Abstract
One proposed mechanism of replication inhibition in iteron-containing plasmids (ICPs) is “handcuffing,” in which the coupling of origins via iteron-bound replication initiator (Rep) protein turns off origin function. In minimal R6K replicons, copy number control requires the interaction of plasmid-encoded π protein with the seven 22-bp iterons of the γ origin of replication. Like other related Rep proteins, π exists as both monomers and dimers. However, the ability of π dimers to bind iterons distinguishes R6K from most other ICPs, where only monomers have been observed to bind iterons. Here, we describe experiments to determine if monomers or dimers of π protein are involved in the formation of handcuffed complexes. Standard ligation enhancement assays were done using π variants with different propensities to bind iterons as monomers or dimers. Consistent with observations from several ICPs, a hyperreplicative variant (π·P106L∧F107S) exhibits deficiencies in handcuffing. Additionally, a novel dimer-biased variant of π protein (π·M36A∧M38A), which lacks initiator function, handcuffs iteron-containing DNA more efficiently than does wild-type π. The data suggest that π dimers mediate handcuffing, supporting our previously proposed model of handcuffing in the γ ori system. Thus, dimers of π appear to possess three distinct inhibitory functions with respect to R6K replication: transcriptional autorepression of π expression, in cis competition (for origin binding) with monomeric activator π, and handcuffing-mediated inhibition of replication in trans.
In a group of related bacterial plasmids (5), initiation of DNA replication occurs at a specific site called the origin of replication (ori) that contains tandemly repeated DNA sequences known as iterons or direct repeats (DRs). Plasmid-encoded replication protein (Rep) binds to these iterons, where it can act as either an initiator of plasmid replication or an inhibitor of overreplication (Fig. 1) (3, 8). “Handcuffing,” or origin pairing, is a generally accepted mechanism of replication inhibition in iteron-containing plasmids (ICPs); this coupling of origins via iteron-bound initiator protein is believed to turn off origin function (20, 27). In minimal R6K replicons, copy number control requires the interaction of the plasmid-encoded π protein with the seven 22-bp iterons of the γ origin. Electron microscopy (EM) studies demonstrated that handcuffing, mediated by the Rep protein π, occurs efficiently between two DNA fragments containing iterons (20, 23, 24) and that the fragments are found in parallel alignment (32). The most suggestive evidence for handcuffing in the copy number control of ICPs comes from plasmids R6K, P1, RK2, and mini-F, where correlations between hyperreplicative (copy-up) phenotypes and handcuffing deficiencies have been demonstrated (2, 20, 22, 26, 31).
FIG. 1.
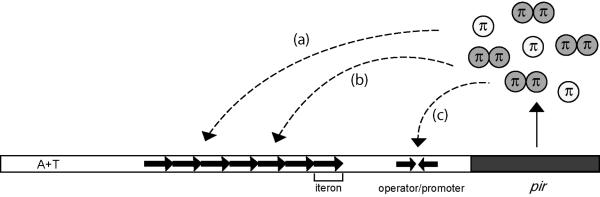
Roles of π binding to γ ori. The pir gene encodes π protein, which exists in two forms: monomers (white) and dimers (gray). The seven direct repeats of γ ori, also called iterons, are indicated by tandem arrows, whereas inverted arrows represent the inverted repeat in the pir gene operator/promoter. π is a multifunctional protein: monomers activate replication (a), dimers inhibit replication (b), and dimers autorepress pir transcription (c).
Although Rep dimers predominate in solution, in most ICPs only Rep monomers, the replication activator form of the protein, have been demonstrated to bind iterons. A variety of data from different systems suggest a relationship between Rep dimers, iteron-mediated plasmid incompatibility (Inc), and handcuffing-based inhibition of replication (reviewed in reference 18). Until recently (4), however, the only Rep protein that had been shown to bind iterons as dimers (in addition to binding as monomers) was the π protein of plasmid R6K (1, 16, 19, 32, 34). Relevant to this is our observation that π dimers bind an individual iteron by using a single subunit, thus leaving the second subunit free to engage a second iteron (32). We have proposed that in the R6K system, negative replication control via handcuffing is mediated by simple dimers of π that can capture iteron sequences in trans or in cis (19). Additionally, another model of handcuffing in R6K has been proposed, in which monomers bound to a single DNA fragment can pair with another DNA fragment also bound by monomers (20). In contrast to R6K, a more elaborate mechanism to handcuff oris has been proposed for plasmid RK2. Although they are iteron binding deficient, dimers of RK2's Rep protein (TrfA) have been proposed to bring together two replication complexes (comprised of monomer-bound iterons), resulting in a tetramer “bridge” (2, 18, 30). Despite the various models for handcuffing, the precise structure(s) of the nucleoprotein complexes involved in the phenomenon has yet to be determined for any ICP. Therefore, we are particularly interested in determining whether π monomer-bound iterons or preformed dimers mediate DNA coupling in R6K.
To address this issue, we have conducted experiments using variants of π protein, His-π·M36A∧M38A and His-π· P106L∧F107S, which differ in their monomer-to-dimer ratios (16). π·M36A∧M38A can inhibit replication dependent on wild-type (wt) π and it does not stimulate γ ori-dependent replication in vivo or in vitro (16; J. Wu and M. Filutowicz, unpublished data), nor can it stimulate “open-complex” formation in the A+T-rich segment of the ori (16). His-π· M36A∧M38A benefited this work due to its property that only dimers of the protein bind iterons (16, 19). Conveniently, the properties of His-π·P106L∧F107S are just the opposite: it is hyperactive in replication (in vivo and in vitro), it can facilitate open-complex formation in vitro, and it binds iterons predominantly as monomers (16, 19). To investigate the handcuffing abilities of these proteins, we conducted standard ligation enhancement assays (20, 21) using wt π as our baseline for activity. The identities of the products of ligation were determined by EM. We show that His-π·M36A∧M38A handcuffs iteron-containing DNA more efficiently than the wt and that His-π·P106L∧F107S is severely impaired in handcuffing. The latter result is consistent with the properties of copy-up Rep proteins from several ICPs (1, 2, 20-22, 26, 31). Taken together, the data support a model of replication inhibition in which preformed π dimers facilitate handcuffing in the R6K system.
MATERIALS AND METHODS
Plasmids and DNA fragment preparation.
A DNA fragment containing seven DRs was released from plasmid pMF34 (9) after digestion with AseI/SnaBI and cloned into the HincII site of pUC9, generating the plasmid pRK4. A PCR-amplified fragment containing the seven DRs from pRK4 digested with XbaI/SalI was cloned into plasmid pBEND5 (13) digested with the same enzymes, generating the plasmid pRK28. Fragments containing seven DRs (321 bp) used for ligation enhancement assays were obtained by digesting pRK28 with EcoRV.
Protein purification and DNA binding characteristics.
His-π·wt, His-π· P106L∧F107S, and His-π·M36A∧M38A were purified as described previously (33). His-tagged proteins retain the characteristics of their nontagged counterparts (15, 16). The monomer:dimer ratios of each protein in complexes with single iteron-containing DNA fragments was previously established; His-π· P106L∧F107S binds iterons preferentially as monomers, while His-π· M36A∧M38A binds exclusively as dimers (16, 19).
Ligation enhancement assay.
DNA containing seven DRs (321-bp EcoRV fragment) was incubated in a 100-μl final reaction volume with His-π·wt or variants of π. The protein amounts used are indicated in the figure legends. The DNA amounts used were 500 pg of [γ-32P]dATP-end-labeled fragment (“limiting” amount of DNA) or 100 ng of unlabeled fragment (“nonlimiting” amount of DNA). All reactions were carried out in 1× ligase buffer (Epicenter) containing 975 ng poly(dI-dC) and 1 mM ATP (Amersham). An aliquot (16 μl) of the sample was removed and subjected to electrophoretic mobility shift assay (EMSA) (4% polyacrylamide gel electrophoresed in 0.5× Tris-borate-EDTA). The remaining samples were further processed by adding 0.5 unit of T4 DNA ligase (Epicenter) and incubated at 37°C for 10 min. Next, samples were extracted with an equal volume of phenol-chloroform and then with chloroform. Ethanol-precipitated DNA was separated on 4% polyacrylamide gels, dried, and scanned with a PhosphorImager using a Storm System (Molecular Dynamics). Radioactive bands were quantified using Image Quant software (Image Quant). For reaction mixtures containing unlabeled DNA, the gels were stained with SYBR green (Sigma) and scanned. Ligation products were also quantified by EM.
Electron microscopy.
EM was done as described previously (7).
RESULTS
π enhances the ligation of fragments containing γ origin iterons.
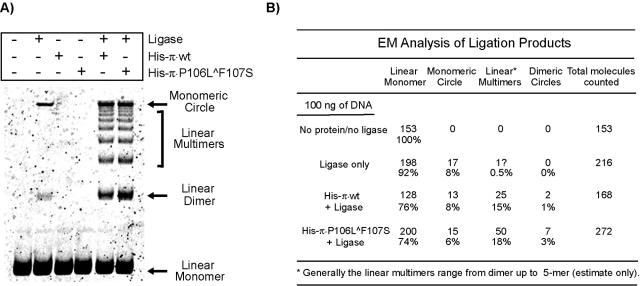
We carried out ligation enhancement assays to characterize the handcuffing ability of π. This commonly used assay is based on the premise that a protein should increase the local concentration of DNA fragments' ends if it can simultaneously bind two DNA molecules. As a result, in ligation reactions, an increase in the rate of intermolecular ligation is observed in the presence of suitable DNA-binding proteins (20, 21, 23). DNA containing seven DRs (100 ng) was incubated with 125 ng of either His-π·wt or His-π·P106L∧F107S and then treated with ligase. As shown in Fig. 2A, the reaction containing DNA and ligase but lacking π (control) predominantly formed a slow-migrating product, and a very small amount of linear dimer was also evident. When His-tagged π protein was added to the ligation reaction mixtures, no significant difference was observed in the amount of ligated products in samples containing the wt protein versus the monomer-biased π·P106L∧F107S. To characterize the products of ligation, we treated ligation samples with Bal 31 nuclease, which digests linear DNA fragments, and again performed gel electrophoresis. We observed that the slowest-migrating band was the sole ligation product surviving Bal 31 digestion, suggesting that the band may contain circular DNA (data not shown).
FIG. 2.
Detection of π-dependent handcuffing in the presence of excess DNA. (A) One hundred nanograms of EcoRV fragment containing seven DRs was incubated in the presence or absence of 125 ng of His-π·wt or His-π·P106L∧F107S and with or without 0.5 unit of ligase as indicated. The samples were processed as described in Materials and Methods. Linear DNA monomers, linear dimers, and monomeric circles are indicated by arrows; linear DNA multimers are indicated by a bracket. (B) Categorization of ligation products by electron microscopy.
To further characterize the products formed in the ligation enhancement assay, aliquots of the ligation mixtures were categorized by EM; the results are summarized in Fig. 2B. EM analysis confirmed that the product of reactions containing DNA plus ligase is circular DNA. Importantly, it is evident from EM as well as gel electrophoresis that multiple ligation products, including linear oligomers (dimers, trimers, etc.) and circles, were formed in the ligation reactions, in a π-dependent manner. Although we do not understand the unusually slow electrophoretic mobility of the DNA circles, similar migration patterns have been described elsewhere (29).
Copy-up variant π is handcuffing deficient under DNA-limiting conditions.
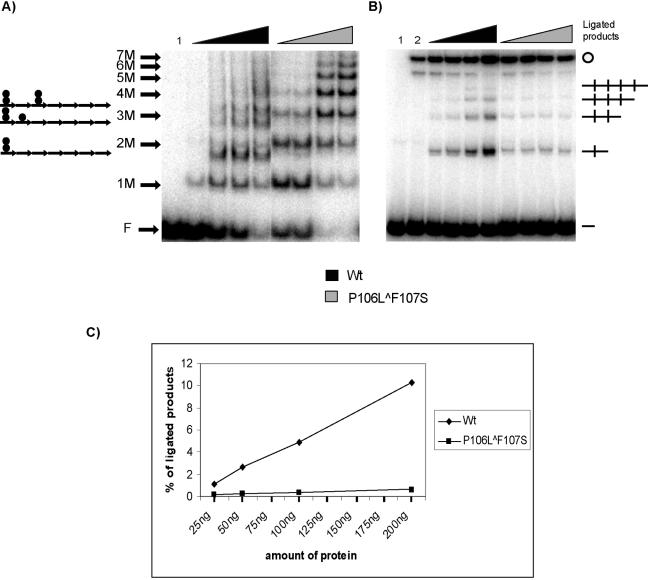
Although there is a known copy-up π protein that does not exhibit a measurable handcuffing deficiency (22), we were somewhat surprised by the outcome of our ligation enhancement assay. We speculated that this assay might be better suited to reveal differences in handcuffing efficiencies if the DNA levels were limiting. Such conditions would be expected to strongly favor π monomer binding in the reactions containing the monomer-biased (16, 19) copy-up variant. After a survey of different levels of DNA, we determined that using 0.5 ng of seven-DR DNA fragment was preferable for discriminating between protein variants (data not shown). This “limiting” DNA level was insufficient for employing EM to analyze ligation products. An example of a ligation enhancement assay with the “limiting” level of DNA and increasing concentrations of π protein is shown in Fig. 3. Before addition of ligase, a fraction of each reaction sample was analyzed by EMSA to visualize nucleoprotein complexes formed under our assay conditions (Fig. 3A). As expected, the monomer-biased copy-up variant (His-π·P106L∧F107S) bound predominantly as a monomer. In comparison, a mixture of monomer and dimer binding was seen in samples containing wt π.
FIG. 3.
Binding of π to the iteron and detection of π-dependent handcuffing by ligation enhancement under limiting DNA conditions. A 0.5-ng amount of 32P-end-labeled EcoRV fragment containing seven DRs was utilized, along with increasing amounts of His-π·wt and His-π· P106L∧F107S. (A) Hypothetical representation of π monomers and π dimers bound to the seven iterons in EMSA is shown (left). F indicates free DNA; 1 M to 7 M indicates the number of π monomer-bound iterons (e.g., 7 M indicates that the seven-DR fragment has each iteron filled by a monomer). Triangles indicate increasing amounts of protein added to the reaction mixtures: 25, 50, 100, and 200 ng. Lane 1 contains DNA without protein. (B) The remaining samples were treated with 0.5 unit of ligase and processed as described in Materials and Methods. Lane 1 contains DNA only (without protein or ligase); lane 2 contains DNA with 0.5 unit of ligase. Handcuffed products (linear DNA dimer, linear trimer, linear tetramer, and monomeric circles) and free DNA (—) are indicated. (C) Quantification of ligated products; results are the averages from three independent experiments.
As was observed using the elevated DNA concentrations in Fig. 2, multiple ligation products were formed in a π-dependent manner (Fig. 3B). One particularly eye-catching result is that only wt π yielded a concentration-dependent increase in the amount of ligated products. Ligation efficiency was insensitive to increasing concentrations of His-π·P106L∧F107S. At the highest concentration of protein, approximately 10 times more ligated product was observed in the presence of wt π in comparison to reactions containing His-π·P106L∧F107S (Fig. 3C). Furthermore, close inspection of the DNA binding patterns generated by the wt and copy-up variant proteins (Fig. 3A) provides a possible explanation for the differences in ligation enhancement. For wt π, increasing protein concentration leads to increasing levels of dimer-bound iteron complexes. In contrast, increasing the concentration of copy-up π simply leads to higher-order monomer-bound complexes. Monomers efficiently bind iterons even though they are less abundant than dimers (16). Thus, our results demonstrate a positive correlation between the levels of dimer-bound iterons and the amount of ligation enhancement observed.
A dimeric variant facilitates π-dependent ligation enhancement under DNA-limiting conditions.
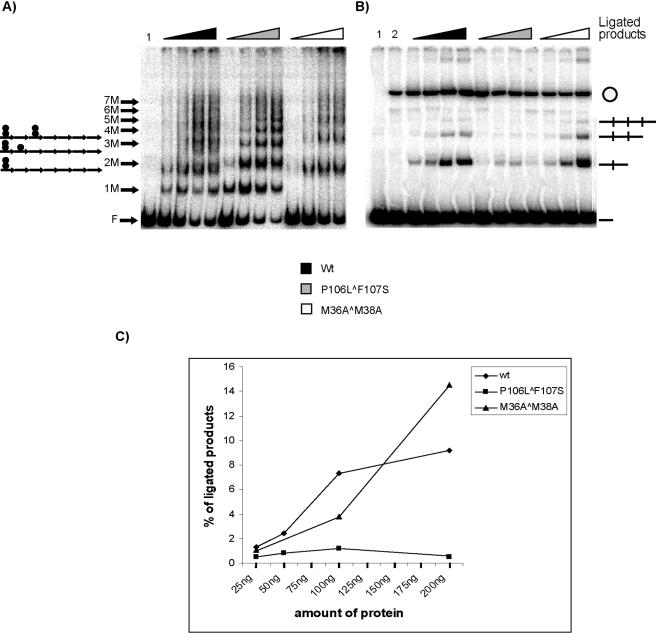
His-π·M36A∧M38A is a novel variant of Repπ available in the R6K system, whose interaction with iterons appears to be solely dimeric (16, 19). We anticipated that this dimer-biased variant would be most helpful in testing our hypothesis that dimers of π protein facilitate handcuffing. As mentioned in the introduction, His-π·M36A∧M38A is unable to activate γ ori (in vivo or in vitro); however, it does inhibit replication (16; Y. Peng, J. Wu, and M. Filutowicz, unpublished data). To investigate whether the inhibition of replication by π dimers might occur via a handcuffing mechanism, we performed assays to compare the ligation enhancement ability of His-π·M36A∧M38A against the abilities of His-π·wt and His-π·P106L∧F107S (monomer biased). DNA fragments were incubated with each π variant, and a fraction of each sample was subjected to EMSA (Fig. 4A). The binding patterns for wt π and the copy-up variant were similar to those observed in the previous experiment. Samples containing the dimeric variant (His-π·M36A∧M38A) showed only four DNA-protein complexes. The remaining samples were treated with ligase and processed (Fig. 4B). In support of our hypothesis that the binding of preformed π dimers stimulates handcuffing, our results showed that 200 ng of His-π· M36A∧M38A yielded 50% more ligation products than His-π·wt (Fig. 4C).
FIG. 4.
Detection of handcuffing using wt and copy-up and dimeric variants of π. A 0.5-ng amount of 32P-labeled EcoRV fragment containing seven DRs was utilized in this experiment with increasing amounts of His-π·wt, His-π·P106L∧F107S, or His-π·M36A∧M38A. (A) Hypothetical representation of π monomers and π dimers bound to the seven iterons in EMSA is shown (left). F indicates free DNA; 1 M to 7 M indicates the number of π monomer-bound iterons (e.g., 7 M indicates that the seven-DR fragment has each iteron filled by a monomer). Triangles indicate increasing amounts of protein added to the reaction mixtures: 25, 50, 100, and 200 ng. (B) The remaining samples were treated with 0.5 unit of ligase and processed as described in Materials and Methods. For the sample containing His-π·M36A∧M38A, only 25-, 100-, and 200-ng samples were analyzed. Handcuffed products (linear DNA dimer, linear trimer, linear tetramer, and monomeric circles) and free DNA (—) are indicated. (C) Quantification of ligated products.
The reduced number of DNA-protein complexes (four) seen with the dimeric variant (His-π·M36A∧M38A) could be explained based on previous EMSA experiments using His-π· M36A∧M38A and a two-DR-containing DNA fragment (17). The results showed a single nucleoprotein complex formed by the dimer-biased protein compared to wt π, which produced very complex binding patterns. These combined data suggest the possibility that two π dimers may be unable to occupy adjacent iterons. Further experiments are under way to test several hypotheses that might explain these observations (e.g., a steric hindrance effect by π dimers or effects of variations in iteron sequences.)
DISCUSSION
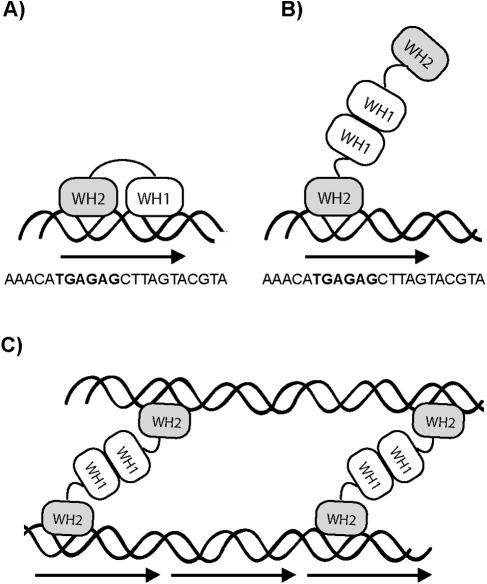
In ICPs, handcuffing of linear DNA fragments containing DRs has been directly demonstrated by EM and suggested by ligation enhancement assay, but the precise structures of nucleoprotein complexes involved are yet to be determined. As illustrated in Fig. 5A, monomers of Rep proteins, including π, contain two DNA-binding domains, an N-terminal WH1 (winged helix 1) motif and a C-terminal WH2 motif, both of which are used for iteron binding (14). Although purified Rep proteins are found predominantly as dimers in solution, with few exceptions such dimers do not appear to be iteron binding proficient. On the other hand, a single layer of Rep protein monomers bound to an ori is expected to be incapable of capturing a second ori to bring about the DNA coupling that is the hallmark of handcuffing. Consequently, a thorough understanding of the mechanism by which Rep proteins pair two iteron-bearing DNA molecules into handcuffed structures has remained elusive.
FIG. 5.
A schematic representation of π bound to iterons. π protein is indicated by two oval winged helix domains connected by a linker, paired wavy lines indicate double-stranded DNA, and an arrow indicates a single iteron, the sequence of which is shown. (A) π monomer bound to a single iteron using both WH1 and WH2 domains. (B) π dimer bound to a single iteron using only the WH2 domain. (C) Proposed model for handcuffing in which both WH2 domains of preformed π dimers contact two iteron-containing DNA fragments. The figure is not drawn to scale.
As an exception to the general rule, dimers of R6K's π protein readily bind γ ori iterons, leading to speculation about whether R6K might employ a novel mechanism for handcuffing. The π dimer apparently uses its C-terminal WH2 to engage the DNA (Fig. 5B); however, this observation is not, in fact, “exceptional” (19, 32). Rather, it coincides well with our general understanding of the three-dimensional structure of Rep protein based on crystallography data generated in two other ICP systems, mini-F plasmid/RepE54 (14) and pPS10/RepA (12). Taken together, the structural analyses suggested that the switch from protein monomer to dimer might involve the remodeling of WH1 (10). Thus, in Rep protein dimers, one of the DNA-binding domains is thought to be occluded due to dimerization. Our own π/iteron contact mapping data (19) support this model, with dimers of π making contacts nearly identical to those of monomers but only in a subset (left half) of the iteron base pairs and DNA backbone (Fig. 5A and B).
Given our current understanding of Rep protein structure and the ability of Rep to handcuff DNA, the ability of π dimers to bind iterons does not make this protein as unusual as it once appeared. It seems likely that the difference between π and other Rep proteins is more subtle, being a matter of different binding affinities for the DNA. π dimers bind strongly enough to be captured by EMSA, perhaps due to a polypeptide loop not found in other Rep proteins (19, 28). In contrast, we believe that other Rep dimers bind DNA more weakly but probably do bind. In fact, evidence for other Rep dimers binding to iterons is beginning to come forward. In plasmid pPS10, for example, UV circular dichroism (spectral analysis) suggests that RepA binds to iteron DNA as dimers, which subsequently dissociate into monomers (6). More recently, Das and Chattoraj demonstrated that dimers of plasmid P1-encoded RepA also bind iterons and participate in handcuffing (4), although the form of dimer employed (preformed, chaperone activated, or both) is uncertain. This diverse collection of data engendered the following question: do preformed dimers of π (and perhaps other Rep proteins) mediate handcuffing?
Here, we present evidence for a correlation between the abundance of π dimer-bound iterons and the ability of the Rep protein to promote handcuffing as measured by ligation enhancement assays. The use of well-characterized variants allowed us to examine the abilities of π monomers and π dimers to participate in DNA coupling. Under limiting DNA concentrations, 200 ng of the dimer-biased π variant proved to be more efficient than the wt in producing ligase-dependent multimerization. In contrast, a monomer-biased copy-up variant exhibited the lowest ligation efficiency, consistent with observations of handcuffing deficiencies in copy-up Rep variants from several ICPs (1, 2, 20-22, 26, 31). We note that at low protein levels (below 200 ng), wt π handcuffed DNA better than the dimeric variant π·M36A∧M38A. One possible reason for this could be that π·M36A∧M38A binds iterons less well than the wt (19). This can be seen by comparing the levels of free DNA in Fig. 4A. Additionally, we observed that compared to reactions containing ligase only, reactions containing ligase plus π (wt or copy-up) show an approximately threefold increase in monomeric circles. This may be a consequence of the known ability of π protein to bend DNA (4, 11, 17, 25). It has been shown that π monomers and dimers bend a single iteron to similar degrees (∼50o) (17).
Notably, when we conducted ligation enhancement assays with nonlimiting DNA concentrations, we were unable to see any significant differences in the amount of ligation between wt and copy-up π. This indicates that care should be taken when characterizing a copy-up protein as having no deficiency in handcuffing; such deficiencies may be manifested or obscured based on reaction conditions. We suspect that when DNA concentrations are low and presumably less favorable for handcuffing, the monomer bias of copy-up π contributes significantly to the binding reaction. On the other hand, high DNA concentrations reduce competition between monomers and dimers for iteron binding, allowing copy-up π to behave similarly to the wt in a handcuffing assay.
There are three fundamental models of handcuffing by Rep protein (reviewed in reference 18), which are distinguished by the perceived roles of Rep monomers and dimers as the DNA-coupling agent. Although the data presented here cannot exclude any of these models, they best support a mode of replication inhibition in the π/γ ori system in which handcuffing is mediated by preformed π dimers bound to iterons (Fig. 5C). Thus, the mechanism of handcuffing used by R6K may be similar if not identical to the mechanism employed by plasmid P1 (4). In fact, based on the structural and functional similarities of all the Rep proteins, we believe that the mechanism of handcuffing in other Rep/iteron systems could also be similar and that the findings described here may provide important insights into a general mechanism of replication inhibition by dimers in iteron/Rep systems.
Acknowledgments
This work was supported by National Institutes of Health grant GM40314 to M.F.
We thank Ricardo Krüger for his support and discussion and Sindhu Chittenipattu for help with EM analysis. We also thank Wilma Ross and Richard L. Gourse for critical reading of the manuscript.
REFERENCES
- 1.Abhyankar, M. M., J. M. Reddy, R. Sharma, E. Bullesbach, and D. Bastia. 2004. Biochemical investigations of control of replication initiation of plasmid R6K. J. Biol. Chem. 279:6711-6719. [DOI] [PubMed] [Google Scholar]
- 2.Blasina, A., B. L. Kittell, A. E. Toukdarian, and D. R. Helinski. 1996. Copy-up mutants of the plasmid RK2 replication initiation protein are defective in coupling RK2 replication origins. Proc. Natl. Acad. Sci. USA 93:3559-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chattoraj, D. K., and T. D. Schneider. 1997. Replication control of plasmid P1 and its host chromosome: the common ground. Prog. Nucleic Acid Res. Mol. Biol. 57:145-186. [DOI] [PubMed] [Google Scholar]
- 4.Das, N., and D. K. Chattoraj. 2004. Origin pairing (‘handcuffing’) and unpairing in the control of P1 plasmid replication. Mol. Microbiol. 54:836-849. [DOI] [PubMed] [Google Scholar]
- 5.del Solar, G., R. Giraldo, M. J. Ruiz-Echevarría, M. Espinosa, and R. Díaz-Orejas. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Díaz-López, T., M. Lages-Gonzalo, A. Serrano-López, C. Alfonso, G. Rivas, R. Díaz-Orejas, and R. Giraldo. 2003. Structural changes in RepA, a plasmid replication initiator, upon binding to origin DNA. J. Biol. Chem. 278:18606-18616. [DOI] [PubMed] [Google Scholar]
- 7.Filutowicz, M., and R. Inman. 1991. A compact nucleoprotein structure is produced by binding of Escherichia coli integration host factor (IHF) to the replication origin of plasmid R6K. J. Biol. Chem. 266:24077-24083. [PubMed] [Google Scholar]
- 8.Filutowicz, M., M. J. McEachern, and D. R. Helinski. 1986. Positive and negative roles of an initiator protein at an origin of replication. Proc. Natl. Acad. Sci. USA 83:9645-9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filutowicz, M., E. Uhlenhopp, and D. R. Helinski. 1986. Binding of purified wild-type and mutant π initiation proteins to a replication origin region of plasmid R6K. J. Mol. Biol. 187:225-239. [DOI] [PubMed] [Google Scholar]
- 10.Forest, K. T., and M. S. Filutowicz. 2003. Remodeling of replication initiator proteins. Nat. Struct. Biol. 10:496-498. [DOI] [PubMed] [Google Scholar]
- 11.Germino, J., and D. Bastia. 1983. The replication initiator protein of plasmid R6K tagged with β-galactosidase shows sequence-specific DNA-binding. Cell 32:131-140. [DOI] [PubMed] [Google Scholar]
- 12.Giraldo, R., C. Fernandez-Tornero, P. R. Evans, R. Díaz-Orejas, and A. Romero. 2003. A conformational switch between transcriptional repression and replication initiation in the RepA dimerization domain. Nat. Struct. Biol. 10:565-571. [DOI] [PubMed] [Google Scholar]
- 13.Kim, J., C. Zwieb, C. Wu, and S. Adhya. 1989. Bending of DNA by gene-regulatory proteins: construction and use of a DNA bending vector. Gene 85:15-23. [DOI] [PubMed] [Google Scholar]
- 14.Komori, H., F. Matsunaga, Y. Higuchi, M. Ishiai, C. Wada, and K. Miki. 1999. Crystal structure of a prokaryotic replication initiator protein bound to DNA at 2.6 Å resolution. EMBO J. 18:4597-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krüger, R., and M. Filutowicz. 2003. Characterization of His-tagged, R6K-encoded π protein variants. Plasmid 50:80-85. [DOI] [PubMed] [Google Scholar]
- 16.Krüger, R., I. Konieczny, and M. Filutowicz. 2001. Monomer/dimer ratios of replication protein modulate the DNA strand-opening in a replication origin. J. Mol. Biol. 306:945-955. [DOI] [PubMed] [Google Scholar]
- 17.Krüger, R., S. A. Rakowski, and M. Filutowicz. 2004. Isomerization and apparent DNA bending by π, the replication protein of plasmid R6K. Biochem. Biophys. Res. Commun. 313:834-840. [DOI] [PubMed] [Google Scholar]
- 18.Krüger, R., S. A. Rakowski, and M. Filutowicz. 2004. Participating elements in the replication of iteron-containing plasmids, p. 25-45. In B. E. Funnell and G. J. Phillips (ed.), Plasmid biology. ASM Press, Washington, D.C.
- 19.Kunnimalaiyaan, S., R. Krüger, W. Ross, S. A. Rakowski, and M. Filutowicz. 2004. Binding modes of the initiator and inhibitor forms of the replication protein π to the γ ori iteron of plasmid R6K. J. Biol. Chem. 279:41058-41066. [DOI] [PubMed] [Google Scholar]
- 20.McEachern, M. J., M. A. Bott, P. A. Tooker, and D. R. Helinski. 1989. Negative control of plasmid R6K replication: possible role of intermolecular coupling of replication origins. Proc. Natl. Acad. Sci. USA 86:7942-7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miron, A., S. Mukherjee, and D. Bastia. 1992. Activation of distant replication origins in vivo by DNA looping as revealed by a novel mutant form of an initiator protein defective in cooperativity at a distance EMBO J. 11:1205-1216. (Erratum, 11:2002.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miron, A., I. Patel, and D. Bastia. 1994. Multiple pathways of copy control of γ replicon of R6K: mechanisms both dependent on and independent of cooperativity of interaction of π protein with DNA affect the copy number. Proc. Natl. Acad. Sci. USA 91:6438-6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukherjee, S., H. Erickson, and D. Bastia. 1988. Detection of DNA looping due to simultaneous interaction of a DNA-binding protein with two spatially separated binding sites on DNA. Proc. Natl. Acad. Sci. USA 85:6287-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee, S., H. Erickson, and D. Bastia. 1988. Enhancer-origin interaction in plasmid R6K involves a DNA loop mediated by initiator protein. Cell 52:375-383. [DOI] [PubMed] [Google Scholar]
- 25.Mukherjee, S., I. Patel, and D. Bastia. 1985. Conformational changes in a replication origin induced by an initiator protein. Cell 43:189-197. [DOI] [PubMed] [Google Scholar]
- 26.Mukhopadhyay, G., S. Sozhamannan, and D. K. Chattoraj. 1994. Relaxation of replication control in chaperone-independent initiator mutants of plasmid P1. EMBO J. 13:2089-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pal, S. K., and D. K. Chattoraj. 1988. P1 plasmid replication: initiator sequestration is inadequate to explain control by initiator-binding sites. J. Bacteriol. 170:3554-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma, S., B. K. Sathyanarayana, J. G. Bird, J. R. Hoskins, B. Lee, and S. Wickner. 2004. Plasmid P1 RepA is homologous to the F plasmid RepE class of initiators. J. Biol. Chem. 279:6027-6034. [DOI] [PubMed] [Google Scholar]
- 29.Sitlani, A., and D. M. Crothers. 1996. Fos and Jun do not bend the AP-1 recognition site. Proc. Natl. Acad. Sci. USA 93:3248-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toukdarian, A. E., and D. R. Helinski. 1998. TrfA dimers play a role in copy-number control of RK2 replication. Gene 223:205-211. [DOI] [PubMed] [Google Scholar]
- 31.Uga, H., F. Matsunaga, and C. Wada. 1999. Regulation of DNA replication by iterons: an interaction between the ori2 and incC regions mediated by RepE-bound iterons inhibits DNA replication of mini-F plasmid in Escherichia coli. EMBO J. 18:3856-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urh, M., J. Wu, K. Forest, R. B. Inman, and M. Filutowicz. 1998. Assemblies of replication initiator protein on symmetric and asymmetric DNA sequences depend on multiple protein oligomerization surfaces. J. Mol. Biol. 283:619-631. [DOI] [PubMed] [Google Scholar]
- 33.Wu, J., and M. Filutowicz. 1999. Hexahistidine (His6)-tag dependent protein dimerization: a cautionary tale. Acta Biochim. Polon. 46:591-599. [PubMed] [Google Scholar]
- 34.Wu, J., M. Sektas, D. Chen, and M. Filutowicz. 1997. Two forms of replication initiator protein: positive and negative controls. Proc. Natl. Acad. Sci. USA 94:13967-13972. [DOI] [PMC free article] [PubMed] [Google Scholar]