Abstract
Abortive infection mechanisms of Lactococcus lactis form a heterogeneous group of phage resistance systems that act after early phage gene expression. One of these systems, AbiK, aborts infection of the three most prevalent lactococcal phage groups of the dairy industry. In this study, it is demonstrated that the antiphage activity depends on the level of expression of the abiK gene and on the presence of a reverse transcriptase (RT) motif in AbiK. The abiK gene was shown to be part of an operon that includes two additional open reading frames, with one of these encoding a phage-related transcriptional repressor named Orf4. Expression of AbiK is driven by two promoters, PabiK and Porf3, the latter being repressed by Orf4 in vivo. Binding of the purified Orf4 to the Porf3 promoter was demonstrated in vitro by gel retardation assays. The N-terminal half of the deduced AbiK protein possesses an RT motif that was modified by site-directed mutagenesis. Conservative mutations in key positions resulted in the complete loss of the resistance phenotype. These data suggest that an RT activity might be involved in the phage resistance activity of AbiK. A model for the mode of action of AbiK is proposed.
Bacteria are used in a variety of fermentation processes because of their ability to convert a wide variety of substrates into complex products or specific molecules. Any process that relies on bacterial fermentation is vulnerable to bacteriophage infections. Lactococcus lactis strains are extensively used by the dairy industry for their fermentation abilities, and they are susceptible to virulent phages, which are ubiquitous in the dairy environment. Three genetically distinct groups of lactococcal phages (936, c2, and P335) are responsible for ruined milk fermentations, and one of the most effective way of controlling them is through the use of phage-resistant strains (49).
One common method to create a phage-resistant starter is to exploit lactococcal native plasmids and to introduce them into industrial phage-sensitive L. lactis strains (61). The genetic determinants of over 50 lactococcal phage resistance mechanisms have been cloned and sequenced (14). They can affect phage multiplication by inhibiting adsorption, blocking DNA entry in the cell, cutting the exogenous DNA (restriction/modification systems), or aborting infection after early phage gene expression. The last group comprises 22 abortive infection (Abi) mechanisms that rarely share amino acid sequence similarity. Although they are effective against similar groups of phages, their mode of action and regulation mechanisms seem quite different, suggesting that they interfere with different viral and/or cellular components (26).
The effects of these antiviral proteins on the course of the phage lytic cycle are usually determined to provide clues regarding their mode of action. Phage DNA replication, early and late transcription, and protein synthesis are modified or inhibited by Abi mechanisms (14). AbiA (20), AbiF (27), AbiK (10), and AbiR (63) rapidly halt phage DNA replication by an unknown mechanism. Recently, it has been shown that AbiP disturbs both phage DNA replication and temporal control of early phage transcription (22). AbiG inhibits mRNA synthesis (54), whereas AbiB triggers the breakdown of phage transcripts through the induction of an RNase (57). AbiD1 likely blocks phage DNA replication or packaging through a decrease of a RuvC-like endonucleolytic activity (4). The transcription of genes encoding AbiA, AbiB, AbiL, AbiP, and AbiT was also found to be constitutive (7, 13, 18, 20, 22). AbiL and AbiT are encoded by two genes that are organized in an operon. Only two lactococcal Abi proteins have been mutated to study the roles of particular motifs in phage resistance. Nonconservative substitutions of regularly spaced leucine residues of AbiA lead to the loss of the phage resistance phenotype (21). Similarly, the substitution of a hydrophobic amino acid by a charged residue or a proline in the C-terminal region of AbiTi abolished the effects of AbiT (7).
AbiK is an abortive infection system encoded by the lactococcal plasmid pSRQ800 (11). The AbiK protein has a predicted size of 599 amino acids and is effective against the 936-, P335-, and c2-like phages (25). It inhibits the DNA replication of phages of the P335 group but affects 936-like phages at a later step of the lytic cycle (10). The phage gene involved in the sensitivity to AbiK has recently been identified in several bacteriophages (9). These genes encode proteins related to single-strand annealing recombinases, but their role in the molecular mechanism of AbiK remains unknown. It has also been reported that the growth temperature and the gene copy number affect the efficacy of AbiK (25). In this study, the transcription and translation of the abiK gene were examined. Conserved residues in the reverse transcriptase (RT) motif of AbiK were also replaced by site-directed mutagenesis.
MATERIALS AND METHODS
Bacterial strains and media.
The strains used in this study are listed in Table 1. Escherichia coli was grown at 37°C in LB medium, and L. lactis was grown at 30°C in M17 (Quélab) supplemented with 0.5% glucose (GM17). When needed, antibiotics were added to the medium at the following concentrations: 50 μg/ml of ampicillin, 20 μg/ml of chloramphenicol, or 10 μg/ml of tetracycline for E. coli and 5 μg/ml of chloramphenicol or erythromycin for L. lactis.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 endA1 gyrA96 thi-1 relA1 | Invitrogen |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| L. lactis strains | ||
| MG1363 | Plasmid-free host for phage p2 | 28 |
| LM0230 | Plasmid-free host for phage p2 | 45 |
| SMQ-20 | LM0230(pSA3, pSRQ800), Emr, Abi+ | 25 |
| SMQ-481 | Plasmid-cured host for phage ul36 | 8 |
| Plasmids | ||
| pBAD-HisB | Expression vector, Apr, 4.1 kb | Invitrogen |
| pBS | Cloning vector, Apr, 2.9 kb | Stratagene |
| pGKH10 | Encodes the lacZ gene of E. coli, Cmr, Emr | 30 |
| pLCF-1 | PCR amplicon of the orf4 gene in pBAD-HisB | This study |
| pMIG3 | Shuttle vector, Cmr, 5.5 kb (intermediate-copy vector for L. lactis) | 67 |
| pNZ123 | High-copy vector for L. lactis, Cmr, 2.8 kb | 19 |
| pSA3 | Shuttle vector, Cmr, Tcr, Emr, 10.2 kb (low-copy vector for L. lactis) | 17 |
| pSRQ800 | Native L. lactis plasmid encoding AbiK, Abi+ | 25 |
| pSRQ817 | 3.7-kb EcoRI-PvuII of pSRQ800 in pMIG3, Cmr, Abi+ | 25 |
| pSRQ818 | 2.2-kb PCR amplicon of the abiK operon in pMIG3, Cmr, Abi+ | This study |
| pSRQ822 | 3.7-kb EcoRI-PvuII in pNZ123, Cmr, Abi+ | This study |
| pSRQ840 | PCR amplicon of lacZ in pMIG3, Cmr | This study |
| pSRQ841 | PCR amplicon of lacZ in pSRQ818 (AbiK-LacZ fusion), Cmr | This study |
| pSRQ842 | 3.2-kb HaeIII-PvuII of pSRQ800 in pMIG3, Cmr, Abi+ | This study |
| pSRQ843 | 3.0-kb NdeI-PvuII of pSRQ800 in pMIG3, Cmr, Abi+ | This study |
| pSRQ851 | 3.7-kb EcoRI-PvuII in pSA3, Cmr, Tcs, Emr, Abi+ | This study |
| pSRQ852 | 3.2-kb HaeIII-PvuII in pSA3, Cmr, Tcs, Emr, Abi+ | This study |
| pSRQ853 | 3.0-kb NdeI-PvuII in pSA3, Cmr, Tcs, Emr, Abi+ | This study |
| pSRQ854 | 0.5-kb EcoRI-HaeIII and 3.0-kb NdeI-PvuII in pSA3, Cmr, Tcs, Emr, Abi+ | This study |
| pSRQ860 | 1.0-kb EcoRI-BglII in pNZ123, Cmr | This study |
| pSRQ871 | 2.2-kb PCR amplicon of the mutated abiK gene (D163E) in pMIG3, Cmr | This study |
| pSRQ872 | 2.2-kb PCR amplicon of the mutated abiK gene (Y168S) in pMIG3, Cmr | This study |
| pSRQ873 | 2.2-kb PCR amplicon of the mutated abiK gene (Y245F) in pMIG3, Cmr | This study |
| pSRQ874 | 2.2-kb PCR amplicon of the mutated abiK gene (Y245S) in pMIG3, Cmr | This study |
| pSRQ875 | 2.2-kb PCR amplicon of the mutated abiK gene (D247E) in pMIG3, Cmr | This study |
| pSRQ876 | 2.2-kb PCR amplicon of the mutated abiK gene (D248E) in pMIG3, Cmr | This study |
Abi+, active Abi mechanism; Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Emr, erythromycin resistance; Tcr, tetracycline resistance; Tcs, sensitive to tetracycline.
Bacteriophage propagation and microbiological assays.
The two virulent bacteriophages used in this study are p2 (L. L. McKay, University of Minnesota), a member of the 936 species, and ul36, a pac-type member of the P335 species (51). They were propagated as described previously (51). The efficiency of plaquing (EOP) was calculated by dividing the phage titer on the tested strain by the titer on the sensitive control strain. One-step growth curves were determined as reported elsewhere (50). The burst size was determined by dividing the average titer after the exponential phase by the average titer before cells began to release virions.
Plasmids, primers, and DNA manipulations.
The plasmids utilized in this study are described in Table 1. All primer sequences cited in this study are available upon request. E. coli plasmid DNA was isolated by the method of Birnboim (5). L. lactis plasmid DNA was obtained as described by O'Sullivan and Klaenhammer (55). All DNA manipulations were carried out as reported by Sambrook and Russell (60). When needed, pBS was first used to clone restriction fragments and to introduce deletions. The abiK operon was amplified using primers SM1 and SM2 to generate pSRQ818. The E. coli lacZ gene was amplified from pGKH10 with primers JB818a and JB818b to construct plasmids pSRQ840 and pSRQ841. The PCR products were digested and purified as specified elsewhere (24). Site-directed mutagenesis of AbiK was made with the megaprimer PCR method (60). The abiK operon was amplified with primers SM1 and SM2, and site-directed mutagenesis was performed with primers Mut1, Mut2, Mut3, Mut4, Mut5, and Mut7. The mutations were confirmed by sequencing the cloned inserts. DNA sequencing was performed using an ABI Prism 3100 apparatus from the service center at the Université Laval or an ABI Prism 3700 apparatus from the genomic platform at the research center of the Centre Hospitalier de l'Université Laval. E. coli and L. lactis were transformed with the Gene Pulser II apparatus as indicated by the manufacturer and by Holo and Nes (32), respectively.
RNA isolation and Northern blot analysis.
RNA of phage-infected L. lactis cells was isolated as described previously (7), and Northern blot analyses were carried out according to Sambrook and Russell (60). The probe corresponding to an internal segment of the abiK gene (coordinates 3988 to 4570 in pSRQ800, accession no. U35629) was amplified with primers IB800.14 and AbiK4. The probe covering the 3′ end of orf3 and the 5′ region of orf4 (coordinates 5405 to 5694) was amplified with primers IB800.12.1 and Orf4. The PCR products were labeled with [α-32P]dATP (Amersham Pharmacia Biotech) by random priming.
5′-RACE.
The transcription initiation site from the abiK promoter was determined by rapid amplification of cDNA ends (RACE) as described by Sambrook and Russell (60). The primer used for cDNA synthesis was PEabiK. PCR amplification was conducted with this primer and the poly(C) primer CB3 (12).
Primer extension analysis.
The transcription initiation site of the 2.6-kb mRNA was confirmed by primer extension analysis using primer LCF-21 as described by Sambrook and Russell (60). Total RNA from L. lactis carrying plasmid pSRQ817 was isolated as reported previously (7).
β-Galactosidase assay.
β-Galactosidase activity was measured as described by Boucher et al. (12). Briefly, cells were grown in LB (E. coli) or GM17 (L. lactis) medium at an optical density at 600 nm of 0.5, washed and resuspended in 0.5 ml of 50 mM sodium phosphate (pH 7.0), and lysed with glass beads. The lysate was cleared by centrifugation, and the protein concentration was measured with the Bio-Rad DC protein assay reagent. The β-galactosidase assay was performed as described by Miller (47).
Cloning into pBAD-HisB and expression of the orf4 gene product.
The orf4 gene was cloned into the pBAD-HisB expression vector (Invitrogen) in order to create a C-terminal fusion of Orf4 with a tail of six histidine residues. The orf4 gene was amplified by PCR from pSRQ800 by using primers LCF-11 and LCF-17. The LCF-17 primer included the codons for the six histidine residues followed by a stop codon. The NcoI-digested PCR product was ligated to the corresponding site in pBAD-HisB and cloned into E. coli TOP10 to create the pLCF-1 construct, which was confirmed by DNA sequencing using primers LCF-14 and LCF-15. Induction of the expression was conducted at 37°C for 2 h in the presence of 0.2% l-(+)-arabinose. The cleared lysate was used for purification of the fusion protein by nickel-chelating nitrilotriacetic acid affinity chromatography (Ni-NTA) as recommended by the manufacturer (Qiagen).
Protein quantification and sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis.
Proteins were quantified using the Bio-Rad protein assay reagent and bovine serum albumin as a standard. The various protein fractions were analyzed by standard 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis run in Tris-glycine buffer (37).
EMSA.
Electrophoretic mobility shift assay (EMSA) with the Ni-NTA-purified Orf4 protein was conducted as described elsewhere (39). The DNA fragments were labeled by incorporation of [α-32P]dATP during PCR amplification and purified with Micro Bio-Spin P-30 chromatography columns (Bio-Rad). The PCR primers used were LCF-3 and LCF-6 for fragment A, LCF-4 and LCF-7 for fragment B, LCF-5 and LCF-8 for fragment C, and LCF-3 and LCF-23 for fragment D. The LCF-24 and LCF-25 complementary oligonucleotides were annealed to create a double-stranded DNA fragment of 41 bp used for competition assays. The binding buffer consisted of 10 mM HEPES (pH 7.9), 10% glycerol, 0.2 mM EDTA, 0.25 mM phenylmethanesulfonyl fluoride, 0.1 μg poly(dI-dC), 50 mM KCl, and 5 mM MgCl2 (39). The purified Orf4 was incubated in the binding buffer with 10,000 cpm of labeled DNA fragment. Binding was allowed to proceed at room temperature for 15 min with or without competitor DNA before analysis on a 5% native polyacrylamide gel (39).
RESULTS
The efficacy of AbiK is enhanced by neighboring genes in pSRQ800.
Previous bioinformatic analyses indicated the presence of a putative promoter (PabiK) and of a rho-independent terminator that were flanking the abiK gene on plasmid pSRQ800 (25). Thus, the abiK gene seemed to be part of a monocistronic unit. In pSRQ800, another putative promoter (Porf3) and two short open reading frames (orf3 and orf4) that have the typical G+C content of L. lactis are located upstream of the abiK operon (Fig. 1A). In this study, we investigated whether or not the presence of this second operon could influence the antiphage activity of AbiK.
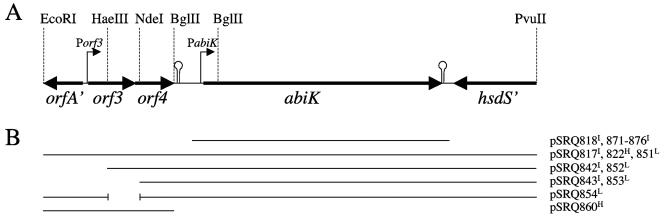
FIG. 1.
Genetic organization of the DNA fragment of pSRQ800 that encodes phage resistance. Panel A: open reading frames, promoters (bent arrows), and rho-independent terminators (stem-loops) identified by bioinformatics analyses. Panel B: representation of the DNA fragments cloned into the different plasmids used in this study. The superscript letter at the end of the plasmid name represents the plasmid copy number in L. lactis (H = high; I = intermediate; L = low).
The native plasmid pSRQ800 and DNA fragments from pSRQ800 subcloned into the shuttle vector pMIG3 confer protection against phages, with constant EOP values of 10−6 (Fig. 1B; Table 2). The antiphage activity is further increased (EOP of ≤1 × 10−9) when the complete operon is cloned into the high-copy vector pNZ123 (Fig. 1B; Table 2). However, when restriction fragments of pSRQ800 are cloned in the low-copy vector pSA3, the antiphage phenotype is weaker and also variable, as EOP values ranged from 10−2 to 10−3 (Fig. 1B; Table 2). The EOP variations with the low-copy vector appear to depend on the absence or the presence of the putative Porf3 promoter on the cloned fragment. Disrupting the orf3-orf4 coding sequences without deleting the Porf3 promoter (pSRQ854) (Table 2) did not alter significantly the EOP (pSRQ851) (Table 2). Thus, the EcoRI-HaeIII fragment harboring the Porf3 promoter is responsible for the increased antiphage activity (EOP from 10−2 to 10−3) with the low-copy vector pSA3 (Table 2). This DNA fragment does not affect phage multiplication (EOP of 1) when cloned without the AbiK system (pSRQ860) (Table 2).
TABLE 2.
Effect of Porf3 and Orf4 on the activity of AbiK against phage p2
| Plasmid | EOPa | Burst sizea | EOP with pSRQ860a |
|---|---|---|---|
| pSRQ800 | (6.8 ± 2.5) × 10−6 | NDb | (3.8 ± 3.2) × 10−2 |
| pSRQ822 | ≤1 × 10−9 | ND | ND |
| pMIG3 | 1.0 | ND | ND |
| pSRQ817 | (1.7 ± 1.7) × 10−6 | ND | ND |
| pSRQ818 | (2.1 ± 1.8) × 10−6 | ND | ND |
| pSRQ842 | (2.6 ± 0.9) × 10−6 | ND | ND |
| pSRQ843 | (3.1 ± 1.2) × 10−6 | ND | ND |
| pSA3 | 1.0 | 83 ± 6 | 1.0 |
| pSRQ851 | (3.1 ± 2.3) × 10−3 | 19 ± 5 | (1.2 ± 0.2) × 10−2 |
| pSRQ852 | (5.5 ± 0.8) × 10−2 | 32 ± 4 | (3.0 ± 1.1) × 10−2 |
| pSRQ853 | (6.1 ± 4.6) × 10−2 | ND | (3.8 ± 2.6) × 10−2 |
| pSRQ854 | (2.0 ± 1.3) × 10−3 | ND | ND |
Means and standard deviations from at least three experiments.
ND, not determined.
Furthermore, when the orfA′-orf3-orf4 operon (Fig. 1A) was provided in trans on the high-copy plasmid pNZ123 (pSRQ860), the enhancement of the antiphage activity was eliminated (Table 2). The presence of plasmid pSRQ860 also drastically reduced the phage resistance phenotype conferred by the native plasmid pSRQ800 (from 10−6 to 10−2) (Table 2). The presence of the high-copy vector pNZ123 (as a control) did not affect the antiphage phenotype of pSRQ800 (EOP of 10−6). Finally, the abortive infection phenotype was not affected when pSRQ860 was provided in trans in L. lactis strains containing low-copy constructs with a deleted Porf3 (pSRQ852 and pSRQ853) (Table 2). Taken altogether, these data suggest that the abiK gene could be expressed from another promoter (Porf3) located upstream from PabiK. Also, a putative protein product encoded by the EcoRI-BglII fragment could interfere with the AbiK antiphage activity. The orf4 gene product is the only possible candidate, since the transcription start site downstream of Porf3 is located inside the start codon of orf3 (see below) and there is no putative ribosome-binding site upstream of this ATG. A series of experiments were carried out to confirm these hypotheses.
Transcription of the abiK gene.
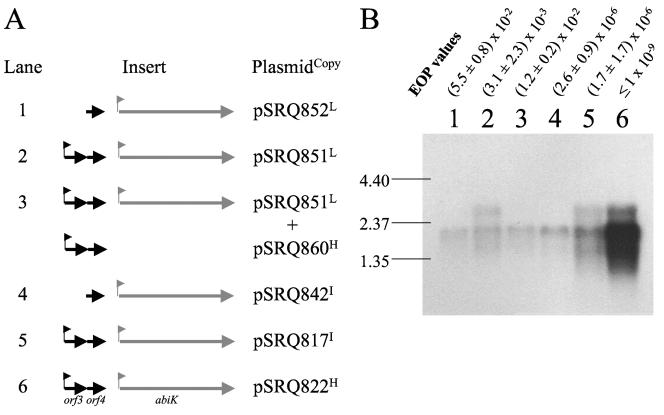
L. lactis MG1363 derivatives carrying different combinations of abiK- and Porf3-containing plasmids were selected for Northern blot experiments (Fig. 2A). Two transcripts (1.9 and 2.6 kb) hybridized with the abiK probe. The smaller mRNA has the size of the predicted abiK operon, and its abundance in the different strains was correlated with the plasmid copy number (Fig. 2). A probe covering orf3 and orf4 also hybridized with the 2.6-kb transcript and with a 0.5-kb mRNA present at a higher concentration (data not shown). Therefore, the 2.6-kb transcript covers the two operons. This also indicates that the putative terminator located downstream of orf4 (Fig. 1A) is partially effective. Interestingly, the 2.6-kb transcript is absent if Porf3 is deleted (Fig. 2, lanes 1 and 4) or if orf4 is provided in trans on pSRQ860 (Fig. 2, lane 3). Judging by the relative concentrations of the 1.9- and 2.6-kb transcripts (Fig. 2, lanes 2, 5, and 6), the orf4 gene product, when provided in a sufficient amount in cis (pSRQ822) (Fig. 2, lane 6) or in trans (pSRQ860) (Fig. 2, lane 3), can repress the expression from Porf3.
FIG. 2.
Transcription of the abiK operon. Panel A shows the plasmids carried by the six strains used, along with a schematic representation of the cloned insert. The corresponding plasmid copy number in L. lactis MG1363 is indicated by a superscript letter at the end of the plasmid name (see the legend to Fig. 1). The pNZ123 vector was cointroduced as a control in addition to the indicated plasmids in lanes 1 and 2. Panel B presents the Northern blot analysis with a probe corresponding to the abiK gene. To facilitate the comparison of the different constructs, some EOP values from Table 2 (without pSRQ860) are shown above the blot. Bands of the molecular weight marker (in kbp) are indicated on the left.
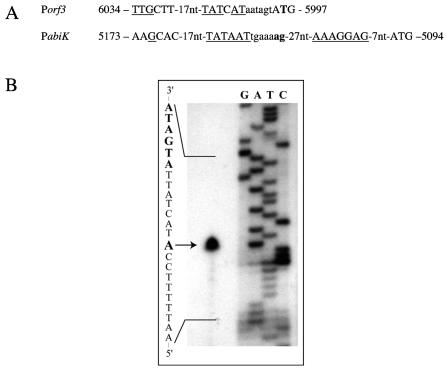
The 5′ ends of the two transcripts were determined by 5′-RACE and primer extension analyses. Sequencing of the amplicon obtained by RACE with a primer downstream of the abiK promoter indicated that there are two possible initiation sites from this promoter (Fig. 3A). The putative Porf3 promoter was analyzed by primer extension analysis of total RNA isolated from strains carrying pSRQ817 and pSRQ860. The extension products confirmed the functionality of the Porf3 promoter and the location of the transcription initiation site (Fig. 3).
FIG. 3.
Promoter determination. Panel A: DNA sequence and transcription initiation sites (in boldface) of the Porf3 and PabiK promoters. The −35 and −10 regions, the ribosome binding site of abiK, and the start codons of orf3 and abiK are in capital letters. Nucleic acids that are identical to the consensus sequence are underlined. The coordinates correspond to the DNA sequence of pSRQ800 (accession no. U35629). Panel B: primer extension analysis of the Porf3 promoter. Total RNA from an L. lactis MG1363 strain carrying the pSRQ817 plasmid was purified, and primer extension was performed as described in Materials and Methods. The lower-strand DNA sequence shown on the left was obtained with the same primer in a T7-based sequencing reaction. The −10 box is highlighted in boldface. The transcription initiation start site is indicated by an arrow.
Binding of Orf4 to the Porf3 promoter.
The Orf4 putative protein possesses a helix-turn-helix (HTH) motif (cd00093, pfam01381.9) and is similar to several regulatory proteins (11). One of its homologues, the repressor (cI) of the coliphage 434, belongs to a group of phage transcriptional regulators with similar structures and low levels of sequence similarity (59, 69). Two structure programs (SAM-T99 and PROF) (34, 56) predicted that Orf4 has the five α-helices and the four turns of this family of proteins. These comparisons strongly suggested that Orf4 could act as a transcriptional repressor.
To test this prediction, the Orf4 gene product was overexpressed in E. coli as a His tag fusion and purified to near homogeneity by Ni-NTA affinity chromatography (see Materials and Methods). This protein preparation was used in a series of EMSA experiments to confirm its DNA-binding activity (see below).
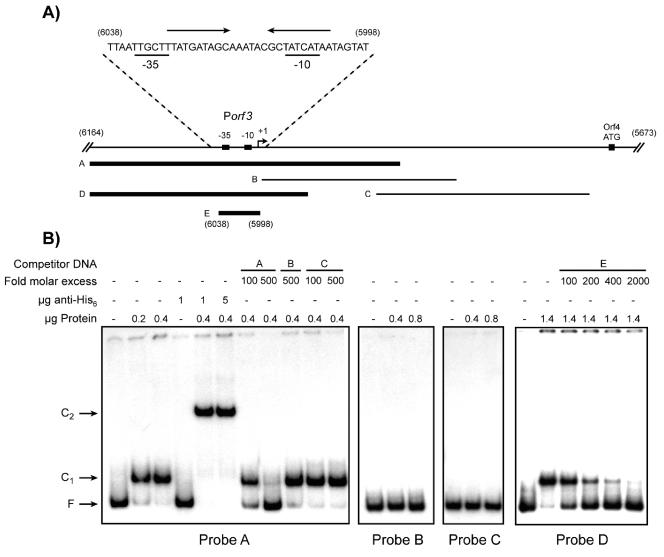
Different promoter fragments were amplified and labeled by PCR (Fig. 4A). Incubation of fragments B (192 bp) and C (210 bp) with the Orf4 protein extract did not show any retardation in EMSA experiments (Fig. 4B). On the other hand, fragments A (306 bp) and D (215 bp) were completely bound and retarded in the presence of Orf4 (Fig. 4B, complex C1). Addition of antihistidine antibodies into the binding reaction mixture resulted in a supershifted complex of higher molecular mass (Fig. 4B, complex C2). This confirmed that the protein causing the retardation of the labeled fragments was indeed Orf4 and not a contaminating protein from E. coli. Up to 2.0 μg protein was tested in EMSA, and even at the highest protein concentration, only one complex was detected (data not shown). This suggests the presence of a single Orf4 binding site in the Porf3 promoter sequence. A 10-bp inverted repeat (10-6-10) was found between the −10 and −35 boxes of the Porf3 promoter and could represent the target of Orf4 (Fig. 4A). A 41-bp DNA fragment covering this region was created by annealing complementary oligonucleotides (Fig. 4A, fragment E). When used in competition assays with fragment D as the labeled probe, the C1 complex gradually disappeared as the amount of unlabeled competitor DNA increased (Fig. 4B). This confirmed that Orf4 binds to the Porf3 sequence and that the inverted repeat might be the targeted structure.
FIG. 4.
Gel retardation assays (EMSA) with recombinant Orf4. Panel A shows the different fragments (indicated by letters A to E) used as labeled probes (10,000 cpm/reaction) or unlabeled competitor DNA. Thick lines represent the fragments retarded by Orf4 in EMSA, whereas thin lines represent unbound fragments. The DNA sequence corresponding to fragment E is shown above, along with the −10 and −35 boxes (underlined characters). The inverted repeats are indicated by arrows above the sequence. Some of the coordinates are shown in parentheses and correspond to the pSRQ800 sequence (GenBank accession no. U35629). The transcription initiation start site (+1) is indicated by a bent arrow. Panel B, EMSA with the various fragments shown in panel A. Unlabeled competitor DNA fragments are indicated by letters (corresponding to fragments in panel A). F. free probe; C1, Orf4-DNA complex; C2, anti-His6-Orf4-DNA complex.
Protein expression analysis with an AbiK-LacZ fusion.
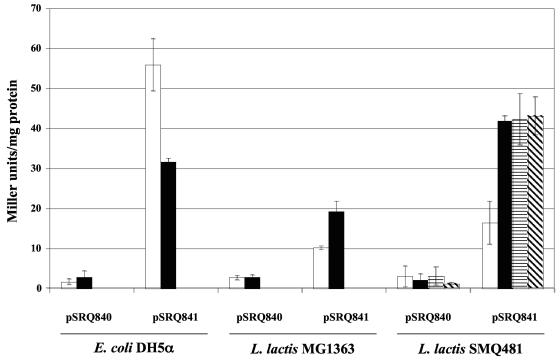
The abiK gene is constitutively transcribed from PabiK and Porf3, but its translation could be regulated. The PabiK promoter, ribosome-binding site and first 178 codons of the abiK gene were fused to the last 1,015 codons of the E. coli lacZ gene and introduced into the shuttle vector pMIG3 to generate pSRQ841 (Table 1). The β-galactosidase activity conferred by pSRQ841 was measured in E. coli and in two L. lactis strains at 30 and 37°C. The promoterless lacZ gene cloned in pMIG3 to generate pSRQ840 was used as a negative control. The fusion protein was constitutively expressed in the three hosts (Fig. 5). In L. lactis, the expression was approximately twice as high at 30°C, which is the optimal growth temperature of this microorganism. This could explain the reduced antiphage activity of AbiK at 37°C (compared to 30°C) and its so-called heat sensitivity (25). The β-galactosidase activity was also measured during infection by phage ul36 and was not found to be significantly different. This result demonstrates that the expression of AbiK is not induced following phage infection (Fig. 5).
FIG. 5.
β-Galactosidase activity in various E. coli and L. lactis strains at 37°C (white bars), at 30°C (black bars), and during infection by phage ul36 (horizontally striped bars, after 15 min; diagonally striped bars, after 30 min). These data represent the means and standard deviations from at least three experiments.
Site-directed mutagenesis of AbiK.
The N-terminal half of the AbiK protein (300 amino acids) contains a motif found in RTs (pfam 00078), as shown by the alignment with AbiA protein, which also possesses a similar motif (Fig. 6). Few amino acids are conserved in all RTs (70). To determine if this RT motif of AbiK is important for phage resistance, five conserved amino acids of the motif were replaced by site-directed mutagenesis and the antiphage activity of mutated AbiK proteins was assayed in L. lactis MG1363 with phage p2. The substitution of an aspartic acid (D163, D247, and D248) by a glutamic acid or of a threonine (Y168 and Y245) by a serine abolished the resistance phenotype (EOP of 1). In some active RTs, the tyrosine in position 245 is replaced by a phenylalanine (70). This substitution reduced slightly the efficacy of AbiK (EOP of 1.4 × 10−5 instead of 2.1 × 10−6).
FIG. 6.
Alignment of AbiK, AbiA, and the RT motifs of Xiong and Eickbush (70). The identical residues in the amino acid sequences of AbiK and AbiA are in boldface. h, hydrophobic residue; p, small polar residue; c, charged residue. The amino acids that were replaced by site-directed mutagenesis are identified by arrows.
DISCUSSION
Effect of the orf4 operon on AbiK expression.
The abiK gene is transcribed from the promoter previously identified by DNA sequence analysis (25) and from another promoter (Porf3) that is not located in the AT-rich region of pSRQ800 that encodes AbiK (this study). Results from Northern blot experiments revealed that the transcription from this second promoter, Porf3, is repressed when Orf4 is provided, in cis or in trans, at high levels relative to AbiK. Thus, Orf4 acts as a repressor by inhibiting transcription from Porf3, resulting in a decreased expression of abiK and a weaker phage resistance phenotype in L. lactis.
The Orf4 protein possesses the HTH motif found in many bacteriophage regulatory proteins and numerous transcriptional regulators (69). Secondary-structure prediction indicated the presence of helices 2 and 3, which bind and recognize DNA, and of helices 4 and 5, which are involved in dimerization (31). Many members of the HTH family of bacterial regulators bind their consensus sequence as homodimers or homotetramers (59). Two putative helices that could play a role in dimer formation were found in the C-terminal part of Orf4. Also, inverted repeats are often part of target promoter/operator regions and can be bound by multimeric forms of transcriptional regulators containing HTH DNA-binding motifs. As examples, two homodimers of the ferric uptake regulator (Fur) from Bacillus subtilis bind to the Fur box composed of a 9-1-9 inverted repeat (2), and four homodimers of the Zn(II)-sensing metal-regulated repressor SmtB from Synechococcus strain PCC7942 bind to a 40-bp oligonucleotide containing a single 12-2-12 inverted repeat (64). Interestingly, the presence of a 10-6-10 inverted repeat inside the 41-bp region bound by the purified Orf4 suggests that this repressor might indeed act as a homodimer. Only one complex was observed in EMSA, over a wide range of Orf4 concentrations, indicating that only one binding site is occupied by a single protein complex. Otherwise, we could have expected retarded bands of intermediate mobility depending on the protein-DNA molar ratio, corresponding to multiple Orf4 complexes composed of monomers or dimers bound to one or more binding sites (41, 58). The nature of the Orf4-binding complex is unknown and remains to be investigated.
Other Abi systems have been shown to be associated with potential regulatory proteins. In L. lactis, the antiphage activity of AbiU is increased (EOP from 10−2 to 10−4) when the putative regulatory gene abiU2 is deleted (16). The abiU2 gene seems to be involved in downregulation of abiU1 or its gene product, AbiU1, which confers phage resistance. This regulation scheme resembles that of PifA, an E. coli protein encoded by plasmid F that aborts T7 infection. The PifC protein negatively controls the expression of the three genes of a polycistronic operon (pifC, pifA, and pifB). Inactivation of pifC increases the expression of the Abi protein (15, 46).
In the case of AbiK, the orf3-orf4 regulatory operon improves the efficacy of the system, since the abiK gene is expressed from an additional promoter. However, the impact of the upstream Porf3 promoter is variable, depending on the copy number of the plasmid carrying the AbiK system. The exact function of AbiK in uninfected cells is yet unknown, but the orf3-orf4 regulatory operon present on pSRQ800 might play a role in proper balancing of the constitutive expression of AbiK and Orf4.
From an evolutionary point of view, the similarity of Orf3 and Orf4 with putative phage proteins suggests that this operon might be of viral origin (11). Associations between Abi systems and foreign regulatory genes or sequences that were not initially associated with these Abi genes have been observed previously. For example, the abiB gene is transcribed from a promoter located in an Iso-ISS1 insertion sequence (13), and the two genes from the AbiT system appear to have been inserted into a preexisting operon (7). Similarly, the orf3-orf4 operon might have been originally associated with other systems before being coupled with the abiK operon. Yet, the association of the orf3-orf4 and abiK operons is undeniable with the native plasmid pSRQ800; when the Porf3 promoter on pSRQ800 is repressed by Orf4 provided in trans from pSRQ860, the antiphage activity is reduced by more than 1000-fold.
Putative RT activity of AbiK.
Despite a complex control of the expression of AbiK, the protein is present in the cell before infection and its expression is not affected by phage infection. The efficacy of AbiK against phages p2 (936 species) and ul36 (P335 species) clearly depends on the intracellular concentration of the protein. The plasmid copy number, the growth temperature that affects the expression of AbiK, or a second promoter can modify the efficacy of the system. At this time, it is not known if AbiK is expressed constitutively in an active form or if it must be activated by a phage component.
Mutagenesis experiments also showed that AbiK has an RT motif that is essential for phage resistance. The three aspartate residues that have been mutated in AbiK are all conserved in other RTs (70). They form a triad that can bind divalent metal ions and are directly involved in catalysis (36). Replacement of one of these aspartate residues by an asparagine, a glutamate, or a glutamine in the human immunodeficiency virus type 1 reverse transcriptase results in 1,000-fold reduction of the polymerase activity (42). This could explain the complete loss of phage resistance when the same conservative mutations were introduced in the AbiK protein. The replacement of one of the two tyrosine residues with a serine also reduces significantly the polymerase activity, probably because both mutations decrease the affinity of the enzyme for dTTPs (42, 44). However, when the tyrosine of the YXDD motif (Y245 in AbiK) is changed to a phenylalanine in human immunodeficiency virus type 1 or murine leukemia virus reverse transcriptases, the processivity of the enzyme is preserved, but several authors reported that the fidelity of DNA synthesis is either increased or reduced (1, 3, 33, 35, 40). The slight reduction of the antiphage activity caused by this mutation in AbiK is consistent with these data. To our knowledge, this is the first time that a link between reverse transcription and antiviral activity has been made.
Previously, it was shown that deletion of the last 46 amino acids of AbiK also eliminated the antiphage activity of AbiK (25). Although we cannot exclude a conformational change in the truncated AbiK to explain the lost of antiphage activity, this result also suggests that the C-terminal domain of the protein (amino acids 302 to 599) might have a second biochemical function that is also involved in resistance. Proteins with an RT domain from viruses and retrotransposons of eukaryotes, but also from E. coli, possess an RNase H domain that cleaves the RNA strand of the DNA-RNA duplex (38, 43). RTs of group II introns, found in bacteria and in organelles of eukaryotes, are associated with maturase and endonuclease domains that seem to be involved in intron mobility (29, 48). The C-terminal region of AbiK does not share similarity with RNase, maturase, or endonuclease domains of proteins in the databases. Deletion and mutation analyses strongly suggest that the abiK gene could encode a novel bacterial reverse transcriptase with low levels of amino acid sequence similarity to other members of this ubiquitous family of proteins. More interestingly, specific mutations in the conserved RT motif suggest that such an activity is essential for phage resistance.
Proposed mode of action of AbiK.
Many well-studied E. coli abortive infection systems involve activation of the Abi mechanism by a phage element which triggers blockage of an essential cellular function before virions are released. Macromolecule synthesis is often the target of the activated Abi system, leading to cell death (23, 52, 62). AbiK does not inhibit DNA and RNA synthesis by the lactococcal phage p2 (936 group) (6, 10). However, some mRNAs may not be translated, because procapsid structures were not observed in infected AbiK+ cells (10). On the other hand, the action of AbiK prevents the DNA replication (10) and the late gene expression (6) of the lactococcal phage ul36 (P335 group). Taken altogether, the data indicate that blockage of protein synthesis is an interesting hypothesis to explain the mode of action of AbiK.
It is proposed that AbiK synthesizes a cDNA strand from a currently unknown template RNA through its RT activity. However, it is still uncertain if the RT activity of AbiK would require a primer (53, 65, 66, 68). If needed, this primer could be of viral or cellular origin. The former would suggest the activation of AbiK upon phage infection, while the latter would imply that the retrotranscription of the target RNA is constitutive and not detrimental to the cell, as noninfected AbiK+ cells behave normally during growth. In these noninfected AbiK+ cells, the RNA strand and the resulting single-stranded cDNA would be degraded by a cellular RNase and an exonuclease, respectively.
It was previously shown that phages can become insensitive to AbiK through a single-nucleotide mutation causing an amino acid substitution in a phage protein (Sak) related to single-strand annealing proteins (9). In infected AbiK+ cells, the single-stranded cDNA released after degradation of the complementary template RNA could be annealed by Sak, thereby protecting it from degradation by cellular exonucleases. Subsequent base pairing of the single-stranded cDNA with a cRNA would sequester this RNA and prevent its further use. Phages harboring a mutation in the sak gene would have an altered activity, resulting in failure of AbiK. Experiments are currently under way to test these hypotheses.
Acknowledgments
We thank S. Zimmerly for fruitful discussion.
J.D.B. is a recipient of graduate scholarships from the Fonds FCAR and the Natural Sciences and Engineering Research Council (NSERC) of Canada. This study was funded by a strategic grant from NSERC to S.M.
REFERENCES
- 1.Avidan, O., M. E. Meer, I. Oz, and A. Hizi. 2002. The processivity and fidelity of DNA synthesis exhibited by the reverse transcriptase of bovine leukemia virus. Eur. J. Biochem. 269:859-867. [DOI] [PubMed] [Google Scholar]
- 2.Baichoo, N., and J. D. Helmann. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184:5826-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakhanashvili, M., O. Avidan, and A. Hizi. 1996. Mutational studies of human immunodeficiency virus type 1 reverse transcriptase: the involvement of residues 183 and 184 in the fidelity of DNA synthesis. FEBS Lett. 391:257-262. [DOI] [PubMed] [Google Scholar]
- 4.Bidnenko, E., S. D. Ehrlich, and M. C. Chopin. 1998. Lactococcus lactis phage operon coding for an endonuclease homologous to RuvC. Mol. Microbiol. 28:823-834. [DOI] [PubMed] [Google Scholar]
- 5.Birnboim, H. C. 1983. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 100:243-255. [DOI] [PubMed] [Google Scholar]
- 6.Bouchard, J. D. 2004. Mode d'action du système de résistance aux phages AbiK de Lactococcus lactis. Ph.D. thesis. Université Laval, Québec, Canada.
- 7.Bouchard, J. D., E. Dion, F. Bissonnette, and S. Moineau. 2002. Characterization of the two-component abortive phage infection mechanism AbiT from Lactococcus lactis. J. Bacteriol. 184:6325-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouchard, J. D., and S. Moineau. 2000. Homologous recombination between a lactococcal bacteriophage and the chromosome of its host strain. Virology 270:65-75. [DOI] [PubMed] [Google Scholar]
- 9.Bouchard, J. D., and S. Moineau. 2004. Lactococcal phage genes involved in sensitivity to AbiK and their relation to single-strand annealing proteins. J. Bacteriol. 186:3649-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boucher, I., E. Emond, E. Dion, D. Montpetit, and S. Moineau. 2000. Microbiological and molecular impacts of AbiK on the lytic cycle of Lactococcus lactis phages of the 936 and P335 species. Microbiology 146:445-453. [DOI] [PubMed] [Google Scholar]
- 11.Boucher, I., E. Emond, M. Parrot, and S. Moineau. 2001. DNA sequence analysis of three Lactococcus lactis plasmids encoding phage resistance mechanisms. J. Dairy Sci. 84:1610-1620. [DOI] [PubMed] [Google Scholar]
- 12.Boucher, I., C. Vadeboncoeur, and S. Moineau. 2003. Characterization of genes involved in the metabolism of alpha-galactosides by Lactococcus raffinolactis. Appl. Environ. Microbiol. 69:4049-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cluzel, P. J., A. Chopin, S. D. Ehrlich, and M. C. Chopin. 1991. Phage abortive infection mechanism from Lactococcus lactis subsp. lactis, expression of which is mediated by an Iso-ISS1 element. Appl. Environ. Microbiol. 57:3547-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffey, A., and R. P. Ross. 2002. Bacteriophage-resistance systems in dairy starter strains: molecular analysis to application. Antonie Leeuwenhoek 82:303-321. [PubMed] [Google Scholar]
- 15.Cram, D., A. Ray, and R. Skurray. 1984. Molecular analysis of F plasmid pif region specifying abortive infection of T7 phage. Mol. Gen. Genet. 197:137-142. [DOI] [PubMed] [Google Scholar]
- 16.Dai, G., P. Su, G. E. Allison, B. L. Geller, P. Zhu, W. S. Kim, and N. W. Dunn. 2001. Molecular characterization of a new abortive infection system (AbiU) from Lactococcus lactis LL51-1. Appl. Environ. Microbiol. 67:5225-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dao, M. L., and J. J. Ferretti. 1985. Streptococcus-Escherichia coli shuttle vector pSA3 and its use in the cloning of streptococcal genes. Appl. Environ. Microbiol. 49:115-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng, Y. M., C. Q. Liu, and N. W. Dunn. 1999. Genetic organization and functional analysis of a novel phage abortive infection system, AbiL, from Lactococcus lactis. J. Biotechnol. 67:135-149. [DOI] [PubMed] [Google Scholar]
- 19.de Vos, W. M. 1987. Gene cloning and expression in lactic streptococci. FEMS Microbiol. Rev. 46:281-295. [Google Scholar]
- 20.Dinsmore, P. K., and T. R. Klaenhammer. 1994. Phenotypic consequences of altering the copy number of abiA, a gene responsible for aborting bacteriophage infections in Lactococcus lactis. Appl. Environ. Microbiol. 60:1129-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dinsmore, P. K., D. J. O'Sullivan, and T. R. Klaenhammer. 1998. A leucine repeat motif in AbiA is required for resistance of Lactococcus lactis to phages representing three species. Gene 212:5-11. [DOI] [PubMed] [Google Scholar]
- 22.Domingues, S., A. Chopin, S. D. Ehrlich, and M. C. Chopin. 2004. The lactococcal abortive phage infection system AbiP prevents both phage DNA replication and temporal transcription switch. J. Bacteriol. 186:713-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duckworth, D. H., J. Glenn, and D. J. McCorquodale. 1981. Inhibition of bacteriophage replication by extrachromosomal genetic elements. Microbiol. Rev. 45:52-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duplessis, M., and S. Moineau. 2001. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol. Microbiol. 41:325-336. [DOI] [PubMed] [Google Scholar]
- 25.Emond, E., B. J. Holler, I. Boucher, P. A. Vandenbergh, E. R. Vedamuthu, J. K. Kondo, and S. Moineau. 1997. Phenotypic and genetic characterization of the bacteriophage abortive infection mechanism AbiK from Lactococcus lactis. Appl. Environ. Microbiol. 63:1274-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forde, A., and G. F. Fitzgerald. 1999. Bacteriophage defence systems in lactic acid bacteria. Antonie Leeuwenhoek. 76:89-113. [PubMed] [Google Scholar]
- 27.Garvey, P., G. F. Fitzgerald, and C. Hill. 1995. Cloning and DNA sequence analysis of two abortive infection phage resistance determinants from the lactococcal plasmid pNP40. Appl. Environ. Microbiol. 61:4321-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorbalenya, A. E. 1994. Self-splicing group I and group II introns encode homologous (putative) DNA endonucleases of a new family. Protein Sci. 3:1117-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haandrikman, A. 1990. Development and use of a broad-host range vector for the simultaneous analysis of divergent promoters. Ph.D. thesis. Rijksuniversiteit Groningen, Groningen, The Netherlands.
- 31.Harrison, S. C., and A. K. Aggarwal. 1990. DNA recognition by proteins with the helix-turn-helix motif. Annu. Rev. Biochem. 59:933-969. [DOI] [PubMed] [Google Scholar]
- 32.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jonckheere, H., E. De Clercq, and J. Anne. 2000. Fidelity analysis of HIV-1 reverse transcriptase mutants with an altered amino-acid sequence at residues Leu74, Glu89, Tyr115, Tyr183 and Met184. Eur. J. Biochem. 267:2658-2665. [DOI] [PubMed] [Google Scholar]
- 34.Karplus, K., C. Barrett, and R. Hughey. 1998. Hidden Markov models for detecting remote protein homologies. Bioinformatics 14:846-856. [DOI] [PubMed] [Google Scholar]
- 35.Kaushik, N., K. Singh, I. Alluru, and M. J. Modak. 1999. Tyrosine 222, a member of the YXDD motif of MuLV RT, is catalytically essential and is a major component of the fidelity center. Biochemistry 38:2617-2627. [DOI] [PubMed] [Google Scholar]
- 36.Kohlstaedt, L. A., J. Wang, J. M. Friedman, P. A. Rice, and T. A. Steitz. 1992. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256:1783-1790. [DOI] [PubMed] [Google Scholar]
- 37.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 38.Lampson, B. C., J. Sun, M. Y. Hsu, J. Vallejo-Ramirez, S. Inouye, and M. Inouye. 1989. Reverse transcriptase in a clinical strain of Escherichia coli: production of branched RNA-linked msDNA. Science 243:1033-1038. [DOI] [PubMed] [Google Scholar]
- 39.Laniel, M. A., A. Béliveau, and S. L. Guérin. 2001. DNA-protein interactions: principles and methods. Methods Mol. Biol. 148:13-30. [DOI] [PubMed] [Google Scholar]
- 40.Larder, B. A., S. D. Kemp, and D. J. Purifoy. 1989. Infectious potential of human immunodeficiency virus type 1 reverse transcriptase mutants with altered inhibitor sensitivity. Proc. Natl. Acad. Sci. USA 86:4803-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, T., S. Nakashima, K. Hirose, M. Shibasaka, M. Katsuhara, B. Ezaki, D. P. Giedroc, and K. Kasamo. 2004. A novel cyanobacterial SmtB/ArsR family repressor regulates the expression of a CPx-ATPase and a metallothionein in response to both Cu(I)/Ag(I) and Zn(II)/Cd(II). J. Biol. Chem. 279:17810-17818. [DOI] [PubMed] [Google Scholar]
- 42.Lowe, D. M., V. Parmar, S. D. Kemp, and B. A. Larder. 1991. Mutational analysis of two conserved sequence motifs in HIV-1 reverse transcriptase. FEBS Lett. 282:231-234. [DOI] [PubMed] [Google Scholar]
- 43.Malik, H. S., and T. H. Eickbush. 2001. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 11:1187-1197. [DOI] [PubMed] [Google Scholar]
- 44.Martin-Hernandez, A. M., E. Domingo, and L. Menendez-Arias. 1996. Human immunodeficiency virus type 1 reverse transcriptase: role of Tyr115 in deoxynucleotide binding and misinsertion fidelity of DNA synthesis. EMBO J. 15:4434-4442. [PMC free article] [PubMed] [Google Scholar]
- 45.McKay, L. L., K. A. Baldwin, and E. A. Zottola. 1972. Loss of lactose metabolism in lactic streptococci. Appl. Microbiol. 23:1090-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller, J. F., and M. H. Malamy. 1983. Identification of the pifC gene and its role in negative control of F factor pif gene expression. J. Bacteriol. 156:338-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Mohr, G., P. S. Perlman, and A. M. Lambowitz. 1993. Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Res. 21:4991-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moineau, S. 1999. Applications of phage resistance in lactic acid bacteria. Antonie Leeuwenhoek 76:377-382. [PubMed] [Google Scholar]
- 50.Moineau, S., E. Durmaz, S. Pandian, and T. R. Klaenhammer. 1993. Differentiation of two abortive mechanisms by using monoclonal antibodies directed toward lactococcal bacteriophage capsid proteins. Appl. Environ. Microbiol. 59:208-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moineau, S., J. Fortier, H.-W. Ackermann, and S. Pandian. 1992. Characterization of lactococcal bacteriophages from Québec cheese plants. Can. J. Microbiol. 38:875-882. [Google Scholar]
- 52.Molineux, I. J. 1991. Host-parasite interactions: recent developments in the genetics of abortive phage infections. New Biol. 3:230-236. [PubMed] [Google Scholar]
- 53.Morozova, T., W. Seo, and S. Zimmerly. 2002. Non-cognate template usage and alternative priming by a group II intron-encoded reverse transcriptase. J. Mol. Biol. 315:951-963. [DOI] [PubMed] [Google Scholar]
- 54.O'Connor, L., M. Tangney, and G. F. Fitzgerald. 1999. Expression, regulation, and mode of action of the AbiG abortive infection system of Lactococcus lactis subsp. cremoris UC653. Appl. Environ. Microbiol. 65:330-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Sullivan, D. J., and T. R. Klaenhammer. 1993. Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus lactis and Lactobacillus spp. Appl. Environ. Microbiol. 59:2730-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ouali, M., and R. D. King. 2000. Cascaded multiple classifiers for secondary structure prediction. Protein Sci. 9:1162-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parreira, R., S. D. Ehrlich, and M. C. Chopin. 1996. Dramatic decay of phage transcripts in lactococcal cells carrying the abortive infection determinant AbiB. Mol. Microbiol. 19:221-230. [DOI] [PubMed] [Google Scholar]
- 58.Rampersaud, A., S. L. Hadockerg, and M. Inouye. 1994. The OmpR protein of Escherichia coli binds to sites in the ompF promoter region in a hierarchical manner determined by its degree of phosphorylation. J. Biol. Chem. 269:12559-12566. [PubMed] [Google Scholar]
- 59.Rosinski, J. A., and W. R. Atchley. 1999. Molecular evolution of helix-turn-helix proteins. J. Mol. Evol. 49:301-309. [DOI] [PubMed] [Google Scholar]
- 60.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 61.Sanders, M. E., P. J. Leonhard, W. D. Sing, and T. R. Klaenhammer. 1986. Conjugal strategy for construction of fast acid-producing, bacteriophage-resistant lactic streptococci for use in dairy fermentations. Appl. Environ. Microbiol. 52:1001-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Snyder, L. 1995. Phage-exclusion enzymes: a bonanza of biochemical and cell biology reagents? Mol. Microbiol. 15:415-420. [DOI] [PubMed] [Google Scholar]
- 63.Twomey, D. P., P. J. De Urraza, L. L. McKay, and D. J. O'Sullivan. 2000. Characterization of AbiR, a novel multicomponent abortive infection mechanism encoded by plasmid pKR223 of Lactococcus lactis subsp. lactis KR2. Appl. Environ. Microbiol. 66:2647-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.VanZile, M. L., X. Chen, and D. P. Giedroc. 2002. Allosteric negative regulation of smt O/P binding of the zinc sensor, SmtB, by metal ions: a coupled equilibrium analysis. Biochemistry 41:9776-9786. [DOI] [PubMed] [Google Scholar]
- 65.Wang, H., J. C. Kennell, M. T. Kuiper, J. R. Sabourin, R. Saldanha, and A. M. Lambowitz. 1992. The Mauriceville plasmid of Neurospora crassa: characterization of a novel reverse transcriptase that begins cDNA synthesis at the 3′ end of template RNA. Mol. Cell. Biol. 12:5131-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, H., and A. M. Lambowitz. 1993. The Mauriceville plasmid reverse transcriptase can initiate cDNA synthesis de novo and may be related to reverse transcriptase and DNA polymerase progenitor. Cell 75:1071-1081. [DOI] [PubMed] [Google Scholar]
- 67.Wells, J. M., P. W. Wilson, and R. W. Le Page. 1993. Improved cloning vectors and transformation procedure for Lactococcus lactis. J. Appl. Bacteriol. 74:629-636. [DOI] [PubMed] [Google Scholar]
- 68.Wilhelm, M., and F. X. Wilhelm. 2001. Reverse transcription of retroviruses and LTR retrotransposons. Cell. Mol. Life Sci. 58:1246-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wintjens, R., and M. Rooman. 1996. Structural classification of HTH DNA-binding domains and protein-DNA interaction modes. J. Mol. Biol. 262:294-313. [DOI] [PubMed] [Google Scholar]
- 70.Xiong, Y., and T. H. Eickbush. 1990. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 9:3353-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]