − | − | − | − | − | − | + | + | + | + | + |
| Tick, 1949, Moscow area, Russia | 257 | GIEM 503/840 | + | − | + | − | − | − | − | − | − | − | − | − | + | + | + | + | + |
| Human, 2000, Pyhäjärvi, Finland | 286 | + | − | + | − | − | − | − | − | − | − | − | − | + | + | + | + | + | |
| Unknown, 1960, Eigelsbach | 014 | FAM SR | + | − | + | − | − | − | − | − | − | − | − | − | + | − | + | + | + |
| Human, 1994, Norway | 158 | CCUG 33391 | + | − | + | − | − | − | − | − | − | − | − | − | ± | − | + | + | + |
| Hare, 1974, Nås (W), Sweden | 074 | SVA T7 | + | − | + | − | − | − | − | − | − | − | − | − | − | ± | + | + | + |
| Passage in small mammals of FSC074 | 069 | SVA T7K | + | − | + | − | − | − | − | − | − | − | − | − | − | + | + | + | + |
| Live vaccine strain, Russia | 155 | LVS | + | − | + | − | − | − | − | − | − | − | − | − | − | − | + | + | D |
| Vaccine strain from Russia | 338 | Strain 015 | + | − | + | − | − | − | − | − | − | − | − | − | − | − | + | + | D |
| Water, 1991, Elista, Kalmykiya, Russia | 118 | 14687 | + | − | + | − | − | − | − | − | − | − | − | − | + | + | + | + | D |
| Water, 1985, Rostov region, Russia | 121 | 12267 | + | − | + | − | − | − | − | − | − | − | − | − | + | + | + | + | D |
| Norwegian rat, 1988, Russia | 150 | 250 | + | − | + | − | − | − | − | − | − | − | − | − | + | + | + | + | D |
| Water, 1988, Rostov, Russia | 151 | 25, A13863 | + | − | + | − | − | − | − | − | − | − | − | − | + | + | + | + | D |
| Human, 1995, Ockelbo, Sweden | 178 | R39/95 | + | − | + | − | − | − | − | − | − | − | − | − | + | + | + | + | D |
| Tick, 1995, Lanzhot, Czech Republic | 180 | T-17 | + | − | + | − | − | − | − | − | − | − | − | − | + | + | + | + | D |
| Mite, 1988, Slovakia | 196 | SE-210/37 | + | − | + | − | − | − | − | − | − | − | − | − | + | + | + | + | D |
| Human, 1998, Ljusdal, Sweden | 200 | + | − | + | − | − | − | − | − | − | − | − | − | + | + | + | + | D | |
| Human, 1995, Äänekoski, Finland | 249 | + | − | + | − | − | − | − | − | − | − | − | − | + | + | + | + | D | |
| Water, 1991, Odessa region, Ukraine | 116 | 14670 | + | − | + | − | − | − | − | − | − | − | − | − | + | + | + | + | T |
| Water, 1990, Odessa region, Ukraine | 124 | 14588 | + | − | + | − | − | − | − | − | − | − | − | − | + | + | + | + | T |
| Human blood, 1994, Örebro, Sweden | 157 | CCUG 33270 | + | − | + | − | − | − | − | − | − | − | − | − | + | + | + | + | T |
| Francisella tularensis subsp. holarctica (from Japan) (4) | |||||||||||||||||||
| Human, 1958, Japan | 021 | Tsuchiya | − | + | + | − | − | + | − | − | + | − | N | − | + | + | + | + | + |
| Human, 1950, Japan | 022 | Ebina | − | + | + | − | − | + | − | − | + | − | N | − | + | + | + | − | + |
| Human lymph node, 1926, Japan | 017 | S-2 | − | + | + | − | − | + | − | − | + | − | N | − | + | + | + | − | + |
| Tick, 1957, Japan | 075 | Jama | − | + | + | − | − | + | − | − | + | − | N | − | + | + | + | − | D |
| Francisella tularensis subsp. novicida (1) | |||||||||||||||||||
| Water, 1950, Utah | 040 | ATCC 15482 | I | I | I | + | + | + | N | + | N | + | N | + | + | + | N | + | + |
The symbol + indicates presence and the symbol − indicates absence of a particular region of difference relative SCHU S4. The symbol ± indicates that the strain is heterogeneous at the particular RD. The letters D or T indicate a duplication or triplication, respectively. N, not detected; I, insertion of a unique sequence (1). Regions used in deletion-based phylogenetic analysis were 1c, 2, 3, 4, 5, 6, 7, 11, and 16. Boldface letters indicate strains subjected to SNV analysis.
A Perl script (detailed in the supplemental text) was devised to search for direct repeats in the, at that time, unfinished genome sequence of F. tularensis strain SCHU S4 (FSC237). The completed genome sequence is assigned accession no. AJ749949 in GenBank (13). More than 70 genomic regions flanked by direct repeats were evaluated by PCR in a subset of five F. tularensis isolates that represented each of the four F. tularensis subspecies and the Japanese F. tularensis subsp. holarctica variant. Genomic regions that exhibited size polymorphism on agarose gels were further analyzed in 45 strains (Table 1). PCR amplicons were sequenced to identify the junctions of the deletion/insertion events. Primer sequences for PCR amplification were posted in the supplemental material in Table S1.
Use of unidirectional deletions for phylogenetic analysis.
The use of RDs for phylogenetic analysis relies on an assumption of unidirectional deletion events that eventually become fixed in bacterial populations (phylogenetic lineages) (2, 14). An RD was used for depicting evolution only when it was found to represent a deletion and not an insertion. Three criteria were used for excluding RDs from use in phylogenetic analysis. (i) The first criterion was whether the corresponding region in the SCHU S4 genome exhibited three or more closely localized direct repeats. We reasoned that the presence of more than two repeats might indicate an evolutionary history of genome amplification. Therefore, the SCHU S4 genome was scrutinized for repeats present in the vicinity of RDs (±10,000 bp). For a review of direct repeat mediated excision or amplification in bacteria, see references 19 and 20. (ii) The second criterion was whether the RD was flanked by multicopy insertion sequence elements. This criterion was based on an assumption that the genomic region is potentially prone to recombination events within and among taxa in phylogenetic lineages. (iii) The third criterion was whether the RD showed polymorphism within a subspecies.
SNVs and calculation of phylogenetic trees.
A subset of 15 F. tularensis strains was selected for analysis of SNVs (Table 1). Additionally, DNA was isolated from a freeze-dried ampoule of the type strain of the tick endosymbiont Wolbachia persica (ATCC VR-331) as described previously (23). A set of seven genes was selected, previously used in multiple locus sequence typing of various bacteria: aroA, parC, pgm, tpiA, trpE, atpA, and uup (28). Internal fragments of the genes were PCR amplified. Primer sequences for PCR amplification were posted in supplemental material in Table S1. Amplification was successful for seven gene fragments from each F. tularensis strain and four gene fragments from W. persica (aroA, parC, tpiA, and atpA). PCR amplicons were sequenced on both strands using the amplification primers and standard protocol on an ABI 377 platform (PE Applied Biosystems). Sequences of gene orthologs in the genomes of Yersinia pestis (CO92) and Agrobacterium tumefaciens (C58) were retrieved from GenBank. Phylogenetic analyses were performed using concatenated nucleotide sequences, individual gene fragments, and concatenated in-frame-translated amino acid sequences. Maximum likelihood trees were reconstructed from DNA sequences by using PAUP v. 4.0b10 (25) as described previously (7). The extent of pairwise congruence between individual gene trees was estimated by a method comparing the likelihood values calculated for single gene trees to the values for other gene trees and to 200 randomly generated trees of the same size (7). Congruence of gene trees indicates a common evolutionary history of investigated genes. Phylogenetic reconstruction from amino acid sequences was performed using PHYLIP (v. 3.6b). Phylogenetic methods are further detailed in the supplemental text.
Analysis of RDs among F. tularensis strains.
PCR analysis of the genomes of 45 F. tularensis isolates identified 17 RDs in F. tularensis (Table 1). Generally, PCR amplification showed the presence of one of two alternate fragment sizes for each RD and isolate. The pattern of amplification allowed an immediate subspecies identification. With very few exceptions, all isolates of a subspecies showed the same fragment size for each RD. A polymorphism was present at RD21-22 among isolates of a subspecies and at RD18-19 among variants of a strain.
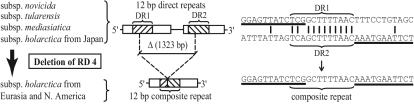
F. tularensis genomes showing alternate PCR fragment sizes were sequenced across each RD. Alignment of sequences for each RD showed the smaller-sized PCR fragments to contain only one direct repeat and to be missing a large genomic segment compared to the SCHU S4 sequence. The boundary of the missing sequence was defined by the two direct repeats (Fig. 1). Nucleotide degeneracy of flanking repeats allowed further analysis. We found each single repeat present in a smaller PCR fragment to be a composite repeat derived from fusion of a left and right flanking repeat (Fig. 1; also Fig. S1 in the supplemental material). The finding is consistent with a deletion mechanism mediated by recombination of flanking repeats followed by excision of the intervening sequence. The deletion will be irreversible if two direct repeats were originally present and a single composite repeat is left after excision. As an alternative, two direct repeats may mediate genome amplification of intervening sequence as demonstrated for RD22 (Table 1). At this locus, sequence analysis showed duplication (12 isolates) or triplication (3 isolates) of the region flanked by repeats (sequence data not shown).
FIG. 1.
Unidirectional repeat-mediated deletion mechanism exemplified by RD4. The symbol Δ represents deleted sequence. Flanking direct repeats (DR1 and DR2) and the resulting composite repeat are depicted as hatched bars. Open bars represent open reading frames that were truncated by the deletion. Partial nucleotide sequences are shown before (DR1 and DR2) and after (composite repeat) the deletion event and allowed analysis of evolutionary direction.
Genes were truncated in isolates with smaller-sized PCR fragments at 16/17 RDs (Table 2; also Table S2 in the supplemental material). Only RD1c represents an intergenic sequence. RD11 and RD16-22 were identified and characterized de novo in this study and found to affect 12 open reading frames in the F. tularensis SCHU S4 genome (Table 2). Gene content of RD1-7 has been previously described (1) and was further characterized in this study (Table S2 in the supplemental material). Among deleted genes, some require discussion with regard to virulence properties of various isolates. The gene pdpD of RD6 was recently suggested to play a role in F. tularensis virulence (15). In concordance to previous analysis of a smaller number of isolates, we confirmed the gene to be deleted in all type B isolates including the live vaccine strain (1, 15). Genes in RD18 and RD19 discriminated among isolates of individual subspecies. Genes in RD19 represent type IV pili building block proteins that were absent in, e.g., the live vaccine strain of F. tularensis. Homologous genes have been implied as virulence determinants in other bacteria (5). Finally, genes in RD18 represent genes of a novel protein family unique to F. tularensis (13). The latter genes were found to be deleted in an avirulent phenotype of strain SCHU (FSC043) and in the live vaccine strain of F. tularensis.
TABLE 2.
Gene content of RDs of F. tularensisa
| RD | Genomic position of deletionb(size in bp) | Gene comprised by deletion | Genome position of flanking direct repeatsb |
|---|---|---|---|
| RD11c | 551874-552722 (849) | FTT0531, ABC transporter, ATP-binding protein, pseudogene | 551856-551879 |
| 552705-552728 | |||
| RD16 | 1664669-1665013 (345) | FTT1598, hypothetical membrane protein | 1664591-1664721 |
| 1664936-1665066 | |||
| RD17 | 1535418-1535717 (300) | aceF, FTT1484, pyruvate dehydrogenase, E2 component | 1535696-1535923 |
| 1535396-1535623 | |||
| RD18c | 928556-930035 (1,480) | FTT0918, hypothetical protein | 928215-928753 |
| FTT0919, hypothetical protein | 929698-930236 | ||
| RD19c | 898418-898956 (539) | FTT0890, type IV pili fiber building block protein | 898901-899020 |
| FTT0889, type IV pili fiber building block protein | 898363-898482 | ||
| RD20 | 546921-547446 (526) | FTT0524, conserved hypothetical protein | 546916-546939 |
| 547442-547465 | |||
| RD21 | 124728-125948 (1,221) | nupC1, FTT0116, nucleoside permease NUP family protein | 123791-125005 |
| nupC2, FTT0115, nucleoside permease NUP family protein | 125009-126233 | ||
| RD22 | 992350-992819 (470) | FTT0980, aminotransferase, class II | 992250-992349 |
| FTT0981, hypothetical protein | 992720-992819 |
Genes comprised by deletion at RD1-RD7 have been described by Broekhuijsen et al. (1). Details of RD1-RD7 have been posted as supplemental material (Table S2).
Position and gene name refer to the genome sequence of F. tularensis subsp. tularensis, strain SCHU S4 (13). Boundaries of repeats were defined by use of a threshold value set at >65% nucleotide identity.
Region of difference independently identified by Samrakandi et al. (21). RD18 and RD19 correspond to L3 and L2, respectively.
A deletion-based phylogeny of F. tularensis.
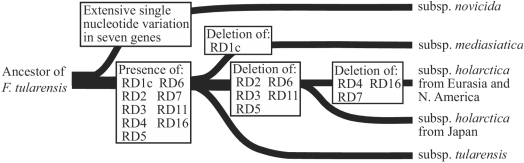
Nine RDs fulfilled our strict selection criteria for a unidirectional genetic event (see above) and were included in the deletion-based phylogenetic analysis (Table 1 and Fig. 2). An evolutionary scenario is suggested where successive losses of genetic material at RDs parallel the taxonomic units of F. tularensis at the subspecies level. The analysis proposes an evolution of F. tularensis where the highly virulent F. tularensis subsp. tularensis (type A) preceded the appearance of the less virulent F. tularensis subsp. holarctica (type B). Our findings partially contrast to the evolution of Y. pestis that apparently became more virulent while losing genetic material in the transition from its ancestor, Yersinia pseudotuberculosis (3). The evolutionary transitions of F. tularensis subspecies (F. tularensis subsp. novicida, subsp. tularensis, and subsp. holarctica) are more complex and apparently involve both acquisition and loss of virulence since F. tularensis subsp. novicida exhibits the lowest virulence and was found to be evolutionarily oldest. Work on other pathogens has shown that the evolution of virulence, in particular the adaptation to living within eukaryotic hosts, may involve both acquisition and loss of genes (16), a fact that may explain the observations.
FIG. 2.
Evolutionary scenario of F. tularensis based on deletion characters assuming a successive loss of genetic material. The scheme reads from left (F. tularensis ancestor) to right. F. tularensis subspecies are indicated. The horizontal distance may not correspond to actual phylogenetic distances calculated by other methods.
All nine RDs were present in all F. tularensis subsp. tularensis isolates. The deletion data show isolates of F. tularensis subsp. mediasiatica to be very similar to isolates of F. tularensis subsp. tularensis. Only RD1c showed a deletion event unique for F. tularensis subsp. mediasiatica isolates. Therefore, the branching order of F. tularensis subsp. mediasiatica and F. tularensis subsp. tularensis remains unclear. Isolates from Japan of F. tularensis subsp. holarctica showed deletion at six RDs. Two additional deletions at RDs were found in Eurasian and North American F. tularensis subsp. holarctica isolates. This suggests that Japanese isolates are evolutionary intermediates between the highly virulent type A (F. tularensis subsp. tularensis) isolates and the less virulent Eurasian/North American type B isolates (F. tularensis subsp. holarctica). The single isolate of F. tularensis subsp. novicida was chosen for rooting, based on the most extensive number of SNVs in seven gene fragments (see below). The deletion analysis per se could not place F. tularensis subsp. novicida due to failure of PCR amplification of three RDs (RD5, RD7, and RD16). Notably, none of 17 RDs showed evidence of deletion events in F. tularensis subsp. novicida. The evolutionary direction might also be inferred from analysis of the gene content of RDs. The finding that 16/17 deletions truncate conserved genes (genes with homologs in other bacteria) implies deletions but not insertions have occurred during evolution. (Table 2; also Table S2 in the supplemental material). Because deletions are expected to represent unidirectional genetic events, RDs of F. tularensis are suggested to be valuable evolutionary markers. In fact, if all RDs with a uniform pattern within a subspecies were used in phylogenetic analysis the depicted evolutionary scenario would not change (data not shown).
A phylogeny inferred from analysis of SNVs.
Compared to other genomic characters, SNVs exhibit slow mutation rates, making them valuable for phylogenetic analysis (9, 17). We analyzed seven housekeeping gene fragments (3,135 bp) in 15 F. tularensis isolates and four gene fragments (1,830 bp) of the tick endosymbiont W. persica. A total of 78 SNVs was detected among 15 F. tularensis isolates (see Fig. S2 in the supplemental material). The single isolate of F. tularensis subsp. novicida was the most divergent, showing 41 unique SNVs. Overall, the sample of worldwide F. tularensis isolates exhibited a low level of average pairwise sequence diversity (π) with values in the range of 0.0026 to 0.0101 for individual genes. The pattern of SNVs allowed a straightforward subspecies recognition, each F. tularensis subspecies being identified by several fixed polymorphisms. There was an extensive sequence similarity among isolates of a F. tularensis subspecies.
Four-gene phylogenetic trees were computed to describe the phylogenetic position of F. tularensis and its four subspecies among the gamma proteobacteria (Fig. 3). To root the phylogeny, we used A. tumefaciens as outgroup, as the α-subgroup is considered ancient among gamma proteobacteria (8). Y. pestis of the γ-subgroup of proteobacteria was included as a second outgroup. In the phylogeny obtained, the tick endosymbiont W. persica is the first and deepest branch, thus representing an early divergence from a common Francisella ancestor. Among the four F. tularensis subspecies, F. tularensis subsp. novicida appeared to represent the first branching. The result is in agreement to findings of a greater amount of genetic divergence among tick endosymbionts or isolates of F. tularensis subsp. novicida compared to isolates of three other F. tularensis subspecies (4, 12, 22). A greater diversity among isolates likely represents a longer evolutionary history. The four-gene trees demonstrated the split between F. tularensis subsp. tularensis/mediasiatica on one hand and F. tularensis subsp. holarctica isolates from Japan/Eurasia/North America on the other hand. Seven-gene trees illustrated the position of Japanese F. tularensis subsp. holarctica isolates close to but distinct from Eurasian/North American F. tularensis subsp. holarctica isolates (Fig. 3). The five F. tularensis subsp. tularensis isolates were grouped into two distinct clades. Overall, the present SNV and deletion-based phylogenies support the genetic relationships of F. tularensis subspecies calculated from VNTR data, including a genetic division of type A isolates into two distinct clades recently designated A.I and A.II, respectively (Fig. 2 and Fig. 3) (11).
FIG. 3.
Phylogenetic analysis of F. tularensis using a maximum-likelihood algorithm and based on concatenated sequences of gene fragments. A. Four-gene tree constructed using amino acid sequences (631 amino acids). The tree was rooted using A. tumefaciens and Y. pestis as outgroups. B. Seven-gene analysis using nucleotide sequences (3,135 bp). The tree was rooted using F. tularensis subsp. novicida as outgroup. F. tularensis subspecies and the division of type A isolates into two clades (A.I, A.II) are indicated. Bootstrap values from 1,000 replications are indicated at interior branch nodes (values of <50% not shown).
The pairwise statistical comparisons of congruence between gene trees indicated that recombination across F. tularensis subspecies occurs rarely or not at all and justifies the use of concatenated gene sequences for tree reconstruction (data not shown). Based on the pattern of deletions and SNVs among isolates, we conclude that the distribution of fixed characters unique to F. tularensis subspecies reflects a genetic separation of subspecies and also that the subspecies have descended clonally.
Deletions that are specific to laboratory strains.
Cell populations within a single F. tularensis strain might carry unique unidirectional deletions, i.e., at RD18-19 (Table 1). The finding of unique deletion characters among bacterial colonies derived from a single bacterial seed stock suggests that regions flanked by direct repeats are prone to mutation in F. tularensis genomes. Under natural evolution, it appears that direct repeat-mediated deletions only rarely became fixed, as illustrated in our analysis across F. tularensis subspecies. According to our results, the majority of observed deletions might be regarded as evolutionary footprints in F. tularensis genomes spanning over wide evolutionary time scales. In contrast, it seems that laboratory culture on artificial media facilitates the fixation of deletion variants. We suggest that deletion at RD18-19 might represent genetic events that occurred at the laboratory during passage or storage of F. tularensis strains. This is supported by the finding of a mix of two distinct cell populations that exhibit these RDs in laboratory stocks of strains FSC074 (RD19) and FSC158 (RD18) (Table 1). Based on sequence analysis, direct repeat-mediated excisions at RD18 and RD19 have occurred also in the genome of the live vaccine strain of F. tularensis. The live vaccine strain was derived during repeated passages on artificial media (26). Thus, it seems plausible that the fixation of a deletion variant (the live vaccine strain) was facilitated by a repeated selection of single colonies during laboratory passages. It remains to be determined if genes located in RD18 and/or RD19 have a role in the attenuation of the live vaccine strain, the Russian vaccine strain 015 (FSC338), and the avirulent phenotype of the SCHU strain (FSC043). All virulence-attenuated strains included in this study showed deletion at RD18 and/or RD19.
Nucleotide sequence accession numbers.
All sequences reported in this paper have been deposited in the GenBank database with accession no. AY794406 to AY794434 (RD2 to RD7, RD11, RD16-22) and AY794435 to AY794543 (SNVs in seven gene fragments). Sequences of RD1 are assigned accession no. AF469614 to AF469619 (1).
Supplementary Material
Acknowledgments
This work was supported by funding from the Swedish MoD, project no. A4854, the Swedish Medical Research Council, Formas, Sweden, and the Medical Faculty, Umeå University, Umeå, Sweden.
We are indebted to numerous scientific colleagues for kindly providing strains to the Francisella Strain Collection in Umeå, Sweden.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Broekhuijsen, M., P. Larsson, A. Johansson, M. Byström, U. Eriksson, E. Larsson, R. G. Prior, A. Sjöstedt, R. W. Titball, and M. Forsman. 2003. Genome-wide DNA microarray analysis of Francisella tularensis strains demonstrates extensive genetic conservation within the species but identifies regions that are unique to the highly virulent F. tularensis subsp. tularensis. J. Clin. Microbiol. 41:2924-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chain, P. S., E. Carniel, F. W. Larimer, J. Lamerdin, P. O. Stoutland, W. M. Regala, A. M. Georgescu, L. M. Vergez, M. L. Land, V. L. Motin, R. R. Brubaker, J. Fowler, J. Hinnebusch, M. Marceau, C. Medigue, M. Simonet, V. Chenal-Francisque, B. Souza, D. Dacheux, J. M. Elliott, A. Derbise, L. J. Hauser, and E. Garcia. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 101:13826-13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarridge, J. E., III, T. J. Raich, A. Sjöstedt, G. Sandström, R. O. Darouiche, R. M. Shawar, P. R. Georghiou, C. Osting, and L. Vo. 1996. Characterization of two unusual clinically significant Francisella strains. J. Clin. Microbiol. 34:1995-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craig, L., M. E. Pique, and J. A. Tainer. 2004. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2:363-378. [DOI] [PubMed] [Google Scholar]
- 6.Dennis, D. T., T. V. Inglesby, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. D. Fine, A. M. Friedlander, J. Hauer, M. Layton, S. R. Lillibridge, J. E. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. M. Perl, P. K. Russell, and K. Tonat. 2001. Tularemia as a biological weapon: medical and public health management. JAMA 285:2763-2773. [DOI] [PubMed] [Google Scholar]
- 7.Feil, E. J., E. C. Holmes, D. E. Bessen, M. S. Chan, N. P. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta, R., and E. Griffiths. 2002. Critical issues in bacterial phylogeny. Theor. Popul. Biol. 61:423. [DOI] [PubMed] [Google Scholar]
- 9.Gutacker, M. M., J. C. Smoot, C. A. Migliaccio, S. M. Ricklefs, S. Hua, D. V. Cousins, E. A. Graviss, E. Shashkina, B. N. Kreiswirth, and J. M. Musser. 2002. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms: resolution of genetic relationships among closely related microbial strains. Genetics 162:1533-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollis, D. G., R. E. Weaver, A. G. Steigerwalt, J. D. Wenger, C. W. Moss, and D. J. Brenner. 1989. Francisella philomiragia comb. nov. (formerly Yersinia philomiragia) and Francisella tularensis biogroup novicida (formerly Francisella novicida) associated with human disease. J. Clin. Microbiol. 27:1601-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johansson, A., J. Farlow, P. Larsson, M. Dukerich, E. Chambers, M. Byström, J. Fox, M. C. Chu, M. Forsman, A. Sjöstedt, and P. Keim. 2004. Worldwide genetic relationships among Francisella tularensis isolates determined by multiple-locus variable-number tandem repeat analysis. J. Bacteriol. 186:5808-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansson, A., A. Ibrahim, I. Göransson, U. Eriksson, D. Gurycova, J. E. Clarridge III, and A. Sjöstedt. 2000. Evaluation of PCR-based methods for discrimination of Francisella species and subspecies and development of a specific PCR that distinguishes the two major subspecies of Francisella tularensis. J. Clin. Microbiol. 38:4180-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsson, P., P. C. Oyston, P. Chain, M. C. Chu, M. Duffield, H. H. Fuxelius, E. Garcia, G. Halltorp, D. Johansson, K. E. Isherwood, P. D. Karp, E. Larsson, Y. Liu, S. Michell, J. Prior, R. Prior, S. Malfatti, A. Sjöstedt, K. Svensson, N. Thompson, L. Vergez, J. K. Wagg, B. W. Wren, L. E. Lindler, S. G. Andersson, M. Forsman, and R. W. Titball. 2005. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37:153-159. [DOI] [PubMed] [Google Scholar]
- 14.Mostowy, S., D. Cousins, J. Brinkman, A. Aranaz, and M. A. Behr. 2002. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J. Infect. Dis. 186:74-80. [DOI] [PubMed] [Google Scholar]
- 15.Nano, F. E., N. Zhang, S. C. Cowley, K. E. Klose, K. K. Cheung, M. J. Roberts, J. S. Ludu, G. W. Letendre, A. I. Meierovics, G. Stephens, and K. L. Elkins. 2004. A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol. 186:6430-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ochman, H., and N. A. Moran. 2001. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292:1096-1099. [DOI] [PubMed] [Google Scholar]
- 17.Pearson, T., J. D. Busch, J. Ravel, T. D. Read, S. D. Rhoton, J. M. U'Ren, T. S. Simonson, S. M. Kachur, R. R. Leadem, M. L. Cardon, M. N. Van Ert, L. Y. Huynh, C. M. Fraser, and P. Keim. 2004. Phylogenetic discovery bias in Bacillus anthracis using single-nucleotide polymorphisms from whole-genome sequencing. Proc. Natl. Acad. Sci. USA 101:13536-13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Penn, R. L. 2005. Francisella tularensis (Tularemia), p. 2674-2685. In G. L. Mandell, J. E. Bennet, and R. Dolin (ed.), Mandell, Douglas and Bennett's principles and practice of infectious diseases, 6th ed., vol. 2. Churchill Livingstone, Ltd., Edinburgh, Scotland.
- 19.Rocha, E. P. 2003. An appraisal of the potential for illegitimate recombination in bacterial genomes and its consequences: from duplications to genome reduction. Genome Res. 13:1123-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romero, D., and R. Palacios. 1997. Gene amplification and genomic plasticity in prokaryotes. Annu. Rev. Genet. 31:91-111. [DOI] [PubMed] [Google Scholar]
- 21.Samrakandi, M. M., C. Zhang, M. Zhang, J. Nietfeldt, J. Kim, P. C. Iwen, M. E. Olson, P. D. Fey, G. E. Duhamel, S. H. Hinrichs, J. D. Cirillo, and A. K. Benson. 2004. Genome diversity among regional populations of Francisella tularensis subspecies tularensis and Francisella tularensis subspecies holarctica isolated from the US. FEMS Microbiol. Lett. 237:9-17. [DOI] [PubMed] [Google Scholar]
- 22.Scoles, G. A. 2004. Phylogenetic analysis of the Francisella-like endosymbionts of Dermacentor ticks. J. Med. Entomol. 41:277-286. [DOI] [PubMed] [Google Scholar]
- 23.Sjöstedt, A., U. Eriksson, L. Berglund, and A. Tärnvik. 1997. Detection of Francisella tularensis in ulcers of patients with tularemia by PCR. J. Clin. Microbiol. 35:1045-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sjöstedt, A. B. 2005. Francisella, p. 200-210. In D. J. Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity (ed.), The Proteobacteria, part B. Bergey's manual of systematic bacteriology, 2nd ed., vol. 2. Springer-Verlag, New York, N.Y.
- 25.Swofford, D. L. 1998. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates Inc., Sunderland, Mass.
- 26.Tigertt, W. D. 1962. Soviet viable Pasteurella tularensis vaccines. Bacteriol. Rev. 26:354-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Titball, R. W., A. Johansson, and M. Forsman. 2003. Will the enigma of Francisella tularensis virulence soon be solved? Trends Microbiol. 11:118-123. [DOI] [PubMed] [Google Scholar]
- 28.Urwin, R., and M. C. Maiden. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11:479-487. [DOI] [PubMed] [Google Scholar]
- 29.Vinogradov, E., W. J. Conlan, J. S. Gunn, and M. B. Perry. 2004. Characterization of the lipopolysaccharide O-antigen of Francisella novicida (U112). Carbohydr. Res. 339:649-654. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.