Abstract
Staphylocoagulase detection is the hallmark of a Staphylococcus aureus infection. Ten different serotypes of staphylocoagulases have been reported to date. We determined the nucleotide sequences of seven staphylocoagulase genes (coa) and their surrounding regions to compare structures of all 10 staphylocoagulase serotypes, and we inferred their derivations. We found that all staphylocoagulases are comprised of six regions: signal sequence, D1 region, D2 region, central region, repeat region, and C-terminal sequence. Amino acids at both ends, 33 amino acids in the N terminal (the signal sequences and the seven N-terminal amino acids in the D1 region) and 5 amino acids in the C terminal, were exactly identical among the 10 serotypes. The central regions were conserved with identities between 80.6 and 94.1% and similarities between 82.8 and 94.6%. Repeat regions comprising tandem repeats of 27 amino acids with a 92% identity on average were polymorphic in the number of repeats. On the other hand, D1 regions other than the seven N-terminal amino acids and D2 regions were less homologous, with diverged identities from 41.5 to 84.5% and 47.0 to 88.9%, respectively, and similarities from 53.5 to 88.7% and 56.8 to 91.9%, respectively, although the predicted prothrombin-binding sites were conserved among them. In contrast, flanking regions of coa were highly homologous, with nucleotide identities of more than 97.1%. Phylogenetic relations among coa did not correlate with those among the flanking regions or housekeeping genes used for multilocus sequence typing. These data indicate that coa could be transmitted to S. aureus, while the less homologous regions in coa presumed to be responsible for different antigenicities might have evolved independently.
Staphylocoagulase is an extracellular protein produced by coagulase-positive staphylococci represented by Staphylococcus aureus. It causes coagulation of plasma. Although some non-staphylocoagulase-producing S. aureus strains have been identified, it has been reported that all strains possess the gene encoding staphylocoagulase (47). Therefore, the production of staphylocoagulase is regarded as a major characteristic that could be used for the identification of S. aureus. It is regarded as the hallmark protein for the classification of S. aureus infections, although it is also produced by three other coagulase-positive species, Staphylococcus intermedius, Staphylococcus delphini, and Staphylococcus scheliferi subsp. coagulans, and a coagulase-variable species, Staphylococcus hyicus subsp. hyicus (7, 37, 46).
Staphylocoagulase has been studied in two ways. One approach was done by defining the structure and function via purification and examination of the characteristics (14, 19, 24, 25). It was reported that the protein binds to prothrombin with a 1:1 stoichiometry to form staphylothrombin, and the complex induces plasma coagulation by converting fibrinogen into fibrin. This would activate prothrombin without the usual proteolytic cleavages (14, 25). Crystal structure analysis of the first 325 amino acids of staphylocoagulase variants bound to α-thrombin or its precursor prethrombin 2 showed that staphylocoagulase inserted its N-terminal peptide into the activation pocket of prethrombin 2 and changed it allosterically to the active form (12). Kinetic studies also confirmed the importance of its N-terminal two amino acids in mature staphylocoagulase (12).
The second approach examines the diversity of staphylocoagulase in antigenicity as well as the gene, both of which are used as epidemiological markers (13, 17, 45). To date, staphylocoagulase has been classified into 10 serotypes, based on the differences in antigenicity via inhibition test of the clotting activity using specific antibodies against each serotype (23, 45).
The structures of three staphylocoagulase serotypes, namely type I, II, and III, have been reported, based on their nucleotide sequences (21, 22, 38). Comparison of the three deduced amino acid sequences and crystal structural analysis indicated that staphylocoagulase is composed of six distinct segments: (i) 26-amino-acid signal sequence, (ii) N-terminal D1 region, (iii) D2 region, (iv) highly conserved central region, (v) 27-amino-acid repeat region, and (vi) 5-amino-acid C-terminal sequence. The D1 and D2 regions consist of α-helices. Through these two domains, staphylocoagulase contacts prothrombin with its two surface sites, the 148 loop and exosite I. The N-terminal amino acids 1 to 270 that are involved in the D1 and D2 regions of the three serotypes are less homologous, with identity of nearly 50%. The repeat regions comprising 27-amino-acid tandem repeats are highly polymorphic in the number of repeats. Although the serotypes do not correlate with the polymorphism of 27-amino-acid tandem repeats (23), both the N-terminal region and the C-terminal region have been used for S. aureus differentiation by analyzing their diversity using amplified DNA fragments and subsequent restriction fragment length polymorphism analysis.
It is very curious that such a large number of serotypes have been found in a singular protein produced by a species. In this study, in order to understand the molecular basis giving rise to these structural variations, we determined the nucleotide sequences of seven coa genes and compared both nucleotide and deduced amino acid sequences of 10 serotypes of staphylocoagulase. Furthermore, aiming to understand the molecular evolution of the serotypes, we sequenced the upstream and downstream regions right after the coa, compared the coa flanking regions of each serotype, and found that they were highly homologous. The reasons a lot of staphylocoagulase serotypes are found in an S. aureus species are discussed.
MATERIALS AND METHODS
Bacterial strains.
Seven reference strains for staphylocoagulase serotypes were used for determination of coa nucleotide sequences and their flanking regions. The seven S. aureus strains were as follows: strain 104 (type I), Stp-28 (type IV), no. 55 (type V), Stp-12 (type VI), Ku (type VIII), 17573 (type IX), and 19 (type X) (23, 45). To investigate the genetic relatedness between serotypes of coa and genetic backgrounds of S. aureus strains, we selected nine strains whose ribotypes had been determined and sequenced their coa. They were as follows: three type III strains of ribotyping group 4, NCTC 10442, 61/6219, and 86/4372; three type IV strains of ribotyping group 4, 85/2082, 85/3907, and 86/961; three type IV strains that did not belong to ribotyping group 4, MR108 (ribotyping group 47), 93/H44 (ribotyping group 48), and 85/2232 (ribotyping group 49) (27). The sources and characteristics of the strains used are listed in Table 1. Seven S. aureus strains whose entire genome sequences have been reported were included in this study: N315 (type II; http://www.bio.nite.go.jp/dogan/DOGAN_html/n315/general_feature.jsp), Mu50 (type II; http://133.5.48.230/microb/VRSA_new/Sequencing_2699SAVs/), MW2 (type VII; http://www.bio.nite.go.jp/dogan/DOGAN_html/mw2/general_feature.jsp), NCTC 8325 (type III; http://www.genome.ou.edu/staph.html), COL (type III; http://www.tigr.org), MRSA252-2 (type IV; http://www.sanger.ac.uk/Projects/S_aureus/), and MSSA476 (type VII; http://www.sanger.ac.uk/Projects/S_aureus/) (2, 18, 28, 48).
TABLE 1.
Characteristics of S. aureus strains in this study
| Staphylocoagulase serotype | S. aureus strain | Clonal complex | Sequence type | Allele (MLST) (arcC-aroE-glpF-gmk-pta-tpi-yqiL) | Reference or source |
|---|---|---|---|---|---|
| I | 104 | Minor group (ST49-ST138) | ST49 | 14-16-11-2-13-12-14 | 45 |
| II | N315 | CC5 | ST5 | 1-4-1-4-12-1-10 | 28 |
| III | NCTC 8325 | CC8 | ST8 | 3-3-1-1-4-4-3 | 18 |
| III | NCTC 10442 | CC8 | ST250 | 3-3-1-1-4-4-16 | 27 |
| III | 61/6219 | CC8 | ST250 | 3-3-1-1-4-4-16 | 27 |
| III | COL | CC8 | ST250 | 3-3-1-1-4-4-16 | TIGRa |
| III | 86/4372 | CC8 | ST8 | 3-3-1-1-4-4-3 | 27 |
| IV | Stp-28 | CC30 | ST30 | 2-2-2-2-6-3-2 | 45 |
| IV | 85/2082 | CC8 | ST239 | 2-3-1-1-4-4-3 | 27 |
| IV | 85/3907 | CC8 | ST239 | 2-3-1-1-4-4-3 | 27 |
| IV | 86/961 | CC8 | ST239 | 2-3-1-1-4-4-3 | 27 |
| IV | MR108 | CC30 | ST30 | 2-2-2-2-6-3-2 | 27 |
| IV | 93/H44 | CC30 | ST30 | 2-2-2-2-6-3-2 | 27 |
| IV | 85/2232 | CC30 | ST30 | 2-2-2-2-6-3-2 | 27 |
| IV | MRSA252-2 | CC30 | ST30 | 2-2-2-2-6-3-2 | 16 |
| V | No. 55 | CC121 | ST95 | 6-5-6-2-7-14-34 | 45 |
| VI | Stp-12 | Minor group (ST96-ST154) | ST96 | 12-1-1-15-11-1-40 | 45 |
| VII | MW2 | CC1 | ST1 | 1-1-1-1-1-1-1 | 2 |
| VII | MSSA476 | CC1 | ST1 | 1-1-1-1-1-1-1 | 16 |
| VIII | Ku | Minor group (ST10-ST145) | ST10 | 8-7-6-2-9-9-7 | 45 |
| IX | 17573 | Minor group (ST130-ST480-ST574) | ST574 | 72-57-81-2-7-58-52 | 23 |
| X | 19 | CC15 | ST15 | 13-13-1-1-12-11-13 | 23 |
TIGR, The Institute for Genomic Research (http://www.tigr.org).
Serological typing of staphylocoagulase.
Staphylocoagulase serotypes were determined by the coagulation inhibition test. Specific antibodies to types I to VIII staphylocoagulases are commercially available as an S. aureus coagulase typing test kit (Denka Seiken Co., Ltd., Tokyo, Japan). A single colony for each test strain grown on brain heart infusion (BHI) agar (Becton Dickinson Co., Ltd.) was suspended in BHI broth (Becton Dickinson Co.) and incubated at 37°C overnight. The culture was then centrifuged. An aliquot (0.1 ml) of the supernatant, which was diluted with diluent (2.0% polypeptone, 1.0% sodium citrate, 0.1% sodium azide) appropriately, was distributed into 11 tubes, and 0.1-ml aliquots of anticoagulase types I to X sera were added to 10 tubes, respectively, from the 1st to the 10th. In the 11th tube, 0.1 ml of 5% normal rabbit serum with diluent was added as control. Following that, the tubes were incubated at 37°C for 1 h. After the incubation, 0.2 ml of diluted rabbit plasma was added to each tube, followed by incubation at 37°C for at least 1 h. The plasma coagulation was judged by visual inspection after 1, 2, 4, 6, and 24 h. Matching of the antiserum and the staphylocoagulase would inhibit the coagulation process. Strains could then be typed according to the type of staphylocoagulase which showed coagulation inhibition.
DNA preparation.
Chromosomal DNA was extracted using Isoprant (Nippon Gene Co., Ltd., Tokyo, Japan) according to protocols recommended by the manufacturer with a slight modification. Briefly, cells were cultured in BHI broth at 37°C overnight. A 1.5-ml aliquot of the culture was poured into a tube, and cells were collected by centrifugation. The pellet was resuspended with 400 μl of SMM buffer (0.5 M sucrose, 0.1 M disodium maleate, 0.002 M MgCl2; pH 6.5), 20 μl of 2 mg/ml lysostaphin (Wako Pure Chemical Industries, Ltd., Osaka, Japan) was added, and the mixture was left at 37°C for more than 20 min until protoplasts were formed. Cells were collected by centrifugation, and their chromosomal DNA was extracted as recommended by the manufacturer.
DNA sequencing.
coa genes and their flanking regions were amplified by PCR using chromosomal DNAs as template. DNA fragments were amplified using primer sets designed based on the S. aureus N315 genome sequence: coa-F (5′-CACAGATGACCCAGCATTAGCTG-3′) and coa-R (5′-CGTTACGGCATGGTTACGATGTG-3′); coaup-F (5′-CTTGCATATGAATATGAGTC-3′) and coaup-R (5′-AAGCTAGATGCAACTGCTAATG-3′); and coadown-F (5′-GACATATGGTCCTAGAGTAACA-3′) and coadown-R (5′-AACTGGTAAGCCATTACGTA-3′). The reaction mixtures consisted of 100-μl total volume solutions containing 10 ng of genomic DNA, 10 pmol of each primer, 0.4 U of Ex Taq DNA polymerase (Takara Bio Inc., Shiga, Japan), 10 μl of 10 Ex Taq buffer, and 20 mM of each deoxynucleoside triphosphate. Amplification was performed with a PCR system 9700 (Applied Biosystems), and the parameters were an initial 1 min at 94°C and 30 cycles of 30 s at 94°C, 1 min at 55°C, and 1 min at 72°C. The PCR products were purified using a High Pure PCR product purification kit (Roche Molecular Systems Inc.). Sequencing reactions were performed with fluorescent dideoxy chain termination chemistry using the BigDye Terminator version 1.1 cycle sequencing kit (Applied Biosystems). DNA sequencing was performed using an ABI Prism 3100 genetic analyzer (Applied Biosystems).
MLST.
Multilocus sequence typing (MLST) was preformed as described previously (11). Briefly, the inner regions of seven housekeeping genes (arcC, aroE, glp, gmk, pta, tpi, and yqiL) in S. aureus were amplified by PCR, and their nucleotide sequences were determined using an ABI Prism 3100 genetic analyzer. Strains were defined by the alleles present at the seven loci (the allelic profile), and each unique allelic profile was assigned a sequence type (ST) based on the MLST website (http://www.mlst.net). STs were then correlated to each other by using the program BURST (based upon related sequence types) in the MLST website.
Genetic analysis.
Multiple alignments were performed with CLUSTAL X software program (version 1.8) (43). The alignment was adjusted manually for any obvious misplacement of gaps on the basis of physiochemical properties of amino acids. The phylogenetic tree was created using the maximum likelihood method in the GCG paupsearch program in the Wisconsin GCG package (Accelrys Inc.) Support for individual nodes on the tree was determined by bootstrap analysis. The phylogenetic trees were visualized with Treeview (34).
Nucleotide sequence accession numbers.
The sequences of the coa genes and their flanking regions of S. aureus strains 104, Stp-28, no. 55, Stp-12, Ku, 17573, and 19 have been deposited in the DDBJ, EMBL, and GenBank databases under accession numbers AB158549, AB158550, AB158551, AB158552, AB158553, AB158554, and AB158555, respectively.
RESULTS
Overall comparison of coa genes among 10 serotypes.
We aimed to compare the genes of all 10 staphylocoagulase serotypes that have been identified by using specific neutralizing antibodies. By the time we started this study, the nucleotide sequences of four types of coa genes had already been reported. Nucleotide sequences of the type I coa of S. aureus BB, type II coa of S. aureus 213, and type III coa of S. aureus 8325-4 were reported individually (21, 22, 38), and nucleotide sequences of type II, III, and VII coa genes were available from the whole genome sequences of S. aureus N315, S. aureus NCTC 8325, and S. aureus MW2 (2, 18, 28). We therefore determined nucleotide sequences of coa in six serotypes (types IV, V, VI, VIII, IX, and X) using the corresponding staphylocoagulase reference strains. In addition to the nucleotide sequence of type I coa of S. aureus strain BB, which has already been reported, we also determined the nucleotide sequence of coa in type I reference strain S. aureus 104 to investigate the phylogenetic relations between coa and other chromosomal regions.
Table 2 shows the characteristics of coa in the 10 serotypes. The lengths of these genes are rather diverse. They ranged from the shortest size of 1,902 bp (type VII) to the longest size of 2,280 bp (type VIII), and the average length was 2,034 bp. The numbers of 81-bp tandem repeats located at the 3′ terminus were diverse, too. They ranged from the smallest number of repeats, five (NCTC 8325, MW2, and 19), to the largest number of repeats, nine (Ku). The differences in nucleotide sequences of the 81-bp tandem repeats and the number of tandem repeats were reported and have been used for epidemiological markers as well (42). When we assigned alleles defined by nucleotide differences in the 81-bp repeat units that constitute the tandem repeat, 36 of 63 were new alleles (V3 to G5 in Table 2). The average GC content was 35.7%, which was slightly higher than that of the whole genome sequence of S. aureus N315 (29%) (28). Table 3 shows overall identities of the nucleotides and their deduced amino acid sequences of 10 coa genes. The nucleotide identity was 77.1% on average, and amino acid identities and similarities were 75.3% and 79.8%, respectively. The highest value of amino acid identity was 88.4% (type IV versus type IX), and the lowest was 69.5% (type V versus type VII).
TABLE 2.
Comparison of 10 staphylocoagulase genes
| Serotype | Strain | Size (bp) | Repeat unitsa | coa repeat profile | Size (R, CT)b | % GC content |
|---|---|---|---|---|---|---|
| I | 104 | 2,148 | 7 | V3W3X3Y3Z3A4B4 | 1,478 | 36.6 |
| II | N315 | 1,977 | 6 | C4BCDEF | 1,473 | 34.7 |
| III | NCTC 8325 | 1,911 | 5 | MNOD4Q | 1,488 | 34.6 |
| IV | Stp-28 | 1,992 | 6 | GHTJKL | 1,488 | 35.3 |
| V | No. 55 | 2,178 | 8 | E4F4G4H4I4J4K4D2 | 1,512 | 36.4 |
| VI | Stp-12 | 1,989 | 6 | E2L4M4N4EF | 1,485 | 35.6 |
| VII | MW2 | 1,902 | 5 | E2O4P4Q4Q | 1,479 | 34.4 |
| VIII | Ku | 2,280 | 9 | R4S4T4U4V4W4X4Y4Z4 | 1,533 | 39.4 |
| IX | 17573 | 1,992 | 6 | YA5B5C5D5E5 | 1,488 | 34.9 |
| X | 19 | 1,935 | 5 | UF5DG5W | 1,512 | 34.6 |
Number of 81-bp tandem repeats.
Size without repeated region and C-terminal region (bp).
TABLE 3.
Nucleotide and amino acid identities among all serotypes
| Serotype | % Identitya with serotype:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | VII | VIII | IX | X | |
| I | 73.7 | 70.8 | 73.9 | 75.4 | 73.9 | 76.3 | 76.4 | 73.2 | 78.5 | |
| II | 78.2 | 81.5 | 75.9 | 78.7 | 79.2 | 75.0 | 78.0 | 77.4 | 73.5 | |
| III | 76.9 | 84.1 | 74.4 | 72.7 | 77.8 | 74.4 | 74.1 | 77.6 | 70.5 | |
| IV | 79.2 | 80.9 | 79.1 | 78.3 | 75.1 | 74.3 | 77.2 | 88.4 | 70.2 | |
| V | 72.4 | 72.4 | 68.4 | 71.4 | 75.2 | 69.5 | 79.9 | 75.2 | 71.3 | |
| VI | 79.8 | 83.7 | 82.0 | 80.6 | 71.3 | 74.9 | 78.2 | 77.2 | 74.3 | |
| VII | 81.9 | 78.8 | 79.5 | 79.0 | 69.4 | 81.2 | 71.3 | 71.9 | 77.6 | |
| VIII | 71.7 | 67.2 | 68.9 | 71.5 | 82.7 | 68.2 | 69.4 | 76.1 | 71.1 | |
| IX | 79.9 | 82.1 | 81.1 | 90.2 | 71.6 | 82.5 | 78.8 | 71.7 | 69.7 | |
| X | 81.4 | 78.6 | 77.6 | 76.1 | 74.9 | 78.1 | 81.9 | 76.7 | 77.4 | |
Identities of nucleotide sequences of coa and their deduced amino acid sequences among 10 serotypes. Nucleotide identities are shown in cells in the bottom left half of the table and amino acid identities are shown in cells in the upper right half of the table. Average nucleotide identity was 77.1% and that of amino acid identities was 75.3%.
Nucleotide sequences of the same serotype were highly conserved. The identities among three type II coa genes of strains 213, N315, and Mu50 (type II) were 99.9 to 100%. Two type III coa genes of 8325 and COL were exactly identical. The identities between the two type IV coa genes of Stp-28 and MRSA252-2 and those between the two type VII coa genes of MW2 and MSSA476 were 97.2% and 99.9%, respectively, although the number of tandem repeats was not identical, since Stp-28 possessed six repeats whereas MRSA252-2 possessed four repeats.
It was reported that staphylocoagulase could bind fibrinogen in addition to prothrombin (6). Cheung et al. purified a protein that binds to fibrinogen by using a fibrinogen column, and they designated it FbpA (9). However, after nucleotide sequence determination of the gene (fbpA), they found that its structure was very similar to coa, and they reported that it should belong to the coa family. When we compared the deduced amino acid sequence of FbpA to that of the 10 staphylocoagulase serotypes, we found that FbpA is nearly identical to type X staphylocoagulase, with an identity of 98.0%. The nucleotide identity between fbpA and type X coa was 98.5%.
Structural comparison of staphylocoagulases among 10 serotypes.
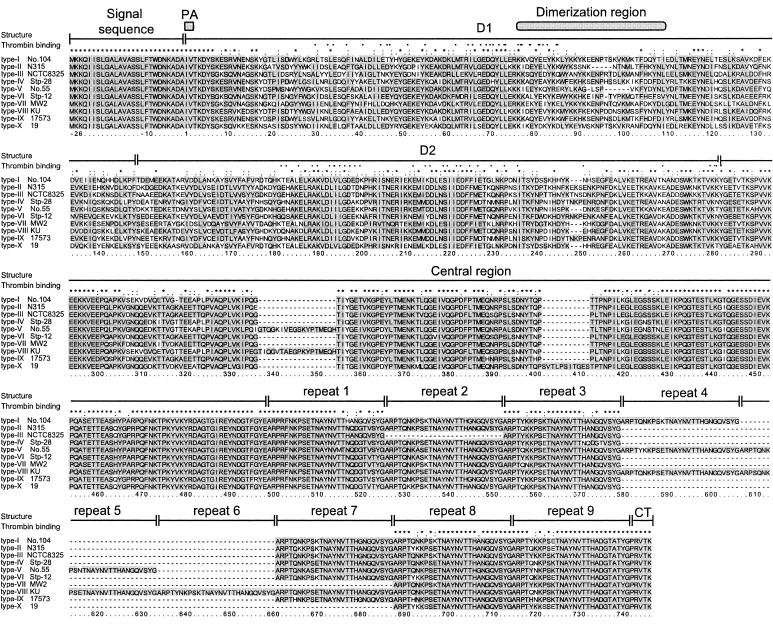
Figure 1 shows the multiple alignments of the deduced amino acid sequences of staphylocoagulases of all 10 serotypes. We found that all deduced amino acids were composed of six regions as previously reported by Fridrich et al. (12). The six regions are as follows: (i) 26-amino-acid signal sequence, (ii) N-terminal D1 region, which contains the prothrombin-activating site and the prothrombin-binding sites, (iii) D2 region, which is required for high-affinity binding of prothrombin through pro-exosite I, (iv) highly conserved central region, (v) 27-amino-acid repeat region, and (vi) 5-amino-acid C-terminal sequence.
FIG. 1.
Deduced amino acid sequences of 10 serotypes of staphylocoagulase compared using the CLUSTAL X program. The corresponding nucleotide sequences were determined using DNA fragments of the following strains: 104 (type I coa), N315 (type II), NCTC 8325 (type III), Stp-28 (type IV), no. 55 (type V), Stp-12 (type VI), MW2 (type VII), Ku (type VIII), 17573 (type IX), and 19 (type X). The highly conserved residues among 10 staphylocoagulases are shown in gray to highlight them. The identical or conserved residues are marked with an asterisk, colon, or period at the top. The residues which are in contact with the 148 loop or exosite I reported by Friedrich et al. (12) are indicated as closed or open triangles, respectively. PA, prothrombin-activated domain; CT, C-terminal sequence.
The N-terminal 26 amino acids of the primary translated products, which are signal sequences, were exactly identical among all serotypes. The first seven amino acids of the secreted mature forms of staphylocoagulase, including two amino acids required for prothrombin activation, were also identical among all serotypes. However, thereafter up to about 300 amino acids the amino acid sequences diverged. The means of amino acid identities and similarities of the D1 regions other than the first seven amino acids among all serotypes were 52.8% and 62.9%, respectively. Those of D2 regions were 60.2% and 69.3%, respectively. Although the identities of amino acid sequences in the D1 and the D2 regions were rather low, amino acid residues that might interact with the surface of prothrombin were highly conserved among them. Interestingly, we found that there were three pairs of serotypes whose amino acid identities in the D2 region were rather high (nearly 90%). They were the pairs of type I and type X (87.1%), type II and type III (88.2%), and type IV and type IX (88.9%).
The central regions located between the D2 region and the tandem repeats were relatively more conserved, and the averages of amino acid identity and similarity were 86.7% and 88.0%. Three strains had insertions that were not carried by other types. They were as follows: 18 amino acids (GTQGKIVGRSKYPTMEQH) in the type V staphylocoagulase of strain no. 55, 18 amino acids (TIQGVTAEGPKYPTMEQH) in the type VIII staphylocoagulase of strain Ku, and 11 amino acids (SVTLPSITGES) in the type X staphylocoagulase of strain 19.
Tandem repeats composed of 27 amino acids were located at the C terminus in all staphylocoagulases we investigated. The number of tandem repeat units varied, but the 27-amino-acid sequences were relatively conserved. The average amino acid identities among the 10 serotypes was 92.9%. The C-terminal 5-amino-acid sequences were completely identical among all serotypes.
Structural relation to vWbp.
S. aureus produces another blood coagulation-causing protein, von Willebrand factor-binding protein (vWbp). Its overall structure is very similar to that of staphylocoagulase, and it is classified into the same zymogen activator and adhesion protein family as staphylocoagulase (3). The deduced amino acids of two regions, called D1 and D2 of vWbp, showed homology to D1 and D2 regions in type I staphylocoagulase, with identities of 30.7% and 28.3%, respectively. Interestingly, vWbp is allelic, too (4). Among seven S. aureus strains whose genome sequences have been published (2, 16, 18, 28), the gene (vwb) is not always intact. It was truncated in three strains, NCTC 8325 (staphylocoagulase type III), MW2 (staphylocoagulase type VII), and MRSA252-2 (staphylocoagulase type IV). When we compared vwb genes in four other strains, two alleles were found. The first allele was found in two strains, COL (staphylocoagulase type III) and MSSA476 (staphylocoagulase type VII), and the second one was found in N315 (staphylocoagulase type II) and Mu50 (staphylocoagulase type II). The vwb gene in strain Newman (3) and two truncated vwb genes in NCTC 8325 and MW2 belonged to the first allele. The amino acid identity and similarity of vWbp between the two alleles represented by S. aureus N315 and S. aureus COL were 70.9% and 76.3%, respectively. The amino acid identities between them are rather low in D1, D2, and the central region, with identities of 56.9%, 53.9%, and 59.7% and similarities of 66.2%, 58.6%, and 70.1%, respectively, whereas the identities and similarities are very high in the vWb binding site and the C-terminal region, with identities of 96.2% and 96.0% and with similarities of 96.2% and 97.6%, respectively. These regions showing low homology are similar to those of staphylocoagulase.
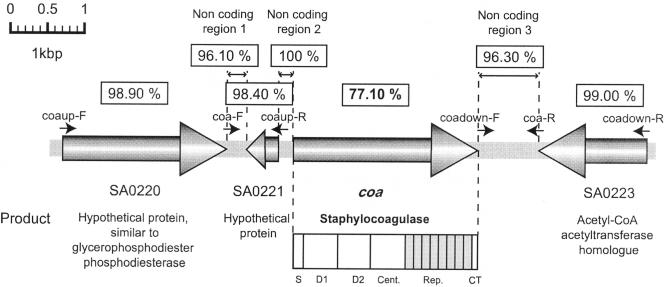
coa flanking regions are highly homologous.
Determination of the entire genome sequences of all seven S. aureus strains showed that coa is located at approximately 1 o'clock on the chromosome. To investigate whether all coa genes are located at the same position, we determined the nucleotide sequences of coa flanking regions in strains whose genomes had not been determined. The strategy and results are summarized in Fig. 2. In the case of S. aureus N315, its coa is located between two open reading frames (ORFs), SA0221 (encoding a hypothetical protein) and SA0223 (encoding an acetyl-coenzyme A acetyltransferase homologue). By using the sets of primers indicated in Fig. 2, two types of DNA fragments, each spanning the upstream and downstream regions of coa, respectively, were amplified by PCR, using chromosomal DNA of the seven reference strains as templates, and sequenced. All 10 strains carried two ORFs corresponding to SA0221 and SA0223 that are located at upstream and downstream of coa. Nucleotide identities of the regions corresponding to the two ORFs are shown in Fig. 2. They were highly homologous in nucleotide identities, with averages of 98.4% and 99.0%, respectively. In addition, noncoding regions flanking coa were well conserved (Fig. 2). Our data showed that the regions flanking coa are nearly identical, while coa sequences located within them are very diverse, especially in the D1 and D2 regions and 81-bp repeat regions.
FIG. 2.
Organization of the region in and around coa and comparison of the nucleotide sequences of coa and surrounding regions. ORFs are shown by bold arrows. The locations of the primers used in this study are indicated by short arrows. The numbers in the boxes show average nucleotide identities of each of the genes or regions among the 10 serotypes.
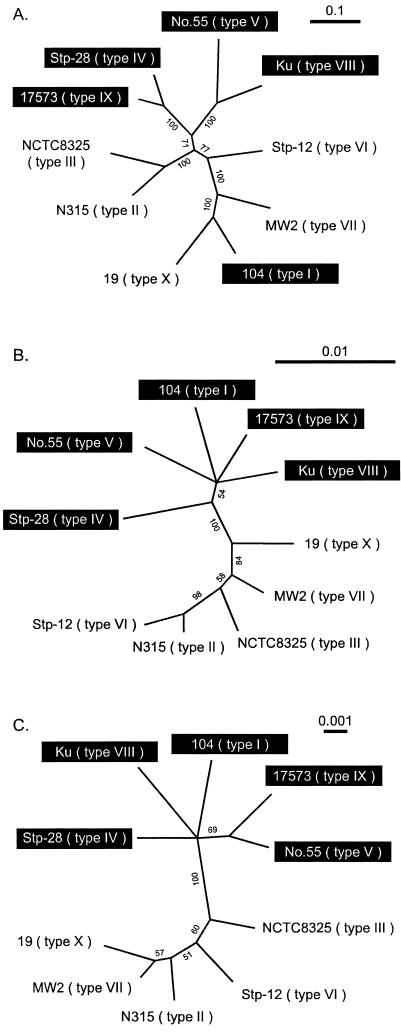
Phylogenetic relationship among coa of 10 serotypes.
To investigate whether allelic differences in coa correlate with the nucleotide differences in the chromosomal region other than coa, we determined nucleotide sequences of seven housekeeping genes used for MLST. STs of 10 strains carrying 10 different serotypes of coa each were different from one another (Table 1). We investigated the phylogenetic relation among the 10 strains by using the nucleotide sequences of the above seven genes. We found that the 10 strains were roughly divided into two groups (Fig. 3C). One group was comprised of five strains carrying type I, IV, V, VIII, and IX coa (group 1), and the other group was comprised of five strains carrying type II, III, VI, VII, and X coa (group 2). When we created a phylogenetic tree using nucleotide sequences of the upstream and downstream regions of coa, it was found that their relative relations were similar to those of the housekeeping genes (Fig. 3B).
FIG. 3.
Maximum likelihood trees constructed using GCG paupsearch and visualized with Treeview software. Nucleotide sequences used for comparison were as follows: A, nucleotide sequencesof coa other than the repeat region and C-terminal sequence; B, regions surrounding coa which were concatenated sequences of SA0220 noncoding region 1, SA0221 noncoding region 2 on the left region flanked by coa and noncoding region 3, and SA0223 on the right region flanked by coa; C, housekeeping genes, which were concatenated parts of seven housekeeping genes used for MLST. Branches with significant bootstrap support (more than 50%) are indicated. Group 1 strains are highlighted by black boxes; the remaining are group 2 strains.
In contrast, the phylogenetic relation among the 10 coa shown in Fig. 3A was not similar to those of housekeeping genes or the flanking region shown in Fig. 3B and C. The most striking difference was the position of the type I staphylocoagulase of strain 104. It was most distantly related to the other four types of coa (types IV, V, VIII, and IX), which constituted group 2 in tree A, whereas it belonged to the same group 1 together with four strains carrying types IV, V, VIII, and IX coa in the trees of Fig. 3B and C. The phylogenetic relation of type III coa in strain NCTC 8325 did not agree with the phylogenetic relation of noncoding regions. In tree B, the region in NCTC 8325 was related to that in MW2 (type VII), more closely than to those in N315 (type II) and Stp-12 (type VI), whereas in trees A and C, the corresponding regions of NCTC 8325 were more closely related to those in N315 than to those in MW2.
Correlations among serotypes of coa and genetic backgrounds.
To determine the correlations between coa alleles and the genetic backgrounds of the strains, we investigated the strains carrying the same serotype of coa but with different genetic backgrounds and the strains carrying different types of coa but with identical genetic backgrounds as determined by ribotyping.
The nucleotide sequences of type IV coa in six S. aureus strains (three of them belonged to ribotype 4 and three of them belonged to ribotypes 47, 48, and 49) were highly homologous, with nucleotide identities of more than 99.9%. The difference in genotypes inferred by ribotyping was also confirmed by MLST. Three ribotype 4 strains, 85/2082, 85/3907, and 86/961, belonged to CC8, and three strains of different ribotypes, MR108, 93/H44, and 85/2232, belonged to CC30. These data suggested that the same coa types are located in S. aureus strains of different genotypes.
Furthermore, we determined nucleotide sequences of type III coa of three strains which belonged to ribotype 4 and CC8. The nucleotide identities of coa of three strains, NCTC 10442, 61/6219, and 86/4372, were highly homologous, with identities of more than 99.9%. The data suggested that different types of coa are located on S. aureus strains of the same genetic background, ribotype 4 and CC8.
DISCUSSION
Structures of staphylocoagulases.
Staphylocoagulase is one of the factors that cause coagulation of plasma. It activates prothrombin without the usual proteolytic cleavages, while other factors such as factor Xa convert prothrombin to thrombin by producing proteolytic cleavage, the classical mechanism of serine protease.
The finding of antigenic variation in staphylocoagulases produced by S. aureus dates as far back as 55 years (10, 39). Since then, 10 serotypes of staphylocoagulases have been identified. Using purified staphylocoagulases as antigens, antibodies specific to each type of staphylocoagulase are manufactured by Denka Seiken in Japan. The method of staphylocoagulase typing was developed based on coagulation inhibition with specific antibodies, and it has been used as a useful method for epidemiology of S. aureus (45). Here we show the genetic basis of those serotypes by nucleotide sequence determinations of staphylocoagulase genes of 10 serotypes.
The 10 staphylocoagulases investigated are composed of common regions with rather diverged regions among 10 serotypes. The N-terminal 33 amino acids comprised of the signal sequence (26 amino acids) and the first 7 amino acids in the D1 region containing the prothrombin-activating domain are exactly identical in all 10 staphylocoagulases. The central regions are relatively conserved. In contrast, D1 regions other than the first 7 amino acids and D2 regions are the most diverged regions among the 10 serotypes, with average amino acid identities of 52.8% and 60.2%, respectively.
Binding of staphylocoagulase to the prothrombin-binding region, which is an essential step for its activity, might be inhibited by antibodies. Purified antibodies that are specific to each serotype might recognize the amino acid sequences of the low-homology region, especially the D1 region rather than the D2 region or their three-dimensional structures, and prevent the binding of staphylocoagulase to the prothrombin-binding region. The fact that there are three pairs of serotypes with rather high amino acid identities (approximately 90%) in D2, namely, type II and type III, type I and type X, and type IV and type VI, also supported the above presumption.
Staphylocoagulase also binds to fibrinogen (6). Other than staphylocoagulase, seven proteins having fibrinogen-binding activity are known to be produced by S. aureus, e.g., clumping factor A (ClfA) (30), clumping factor B (ClfB) (32), major histocompatibility complex class II analog protein (Map) (31), extracellular adherence protein (Eap) (35), fibronectin-binding proteins A and B (FnbpA and FnbpB) (48), and extracellular fibrinogen-binding protein (Efb) (5). Among these proteins, Efb showed the highest similarity to staphylocoagulase. An identical sequence of 22 amino acids between a spacing of 9 amino acids in the N-terminal part of the mature protein showed homology to the tandem repeats of staphylocoagulase at the C terminus. Although Efb binds fibrinogen at two sites, which are the repeat regions at the N terminus and C terminus, only the N-terminal segment of the protein, containing the two 22-amino-acid repeats, could bind to soluble fibrinogen. The fact that Efb and staphylocoagulase compete for the same binding domain on fibrinogen might be due to their structural similarity (36). Staphylothrombin (a complex of staphylocoagulase and prothrombin) binds fibrinogen at the central region and repeat region. The staphylothrombin plays an important role in plasma coagulation by converting fibrinogen to fibrin.
Differences between staphylocoagulases produced by S. aureus and S. intermedius.
Staphylocoagulase produced by S. intermedius is different from that of S. aureus. It contains sugar, fucose, and galactose, which are not found in the staphylocoagulase of S. aureus (26). They are different in antigenicity (20). The three amino acids at the N terminus of S. intermedius are Ser-Glu-Pro, whereas those of S. aureus are Ile-Val-Thr. Since the N terminus, including the three amino acids, is essential for prothrombin activation, this may explain the observation that clotting activities of these two coagulases are not identical (26).
Why are there so many serotypes of proteins that cause blood coagulation?
From our results, we presume that antigenic variation in staphylocoagulases might be useful to evade the host immune response and/or to adapt to the different staphylocoagulase-prothrombin binding sites of mammalian species. Vaccines against the type b capsular polysaccharides of Haemophilus influenzae (Hib) have been used to reduce the incidence of invasive diseases caused by H. influenzae in industrial countries. However, as the use of the vaccines increased, the number of non-Hib strains in clinical isolates has also increased (29, 33, 49). The possibility of “serotype replacement” has been raising concerns. It can be postulated that staphylocoagulases changed their antigenicities by altering amino acids to evade the host defense system.
S. aureus could colonize and be pathogenic not only to human beings, but also to other warm-blooded animals. In mammals, prothrombin is one of the conserved proteins. However, its structure is not identical among various species; rather, it differs from species to species. The amino acid identity between human prothrombin and rat prothrombin is 79.7%, and that between human and cow prothrombin is 81.5%. Type IX staphylocoagulase of strain 17573 was isolated from a cow. Type I to VIII staphylocoagulases were carried in strains isolated from humans. Carter et al. reported that the 5′-terminal sequence of the strain AN3 staphylocoagulase was isolated from a lamb and that the HinfI restriction pattern of it was classified as type 3 (8). When we compared the two 5′-terminal nucleotide sequences of AN3 staphylocoagulase and type IX staphylocoagulase, we found they were exactly identical. Therefore, it might be speculated that type IX staphylocoagulase strains may be inclined to colonize animals. Other 5′-terminal sequences of coa isolated from animals were slightly different from coa types I to X. The nucleotide sequences were similar between HinfI type 1 and type I coa, HinfI type 2 and type VI coa, HinfI type 4 and type V, HinfI type 5 and type VI coa, and HinfI type 6 and type V coa. The amino acid identities of the D1 region between each pair were 98.6%, 99.3%, 94.1%, 94.3%, and 94.8%, respectively.
Correlations between staphylocoagulase serotype and genetic background of S. aureus.
The average coa nucleotide identities among 10 serotypes was 77.1%. It was lower than the identities between the whole genomes of S. aureus, since three methicillin-resistant S. aureus strains possess high nucleotide sequence identities: 99.7% between N315 and Mu50, 94.8% between MW2 and N315, and 94.7% between MW2 and Mu50 (2). Most of the nucleotide differences in the S. aureus chromosome are due to the insertion of some DNA regions, such as genomic islands (Gislands) and genomic islets (Gislets) (1, 2, 15). The Gisland is defined as a relatively large chromosomal region (10 to 200 kbp) whose GC content differs from the chromosome and which was acquired from other bacteria by lateral gene transfer. It encodes most of the genetic information involved in virulence and drug resistance (41, 44). In contrast to Gisland, Gislet is composed of a single gene, which is allelic, and its flanking regions are highly conserved (15). No recombinase-like protein was encoded in Gislet, and so an insertion sequence is not involved in this category. In addition to the whole genome sequences of seven S. aureus strains, data from this study indicated that coa should be regarded as a member of Gislets, since all 10 coa serotypes were found to be located at the same position on the chromosome between the highly conserved regions. Carter et al. previously reported that some strains with different pulsed-field gel electrophoresis patterns and MLST have the same types of staphylocoagulase, judging from the HinfI-random fragment length polymorphism pattern of the N-terminal fragment of coa (8). We have shown that seven strains of two different STs, CC8 and CC30, carried the same type IV coa. Furthermore, different types of staphylocoagulase, namely type III and type IV, were carried by strains of the same genotypes, CC8. The large chromosomal replacement in the S. aureus chromosome was reported by Robinson et al. (40). Since the size of the replaced region is at least ∼557 kbp and the region involves coa, this might explain the reason why different coa are carried in the same genotypes. However, further studies are needed to conclude this.
When we examined phylogenetic relatedness between coa and other chromosomal regions, we found small discrepancies between them, i.e., the type I coa of strain 104 belonged to group 2, whereas the flanking regions of coa and the housekeeping genes in the chromosome belonged to group 1 in their phylogenetic trees. Similarly, coa of NCTC 8325 is located at a slightly different position from the flanking regions of coa in the phylogenetic tree. These data strongly suggest that some S. aureus strains acquired coa by recombination, although its mechanism remains unknown. S. aureus is a part of our normal flora, and it persists in the human body by altering its genetic traits successfully. Hence, it could be possible that S. aureus strains might alter their staphylocoagulase type to adapt to host immunity and/or interspecies differences of prothrombin.
Acknowledgments
We thank Tsai-Ling Yang and Bruce E. Allen for their kind help in manuscript preparation. We thank Mark C. Enright for his kind help with MLST.
This work was supported by a Grant-in-Aid for 21st Century COE Research and Grant-in-Aid for Scientific Research on Priority Areas (13226114) from The Ministry of Education, Science, Sports, Culture and Technology of Japan.
REFERENCES
- 1.Baba, T., F. Takeuchi, M. Kuroda, T. Ito, H. Yuzawa, and K. Hiramatsu. 2004. The Staphylococcus aureus genome. Horwood Publishing, Chichester, United Kingdom.
- 2.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 3.Bjerketorp, J., K. Jacobsson, and L. Frykberg. 2004. The von Willebrand factor-binding protein (vWbp) of Staphylococcus aureus is a coagulase. FEMS Microbiol. Lett. 234:309-314. [DOI] [PubMed] [Google Scholar]
- 4.Bjerketorp, J., M. Nilsson, A. Ljungh, J. I. Flock, K. Jacobsson, and L. Frykberg. 2002. A novel von Willebrand factor binding protein expressed by Staphylococcus aureus. Microbiology 148:2037-2044. [DOI] [PubMed] [Google Scholar]
- 5.Boden, M. K., and J. I. Flock. 1994. Cloning and characterization of a gene for a 19 kDa fibrinogen-binding protein from Staphylococcus aureus. Mol. Microbiol. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 6.Boden, M. K., and J. I. Flock. 1992. Evidence for three different fibrinogen-binding proteins with unique properties from Staphylococcus aureus strain Newman. Microb. Pathog. 12:289-298. [DOI] [PubMed] [Google Scholar]
- 7.Capurro, A., C. Concha, L. Nilsson, and K. Ostensson. 1999. Identification of coagulase-positive staphylococci isolated from bovine milk. Acta Vet. Scand. 40:315-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter, P. E., K. Begbie, and F. M. Thomson-Carter. 2003. Coagulase gene variants associated with distinct populations of Staphylococcus aureus. Epidemiol. Infect. 130:207-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung, A. I., S. J. Projan, R. E. Edelstein, and V. A. Fischetti. 1995. Cloning, expression, and nucleotide sequence of a Staphylococcus aureus gene (fbpA) encoding a fibrinogen-binding protein. Infect. Immun. 63:1914-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duthie, E. S., and L. L. Lorenz. 1952. Staphylococcal coagulase: mode of action and antigenicity. J. Gen. Microbiol. 6:95-107. [DOI] [PubMed] [Google Scholar]
- 11.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedrich, R., P. Panizzi, P. Fuentes-Prior, K. Richter, I. Verhamme, P. J. Anderson, S. Kawabata, R. Huber, W. Bode, and P. E. Bock. 2003. Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation. Nature 425:535-539. [DOI] [PubMed] [Google Scholar]
- 13.Goh, S. H., S. K. Byrne, J. L. Zhang, and A. W. Chow. 1992. Molecular typing of Staphylococcus aureus on the basis of coagulase gene polymorphisms. J. Clin. Microbiol. 30:1642-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemker, H. C., B. M. Bas, and A. D. Muller. 1975. Activation of a pro-enzyme by a stoichiometric reaction with another protein. The reaction between prothrombin and staphylocoagulase. Biochim. Biophys. Acta 379:180-188. [DOI] [PubMed] [Google Scholar]
- 15.Hiramatsu, K., S. Watanabe, F. Takeuchi, T. Ito, and T. Baba. 2004. Genetic characterization of methicillin-resistant Staphylococcus aureus. Vaccine 22(Suppl. 1):S5-S8. [DOI] [PubMed] [Google Scholar]
- 16.Holden, M. T., E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 101:9786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hookey, J. V., J. F. Richardson, and B. D. Cookson. 1998. Molecular typing of Staphylococcus aureus based on PCR restriction fragment length polymorphism and DNA sequence analysis of the coagulase gene. J. Clin. Microbiol. 36:1083-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iandolo, J. J., V. Worrell, K. H. Groicher, Y. Qian, R. Tian, S. Kenton, A. Dorman, H. Ji, S. Lin, P. Loh, S. Qi, H. Zhu, and B. A. Roe. 2002. Comparative analysis of the genomes of the temperate bacteriophages phi 11, phi 12 and phi 13 of Staphylococcus aureus 8325. Gene 289:109-118. [DOI] [PubMed] [Google Scholar]
- 19.Igarashi, H., T. Morita, and S. Iwanaga. 1979. A new method for purification of staphylocoagulase by a bovine prothrombin-Sepharose column. J. Biochem. (Tokyo) 86:1615-1618. [DOI] [PubMed] [Google Scholar]
- 20.Igarashi, H., M. Shingaki, H. Ushioda, H. Fujikawa, T. Terayama, J. Kawano, and A. Shimizu. 1985. Immunological differentiation of staphylocoagulase produced by Staphylococcus aureus and Staphylococcus intermedius. Gustav Fischer Verlag, Stuttgart, Germany.
- 21.Kaida, S., T. Miyata, Y. Yoshizawa, H. Igarashi, and S. Iwanaga. 1989. Nucleotide and deduced amino acid sequences of staphylocoagulase gene from Staphylococcus aureus strain 213. Nucleic Acids Res. 17:8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaida, S., T. Miyata, Y. Yoshizawa, S. Kawabata, T. Morita, H. Igarashi, and S. Iwanaga. 1987. Nucleotide sequence of the staphylocoagulase gene: its unique COOH-terminal 8 tandem repeats. J. Biochem. (Tokyo) 102:1177-1186. [DOI] [PubMed] [Google Scholar]
- 23.Kanemitsu, K., H. Yamamoto, H. Takemura, M. Kaku, and J. Shimada. 2001. Relatedness between the coagulase gene 3′-end region and coagulase serotypes among Staphylococcus aureus strains. Microbiol. Immunol. 45:23-27. [DOI] [PubMed] [Google Scholar]
- 24.Kawabata, S., and S. Iwanaga. 1994. Structure and function of staphylothrombin. Semin. Thromb. Hemost. 20:345-350. [DOI] [PubMed] [Google Scholar]
- 25.Kawabata, S., T. Morita, T. Miyata, S. Iwanaga, and H. Igarashi. 1986. Isolation and characterization of staphylocoagulase chymotryptic fragment. Localization of the procoagulant- and prothrombin-binding domain of this protein. J. Biol. Chem. 261:1427-1433. [PubMed] [Google Scholar]
- 26.Komori, Y., N. Iimura, R. Yamashita, H. Sugihara, and T. Nikai. 2001. Characterization of coagulase from Staphylococcus intermedius. J. Nat. Toxins 10:111-118. [PubMed] [Google Scholar]
- 27.Kondo, N., T. Ito, and K. Hiramatsu. 1997. Genetic basis for molecular epidemiology of MRSA. Nippon Saikingaku Zasshi 52:417-434. [DOI] [PubMed] [Google Scholar]
- 28.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 29.Lipsitch, M. 1999. Bacterial vaccines and serotype replacement: lessons from Haemophilus influenzae and prospects for Streptococcus pneumoniae. Emerg. Infect. Dis. 5:336-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDevitt, D., P. Francois, P. Vaudaux, and T. J. Foster. 1994. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol. 11:237-248. [DOI] [PubMed] [Google Scholar]
- 31.McGavin, M. H., D. Krajewska-Pietrasik, C. Ryden, and M. Hook. 1993. Identification of a Staphylococcus aureus extracellular matrix-binding protein with broad specificity. Infect. Immun. 61:2479-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ni Eidhin, D., S. Perkins, P. Francois, P. Vaudaux, M. Hook, and T. J. Foster. 1998. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol. Microbiol. 30:245-257. [DOI] [PubMed] [Google Scholar]
- 33.Nitta, D. M., M. A. Jackson, V. F. Burry, and L. C. Olson. 1995. Invasive Haemophilus influenzae type f disease. Pediatr. Infect. Dis. J. 14:157-160. [PubMed] [Google Scholar]
- 34.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 35.Palma, M., A. Haggar, and J. I. Flock. 1999. Adherence of Staphylococcus aureus is enhanced by an endogenous secreted protein with broad binding activity. J. Bacteriol. 181:2840-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palma, M., O. Shannon, H. C. Quezada, A. Berg, and J. I. Flock. 2001. Extracellular fibrinogen-binding protein, Efb, from Staphylococcus aureus blocks platelet aggregation due to its binding to the alpha-chain. J. Biol. Chem. 276:31691-31697. [DOI] [PubMed] [Google Scholar]
- 37.Pantucek, R., F. Gotz, J. Doskar, and S. Rosypal. 1996. Genomic variability of Staphylococcus aureus and the other coagulase-positive Staphylococcus species estimated by macrorestriction analysis using pulsed-field gel electrophoresis. Int. J. Syst. Bacteriol. 46:216-222. [DOI] [PubMed] [Google Scholar]
- 38.Phonimdaeng, P., M. O'Reilly, P. Nowlan, A. J. Bramley, and T. J. Foster. 1990. The coagulase of Staphylococcus aureus 8325-4. Sequence analysis and virulence of site-specific coagulase-deficient mutants. Mol. Microbiol. 4:393-404. [DOI] [PubMed] [Google Scholar]
- 39.Rammelkamp, C. H., Jr., and G. F. Badger. 1950. Antigenicity of cell-free staphylococcal coagulase. J. Infect. Dis. 86:159-163. [DOI] [PubMed] [Google Scholar]
- 40.Robinson, D. A., and M. C. Enright. 2004. Evolution of Staphylococcus aureus by large chromosomal replacements. J. Bacteriol. 186:1060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruzin, A., J. Lindsay, and R. P. Novick. 2001. Molecular genetics of SaPI1—a mobile pathogenicity island in Staphylococcus aureus. Mol. Microbiol. 41:365-377. [DOI] [PubMed] [Google Scholar]
- 42.Shopsin, B., M. Gomez, M. Waddington, M. Riehman, and B. N. Kreiswirth. 2000. Use of coagulase gene (coa) repeat region nucleotide sequences for typing of methicillin-resistant Staphylococcus aureus strains. J. Clin. Microbiol. 38:3453-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ubeda, C., M. A. Tormo, C. Cucarella, P. Trotonda, T. J. Foster, I. Lasa, and J. R. Penades. 2003. Sip, an integrase protein with excision, circularization and integration activities, defines a new family of mobile Staphylococcus aureus pathogenicity islands. Mol. Microbiol. 49:193-210. [DOI] [PubMed] [Google Scholar]
- 45.Ushioda, H., T. Terayama, S. Sakai, H. Zen-Yoji, M. Nishiwaki, and A. Hidano. 1981. Coagulase typing of Staphylococcus aureus and its application in routine work. Gustav Fischer Verlag, Stuttgart, Germany.
- 46.Vandenesch, F., C. Lebeau, M. Bes, G. Lina, B. Lina, T. Greenland, Y. Benito, Y. Brun, J. Fleurette, and J. Etienne. 1994. Clotting activity in Staphylococcus schleiferi subspecies from human patients. J. Clin. Microbiol. 32:388-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vandenesch, F., C. Lebeau, M. Bes, D. McDevitt, T. Greenland, R. P. Novick, and J. Etienne. 1994. Coagulase deficiency in clinical isolates of Staphylococcus aureus involves both transcriptional and post-transcriptional defects. J. Med. Microbiol. 40:344-349. [DOI] [PubMed] [Google Scholar]
- 48.Wann, E. R., S. Gurusiddappa, and M. Hook. 2000. The fibronectin-binding MSCRAMM FnbpA of Staphylococcus aureus is a bifunctional protein that also binds to fibrinogen. J. Biol. Chem. 275:13863-13871. [DOI] [PubMed] [Google Scholar]
- 49.Wenger, J. D., R. Pierce, K. Deaver, R. Franklin, G. Bosley, N. Pigott, C. V. Broome, et al. 1992. Invasive Haemophilus influenzae disease: a population-based evaluation of the role of capsular polysaccharide serotype. J. Infect. Dis. 165(Suppl. 1):S34-S35. [DOI] [PubMed] [Google Scholar]