Abstract
Bordetella pertussis and Bordetella bronchiseptica, which are respiratory mucosal pathogens of mammals, produce and utilize the siderophore alcaligin to acquire iron in response to iron starvation. A predicted permease of the major facilitator superfamily class of membrane efflux pumps, AlcS (synonyms, OrfX and Bcr), was reported to be encoded within the alcaligin gene cluster. In this study, alcS null mutants were found to be defective in growth under iron starvation conditions, in iron source utilization, and in alcaligin export. trans complementation using cloned alcS genes of B. pertussis or B. bronchiseptica restored the wild-type phenotype to the alcS mutants. Although the levels of extracellular alcaligin measured in alcS strain culture fluids were severely reduced compared with the wild-type levels, alcS mutants had elevated levels of cell-associated alcaligin, implicating AlcS in alcaligin export. Interestingly, a ΔalcA mutation that eliminated alcaligin production suppressed the growth defects of alcS mutants. This suppression and the alcaligin production defect were reversed by trans complementation of the ΔalcA mutation in the double-mutant strain, confirming that the growth-defective phenotype of alcS mutants is associated with alcaligin production. In an alcA::mini-Tn5 lacZ1 operon fusion strain background, an alcS null mutation resulted in enhanced AlcR-dependent transcriptional responsiveness to alcaligin inducer; conversely, AlcS overproduction blunted the transcriptional response to alcaligin. These transcription studies indicate that the alcaligin exporter activity of AlcS is required to maintain appropriate intracellular alcaligin levels for normal inducer sensing and responsiveness necessary for positive regulation of alcaligin system gene expression.
Bordetella pertussis, Bordetella parapertussis, and Bordetella bronchiseptica are mucosal respiratory pathogens of mammals. The etiologic agent of whooping cough or pertussis, B. pertussis, is an obligate human pathogen that has no known environmental or nonhuman reservoirs; the closely related species B. parapertussis causes a pertussis-like syndrome in humans, as well as respiratory infections in sheep. B. bronchiseptica infects nonhuman mammals primarily and is responsible for swine atrophic rhinitis, canine kennel cough, and snuffles in rabbits. Bordetellae are remarkably versatile in their ability to acquire the essential biometal iron; the multiplicity of their iron transport systems underscores the fundamental biological importance of iron for these microbes, which is reflected in the diverse metabolic and enzymatic activities in which iron plays a central role. It is well established that B. pertussis and B. bronchiseptica produce and utilize the siderophore alcaligin (20, 46) to scavenge iron for biological assimilation. B. pertussis and B. bronchiseptica are also capable of utilizing certain siderophores produced by other microbial species, such as enterobactin (2, 7), ferrichrome (6), desferrioxamine B (6), and certain pyoverdins (52), to satisfy their iron requirement. Apart from these iron sources of microbial origin, eukaryotic host-derived iron sources, such as hemin and hemoglobin, are also readily assimilated by B. pertussis and B. bronchiseptica cells by way of a specific heme-iron transport system (60). Iron acquisition by Bordetella is a highly regulated process. Expression of the known iron transport systems of Bordetella species, specifically those for assimilation of ferric alcaligin, ferric enterobactin, and heme iron, is repressed by the Fur protein and iron when intracellular iron levels are sufficient (24). Maximal expression of these iron transport systems under iron starvation conditions is orchestrated by priority regulation mechanisms that depend on iron source sensing, in that expression of each iron system is maximized by a specific positive transcriptional regulator that is responsive to the cognate iron source acting as the inducer (2, 21, 61). Previous studies have established that under iron starvation growth conditions, transcription of the alcABCDER operon promoter and the fauA promoter is activated by the AraC family regulator protein AlcR in response to the alcaligin inducer (18, 21). In this way, alcaligin participates directly in the control of its own production and transport. Induction of AlcR-mediated transcriptional activation is responsive to nanomolar concentrations of alcaligin (21). This iron source sensing for priority regulation may be important for effective adaptation to changes in the host environment that affect iron source availability during the course of an infection (24). The full iron-scavenging potential of B. pertussis and B. bronchiseptica is undetermined, but in addition to the three known outer membrane receptor proteins required for utilization of ferric alcaligin (FauA), ferric enterobactin (BfeA), and heme iron (BhuR), at least nine more genes encoding siderophore receptor homologs of unknown specificity can be identified in the B. pertussis genome (50).
The alcaligin siderophore gene cluster encodes activities required for the biosynthesis and transport of alcaligin, and this system is the most thoroughly characterized iron acquisition system of the bordetellae. The organization of the alcaligin genes is conserved among the related species B. pertussis, B. bronchiseptica, and B. parapertussis (50). Significant progress has been made in understanding the production of alcaligin (4, 19, 34, 41) and its chemistry of iron chelation (12, 13, 20, 38, 39, 58), iron uptake by way of the ferric alcaligin receptor protein FauA (18), and the negative and positive transcriptional regulation of alcaligin biosynthesis and transport genes (8, 9, 14, 15, 16, 21, 40, 51). However, as with most siderophore systems, elucidation of the process of alcaligin siderophore export or release from the producing cell has lagged. Relatively recently, several bacterial siderophore gene clusters have been reported to encode major facilitator superfamily (MFS) proteins with known or postulated roles in either siderophore export or siderophore uptake. Experimental evidence has established that the Escherichia coli enterobactin system gene product EntS (synonyms, YbdA and P43) is a MFS transporter that is required for enterobactin export (33). Likewise, the csbX gene adjacent to the catecholate siderophore biosynthesis gene cluster of Azotobacter vinelandii has been shown to be involved in export of catecholates related to the siderophore protochelin (49), and in the plant pathogen Erwinia chrysanthemi, the yhcA gene of the achromobactin siderophore gene cluster encodes an MFS protein involved in export or release of newly synthesized achromobactin (31). Other siderophore system genes, for example, ybtX of the Yersinia pestis yersiniabactin system (29) and pvsC of the Vibrio parahemolyticus vibrioferrin system (59), also encode putative MFS proteins. However, although it has been reported that pvsC mutants are defective in vibrioferrrin export (59), ybtX appears to be dispensable for growth under iron-deficient culture conditions in vitro, and no role for ybtX in yersiniabactin export has been identified (29). In contrast to the siderophore export functions of some MFS proteins, the MFS transporter RhtX of Sinorhizobium meliloti has been reported to be required for uptake of the siderophores rhizobactin and schizokinen (27). Uptake of ferrioxamine-type siderophores by Saccharomyces cerevisiae is known to involve an MFS transporter (44).
Prior to the determination of the full genome sequences of B. pertussis and B. bronchiseptica by Parkhill and colleagues (50), an open reading frame encoding a putative MFS inner membrane translocase of unknown specificity, originally named orfX (now locus tag BP2462) but having the newly proposed designation alcS, was identified by analysis of nucleotide sequences flanking the alcR regulator gene of B. pertussis strain UT25 (9). The B. bronchiseptica alcS ortholog, dubbed bcr (locus tag BB3899) because of the amino acid sequence similarity of its product to the E. coli Bcr bicyclomycin resistance protein, was independently identified by Pradel and coworkers (51) by nucleotide sequencing of the alcR region of B. bronchiseptica. Because alcS resides within the alcaligin gene cluster and the AlcS protein exhibits similarity with other MFS proteins associated with siderophore systems, AlcS was hypothesized to function in alcaligin-mediated iron assimilation.
This study demonstrates that AlcS is involved in alcaligin siderophore efflux or release by Bordetella cells. Furthermore, the results indicate that AlcS function is integral to the process of inducer sensing for AlcR-mediated transcriptional control of alcaligin system gene expression.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bordetella strains and plasmids used in this study are listed in Table 1. B. bronchiseptica strains were maintained on Luria-Bertani agar plates, and modified Stainer-Scholte medium (SS) (56) was used for broth cultures. Iron-replete SS was supplemented with 36 μM ferrous sulfate as described previously (4). This concentration of iron is sufficient to support growth and repress siderophore production, but it is not toxic or inhibitory to growth. For iron-depleted SS cultures, SS was deferrated by treatment with Chelex 100 resin (Bio-Rad Laboratories, Richmond, CA). Batch cultures using iron-depleted (deferrated) SS and acid-cleaned glassware contained only trace amounts of iron, which limited the growth of all B. bronchiseptica strains. The final concentrations of antibiotics used for plasmid marker selection were as follows: tetracycline, 15 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 30 μg/ml; and ampicillin, 100 μg/ml. In analyses of induction of alcA::mini-Tn5 lacZ1 transcription by alcaligin, iron-depleted SS was amended as indicated below using purified alcaligin siderophore. Bacterial growth yields from broth cultures were estimated spectrophotometrically at 600 nm, and the numbers of CFU/ml were determined by using standard plate counting methods. All glassware was acid cleaned and rinsed repeatedly in distilled deionized water prior to use.
TABLE 1.
Bordetella strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Reference(s) or source |
|---|---|---|
| B. bronchiseptica strains | ||
| B013N | Wild-type, nalidixic acid-resistant derivative of swine isolate strain B | 4 |
| BRM18 | ΔfauA2 mutant derivative of B013N, with a nonpolar in-frame fauA deletion mutation, alcaligin transport deficient | 18 |
| BRM16 | alcS::Kanr mutant derivative of B013N, with a kanamycin resistance cassette insertion at the unique PstI restriction site in alcS | This study |
| BRM26 | ΔalcA1 mutant derivative of B013N, with a nonpolar in-frame alcA deletion mutation, alcaligin deficient | This study |
| BRM27 | ΔalcA1 alcS::Kanr mutant derived from BRM16 | This study |
| BRM1 | alcA::mini-Tn5 lacZ1 mutant derivative of B013N, alcaligin deficient | 4 |
| BRM30 | ΔalcS1 alcA::mini-Tn5 lacZ1 mutant derived from BRM1 | This study |
| B. pertussis UT25 | Wild-type, virulent phase isolate | 30 |
| Recombinant plasmids | ||
| pBRM6 | 20-kb alcS+ SalI DNA fragment of B. bronchiseptica alcC mutant BRM6 cloned into pGEM3Z, Apr | 4 |
| pBB21 | 2.3-kb alcA+ BamHI-SphI DNA fragment of B. pertussis UT25 cloned into pBBR1MCS, Cmr | Kang and Armstrong, unpublished |
| p3Z/4.6S | 4.6-kb alcABC+ SmaI DNA fragment of B. bronchiseptica B013N cloned into pGEM3Z, Apr | This study |
| pRK/alcSBb | 1.4-kb alcS+ PCR-generated DNA fragment of B. bronchiseptica B013N cloned into pRK415, Tcr | This study |
| pRK/alcSBp | 1.4-kb alcS+ PCR-generated DNA fragment of B. pertussis UT25 cloned into pRK415, Tcr | This study |
| Other plasmids | ||
| pGEM3Z | 2.7-kb cloning vector, Apr | Promega |
| pBluescript II KS(+) | 2.9-kb cloning vector, Apr | Stratagene |
| pRK415 | 10.5-kb broad-host-range cloning vector, Tcr | 42 |
| pBBR1MCS | 4.7-kb broad-host-range cloning vector, Cmr | 3, 43 |
| pEG7 | 8.0-kb allelic exchange vector, Apr Gmr | 26 |
| pEG18.3 | 11.8-kb allelic exchange vector, Apr Gmr Kmr SacBR+ | 26 |
| pBSL86 | 3.6-kb vector source of kanamycin resistance cassette used in construction of alcS::Kanr allele of BRM16 | 1 |
Routine DNA procedures.
DNA cloning procedures used standard methods described previously (55). For conjugal transfer of pRK415 and pBBR1MCS plasmid derivatives to Bordetella strains we used triparental mating as described previously (19), with E. coli DH5α as the plasmid donor strain and DH5α(pRK2013) as the source of mobilization functions. Transconjugants were selected on agar plates containing the appropriate antibiotics and crude colicin B (17).
Nucleotide sequence and protein analysis.
Bordetella genomic DNA sequence data were produced by the Bordetella Sequencing Group at the Wellcome Trust Sanger Institute. Annotated Bordetella genome sequences were accessed at the GeneDB website (http://www.genedb.org/), developed and maintained by the Sanger Institute's Pathogen Sequencing Unit; genome sequence features were visualized using the Artemis (54) genome sequence viewer and annotation tool (http://www.sanger.ac.uk/Software/Artemis/). AlcS protein motifs were predicted using bioinformatics tools available online through the ExPASy proteomics server (http://us.expasy.org/tools/scanprosite/) of the Swiss Institute of Bioinformatics and tools available at the Transport Classification Database (http://tcdb.ucsd.edu/) operated by Milton Saier's bioinformatics group at the University of California-San Diego. Multiple-protein alignment was performed using the online Clustal W multiple sequence alignment program (25) provided by the European Bioinformatics Institute of the European Molecular Biology Laboratory (http://www.ebi.ac.uk/clustalw/index.html). In other DNA and protein analyses for sequence data management and PCR primer selection we used the Lasergene software package, version 5.53 for Mac OS X (DNASTAR, Inc., Madison, WI).
Bordetella alcaligin siderophore purification and detection.
Alcaligin used in quantitative siderophore detection assays, in growth stimulation bioassays, and as the inducer in transcriptional analyses was purified from B. bronchiseptica culture supernatant fluids by a modification of the benzyl alcohol-ether extraction method (48), as previously described (20), and was recrystallized multiple times from ethanolic solutions. The chrome azurol S (CAS) universal siderophore detection assay (57) was used to quantitate alcaligin produced by B. bronchiseptica grown in SS as reported previously (4). Alcaligin concentrations in test samples were determined by linear regression analysis using standard curves generated using aqueous solutions of highly purified alcaligin (0.00, 0.39, 0.78, 1.56, 3.13, 6.25, 12.50, 25.00, and 50.00 μM) in the CAS assay. Alcaligin standard solutions were assayed in triplicate in parallel with test samples and routinely yielded correlation coefficients (r) of ≥0.995.
For quantitation of cell-associated alcaligin in B. bronchiseptica cells, bacteria were recovered from 50-ml iron-depleted SS cultures by centrifugation, washed twice using 25 ml of cold 10 mM Tris HCl-0.9% NaCl (TBS), and resuspended in 10 ml TBS, and cell densities (A600) of the suspensions were measured spectrophotometrically. Suspensions were diluted to 1 U of optical density at 600 nm (OD600)/ml in TBS, and portions were reserved for CFU/ml determinations. Twenty-milliliter portions of each suspension (20 U of optical density at 600 nm [ODU600]) were supplemented with 50 mg lysozyme and then subjected to three cycles of freezing in a dry ice-ethanol bath, followed by thawing at 37°C. Each lysate was vigorously extracted using 5 ml phenol:chloroform (1:1), and the organic phase containing alcaligin was recovered after brief centrifugation for phase separation. Two milliliters of water was added to the phenol:chloroform extract, and alcaligin was driven to the aqueous phase by addition of 30 ml diethyl ether. The aqueous phase was recovered and evaporated to dryness using a rotary evaporator. The dried residue was redissolved in 500 μl water, and alcaligin concentrations were determined by the CAS method using purified alcaligin as the standard.
TLC.
For thin-layer chromatography (TLC) analysis of alcaligin exported by B. bronchiseptica cultured in iron-depleted SS, 10-ml portions of cell-free culture supernatant fluids were extracted using 1 ml phenol:chloroform (1:1). The organic phase was recovered, and 0.5 ml water was added, followed by 3 ml diethyl ether. The aqueous phase containing alcaligin was evaporated to dryness, and the residue was redissolved in 20 μl methanol. The methanolic solution was spotted onto glass TLC plates (20 by 20 cm) precoated with a 250-μm layer of silica gel (Sigma Chemical Co., St. Louis, MO) and developed with n-butanol:acetic acid:water (4:1:5) (upper phase). Ferric iron-reactive alcaligin was localized by spraying the dried TLC plates with freshly prepared 1% ferric chloride in 1 mM HCl.
Iron source feeding assays.
Utilization of various iron sources by B. bronchiseptica was assessed by using agar plate bioassays as described previously (9, 20), with iron-restricted Luria-Bertani agar plates containing the nonutilizable iron chelator ethylenediaminedi-[(o-hydroxyphenyl)acetic acid] (EDDA) at a final concentration of 100 μg/ml. Iron restriction by EDDA limits growth of the indicator bacteria seeded into the agar, allowing growth stimulation by iron sources to be observed. The diameters of bacterial growth zones surrounding the 6-mm-diameter sample wells were measured after 24 h of incubation at 37°C.
β-Galactosidase (LacZ) assays.
B. bronchiseptica alcA::mini-Tn5 lacZ1 gene fusion strains were assayed for LacZ activity by the Miller method (45) modified as described previously (23), after culture in iron-replete or iron-depleted SS. In experiments involving induction of alcA::mini-Tn5 lacZ1 transcription by the alcaligin siderophore, SS cultures were amended with purified alcaligin at different final concentrations. The LacZ activities presented below are means ± standard deviations for triplicate assays.
Construction of B. bronchiseptica and B. pertussis alcS+ plasmids for genetic complementation analysis.
Chromosomal DNA regions (1.4 kb) of B. bronchiseptica B013N and B. pertussis UT25, encompassing the alcS coding sequence (CDS) with 112-bp upstream and 40-bp downstream DNA sequences, were amplified by PCR using primer/adapters incorporating synthetic BamHI (5′-GGCCGGATCCTTGCGGCGGCAGGCATTGACATT-3′) and EcoRI (5′-GGCCGAATTCTCGGCGATGACGGAGTGCGTATGG-3′) restriction sites for cloning of the products. The adapters were trimmed by digestion of the PCR products using BamHI and EcoRI, and the trimmed DNA fragments were cloned into plasmid vectors pRK415 and pBluescript II KS(+).
Construction of B. bronchiseptica alcS mutant strains BRM16 and BRM30.
A 2.7-kb KpnI-EcoRV B. bronchiseptica DNA fragment was subcloned from plasmid pBRM6 into the plasmid cloning vector pBluescript II KS(+), and the PstI kanamycin resistance gene cassette of pBSL86 was ligated at the unique PstI restriction site within the alcS coding region. This alcS::Kanr recombinant plasmid was delivered to B. bronchiseptica strain B013N by electroporation, and plasmid integrants were selected on the basis of ampicillin resistance conferred by the cloning vector marker and kanamycin resistance associated with the insertion. Several integrant clones were passaged without selection in Luria-Bertani broth, and then bacteria were replica plated and scored for ampicillin sensitivity and kanamycin resistance, which yielded presumptive B013N alcS::Kanr derivatives. Allelic exchange resulting in transfer of the alcS::Kanr allele to the B. bronchiseptica chromosome, yielding alcS::Kanr mutant strain BRM16, was confirmed by Southern blot DNA hybridization analysis.
The ΔalcS1 mutant allele with an in-frame coding sequence deletion was generated by PCR splicing by overlap extension (37). DNA oligonucleotide primer A (5′-GGGGGAATTCGGGGCGCCGGCATTGAGAACCTT-3′) with primer A′ (5′-ATGCTGCAACCACGCCCACAGCAGCCCCAGCATTACCATCAT-3′) and primer B (5′-GGGGGGATCCCGGGCCCAGGGAGCGTTGTTG-3′) with primer B′ (5′-GTAATGCTGGGGCTGCTGTGGGCGTGGTTGCAGCATTTATCG-3′) were used to generate two B. bronchiseptica DNA amplimers with overlapping termini spanning the alcS deletion junction. The amplimers were spliced together at their overlapping termini by combining them in equimolar ratios and joining them together by PCR using the outside primers, primers A and B. The approximately 2-kb spliced product was trimmed at the synthetic primer-directed EcoRI and BamHI adapter sequences, and the product was cloned into allelic exchange plasmid pEG7. The ΔalcS1 mutant allele, deleted for all of the alcS CDS except the N-terminal 20 codons and the C-terminal 17 codons, was transferred to the chromosome of B. bronchiseptica strain BRM1 by allelic exchange using the suicide plasmid pEG7 sacBR-based positive selection system (26) to produce mutant strain BRM30 (ΔalcS1 alcA::mini-Tn5 lacZ1). Deletion of the chromosomal alcS sequences was confirmed by PCR with alcS-specific primers and chromosomal DNA templates.
Construction of B. bronchiseptica ΔalcA1 mutant strains BRM26 and BRM27.
A pGEM3Z-based recombinant plasmid carrying a 4.6-kb alcABC+ B. bronchiseptica SmaI DNA fragment was digested to completion using BglII, and the plasmid was religated to produce an in-frame 297-bp deletion in the alcA CDS. The ΔBglII DNA insert fragment was excised using BamHI and EcoRI, subcloned into the allelic exchange vector pEG7, and transferred to the chromosome of B. bronchiseptica strains B013N and BRM16 (alcS::Kanr) by allelic exchange using the suicide plasmid pEG7 sacBR-based positive selection system, yielding the ΔalcA1 mutant strain BRM26 and the ΔalcA1 alcS::Kanr double-mutant strain BRM27. Deletion of the chromosomal alcA sequences was confirmed by using PCR with alcA-specific primers and chromosomal DNA templates. The ΔalcA1 mutation eliminated alcaligin production by BRM26 and BRM27, and alcaligin production was restored to both strains by trans complementation using cloned copies of the alcA gene alone as plasmid pBB21 (H. Y. Kang and S. K. Armstrong, unpublished data), confirming that the ΔalcA1 mutation was not polar on the downstream, cotranscribed alcaligin biosynthesis genes.
RESULTS
alcS encodes a predicted MFS transporter.
B. pertussis and B. bronchiseptica produce and utilize the macrocyclic dihydroxamate siderophore alcaligin (Fig. 1A) (20, 46). The organization of the alcaligin siderophore gene cluster (Fig. 1B) is conserved among the related species B. pertussis, B. bronchiseptica, and B. parapertussis (41, 50). The alcS gene maps between alcR, encoding the alcaligin-responsive AraC-family transcriptional regulator, and the fauA ferric alcaligin outer membrane receptor gene. Previous transcription studies and mutational analyses determined that alcS transcription is not iron repressible and is independent of the distal upstream alcABCDER operon promoter and the iron-regulated secondary promoter upstream of alcR (9, 40). The predicted alcS gene product, a 407-amino-acid protein with a molecular mass of approximately 43 kDa, is a putative transporter of the MFS class of membrane efflux pumps (PROSITE:PS50850). AlcS structural analysis predicts 12 transmembrane α-helical spanners (TMS) (Fig. 1C) and a potential prokaryotic membrane lipoprotein lipid attachment site (amino acid residues 12 to 22; PROSITE:PS00013). AlcS exhibits similarity with MFS proteins with known functions; in BLAST database searches, high-scoring segment pairs were identified for AlcS and the bicyclomycin resistance protein (sulfonamide resistance protein) Bcr of E. coli (accession no. P28246) (10), the chloramphenicol resistance protein CmlA of Pseudomonas aeruginosa (accession no. P32482) (11), and the multidrug resistance proteins EmrD (accession no. P31442) (47) and MdfA (accession no. CAA69997) (28) of E. coli. A multiple-protein sequence alignment of AlcS with these known MFS transporters is shown in Fig. 1C. AlcS has a protein motif that is conserved among the 12-TMS subfamily of MFS proteins, motif A (GxLaDrxGrkxx[x]l]; motif consensus sequences as described in reference 53, where x is any amino acid, uppercase letters show amino acids occurring in >70% of 12- and 14-TMS MFS proteins examined, lowercase letters show amino acids occurring in <40% of 12- and 14-TMS MFS proteins examined, and (x) is an amino acid not always present) (AlcS amino acid residues 68 to 80 [Fig. 1C]), found in the cytoplasmic loop between TMS2 and TMS3. Motif A is thought to impart a β-turn involved in a conformational change of the protein that opens and closes a transport channel (53). AlcS also has the conserved basic arginine residue found in motif B of TMS4 (AlcS residues 108 to 126) (Fig. 1C), which has been suggested to function in proton transfer by MFS transporters (53), and features of motif C in TMS5 (AlcS residues 138 to 156) (Fig. 1C) found in multidrug transporters and drug efflux pumps but not in MFS symporter proteins; motif C determinants are thought to dictate the direction of substrate transport (53). Among the MFS transporters with demonstrated involvement in bacterial siderophore export, AlcS exhibits 26% amino acid sequence identity with the E. coli enterobactin exporter EntS (33) over 324 amino acids and 26% identity over 340 amino acids with the Azotobacter vinelandii catecholate siderophore exporter CsbX (49).
FIG. 1.
AlcS is a predicted MFS transporter encoded within the Bordetella alcaligin siderophore gene cluster. (A) Molecular structure of alcaligin siderophore [1,8(S),11,18(S)-tetrahydroxy-1,6,11,16-tetraazacycloeicosane-2,5,12,15-tetrone] produced by Bordetella species. (B) Spatial organization of the Bordetella alcaligin siderophore gene cluster. The linear genetic map represents an approximately 12-kb BamHI-PstI chromosomal DNA region of B. bronchiseptica and B. pertussis (41). The arrows indicate the transcriptional orientations of genes, and the open rectangles upstream from alcA, alcR, and fauA represent the locations of known Fur-regulated promoter-operator regions. The arrow representing the alcS gene is shaded. (C) Multiple-protein sequence alignment of AlcS with representative bacterial MFS transporters with known functions. The proteins and GenBank accession numbers are as follows: AlcS, the Bordetella alcaligin exporter protein (B. bronchiseptica accession no. NP_890434, B. pertussis accession no. NP_881089); E. coli Bcr bicyclomycin resistance protein (accession no. P28246); Pseudomonas aeruginosa chloramphenicol resistance protein CmlA (accession no. P32482); E. coli multidrug resistance protein EmrD (accession no. P31442); and E. coli multidrug resistance protein MdfA (accession no. CAA69997). In the ClustalW alignment consensussequence, asterisks indicate residues that are identical in all sequences, colons indicate conserved substitutions, and periods indicate semiconserved substitutions. Predicted transmembrane segments of AlcS are overlined, and motif A conserved among 12-transmembrane segment MFS proteins is shaded.
alcS is required for maximal growth yields under iron starvation conditions.
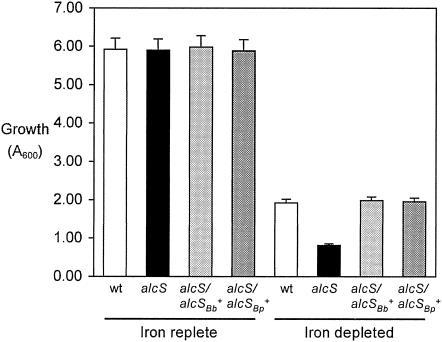
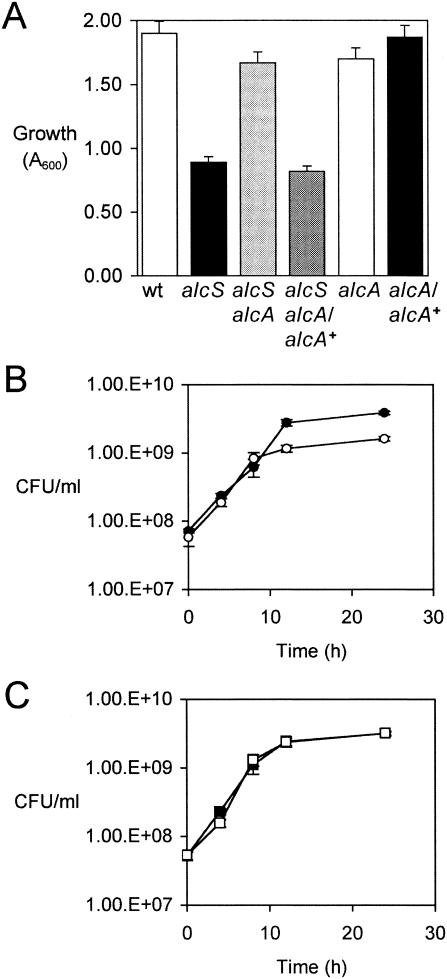
AlcS was hypothesized to function in iron assimilation because of the genomic location of alcS within the alcaligin gene cluster and its similarity with other MFS proteins associated with siderophore systems. As a preliminary test of this hypothesis, iron starvation growth studies of a B. bronchiseptica alcS null mutant were performed. B. bronchiseptica strains were cultured to the early stationary growth phase (24 h) in iron-replete or iron-depleted SS, and the growth yields were measured. As typically observed, growth of wild-type B. bronchiseptica strain B013N was reduced by iron starvation more than threefold compared with iron-replete SS cultures (Fig. 2). Growth limitation for Bordetella in iron-depleted SS batch cultures, which were stringently deferrated, was imposed by the limited availability of iron in the culture system, and alcaligin production was maximized. Compared with B013N, isogenic alcS::Kanr strain BRM16 exhibited a marked further reduction in growth yield in iron-depleted cultures. Growth of the alcS mutant strain was reduced over sevenfold by iron starvation, yet no significant difference in growth yield between the wild type and the alcS mutant strain was observed after culture in iron-replete SS, which contained sufficient iron (36 μM) to satisfy nutritional requirements of Bordetella and to repress alcaligin production. Remarkably, the growth yields of the alcS mutant in iron-depleted SS were even less than those commonly achieved with non-alcaligin-producing mutant strains (4; unpublished data). trans complementation of the BRM16 alcS::Kanr mutation by plasmid-borne copies of the B. bronchiseptica and B. pertussis alcS genes alone (in plasmids pRK/alcSBb and pRK/alcSBp, respectively) (Table 1) restored growth in iron-depleted SS to wild-type levels.
FIG. 2.
Growth yields of B. bronchiseptica strains. The bars indicate growth yields of B. bronchiseptica strains after 24 h of culture in iron-replete batch cultures (36 μM iron; iron-sufficient growth conditions that repress alcaligin production) and iron-depleted SS batch cultures, expressed as A600 (means ± standard deviations; n = 3). In iron-depleted SS batch cultures, growth of all strains was limited by the presence of only trace amounts of iron in the system. Cultures were initiated at a cell density of 0.05 ODU600l using washed cells from 24-h iron-replete SS seed cultures. Abbreviations for strains used are as follows: wt, B103N; alcS, BRM16; alcS/alcSBb+, BRM16 (pRK/alcSBb); alcS/alcSBp+, BRM16 (pRK/alcSBp)
AlcS functions in alcaligin export.
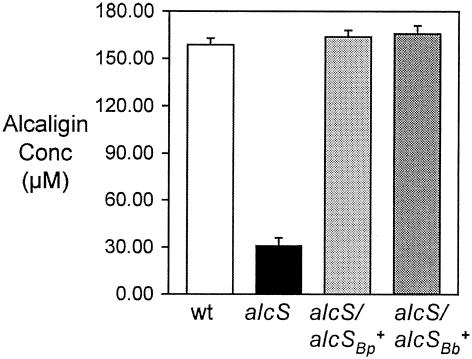
Maximal growth of B. bronchiseptica strains in iron-depleted SS requires the production and utilization of alcaligin. Since alcS mutant strain BRM16 displayed a growth defect under iron starvation conditions, it was hypothesized that alcS might be required for maximal alcaligin production. B. bronchiseptica strains were cultured for 24 h in iron-depleted SS, and alcaligin concentrations in cell-free culture supernatant fluids were measured by a quantitative CAS siderophore assay method. The alcaligin levels of the alcS strain were normalized to wild-type cell numbers (CFU) based on standard plate counting (Fig. 3). The alcaligin levels in BRM16 culture supernatant fluids were severely reduced to only 20% of the wild-type levels. trans complementation of the alcS::Kanr mutation by alcSBb+ or alcSBp+ plasmids restored alcaligin concentrations to wild-type levels. Using feeding assays, the presence of authentic alcaligin in cell-free alcS culture fluids was verified by demonstrating the ability of those fluids to stimulate the growth of wild-type B. bronchiseptica but not the growth of ΔfauA2 mutant strain BRM18 (18), which does not produce the ferric alcaligin receptor protein (data not shown).
FIG. 3.
Alcaligin concentrations in cell-free culture supernatant fluids. Alcaligin concentrations in cell-free culture supernatant fluids after 24 h of growth in iron-depleted SS, expressed as alcaligin monomer concentrations (means ± standard deviations; n = 3), were measured by the quantitative CAS method. The concentrations shown were normalized to growth yields based on CFU/ml determinations. Abbreviations for strains used are as follows: wt, B013N; alcS, BRM16; alcS/alcSBb+, BRM16 (pRK/alcSBb); alcS/alcSBp+, BRM16 (pRK/alcSBp).
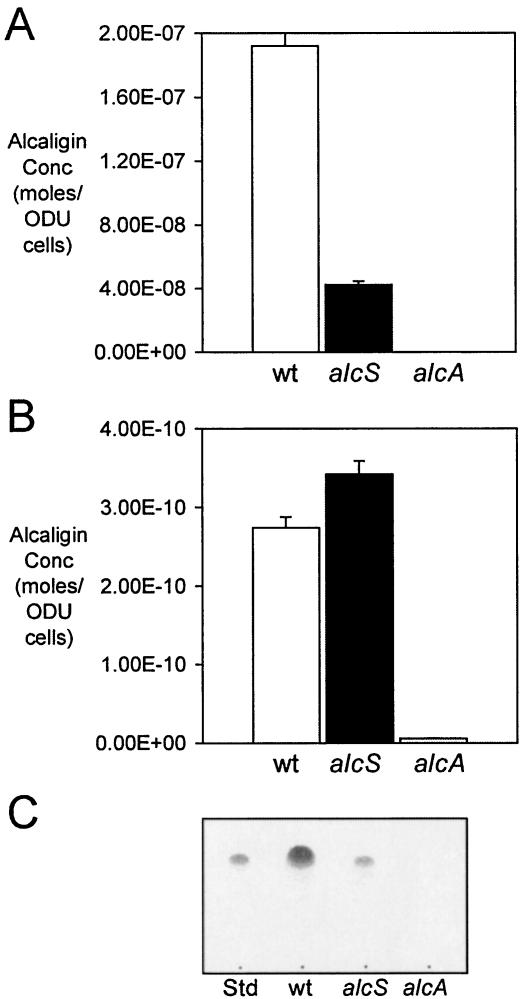
Given the similarity of AlcS with known solute efflux proteins, the reduced alcaligin levels measured in alcS culture supernatant fluids suggested that alcS mutants might be defective in export or release of the alcaligin siderophore from the bacterial cell. To determine whether alcaligin levels were altered in alcS cells, B. bronchiseptica B013N and the isogenic alcS::Kanr strain BRM16 were cultured for 24 h in iron-depleted SS, and alcaligin siderophore concentrations in extracellular and cell-associated fractions were quantified and normalized to cell numbers. As observed in previous experiments, the levels of alcaligin export by alcS cells measured in culture supernatant fluids were only 22% of wild-type levels (Fig. 4A). In contrast, the levels of alcS cell-associated alcaligin were 125% of the wild-type levels (Fig. 4B). Control fractions from the alcaligin biosynthesis mutant BRM26 contained no significant amounts of the siderophore. The disparity between the acute reduction in extracellular alcaligin levels and the significant elevation in cell-associated levels of alcaligin in alcS cells compared with wild-type cells is consistent with the hypothesized role for AlcS in siderophore release or export from Bordetella cells. Using TLC analysis, the siderophore activity in alcS culture fluids was identified as alcaligin (Fig. 4C), based on alcaligin's known retention factor constant (Rf) (20) and comigration with purified alcaligin.
FIG. 4.
Extracellular and cell-associated alcaligin siderophore. Alcaligin production by B. bronchiseptica strains cultured for 24 h in iron-depleted SS, expressed as alcaligin monomer concentrations (means ± standard deviations; n = 3), was normalized to cell numbers (1 U of OD600 [ODU] = ∼2 × 109 CFU). (A) Exported alcaligin as measured in cell-free culture supernatant fluids. (B) Cell-associated alcaligin production as measured in cell extracts. Abbreviations for strains used are as follows: wt, B013N; alcS, BRM16; alcA, BRM26. (C) Thin-layer chromatography of alcaligin extracted from cell-free culture supernatant fuids after 24 h growth in iron-depleted SS. Abbreviations for samples used; Std, 50 μg purified alcaligin; wt. B013N extract; alcS, BRM16 extract; alcA, BRM26 extract.
Alcaligin production in an alcS mutant is inhibitory to growth.
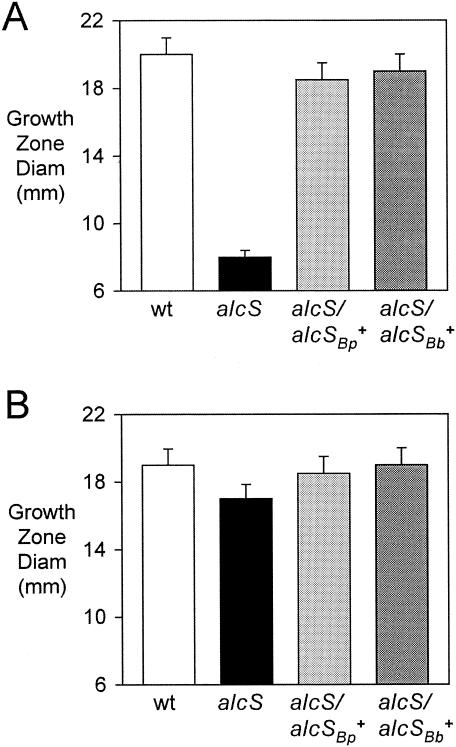
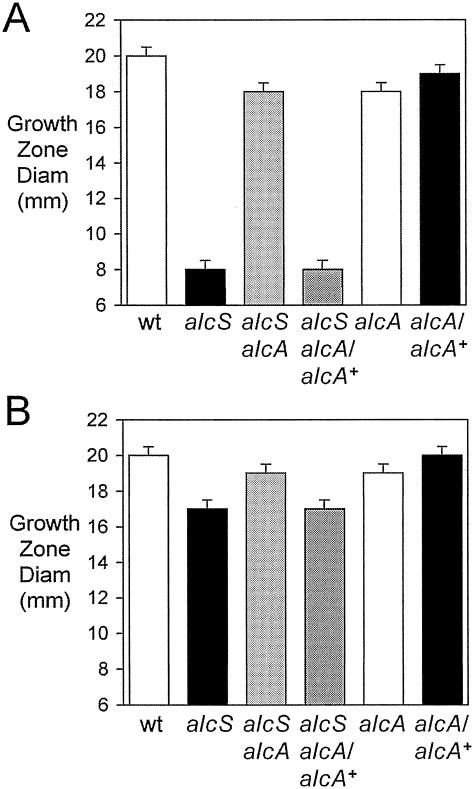
Feeding assays revealed that BRM16 had an acute defect in growth stimulation by ferric chloride relative to the wild-type strain (Fig. 5A). A qualitatively similar alcS growth stimulation defect was also observed in feeding assays using iron-restricted agar and ferric nitrate, ferrous sulfate, and ferrous ammonium sulfate as iron sources (data not shown). The unexpected observation that the alcS strain was severely defective in growth stimulation by iron salts at millimolar concentrations was remarkable. Ferric chloride is routinely used as the positive control iron source in growth stimulation feeding assays because at millimolar concentrations far exceeding the iron-chelating capacity of the 100-μg/ml (approximately 277 μM) EDDA used to restrict iron availability, iron assimilation does not require the involvement of any known high-affinity uptake system. Iron chelation by EDDA in the feeding assay agar results in iron starvation and subsequent alcaligin production by the bacteria seeded into the medium. This iron restriction is necessary to allow observation of any growth stimulation by iron sources, which is the purpose of the feeding assay. Even though the alcS::Kanr mutation is not polar on any known alcaligin utilization genes, BRM16 also exhibited a small, yet reproducible reduction in growth stimulation by alcaligin (Fig. 5B); reduced growth stimulation was also observed using hemin chloride and the xenosiderophore enterobactin in these feeding studies (data not shown), both of which are dependent on other Bordetella iron transport systems. As determined for the reduced growth and alcaligin production phenotypes of the alcS strain, genetic complementation using B. bronchiseptica or B. pertussis alcS+ plasmids also restored nearly wild-type levels of growth stimulation by both ferric chloride (Fig. 5A) and alcaligin (Fig. 5B).
FIG. 5.
Feeding by nutritional iron sources. Feeding assays with B. bronchiseptica indicator strains were performed using iron-restricted EDDA agar plates as described in Materials and Methods. The bars indicate the mean diameters of growth stimulation zones, including the 6-mm diameter of the sample well, and the error bars indicate standard deviations (n =3). (A) Growth stimulation by 1.25 mM ferric chloride. (B) Growth stimulation by 300 μM alcaligin. Abbreviations for strains used are as follows: wt, B013N; alcS, BRM16; alcS/alcSBb+, BRM16 (pRK/alcSBp); alcS/alcSBp+, BRM16 (pRKalcSBp).
The iron starvation growth defect, the reduced extracellular alcaligin levels, and the elevated cell-associated levels of alcaligin in the alcS mutant strain were consistent with an alcaligin export defect. The alcS phenotype of poor growth stimulation by iron salts in feeding assays was unusual; reduced growth stimulation by iron salts is a trait evocative of fur mutants, for which deregulated iron transport under iron-replete conditions can cause toxic iron overload (32, 35, 36). Furthermore, the initial observations that the alcS mutant growth yields in iron-depleted SS were even less than those of non-alcaligin-producing strains suggested that alcaligin production itself might be deleterious to the growth of alcS mutants. To further address the hypothesis that alcaligin production and intracellular accumulation contribute to the defective alcS growth and iron source feeding phenotype, a ΔalcA1 null mutation abrogating alcaligin production was crossed into alcS::Kanr strain BRM16, and the phenotype of the double-mutant strain, BRM27, was examined. Similarly, BRM26, an isogenic alcaligin-deficient ΔalcA1 mutant derivative of AlcS+ parent strain B013N, was generated for use as a control strain.
Growth studies revealed that the ΔalcA1 mutation, which abrogated alcaligin production, strongly suppressed the alcS-related growth defect in iron-depleted SS (BRM27 [alcS alcA] versus BRM16 [alcS]) (Fig. 6A); the BRM27 growth yields were similar to those of the isogenic non-alcaligin-producing ΔalcA1 AlcS+ strain BRM26. Furthermore, trans complementation of BRM27 using alcA+ plasmid pBB21 restored alcaligin production and reversed the suppression, resulting in poor growth yields approximating those of the alcaligin-producing alcS::Kanr strain BRM16 (Fig. 6A). When bacterial multiplication in iron-depleted SS was monitored by viable cell counting, alcS::Kanr strain BRM16 was found to have an exponential growth rate similar to that of wild-type strain B013N, but BRM16 entered the stationary phase sooner and the overall growth yield was reduced (Fig. 6B). In contrast, in an alcaligin-deficient ΔalcA1 strain background, the alcS null mutation was found to have no effect on the growth kinetics and growth yield (BRM26 versus BRM27) (Fig. 6C). In these experiments, the growth yield difference between alcaligin-deficient and alcaligin-producing strains in iron-depleted SS batch cultures was modest but significant (P < 0.0005, as determined by Student's t test). It is important to note that the growth of all strains tested was limited by the minimal iron content of this closed system, so any growth advantage that would be conferred by alcaligin production was minimal. This culture system circumvented any repressing influence of iron on alcaligin system gene expression.
FIG. 6.
Iron starvation growth defect of alcS mutants is related to alcaligin production. (A) The bars indicate the mean growth yields of B. bronchiseptica strains, expressed as A600, and the error bars indicate standard deviations (n = 3) after 24 h of culture in iron-depleted SS batch cultures. Note that the growth of all strains was limited by the availability of only trace amounts of iron in this culture system, but alcaligin production by alcaligin-positive strains was maximized. Abbreviations for strains used are as follows: wt, B013N; alcS, BRM16; alcS alcA, BRM27; alcS alcA/alcA+, BRM27(pBB21); alcA, BRM26; alcA/alcA+, BRM26(pBB21). (B) Growth curves for B. bronchiseptica strains cultured in iron-depleted SS. Viable cell counts, expressed as CFU/ml (means ± standard deviations; n = 3), are shown. •, B013N (wild type); ○, BRM16 (alcS). (C) Growth curves for B. bronchiseptica strains cultured in iron-depleted SS. ▪, BRM26 (alcA); □, BRM27 (alcS alcA).
The defects in growth stimulation of alcS mutants by iron salts and alcaligin in feeding assays (Fig. 7A and B, respectively) were also strongly suppressed by the alcA mutation. As shown for ΔalcA1 alcS::Kanr double-mutant strain BRM27 in Fig. 7A, the extreme reduction in growth stimulation by ferric chloride typical of alcS mutants was not seen in an alcaligin-deficient strain background. In fact, the growth stimulation of ΔalcA1 alcS::Kanr strain BRM27 was comparable to that of ΔalcA1 strain BRM26. Accordingly, suppression of the alcS mutation was reversed, and the severely defective growth phenotype was restored to BRM27 by complementation of the ΔalcA1 mutation by alcA+ plasmid pBB21, which restored alcaligin production. Likewise, suppression of the defective alcS growth stimulation by alcaligin was also accomplished by eliminating alcaligin production (Fig. 7B). Together, these results indicate that alcaligin production is detrimental for growth of alcS strains, presumably because the export defect results in abnormal intracellular accumulation of alcaligin that is inhibitory or toxic for the bacterium.
FIG. 7.
Feeding defects of alcS mutants are related to alcaligin production. The bars indicate the mean diameters of growth stimulation zones in iron-restricted EDDA agar plates, including the 6-mm diameter of the sample wells, and the error bars indicate standard deviations (n =3). Iron restriction by EDDA chelation resulted in alcaligin production by alcaligin-positive indicator bacteria seeded into the agar. (A) Growth stimulation by 1.25 mM ferric chloride. (B) Growth stimulation by 300 μM alcaligin. Abbreviations for strains used are as follows: wt, B013N; alcS, BRM16; alcS alcA, BRM27; alcS alcA/alcA+, BRM27(pBB21); alcA, BRM26; alcA/alcA+, BRM26(pBB21).
alcS expression level is important for inducer sensing required for positive regulation of alcaligin system genes.
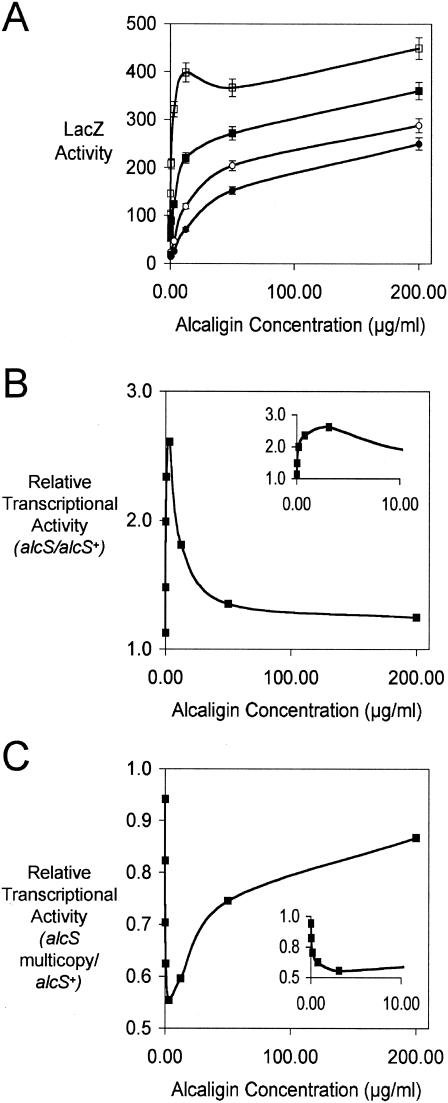
It is known that maximal expression of Bordetella alcaligin biosynthesis and transport genes involves an AlcR-dependent positive regulatory circuit that is responsive to alcaligin as the inducer and that within the normal physiologic range of alcaligin concentrations produced by Bordetella cells, transcriptional activity of the alcABCDER operon promoter shows an inducer dose-dependent relationship (21). Since AlcS was shown in these studies to be required for export of alcaligin from the producing bacterial cell, it was inferred that AlcS activity was integral to the normal inducer-sensing process. Hence, inducer sensing and responsiveness should vary with AlcS production levels. Transcriptional responses to different alcaligin inducer levels were determined for a set of isogenic alcA::mini-Tn5 lacZ1 fusion strains derived from the well-characterized alcaligin-deficient fusion strain BRM1 (4, 21, 41). Inducer responses were compared for AlcS+, AlcS−, and AlcS-overproducing alcA-lacZ fusion strains. Since the mini-Tn5 lacZ1 reporter element disrupts the alcaligin biosynthesis gene alcA in these strains, inducer sensing and transcriptional activation could be assessed in response to known levels of purified inducer supplied exogenously, thus circumventing the involvement of autoinduction. Strains were cultured in iron-depleted SS supplemented with alcaligin, and LacZ activities were measured as a reporter of AlcR-dependent activation of the alcABCDER operon promoter (Fig. 8A). Transcription was found to be inducible irrespective of AlcS production; however, the AlcS− derivative BRM30 and AlcS-overproducing strain BRM1(pRK/alcSBb+) showed significant deviations from the normal induction curves observed for AlcS+ strains BRM1 and BRM1(pRK415). In particular, the inducer responsiveness of ΔalcS1 strain BRM30 was significantly elevated at the lower range of alcaligin concentrations tested compared with BRM1 (Fig. 8B); maximal enhancement, resulting in a 2.6-fold increase in transcriptional activity in the ΔalcS1 strain compared with the AlcS+ strain, occurred at approximately 3 μg/ml (7.5 μM) inducer. Conversely, AlcS overproduction by multicopy gene dosage in BRM1(pRK/alcSBb+) blunted the transcriptional responses compared with the AlcS+ control strain BRM1 carrying the plasmid vector pRK415 that has only the chromosomal copy of alcS, and the diminution was also greatest at the lower inducer concentrations used (Fig. 8C), with the maximal reduction (0.6-fold) also occurring at approximately 3 μg/ml. Thus, the fusion strains showed inducer sensitivities that correlated with their levels of AlcS production and exporter activity. These results indicate that the alcaligin export activity of AlcS is important for maintenance of intracellular alcaligin levels within the range required for appropriate inducer sensing and transcriptional regulation.
FIG. 8.
alcS expression affects inducer sensing and responsiveness in positive regulation of alcaligin gene transcription. (A) Transcriptional activity of alcA::mini-Tn5 lacZ1 fusion reporter strains, expressed in Miller units (means ± standard deviations; n = 3), after 24 h of culture in iron-depleted SS supplemented with alcaligin inducer at 0.00, 0.04, 0.20, 0.78, 3.13, 12.50, 50.00, and 200.00 μg/ml. The strains used were strains BRM1 (AlcS+) (▪), BRM30 (AlcS−) (□), BRM1(pRK415) (AlcS+) (○), and BRM1(pRK/alcSBb+) (B. bronchiseptica AlcS overproducer) (•). (B) Relative transcriptional activity (mean LacZ activity of AlcS− strain BRM30/mean LacZ activity of AlcS+ strain BRM1), plotted as a function of alcaligin inducer concentration. (Inset) Expanded portion of the graph, showing relative transcriptional activity as a function of inducer concentrations ranging from 0.00 to 10.00 μg/ml. (C) Relative transcriptional activity [mean LacZ activity of AlcS-overproducing strain BRM1(pRK/alcSBb+)/mean LacZ activity of AlcS+ strain BRM1(pRK415)], plotted as a function of alcaligin inducer concentration. (Inset) Expanded portion of the graph, showing relative transcriptional activity as a function of inducer concentrations ranging from 0.00 to 10.00 μg/ml.
DISCUSSION
Early studies noted a gene within the B. pertussis and B. bronchiseptica alcaligin siderophore gene clusters encoding a protein that exhibited protein sequence similarity with bacterial multidrug resistance efflux proteins, a subset of the MFS transporters (9, 51). The orthologous Bordetella MFS transporter genes were provisionally named orfX and bcr, but no specific function could be ascribed to their products in the absence of any data other than the partial nucleotide sequence. Completion of the Bordetella genome sequencing projects at the Sanger Institute yielded the full nucleotide sequences of orfX and bcr, and the products were annotated as putative drug resistance translocases (MFS transporters). Although MFS transporter substrate specificity cannot be reliably predicted from the protein sequence, the genetic location of the putative MFS transporter within the alcaligin gene cluster led to the hypothesis that its function might be related to alcaligin flux across the cytoplasmic membrane. This report provides the first evidence that orfX and bcr are involved in alcaligin export or release from Bordetella cells. We propose the revised genetic designation alcS to reflect the newly described alcaligin efflux function of the Bordetella gene product.
As in most organisms, nutritional iron limitation restricts growth of Bordetella. In Bordetella cells, iron starvation results in increased alcaligin biosynthesis and transport gene expression. Generally, an iron starvation growth defect in a mutant strain that can be reversed by supplementation of the culture medium with iron is presumptive evidence that the mutated gene is involved in iron assimilation. In this study, alcS mutant strain BRM16 showed significantly reduced growth yields in iron-depleted SS, exhibiting only about 40% of the wild-type growth; however, the growth of the mutant in iron-replete medium was similar to that of wild-type cells. Growth of the mutant in iron-depleted SS was restored to wild-type levels by complementation with the alcS gene of either B. bronchiseptica or B. pertussis. These initial observations suggested that AlcS was important for coping with iron starvation stress. Previous studies demonstrating the maximal growth difference between alcaligin-deficient and alcaligin-producing strains (20) used culture medium supplemented with EDDA-chelated iron that could be assimilated only via an alcaligin-dependent mechanism. In iron-depleted SS alone, growth of even alcaligin-producing strains is limited by the negligible iron content of the batch culture system. In this study, the finding that an alcaligin-producing alcS mutant had a lower growth yield in iron-depleted SS than alcaligin biosynthesis null mutants was unexpected and suggested that alcaligin production compounded the iron starvation growth defect of the alcS strain.
The alcaligin concentrations in wild-type B. bronchiseptica culture supernatant fluids after 24 h of growth normally range from 100 to 300 μM (40 to 120 μg/ml) (14, 20). The alcaligin levels in culture supernatant fluids of alcS strain BRM16 were reduced to approximately 30 μM after normalization for cell number. As predicted for an efflux mutant phenotype, the cell-associated alcaligin levels were found to be significantly elevated in the alcS mutant. The increase in the amount of cell-associated alcaligin in alcS cells compared with wild-type cells does not fully account for the difference in extracellular levels between the two strains, suggesting that any intracellular accumulation of alcaligin in alcS cells is limited, perhaps because elevated intracellular levels of alcaligin inhibit growth or siderophore production. A deleterious effect of alcaligin accumulation in the bacterial cells might explain the fact that the growth of alcS strain BRM16 was reduced compared with that of an alcA mutant that produces no alcaligin. Furthermore, mutation of the alcA gene of BRM16 relieved the growth inhibition caused by alcS mutation. It also seems plausible that alcaligin that cannot exit the cell may be degraded or its biosynthesis may be restricted by a metabolic feedback mechanism resulting from product accumulation. Altogether, the disparity between the cell distribution of alcaligin in wild-type and alcS cells indicated that AlcS is involved in alcaligin export from the bacterial cell.
Because AlcS was not predicted to be directly involved in iron source utilization, feeding assays produced some unanticipated results. Most notably, the alcS mutant strain was severely defective for growth in iron-restricted EDDA agar with iron salt solutions supplied in wells cut into the agar. Wild-type growth stimulation was restored by either complementing the mutant with alcS or mutating the alcA alcaligin biosynthesis gene of the alcS mutant. Therefore, this significant growth impairment phenotype suggested that iron supplied at high concentrations was deleterious for growth of alcS mutants under conditions that caused siderophore production leading to excessive levels of intracellular alcaligin. Although the mechanistic basis of this iron effect is unknown, the phenotype is reminiscent of previously described fur mutants having deregulated iron uptake systems (5, 36), where, in the presence of molecular oxygen, bacterial iron overload can result in oxygen radical damage via Haber-Weiss Fenton chemistry (32, 35). The aberrantly elevated intracellular concentrations of alcaligin (and conceivably, its ferric complex) in alcS cells might account, at least in part, for the similarity between the growth phenotypes of alcS and fur strains when they are supplied with high concentrations of iron. In addition, it is possible that elevated intracellular alcaligin levels in alcS cells could remove iron from key iron enzymes and proteins, inactivating them and causing growth inhibition. In alcS cells, accumulated alcaligin may even be able to remove iron from the Fur holorepressor, preventing repression of alcaligin production.
Mutation of alcA, resulting in elimination of alcaligin production, suppressed all of the growth defects resulting from the alcS mutation, and this suppression could be reversed by restoring alcaligin production. These observations are consistent with reports by other investigators that specific transporters may be required to export siderophores in part to prevent toxicity associated with their intracellular accumulation (33, 49). In a study of the enterobactin EntS efflux protein of E. coli, attempts to construct an entS fes double-mutant strain by P1 transduction yielded transductants at a low frequency, and the surviving transductants had severe growth defects under iron-restricted conditions (33). The cytoplasmic Fes esterase is required for iron removal from ferric enterobactin (22), and ferric enterobactin accumulates in fes cells to levels that turn them visibly pink. In addition, attempts in E. chrysanthemi to construct a nonpolar yhcA achromobactin MFS exporter null mutant were unsuccessful; only yhcA mutations that were polar on achromobactin biosynthesis genes were tolerated, and even then, they were tolerated only in a strain that did not produce another E. chysanthemi siderophore, chrysobactin (31).
MFS transporters are involved in the export of a broad range of compounds. In addition to the role of MFS transporters in export of newly synthesized siderophores, it has been proposed that they might also be involved in siderophore recycling once the iron has been stripped from the ferric complex or in ferric siderophore extrusion if iron removal is delayed within the bacterial cell (31). MFS transporters may be only one component of the siderophore export machinery; in gram-negative bacteria, most multidrug transporters have three components, an inner membrane transporter, an outer membrane channel, and a periplasmic accessory protein to juxtapose the inner and outer membranes for solute expulsion (62).
Maximal expression of Bordetella alcaligin system genes requires the regulatory protein AlcR, with alcaligin acting as the inducer, and transcriptional activation is proportional to the siderophore concentration (21) up to some maximal level of induction. Previous studies have determined that the active inducer is the iron-free form of alcaligin rather than the ferric alcaligin complex (Brickman and Armstrong, unpublished). The mechanism detecting the alcaligin inducer is very sensitive; the normal threshold concentration of alcaligin required for measurable induction of transcription in B. bronchiseptica is more than 1,000-fold less than the concentration required for measurable growth stimulation (20, 21). At low alcaligin inducer concentrations, an alcS mutant strain showed increased transcriptional responsiveness compared with the isogenic AlcS+ strain. Thus, inducer sensing was perturbed because intracellular alcaligin levels were elevated when alcaligin export was abrogated. Conversely, in strains carrying multiple copies of alcS, induction was blunted, presumably because of diminished intracellular inducer levels resulting from abnormally increased alcaligin export activity. The findings that AlcS is an alcaligin exporter and that AlcS levels can influence induction of transcription by alcaligin are consistent with the hypothesis that inducer levels are perceived intracellularly, possibly by direct interaction of alcaligin with AlcR.
Expression of Bordetella alcaligin biosynthesis and transport genes is negatively regulated at the transcriptional level by the Fur repressor using ferrous iron as a corepressor, and maximal expression during iron starvation is dependent on AlcR in response to alcaligin, which acts as the inducer. Through the action of Fur and AlcR, iron participates directly in the genetic control of its own assimilation, and alcaligin is directly involved in control of its own production and uptake. Since the regulators are responsive to various levels of iron and alcaligin, it follows that these regulatory mechanisms have evolved to function in the context of related activities that affect levels of the corepressor and inducer. A model for AlcS function proposes that AlcS is produced constitutively and is localized to the cytoplasmic membrane, where it is involved in the export of alcaligin from the bacterial cell. Exported alcaligin can solubilize iron from various extracellular sources and make it bioavailable for assimilation by Bordetella cells. There are at least three potential alcaligin substrate sources for AlcS: newly synthesized alcaligin, deferrated alcaligin after iron delivery via the FauA-dependent high-affinity uptake system, and alcaligin that functions intracellularly as an inducer with AlcR. If AlcS function is abrogated by mutation, export of alcaligin is reduced, with concomitant increases in intracellular alcaligin concentrations. Elevated intracellular concentrations of alcaligin are deleterious for growth of the cell. In addition, the export defect perturbs the alcaligin pool that participates in regulation of its own synthesis, further upregulating siderophore production. It is reasonable to propose that the inducer sensing process evolved to function in the context of the AlcS siderophore export mechanism and that those processes must be in balance. In that respect, the alcaligin export activity of AlcS is part of the complex regulatory process that controls alcaligin gene expression.
Acknowledgments
We thank Chantel Sabus for technical assistance in the construction of B. bronchiseptica alcS::Kanr strain BRM16.
This work was supported by University of Minnesota grant-in-aid 19473 and by Public Health Service grant AI-31088 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Alexeyev, M. F. 1995. Three kanamycin resistance gene cassettes with different polylinkers. BioTechniques 18:52, 54, 56. [PubMed] [Google Scholar]
- 2.Anderson, M. T., and S. K. Armstrong. 2004. The BfeR regulator mediates enterobactin-inducible expression of Bordetella enterobactin utilization genes. J. Bacteriol. 186:7302-7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antoine, R., and C. Locht. 1992. Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from gram-positive organisms. Mol. Microbiol. 6:1785-1799. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong, S. K., and M. O. Clements. 1993. Isolation and characterization of Bordetella bronchiseptica mutants deficient in siderophore activity. J. Bacteriol. 175:1144-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagg, A., and J. B. Neilands. 1987. Ferric uptake regulation protein acts as a repressor, employing iron(II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471-5477. [DOI] [PubMed] [Google Scholar]
- 6.Beall, B., and T. Hoenes. 1997. An iron-regulated outer-membrane protein specific to Bordetella bronchiseptica and homologous to ferric siderophore receptors. Microbiology 143:135-145. [DOI] [PubMed] [Google Scholar]
- 7.Beall, B., and G. N. Sanden. 1995. A Bordetella pertussis fepA homologue required for utilization of exogenous ferric enterobactin. Microbiology 141:3193-3205. [DOI] [PubMed] [Google Scholar]
- 8.Beall, B. W., and G. N. Sanden. 1995. Cloning and initial characterization of the Bordetella pertussis fur gene. Curr. Microbiol. 30:223-226. [DOI] [PubMed] [Google Scholar]
- 9.Beaumont, F. C., H. Y. Kang, T. J. Brickman, and S. K. Armstrong. 1998. Identification and characterization of alcR, a gene encoding an AraC-like regulator of alcaligin siderophore biosynthesis and transport in Bordetella pertussis and Bordetella bronchiseptica. J. Bacteriol. 180:862-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bentley, J., L. S. Hyatt, K. Ainley, J. H. Parish, R. B. Herbert, and G. R. White. 1993. Cloning and sequence analysis of an Escherichia coli gene conferring bicyclomycin resistance. Gene 127:117-120. [DOI] [PubMed] [Google Scholar]
- 11.Bissonnette, L., S. Champetier, J. P. Buisson, and P. H. Roy. 1991. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J. Bacteriol. 173:4493-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boukhalfa, H., T. J. Brickman, S. K. Armstrong, and A. L. Crumbliss. 2000. Kinetics and mechanism of iron(III) dissociation from the dihydroxamate siderophores alcaligin and rhodotorulic acid. Inorg. Chem. 39:5591-5602. [DOI] [PubMed] [Google Scholar]
- 13.Boukhalfa, H., and A. L. Crumbliss. 2000. Multiple-path dissociation mechanism for mono- and dinuclear tris(hydroxamato)iron(III) complexes with dihydroxamic acid ligands in aqueous solution. Inorg. Chem. 39:4318-4331. [DOI] [PubMed] [Google Scholar]
- 14.Brickman, T. J., and S. K. Armstrong. 2002. Alcaligin siderophore production by Bordetella bronchiseptica strain RB50 is not repressed by the BvgAS virulence control system. J. Bacteriol. 184:7055-7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brickman, T. J., and S. K. Armstrong. 2002. Bordetella interspecies allelic variation in AlcR inducer requirements: identification of a critical determinant of AlcR inducer responsiveness and construction of an alcR(Con) mutant allele. J. Bacteriol. 184:1530-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brickman, T. J., and S. K. Armstrong. 1995. Bordetella pertussis fur gene restores iron repressibility of siderophore and protein expression to deregulated Bordetella bronchiseptica mutants. J. Bacteriol. 177:268-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brickman, T. J., and S. K. Armstrong. 1996. Colicins B and Ia as novel counterselective agents in interspecies conjugal DNA transfers from colicin-sensitive Escherichia coli donors to other gram-negative recipient species. Gene 178:39-42. [DOI] [PubMed] [Google Scholar]
- 18.Brickman, T. J., and S. K. Armstrong. 1999. Essential role of the iron-regulated outer membrane receptor FauA in alcaligin siderophore-mediated iron uptake in Bordetella species. J. Bacteriol. 181:5958-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brickman, T. J., and S. K. Armstrong. 1996. The ornithine decarboxylase gene odc is required for alcaligin siderophore biosynthesis in Bordetella spp.: putrescine is a precursor of alcaligin. J. Bacteriol. 178:54-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brickman, T. J., J. G. Hansel, M. J. Miller, and S. K. Armstrong. 1996. Purification, spectroscopic analysis and biological activity of the macrocyclic dihydroxamate siderophore alcaligin produced by Bordetella pertussis and Bordetella bronchiseptica. Biometals 9:191-203. [DOI] [PubMed] [Google Scholar]
- 21.Brickman, T. J., H. Y. Kang, and S. K. Armstrong. 2001. Transcriptional activation of Bordetella alcaligin siderophore genes requires the AlcR regulator with alcaligin as inducer. J. Bacteriol. 183:483-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brickman, T. J., and M. A. McIntosh. 1992. Overexpression and purification of ferric enterobactin esterase from Escherichia coli. Demonstration of enzymatic hydrolysis of enterobactin and its iron complex. J. Biol. Chem. 267:12350-12355. [PubMed] [Google Scholar]
- 23.Brickman, T. J., B. A. Ozenberger, and M. A. McIntosh. 1990. Regulation of divergent transcription from the iron-responsive fepB-entC promoter-operator regions in Escherichia coli. J. Mol. Biol. 212:669-682. [DOI] [PubMed] [Google Scholar]
- 24.Brickman, T. J., C. K. Vanderpool, and S. K. Armstrong. 2004. Bordetella, p. 311-328. In J. H. Crosa, A. R. Mey, and S. M. Payne (ed.), Iron transport in bacteria. ASM Press, Washington, D.C.
- 25.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cotter, P. A., and J. F. Miller. 1997. A mutation in the Bordetella bronchiseptica bvgS gene results in reduced virulence and increased resistance to starvation, and identifies a new class of Bvg-regulated antigens. Mol. Microbiol. 24:671-685. [DOI] [PubMed] [Google Scholar]
- 27.Cuiv, P. O., P. Clarke, D. Lynch, and M. O'Connell. 2004. Identification of rhtX and fptX, novel genes encoding proteins that show homology and function in the utilization of the siderophores rhizobactin 1021 by Sinorhizobium meliloti and pyochelin by Pseudomonas aeruginosa, respectively. J. Bacteriol. 186:2996-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar, R., and E. Bibi. 1997. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J. Bacteriol. 179:2274-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fetherston, J. D., V. J. Bertolino, and R. D. Perry. 1999. YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol. Microbiol. 32:289-299. [DOI] [PubMed] [Google Scholar]
- 30.Field, L. H., and C. D. Parker. 1979. Differences observed between fresh isolates of Bordetella pertussis and their laboratory-passaged derivatives, p. 124-132. In C. R. Manclark and J. C. Hill (ed.), International Symposium on Pertussis. U.S. Department of Health, Education and Welfare, Public Health Service, Washington, D.C.
- 31.Franza, T., Mahé, B., and D. Expert. 2005. Erwinia chrysanthemi requires a second iron transport route dependent of the siderophore achromobactin for extracellular growth and plant infection. Mol. Microbiol. 55:261-275. [DOI] [PubMed] [Google Scholar]
- 32.Fridovich, I. 1978. The biology of oxygen radicals. Science 201:875-880. [DOI] [PubMed] [Google Scholar]
- 33.Furrer, J. L., D. N. Sanders, I. G. Hook-Barnard, and M. A. McIntosh. 2002. Export of the siderophore enterobactin in Escherichia coli: involvement of a 43 kDa membrane exporter. Mol. Microbiol. 44:1225-1234. [DOI] [PubMed] [Google Scholar]
- 34.Giardina, P. C., L. A. Foster, S. I. Toth, B. A. Roe, and D. W. Dyer. 1995. Identification of alcA, a Bordetella bronchiseptica gene necessary for alcaligin production. Gene 167:133-136. [DOI] [PubMed] [Google Scholar]
- 35.Halliwell, B., and J. M. Gutteridge. 1984. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 219:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hantke, K. 1981. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol. Gen. Genet. 182:288-292. [DOI] [PubMed] [Google Scholar]
- 37.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 38.Hou, Z., K. N. Raymond, B. O'Sullivan, T. W. Esker, and T. Nishio. 1998. A preorganized siderophore: thermodynamic and structural characterization of alcaligin and bisucaberin, microbial macrocyclic dihydroxamate chelating agents. Inorg. Chem. 37:6630-6637. [DOI] [PubMed] [Google Scholar]
- 39.Hou, Z. G., C. J. Sunderland, T. Nishio, and K. N. Raymond. 1996. Preorganization of ferric alcaligin, Fe(2)L(3). The first structure of a ferric dihydroxamate siderophore. J. Am. Chem Soc. 118:5148-5149. [Google Scholar]
- 40.Kang, H. Y., and S. K. Armstrong. 1998. Transcriptional analysis of the Bordetella alcaligin siderophore biosynthesis operon. J. Bacteriol. 180:855-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang, H. Y., T. J. Brickman, F. C. Beaumont, and S. K. Armstrong. 1996. Identification and characterization of iron-regulated Bordetella pertussis alcaligin siderophore biosynthesis genes. J. Bacteriol. 178:4877-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 43.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop, 2nd, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 44.Lesuisse, E., M. Simon-Casteras, and P. Labbe. 1998. Siderophore-mediated iron uptake in Saccharomyces cerevisiae: the SIT1 gene encodes a ferrioxamine B permease that belongs to the major facilitator superfamily. Microbiology 144:3455-3462. [DOI] [PubMed] [Google Scholar]
- 45.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 46.Moore, C. H., L. A. Foster, D. G. Gerbig, Jr., D. W. Dyer, and B. W. Gibson. 1995. Identification of alcaligin as the siderophore produced by Bordetella pertussis and B. bronchiseptica. J. Bacteriol. 177:1116-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naroditskaya, V., M. J. Schlosser, N. Y. Fang, and K. Lewis. 1993. An E. coli gene emrD is involved in adaptation to low energy shock. Biochem. Biophys. Res Commun. 196:803-809. [DOI] [PubMed] [Google Scholar]
- 48.Neilands, J. B. 1952. A crystalline organo-iron pigment from a rust fungus (Ustilago sphaerogena). J. Am. Chem. Soc. 74:4846-4847. [Google Scholar]
- 49.Page, W. J., E. Kwon, A. S. Cornish, and A. E. Tindale. 2003. The csbX gene of Azotobacter vinelandii encodes an MFS efflux pump required for catecholate siderophore export. FEMS Microbiol. Lett. 228:211-216. [DOI] [PubMed] [Google Scholar]
- 50.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. Holden, C. M. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeno-Tarraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35:32-40. [DOI] [PubMed] [Google Scholar]
- 51.Pradel, E., N. Guiso, and C. Locht. 1998. Identification of AlcR, an AraC-type regulator of alcaligin siderophore synthesis in Bordetella bronchiseptica and Bordetella pertussis. J. Bacteriol. 180:871-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pradel, E., and C. Locht. 2001. Expression of the putative siderophore receptor gene bfrZ is controlled by the extracytoplasmic-function sigma factor BupI in Bordetella bronchiseptica. J. Bacteriol. 183:2910-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 55.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 56.Schneider, D. R., and C. D. Parker. 1982. Effect of pyridines on phenotypic properties of Bordetella pertussis. Infect. Immun. 38:548-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 58.Spasojevic, I., S. K. Armstrong, T. J. Brickman, and A. L. Crumbliss. 1999. Electrochemical behavior of the Fe(III) complexes of the cyclic hydroxamate siderophores alcaligin and desferrioxamine E. Inorg. Chem. 38:449-454. [DOI] [PubMed] [Google Scholar]
- 59.Tanabe, T., T. Funahashi, H. Nakao, S. Miyoshi, S. Shinoda, and S. Yamamoto. 2003. Identification and characterization of genes required for biosynthesis and transport of the siderophore vibrioferrin in Vibrio parahaemolyticus. J. Bacteriol. 185:6938-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vanderpool, C. K., and S. K. Armstrong. 2001. The Bordetella bhu locus is required for heme iron utilization. J. Bacteriol. 183:4278-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vanderpool, C. K., and S. K. Armstrong. 2003. Heme-responsive transcriptional activation of Bordetella bhu genes. J. Bacteriol. 185:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zgurskaya, H. I., and H. Nikaido. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37:219-225. [DOI] [PubMed] [Google Scholar]