Abstract
AtzF, allophanate hydrolase, is a recently discovered member of the amidase signature family that catalyzes the terminal reaction during metabolism of s-triazine ring compounds by bacteria. In the present study, the atzF gene from Pseudomonas sp. strain ADP was cloned and expressed as a His-tagged protein, and the protein was purified and characterized. AtzF had a deduced subunit molecular mass of 66,223, based on the gene sequence, and an estimated holoenzyme molecular mass of 260,000. The active protein did not contain detectable metals or organic cofactors. Purified AtzF hydrolyzed allophanate with a kcat/Km of 1.1 × 104 s−1 M−1, and 2 mol of ammonia was released per mol allophanate. The substrate range of AtzF was very narrow. Urea, biuret, hydroxyurea, methylcarbamate, and other structurally analogous compounds were not substrates for AtzF. Only malonamate, which strongly inhibited allophanate hydrolysis, was an alternative substrate, with a greatly reduced kcat/Km of 21 s−1 M−1. Data suggested that the AtzF catalytic cycle proceeds through a covalent substrate-enzyme intermediate. AtzF reacts with malonamate and hydroxylamine to generate malonohydroxamate, potentially derived from hydroxylamine capture of an enzyme-tethered acyl group. Three putative catalytically important residues, one lysine and two serines, were altered by site-directed mutagenesis, each with complete loss of enzyme activity. The identity of a putative serine nucleophile was probed using phenyl phosphorodiamidate that was shown to be a time-dependent inhibitor of AtzF. Inhibition was due to phosphoroamidation of Ser189 as shown by liquid chromatography/matrix-assisted laser desorption ionization mass spectrometry. The modified residue corresponds in sequence alignments to the nucleophilic serine previously identified in other members of the amidase signature family. Thus, AtzF affects the cleavage of three carbon-to-nitrogen bonds via a mechanism similar to that of enzymes catalyzing single-amide-bond cleavage reactions. AtzF orthologs appear to be widespread among bacteria.
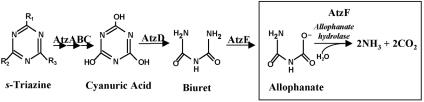
Microorganisms respond to new chemical inputs in the environment by evolving new enzymes and pathways (30). s-Triazine herbicides such as atrazine are anthropogenic chemicals that have recently been observed to be metabolized by soil bacteria (21, 26, 28, 29, 31). Atrazine metabolism has been most well studied using Pseudomonas sp. strain ADP. In that bacterium, all six genes encoding atrazine catabolism to carbon dioxide and ammonia have been localized to a self-transmissible plasmid, pADP-1, which has been completely sequenced (19). AtzA, AtzB, and AtzC metabolize atrazine to cyanuric acid via three hydrolytic reactions (Fig. 1). Cyanuric acid is further hydrolyzed by enzymes encoded by the contiguous, transcriptionally linked genes atzD, atzE, and atzF. AtzF is the terminal enzyme in the atrazine catabolic pathway. Recombinant AtzF activity in crude protein extracts from Escherichia coli transformed allophanate to carbon dioxide and ammonia (19) (Fig. 1). Previous studies on cyanuric acid metabolism had concluded that the terminal pathway intermediate was urea (6, 7). However, this product was identified after acidifying extracts to pH 2.0, which would cause decarboxylation of allophanate to urea. In this context, the metabolic intermediate penultimate to ammonia and carbon dioxide is still open to experimental verification, and the purification of AtzF as described here contributes to that determination.
FIG. 1.
Atrazine catabolic pathway in Pseudomonas sp. strain ADP.
In a different context, allophanate is formed by the activity of urea carboxylase that has been purified from the prokaryote Oleomonas sagaranensis (12). In this organism, urea carboxylase is proposed to be functional in the catabolism of urea via a two-reaction sequence consisting of an ATP-dependent urea carboxylation followed by allophanate hydrolysis, ultimately liberating 2 moles of ammonia from urea. In yeast, this reaction sequence is carried out by a two-domain protein consisting of separate biotin carboxylase and allophanate hydrolase domains (27). However, an allophanate hydrolase enzyme has yet to be purified from prokaryotic sources. This would help reveal the different potential functions that this protein might serve in bacterial systems (10).
Translation of the allophanate hydrolase gene, atzF, from Pseudomonas sp. strain ADP reveals that it is in the amidase signature family, a large class of hydrolytic enzymes found in bacteria, plants, and mammals (20, 24, 25). Members of the amidase signature family are characterized by a serine- and glycine-rich motif that is also found in AtzF (19). X-ray structures are now available for several members of the family: fatty acid amidase from rat (4), the C-terminal peptide amidase from the bacterium Stenotrophomonas maltophila (17), and malonamidase E2 from Bradyrhizobium japonicum (24). Interestingly, malonamidase and AtzF work on structurally analogous compounds, malonamate and allophanate, respectively. Malonamate differs from allophanate only by having a methylene carbon in place of the secondary nitrogen atom. However, AtzF and malonamidase share only modest (30%) amino acid sequence identity.
The determined X-ray structures, along with site-directed mutagenesis experiments (8, 16, 20, 25), have revealed a unique Ser-cisSer-Lys catalytic triad in the amidase signature family. One of the serine residues has been implicated as functioning in nucleophilic attack on the substrate carbonyl center in each respective substrate. It is proposed that there are two distinct classes of enzymes in the amidase signature family. One distinguishing feature is the susceptibility to inhibition by serine-modifying reagents such as phenylmethanesulfonyl fluoride (8, 17).
In this study, we describe the purification and characterization of AtzF. We confirmed that allophanate is transformed to 2 mol of ammonia by a single enzyme. AtzF was shown to be highly specific for allophanate but hydrolyzed malonamate at rates which are likely not to be physiologically relevant. Acyl group capture, site-directed mutagenesis, and time-dependent inhibitor studies were consistent with serine nucleophilic catalysis. AtzF appears to be one of an emerging class of prokaryotic amidase signature proteins with a high degree of substrate selectivity for allophanate.
MATERIALS AND METHODS
Chemicals.
Potassium allophanate was prepared by hydrolyzing diethyl allophanate (Acros Organics, Fisher Scientific, Pittsburgh, PA) as described previously by Whitney and Cooper (34). Malonohydroxamate was synthesized as described previously by Bernheim (3). The product was analyzed by nuclear magnetic resonance, mass spectrometry, and high-performance liquid chromatography using a reverse-phase C18 column. Acetylurea was purchased from Lancaster Synthesis, Inc. (Pelham, NH). All other commercially available compounds were purchased from Aldrich Chemical Co. (Milwaukee, WI).
Enzymatic assays.
Ammonia released from enzymatic assays was determined using the Berthelot reaction as described previously by Weatherburn (33). One unit of enzyme activity was defined as the amount of enzyme converting 1 μmol of allophanate to 2 μmol of ammonia in 1 min.
Construction of recombinant plasmid for the expression of AtzF with a His tag.
The atzF gene from Pseudomonas sp. strain ADP was amplified, without its native promoter, by PCR using primers atzFf-His (5′-GCCATATGAATGACCGCGCG-3′) and atzFr-His (5′-TAGAATTCCGGAGCGGCTTG-3′). The primers contained NdeI and EcoRI restriction enzyme cloning sites in their 5′ and 3′ regions, respectively. The gene was cloned downstream of the lac promoter and His tag coding sequence in vector pET28b+ (Novagen, Madison, WI) and transformed into E. coli BLR(DE3) (Novagen, Madison, WI), and its sequence was verified.
Site-directed mutagenesis.
Suitable primer pairs (Table 1) were designed to generate mutated AtzF by PCR using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). Plasmid pET28b+::atzF was used as a template for mutagenic reactions. Following PCR, the parental plasmid was digested with DpnI, and the nicked mutated plasmid was transformed into XL10-Gold competent cells (Stratagene, La Jolla, CA). The plasmid was extracted and transformed into the expression His-tagged vector host E. coli BLR(DE3) (Novagen, Madison, WI). Mutations were confirmed by complete sequencing of the resultant plasmids.
TABLE 1.
Oligonucleotides used for site-directed mutagenesis
| AtzF gene allele | Mutagenic primer setsa |
|---|---|
| K91A | CTTCGGCGTCGCGGACAACATCG |
| CGATGTTGTCCGCGACGCCGAAG | |
| S165A | CGTATCCGGCGGCGCAAGCAGTGGCTCC |
| GGAGCCACTGCTTGCGCCGCCGGATACG | |
| S189A | CGGACACTGCCGGTGCTGGCCGCATTCCTGCTG |
| CAGCAGGAATGCGGCCAGCACCGGCAGTGTCCG |
Nucleotide codons in boldface type encode the mutated amino acids.
Expression and purification of AtzF wild-type and mutagenized enzymes.
E. coli BLR(DE3) (pET28b+::atzF) was grown in LB medium (22) containing 50 μg of kanamycin per ml at 30°C, with shaking at 150 rpm. When the culture reached an optical density at 600 nm of 0.5, 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added and the induced cells were grown overnight under the same conditions. Cultures were centrifuged at 10,000 × g for 10 min at 4°C and washed three times in 0.85% NaCl, and cell pellets (2.5 g [wet weight]) were resuspended in 20 ml of 25 mM 3-(N-morpholino)propanesulfonate (MOPS) buffer, pH 6.9. Cells were broken by four sonication cycles (2 min at 70% intensity and a 50% duty cycle), with intermittent cooling on ice for 5 min, using a Heat Systems-Ultrasonics (Farmingdale, NY) model W-225R sonicator and a microtip. Cell extracts were obtained by centrifugation at 18,000 × g for 90 min at 4°C, and lysates were applied to a 5-ml HiTrap chelating column (Amersham Pharmacia Biotech, Piscataway, NJ) complexed with Ni2+ according to the procedure recommended by the manufacturer. The column was washed with 15 ml 25 mM MOPS buffer, pH 6.9, followed by a second wash with the same buffer supplemented with 0.05 M imidazole. The enzyme was eluted from the column with 15 ml of 0.25 M imidazole in MOPS buffer, pH 6.9. All buffers contained 10% glycerol. The pure enzyme was concentrated using a Centricon-10 filtration unit (Amicon, Beverly, MA), and enzyme purity and subunit molecular mass were estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Gel filtration chromatography.
The holoenzyme molecular mass was estimated by gel filtration chromatography on a Superose 12 HR column (Pharmacia, Uppsala, Sweden), using an FPLC system (Pharmacia). The column was equilibrated with 25 mM MOPS buffer, pH 6.9, containing 0.15 M KCl, at a flow rate of 0.1 ml min−1. The molecular mass of AtzF was estimated by comparing its elution to that of standards with well-defined molecular masses: thyroglobulin (670,000), gamma globulin (158,000), chicken ovalbumin (44,000), horse myoglobin (17,000), and vitamin B12 (1,350).
Testing for transition metal cofactors.
Purified AtzF (2.0 mg) was dissolved in 4 ml 10% HCl in 25 mM MOPS buffer, pH 6.9. The sample was incubated overnight at 90°C, and the metal content was determined by inductively coupled plasma emission spectroscopy at the University of Minnesota Soils Analytical Laboratory (St. Paul, MN). To test for metal activation, purified AtzF was incubated for 30 min with 100 μM of the divalent metal Ni2+, Co2+, Fe2+, or Mn2+, and the activity of the enzyme was measured as described above. To test for the effect of metal chelators, purified AtzF was incubated for 30 min with 5 mM 1,10-phenanthroline, 8-hydroxyquinoline-5-sulfonic acid, or EDTA and assayed for activity as described above.
Substrate specificity and kinetic studies.
Enzyme activity was measured in two types of basic buffers, 0.1 M potassium phosphate and 0.1 M sodium bicarbonate. The pH range tested was from 8 to 11.3. Neutral and acidic pH buffers were not tested since the enzyme substrate, allophanate, spontaneously decarboxylates at an appreciable rate at those pH values. All substrate assays and kinetic studies were conducted at the pH optimum determined for allophanate. Solutions containing various allophanate analogs, cyclic amides, and thioamide compounds were prepared in 0.1 M sodium bicarbonate buffer at pH 9.0. The allophanate analogs were incubated with purified AtzF for 30 min, and ammonia release was measured as described above. The remainder of the tested compounds that had defined UV spectra were incubated with enzyme, and potential changes in UV absorbance were followed over time. In cases where enzyme activity with a compound was detected, steady-state kinetic parameters were determined.
Kinetic parameters were calculated from the initial hydrolysis rates at different concentrations using the Hanes-Woolf equation: [S]/V0 = [S]/Vmax + Km/Vmax (23). Linear regression of the plot [S]/V0 versus [S] was used to determine the Vmax and Km parameters. The kcat was calculated by dividing Vmax by the moles of AtzF based on the subunit molecular mass. Substrate concentrations used ranged from 200 μM to 10 mM. Control analyses without enzyme were conducted in parallel. Compounds that were not hydrolyzed were examined as inhibitors of AtzF. The influence of the serine protease inhibitor phenylmethylsulfonyl fluoride (PMSF) on enzyme activity was tested by incubating 2 mM PMSF with 17 μM AtzF, and allophanate hydrolase activity was measured at 25°C as described above.
Acyl group trapping with hydroxylamine to make malonohydroxamate.
AtzF, at a final concentration of 12 μM, was incubated at room temperature with 9.3 mM malonamate and 93 mM hydroxylamine in 0.1 M sodium bicarbonate buffer at pH 9.0. Control samples without each one of the components or the S189A AtzF mutant enzyme was also analyzed. At selected time points, 100-μl aliquots were taken for detection of malonohydroxamate using the method described previously by Kim and Kang (15), with several modifications. Reaction mixtures were diluted with 800 μl of water, followed by the addition of 100 μl 15% FeCl3 in 0.66 N HCl solution. The protein precipitate was removed by centrifugation, and the color intensity was monitored spectrophotometrically at 540 nm. The monitored red-brown color developed from the formation of Fe(III) complexed with malonohydroxamate. The concentration was calculated according to a standard curve constructed using synthetic malonohydroxamate as the standard.
A ThermoFinnigan LCQ Classic electrospray-ion trap mass spectrometer (MS) (Thermo Electron, San Jose, CA), operated in a negative-ion mode, was used to verify the formation of malonohydroxamate. All enzymatic reactions monitored by mass spectrometry were done in 20 mM ammonium acetate buffer, pH 8.0, and were filtered before use with Nanosep 3K (Pall Filtron Corporation, Northborough, MA). Subsamples were directly injected into a 5-μl loop with ammonium acetate as makeup solvent at a flow rate of 50 μl per min. The instrument source voltage was 5 kV, the sheath gas flow rate was 70, the capillary was heated to 200°C, and its voltage was −9.52 V. Full scans from 100 to 300 m/z were taken, targeting the ions with [M − 1H]−1 of 118 m/z (malonohydroxamate) and 102 m/z (malonamate). Standards of the two compounds were also analyzed.
Inhibition of AtzF by phenyl phosphorodiamidate.
AtzF (50 μM subunit concentration) was incubated at room temperature with 0.25, 1.25, 2.5, or 5 mM phenyl phosphorodiamidate in 0.1 M sodium bicarbonate buffer, pH 9.0. At selected time points, subsamples were withdrawn and allophanate degradation activity was measured. To test for reversibility of inhibition, AtzF was incubated with 5 mM phenyl phosphorodiamidate, and the reaction was allowed to proceed until there was greater than 95% inhibition. The mixture was dialyzed twice against buffer containing no phenyl phosphorodiamidate, and the activity was measured.
Mass spectrometric analysis of irreversibly inhibited AtzF was used to identify modified residue(s). Twenty micrograms of AtzF was incubated with 2.5 mM phenyl phosphorodiamidate for 40 min at 25°C, and inactive enzyme was digested for 22 h with 0.2 μg chymotrypsin (Roche Applied Science, Penzberg, Germany). A control sample of unmodified digested AtzF was also prepared under the same conditions. Prior to matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis, a portion of the peptide mixture was desalted using Millipore C18 ZipTips (Billerica, MA) according to the protocol described by the manufacturer. Full scans of the peptide mixture, from 500 to 3,500 m/z, and tandem mass spectral data of select ions were collected with a QSTAR XL (Applied Biosystems Inc., Foster City, California) quadrupole time of flight mass spectrometer with an orthogonal MALDI source using α-cyano-4-hydroxycinnamic acid as the matrix (Sigma-Aldrich, St. Louis, MO). The instrument was operated under the same conditions described previously by Kapphahn et al. (13). Mass spectra were the averages of approximately 50 laser shots collected in a positive mode. External calibration was performed using human angiotensin II (monoisotopic [MH+] m/z 1,046.5417) and adrenocorticotropin hormone fragment 18-39 (monoisotopic [MH+] m/z 2,465.1989).
For liquid chromatography-electrospray ionization (LC-ESI)-tandem mass spectrometry (MS/MS), samples were prepared as described below. AtzF was digested and desalted as described above and diluted 1:20 using a 98:2 ratio of water to acetonitrile and 0.1% trifluoroacetic acid in a total volume of 30 μl. An LC Packings (Sunnyvale, CA) Famos autosampler aspirated 27.5 μl of the peptide dilution mixture into a 100-μl sample loop using the Famos μl-pick-up injection mode and a 98:2 ratio of water to acetonitrile and 0.1% formic acid as the transfer reagent. An LCP Switchos pump was used to concentrate and desalt the sample on an LCP C18 nano-precolumn (0.3-mm internal diameter by 5-mm length) with load buffer. The precolumn was switched in-line with a capillary column, and peptides were eluted at 350 nl per min using an LCP Ultimate LC system. The capillary column (100-μm internal diameter) was packed in-house to 12 cm in length with 200-Å, 5-μm-pore-size C18 particles (Michrom BioResources, Auburn, CA) as described previously by Hernandez et al. (11). Peptides were eluted from the column and introduced into a QSTAR Pulsar quadrupole-TOF MS which was equipped with Protana's nano-electrospray source under the same conditions described previously by Kapphahn et al. (13). The [M + 3H]3+ monoisotopic peak at 586.9830 m/z and [M + 2H]2+ monoisotopic peak at 879.9705 m/z from human renin substrate tetradecapeptide (Sigma-Aldrich, St. Louis, MO) were used for external calibration. For protein identification by MS/MS, the eluted peptides from the column were focused into the mass spectrometer, where product ion spectra were collected in an information-dependent acquisition (IDA) mode. IDA mode settings included continuous cycles of one full-scan TOF MS from 400 to 1,100 m/z (1.5 seconds) plus three product ion scans from m/z 50 to 2,500 (3 seconds each). Precursor m/z values were selected from a peak list automatically generated by Analyst QS software (ABI) from the TOF MS scan during acquisition, starting with the most intense ion. Product ion mass spectra were searched using ProID (ABI) against the National Center for Biotechnological Information (NCBI) database.
For peptide sequence analysis, samples were prepared as described above and analyzed by MS/MS. Selected ion monitoring scan mode was performed for fragmentation of active-site-containing peptides (181SLGTDTAGSGRIPAAF). Settings included full-scan TOF MS from 400 to 1,100 m/z (1.5 seconds) plus 10-second averaging for ions in selected ion monitoring list. Product ion mass spectra were inspected manually using the ABI (Foster City, CA) BioAnalyst software package to view the theoretical fragment ion table and compare ions to experimental data.
RESULTS
AtzF purification and stability.
His-tagged AtzF protein was eluted from a nickel column to yield a homogeneous protein with a 3.1-fold purification (Table 2). Approximately one-half of the activity found in the crude extract was retained in the final purified material. AtzF was found to rapidly lose activity when stored in 25 mM MOPS buffer at 4°C or −80°C. However, the addition of 10% glycerol prior to freezing maintained AtzF activity for up to 1 month with only marginal losses.
TABLE 2.
Purification of AtzF
| Fraction | Total protein (mg) | Total act (U) | Sp act (U mg−1) | Yield (%) | Purifi- cation (fold) |
|---|---|---|---|---|---|
| Crude extract | 183 | 606a | 3.32 | 100 | |
| Eluate from nickel column | 26.9 | 277 | 10.3 | 45.8 | 3.10 |
One unit of enzyme activity is defined as the conversion of 1 μmol of allophanate to 2 μmol of ammonia in 1 min.
AtzF subunit structure and analysis for a possible metal cofactor.
The AtzF protein had a subunit molecular mass of 66,223 as determined from translation of the gene sequence. Gel filtration gave a holoenzyme molecular mass of 260,000, consistent with an α4 subunit structure. UV/visible spectroscopy showed no discernible absorbance above 290 nm. Metal addition or incubation with metal chelators did not give any appreciable stimulation or diminution of activity. Moreover, only trace levels of metals, likely to be contaminants, were detected by inductively coupled plasma emission spectroscopy; Co, Cu, Ni, Fe, and Zn were present at less then 0.1 mol per mol subunit.
Product formation and kinetics with allophanate.
AtzF was incubated with potassium allophanate at pH values ranging from 8.0 to 11.3. Activity was maximal at pH 9.5, where the specific activity was 10.3 μmol per min per mg protein. The kcat was 16.4 s−1, the Km was 1.5 mM, and the kcat/Km was 1.1 × 104 s−1 M−1. With limiting concentrations of allophanate, and allowing the reaction to proceed to completion, 2.0 mol of ammonia was detected per mol of substrate consumed.
AtzF unreactive with compounds structurally analogous to allophanate.
Biuret, a metabolite in the atrazine degradation pathway, and urea were not substrates for AtzF. The following allophanate analogs were not detectably reactive with AtzF: methyl allophanate, hydantoic acid, oxamic acid, hydroxyurea, methyl carbamate, N-methylurea, acetylurea, 1-acetyl-2-thiourea, and semicarbazide. No activity was observed with the following cyclic amide or thioamide compounds: rhodanine, rhodanine-3-acetic acid, 3-aminorhodanine, (4R)-(−)-2-thioxo-4-thiazolidinecarboxylic acid, (−)-2-oxo-4-thiazolinecarboxylic acid, and 2-amino-5-bromothiazole.
Malonamate is a poor substrate for AtzF.
Malonamate was hydrolyzed by AtzF to yield 1 mol of ammonia per mol of substrate consumed, but it was hydrolyzed very slowly. The specific activity was 53 nmol per min per mg protein. The kcat was 0.1 s−1, which was 0.7% of the rate seen with allophanate. The Km was 5.3 mM. The kcat/Km was 21 s−1 M−1, almost 3 orders of magnitude lower than when allophanate was the substrate.
Despite the much lower kcat/Km for malonamate than for allophanate, malonamate strongly inhibited the release of ammonia from allophanate by AtzF. When both malonamate and allophanate were added to reaction mixtures at an equimolar concentration (8.6 mM), the activity, measured as ammonia release, was 14% of that found with the uninhibited enzyme. The relative activity was reduced to 1% when the malonamate concentration was increased fourfold. This was consistent with the hypothesis that a putative malonyl-enzyme intermediate might be generated and is subsequently slowly hydrolytically released from the enzyme.
Hydroxylamine trapping experiments.
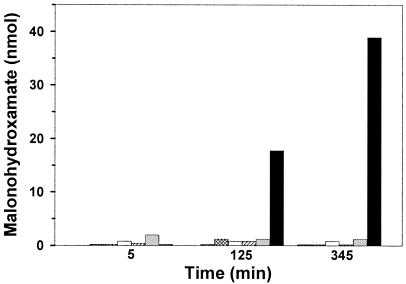
Malonohydroxamate coordinates Fe(III) to yield a hydroxamate-metal chelate that absorbs in the visible range, and its formation was used to obtain further evidence for the intermediacy of an acyl-enzyme species. Malonamate (9.3 mM) was incubated with 12 μM AtzF and 93 mM hydroxylamine. In 125 min, 17 nmol of malonohydroxamate was detected; and at 345 min, 39 nmol was detected (Fig. 2). Mass spectrometry of the reaction mixtures yielded a compound with a [M − 1H]−1 parent ion of 118 m/z, consistent with the formation of malonohydroxamate. This assignment was confirmed by comparison with a synthetic standard. Controls with malonamate and hydroxylamine, but without enzyme, or with a Ser189Ala mutant (described below), as well as enzyme with either compound alone yielded negligible product as determined using the colorimetric assay. Moreover, mass spectrometric analysis of control samples showed only malonamate.
FIG. 2.
Malonohydroxamate formation by wild-type AtzF and mutant AtzF; vertical striped bars, malonamate plus hydroxylamine; hatched bars, enzyme buffer plus malonamate plus hydroxylamine; open bars, AtzF plus hydroxylamine; diagonal striped bars, AtzF plus malonamate; dotted bars, S189A AtzF mutant plus malonamate plus hydroxylamine; solid bars, AtzF plus malonamate plus hydroxylamine.
Site-directed mutagenesis.
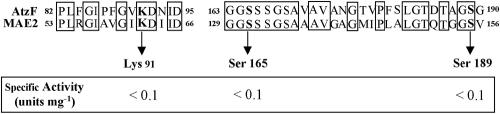
The malonamate inhibition and hydroxylamine trapping experiments suggested the intermediacy of an active-site nucleophile. Based on sequence alignments with other amidase signature family members (16, 24), a potential serine nucloeophile and other putative mechanistically relevant residues were subjected to site-directed mutagenesis (Fig. 3). There was no detectable AtzF activity following mutagenesis of Lys91, Ser165, and Ser189 to alanine in AtzF, separately. A total of 0.5% of the wild-type activity could have been readily detected via the assay methodology that was used.
FIG. 3.
Sequence alignment of AtzF and malonamidase at regions known to be most highly conserved in the amidase signature proteins. Arrows indicates AtzF amino acids that had been mutated to alanine. The specific activity of each mutated enzyme is listed below.
Phenyl phosphorodiamidate is a time-dependent inhibitor of AtzF.
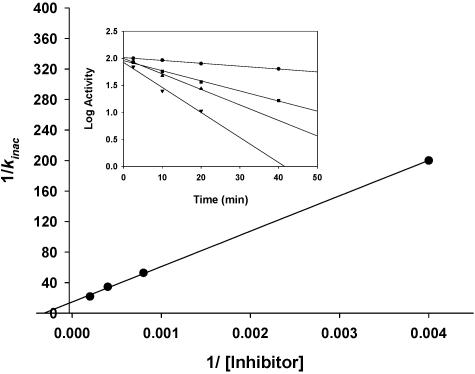
Known inhibitors for amidase signature family members were tested for their influence on allophanate turnover. PMSF was not observed to be an inhibitor of AtzF. However, phenyl phosphorodiamidate was found to inhibit AtzF activity in a time-dependent manner (Fig. 4). Activity loss followed first-order kinetics, with a Ki of 3.2 mM. Activity was not observed after treatment with excess inhibitor. However, enzyme activity could be recovered over time; 50% of the initial activity was recovered after completely inhibited enzyme was dialyzed twice against 0.1 M sodium bicarbonate buffer. These data are consistent with the covalent modification of a single amino acid residue followed by slow hydrolysis of an acyl-enzyme intermediate that regenerates active AtzF.
FIG. 4.
Effect of phenyl phosphorodiamidate concentration on the rate of inactivation of AtzF, yielding Ki (1/Kinac). Insert, time-dependent inhibitory effect of different concentrations of phenyl phosphorodiamidate on AtzF activity. Legend: 0.25 mM (•), 1.25 mM (▪), 2.5 mM (▴), and 5 mM (▾) phenyl phosphorodiamidate.
Identification of the modified amino acid.
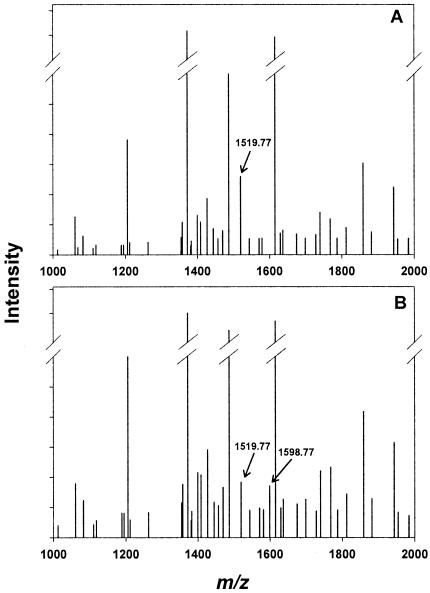
MALDI-XL spectrometry (Applied Biosystems, Foster City, CA) was initially used to identify the modified peptide(s) formed after incubation of AtzF with phenyl phosphorodiamidate and digestion with chymotrypsin. Overall, 18 peptides were identified within a 60-ppm mass tolerance, giving a coverage of 35% of the protein. Two ions differed when peptide fragments were analyzed in comparison with digested uninhibited enzyme (Fig. 5). In the inhibited AtzF sample, the peptide profile showed the appearance of a new peak with a mass of 1,598.77 Da and the concomitant diminution of an ion with a mass of 1,519.77 Da. MS/MS on the 1,519.77-Da fragment confirmed that the sequence of this peptide was 181SLGTDTAGSGRIPAAF, which contains the putative nucleophilic serine (S189). The increase of 79 mass units is consistent with the addition of a phosphonamide group (PO2NH3), which could arise by nucleophilic capture by serine and hydrolytic release of an amino group while tethered to the enzyme. No significant mass changes were detected in any other peptides.
FIG. 5.
Mass spectrometric analysis of chymotrypsin-digested AtzF peptides. Spectra were plotted as peak intensity versus m/z (centroid mass) and represented as a zoom scan of the region that corresponds to the peptide which was modified by the inhibitor phenyl phosphorodiamidate. (A) Spectrum of digested unmodified enzyme; (B) spectrum of digested modified enzyme. Arrows indicate the peptides that include the catalytic serine (Ser189).
To more definitively determine which amino acid(s) was modified, the digested AtzF peptides were analyzed by LC-ESI-MS/MS. When the sample was run under IDA mode and product ion mass spectra were searched using ProID (ABI) against the NCBI database, 42% of the protein sequence was covered. To target specific peptides, the sample was rerun under a different scheme in which the mass spectrometer switched automatically to MS/MS mode when the ion masses were of the peptide of interest. The selected peptide, 181SLGTDTAGSGRIPAAF, which contained the putative catalytic serine (Ser189), was analyzed by the masses 760.88 m/z or 800.38 m/z ([M + 2H]2+, unmodified or modified peptide, respectively) and 507.59 m/z or 533.92 m/z ([M + 3H]3+, unmodified or modified peptide, respectively). The collision-induced dissociation spectrum of the doubly charged ion of the inhibited peptide corresponded well with 181SLGTDTAGSGRIPAAF, in which Ser189 was modified. The y11 ion at m/z 1,126.5 supports this, since it contains the correct mass change on Ser189. This conclusion was supported by the identification of other y- and b-type ions. Moreover, a few y and b ions and other internal fragments, which were consistent with the loss of PO2NH3 from Ser189, were found. It is well known that a bond between the serine oxygen and phosphorus is not stable under MS/MS analysis (1, 2, 35).
DISCUSSION
The present study was conducted to (i) confirm that AtzF catalyzes the terminal step in the s-triazine catabolic pathway using allophanate as its substrate, (ii) determine the substrate range of AtzF and whether it is involved in multiple physiological processes, and (iii) investigate the mechanism of AtzF as a putative member of the amidase signature family. First, purified AtzF was shown to catalyze the hydrolysis of allophanate to 2 mol of ammonia, and it was not active with biuret or urea. This is consistent with the observation that allophanate hydrolase in Pseudomonas sp. strain ADP catalyzes the last metabolic step of bacterial cyanuric acid metabolism, thus releasing nitrogen from s-triazine ring herbicides. Previously, bacteria were shown to metabolize s-triazine herbicides as their sole source of nitrogen to support growth (7, 21, 28, 29). Moreover, the data are consistent with the idea that Pseudomonas sp. strain ADP metabolizes allophanate, and not urea, as an intermediate during the catabolism of cyanuric acid. We have shown that other cyanuric acid-metabolizing bacteria contain genes with high sequence identity to AtzF (18). These data suggest that a number of bacteria metabolize cyanuric acid via allophanate.
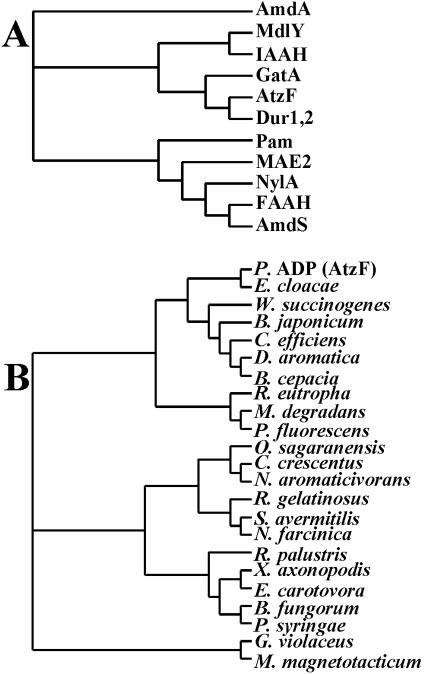
Allophanate hydrolase activity is also suggested to function in some bacteria, in concert with urea carboxylase, for the purpose of transforming urea to 2 mol of ammonia as an alternative to direct hydrolysis by urease (10, 12). That metabolic role is based on the stability of urea and the relative instability of allophanate; urea carboxylation renders the amide bonds more susceptible to hydrolysis. This alternative pathway for urea hydrolysis may be important because nickel or bicarbonate might be limiting in some environments, and thus, urease activity may not be expressed. Moreover, the urease enzyme system consists of multiple components and requires ancillary proteins for proper folding and elaboration of the binuclear nickel center that functions in urea hydrolysis (5, 10). In this context, the urea carboxylase/allophanate hydrolase couple, which requires an ATP to drive carboxylation at every cycle, might be a viable alternative for biological harvest of two ammonia molecules from one urea. Allophanate hydrolase had previously been identified in yeast, algae, and protozoa, but it now appears to be fairly widespread in prokaryotes also (Fig. 6). This recent discovery highlighted the importance of purifying and characterizing allophanate hydrolase here, which had not previously been obtained from a prokaryotic source.
FIG. 6.
Dendrograms showing amino acid sequence relatedness between AtzF and (A) members of amidase signature family proteins from diverse organisms that have been structurally or mechanistically characterized and (B) putative allophanate hydrolases found in different bacteria that contain a urea carboxylase or s-triazine biodegradation genes. Part A gives the protein designations, and part B shows the organism name with the GenBank accession numbers for the relevant sequence given in parentheses. (A) The amidase signature family members are as follows: AmdA, amidase from Rhodococcus erythropolis, P22984; MdlY, mandelamide hydrolase from Pseudomonas putida, AAO23019; IAAH, indoleacetamide hydrolase from Pseudomonas syringae, P52831; GatA, glutamyl-tRNA amidotransferase subunit A from Streptococcus pyogenes, Q99YC0; AtzF, allophanate hydrolase from Pseudomonas sp. strain ADP (P. ADP), NP_862539; Dur1,2, urea amidolyase from Saccharomyces cerevisiae, AAC41643; Pam, peptide amidase from Stenotrophomonas maltophilia, CAC93616; MAE2, malonamidase E2 from Bradyrhizobium japonicum, AF012735; NylA, 6-aminohexanoate cyclic dimer hydrolase from Flavobacterium sp. strain K172, P13397; FAAH, fatty-acid amide hydrolase from Rattus norvegicus, P97612; and AmdS, acetamidase from Emericella nidulans, P08158. (B) Putative allophanate hydrolases from genome projects which share significant sequence identity (>39%) with AtzF: Pseudomonas sp. strain ADP AtzF (NP_862539), Enterobacter cloacae (AAK11683), Wolinella succinogenes (NP_907309), Bradyrhizobium japonicum (NP_772446), Corynebacterium efficiens (NP_737955.1), Dechloromonas aromatica (ZP_00152874), Burkholderia cepacia (ZP_00211646), Ralstonia eutropha (ZP_00202612), Microbulbifer degradans (ZP_00314767), Pseudomonas fluorescens (ZP_00266691), Oleomonas sagaranensis (BAD16655), Caulobacter crescentus (NP_420637), Novosphingobium aromaticivorans (ZP_00304984), Rubrivivax gelatinosus (ZP_00241447), Streptomyces avermitilis (NP_827873), Nocardia farcinica (YP_118429), Rhodopseudomonas palustris (CAE26847), Xanthomonas axonopodis (NP_644621), Erwinia carotovora (yp_050236), Burkholderia fungorum (ZP_00282169), Pseudomonas syringae (NP_791188), Gloeobacter violaceus (NP_923907), and Magnetospirillum magnetotacticum (ZP_00208237).
There is no evidence for allophanate hydrolase in Pseudomonas sp. strain ADP acting in urea metabolism. Pseudomonas sp. strain ADP shows ATP-independent urease activity which does not function in cyanuric acid metabolism (G. Cheng, unpublished data). In addition, the atzF gene is clustered with atzD and atzE, and it is cotranscribed with those genes. The atzDEF genes are found on a catabolic plasmid, and no urea carboxylase gene has been identified on that genetic element. It has been suggested that the allophanate hydrolase that is active in urea metabolism might act on a wider range of amides (10), but this was shown not to be the case for AtzF. In fact, the substrate range of allophanate hydrolase from Pseudomonas sp. strain ADP is very narrow. Of 20 compounds tested, only malonamate was an alternative substrate for AtzF. Moreover, the kcat/Km for malonamate was almost 3 orders of magnitude lower than that for allophanate, suggesting that allophanate is the physiological substrate. Allophanate was previously shown to be the product of the AtzE-catalyzed reaction in crude protein extracts prepared using recombinant E. coli cells containing the atzE gene (19). This observation also supports the idea that allophanate is the physiologically relevant substrate for AtzF.
AtzF is a member of the amidase signature family, with significant sequence identity to structurally and mechanistically well-defined members of that family (Fig. 6A). The AtzF sequence clustered most closely with the sequence of the allophanate hydrolase domain of the urea carboxylase/allophanate hydrolase multifunctional protein from Saccharomyces cerevisiae (Dur1,2) (34). Other well-characterized amidase signature family members with the highest sequence identity to AtzF (>40%) include glutamyl-tRNA amidotransferases (GatA) (9) and mandelamide hydrolase (MdlY) (8). More distantly related proteins for which X-ray structures are available include malonamidase (MAE2) (24), peptide hydrolase (Pam) (17), and fatty acid amidase (FAAH) (4). Malonamidase is of particular interest because malonamate is structurally analogous to allophanate and AtzF showed some, albeit low, activity with malonamate in the present study.
Interestingly, AtzF clusters with what we consider here to be putative allophanate hydrolases identified in bacterial gene cluster and genome sequencing projects (Fig. 6B). For example, the most closely related protein (67% sequence identity) to AtzF from Pseudomonas sp. strain ADP is a protein with undefined function from Enterobacter cloacae. The latter was identified as an open reading frame on a plasmid DNA region that was sequenced because of its perceived importance in metabolizing s-triazine ring compounds (GenBank accession no. AF342826). We predict here that this protein will have allophanate hydrolase activity, and experiments are in progress to test this hypothesis. The other genes encoding the proteins shown in Fig. 6B were found in the same genome as a putative or demonstrated urea carboxylase gene, further suggesting that these are yet-to-be-characterized allophanate hydrolases. In Oleomonas sagaranensis, the urea carboxylase and allophanate hydrolase genes were contiguous, and urea carboxylase activity was demonstrated with a purified protein (12). In Caulobacter crescentus CB15, a gene encoding an amidase signature protein with 53% amino acid identity to AtzF was adjacent to a gene encoding a putative urea carboxylase protein with 58% identity to the urea carboxylase from Oleomonas. Overall, 21 putative allophanate hydrolase proteins were identified based on (i) significant (>39%) sequence identity to AtzF and (ii) linkage in the same genome to a demonstrated or putative urea carboxylase. There are additional proteins not shown that have been annotated as allophanate hydrolases, for example, in Bacillus subtilis strain 168 (14) and Fusobacterium nucleatum strain ATCC 25586 (GenBank accession no. NP_603337) genomes. These were not included here because the sequence identity with AtzF was only about 30%, and so their potential function is more tenuous. Yet those genes are also linked to urea carboxylases and thus may turn out to be true allophanate hydrolases. The bacteria included in the analysis of Fig. 6B are found in the alpha, beta, gamma, and epsilon Proteobacteria, the Actinobacteria, and the Cyanobacteria. Thus, members of the amidase signature family that putatively display allophanate hydrolase activity are widely distributed throughout the bacterial kingdom.
Sequence alignment with proteins of well-defined structure and mechanism (like malonamidase) (Fig. 3) suggests that the unusual cleavage of three carbon-to-nitrogen bonds carried out by AtzF could be explained by a simple amidase mechanism. The activity of AtzF with malonamate in which hydroxylamine is exchanged for the terminal amino group argues for a cleavage of the terminal C-N bond of amide substrates by AtzF. Other data reported here support the role of Ser189 as the active-site nucleophile that would capture the imidodicarbonate fragment after displacing ammonia from allophanate. Several efforts to demonstrate the appearance of free imidodicarbonate in solution have been unsuccessful. However, we have chemically hydrolyzed synthetic diethylimidodicarbonate in buffer and shown that this is extremely unstable and difficult to trap, even with large starting concentrations (data not shown). In this context, we cannot presently discern whether a likely serine-imidodicarbonate species decomposes while tethered to the enzyme or whether it is released off of the enzyme surface and then decomposes in solution.
Serine hydrolases, including members of the amidase signature family, have often been probed with reactive reagents that form a stable, covalent bond with the nucleophilic serine residue (32). Interestingly, members of the amidase signature family have shown differential sensitivity to these agents. Malonamidase, peptide amidase, and mandelamide hydrolase were unaffected by PMSF, whereas fatty acid amidase was sensitive (8). AtzF was not significantly inhibited by PMSF at the concentrations tested. However, AtzF was inhibited, in a first-order, time-dependent manner, by phenyl phosphorodiamidate. The inhibitor was shown to form a covalently modified Ser189. The modification was shown to consist of a mass consistent with that of the serine containing a phosphoromonoamidate group. Shin et al. (24) showed that phenyl phosporodiamidate reacted with crystals of malonamidase to yield a covalently bonded adduct. By interpretation of the electron density maps, they concluded that a pyrophosphate group was appended to the nucleophilic serine. The Ser155 residue in malonamidase corresponds to Ser189 in AtzF. It was not explained in their study how a pyrophosphate molecule was formed. In the present study with AtzF, the phosphoromonoamidate modification may have arisen from serine displacement of phenol to yield serine-O-phosphorodiamidate which then underwent hydrolytic displacement of ammonia to yield the more stable serine-O-phosphoromonoamidate. Shin et al. (24) proposed that Ser131, corresponding to Ser165 in AtzF, acts as a general base in malonamidase to activate water for attack on the serine ester intermediate. The activated water may also catalyze the conversion of AtzF-serine-O-phosphorodiamidate to serine-O-phosphoromonoamidate. Since Ser165 was shown in this study to be required for AtzF activity, it may be playing a similar role in AtzF as in malonamidase.
Acknowledgments
This work was supported, in part, by grant USDA/CSREES/NRI 2202-35107-12508 and by the Department of Energy Office of Biological and Environmental Research grant no. DE-FG02-01ER63268.
We also thank Jack Richman for the help with malonohydroxamate synthesis and analysis, Gilbert Johnson for synthesis of allophanate, and LeeAnn Higgins and Sudha Marimanikkuppam for their help with mass spectrometric analyses and helpful discussions.
REFERENCES
- 1.Affolter, M., J. D. Watts, D. L. Krebs, and R. Aebersold. 1994. Evaluation of two-dimensional phosphopeptide maps by electrospray ionization mass spectrometry of recovered peptides. Anal. Biochem. 223:74-81. [DOI] [PubMed] [Google Scholar]
- 2.Annan, D. S., and S. A. Carr. 1996. Phosphopeptide analysis by matrix-assisted laser desorption time-of-flight mass spectrometry. Anal. Chem. 68:3413-3421. [DOI] [PubMed] [Google Scholar]
- 3.Bernheim, M. L. C. 1964. The enzymic hydrolysis of certain hydroxamic acids. Arch. Biochem. Biophys. 107:313-318. [DOI] [PubMed] [Google Scholar]
- 4.Bracey, M. H., M. A. Hanson, K. R. Masuda, R. C. Stevens, and B. F. Cravatt. 2002. Structural adaptations in a membrane enzyme that terminates endocannabinoid signaling. Science 298:1793-1796. [DOI] [PubMed] [Google Scholar]
- 5.Chang, Z., J. Kuchar, and R. P. Hausinger. 2004. Chemical cross-linking and mass spectrometric identification of sites of interaction for UreD, UreF, and urease. J. Biol. Chem. 279:15305-15313. [DOI] [PubMed] [Google Scholar]
- 6.Cook, A. M., P. Beilstein, H. Grossenbacher, and R. Hütter. 1985. Ring cleavage and degradative pathway of cyanuric acid in bacteria. Biochem. J. 231:25-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook, A. M. 1987. Biodegradation of s-triazine xenobiotics. FEMS Microbiol. Rev. 46:93-116. [Google Scholar]
- 8.Gopalakrishna, K. N., B. H. Stewart, M. M. Kneen, A. D. Andricopulo, G. L. Kenyon, and M. J. McLeish. 2004. Mandelamide hydrolase from Pseudomonas putida: characterization of a new member of the amidase signature family. Biochemistry 43:7725-7735. [DOI] [PubMed] [Google Scholar]
- 9.Harpel, M. R., K. Y. Horiuchi, Y. Luo, L. Shen, W. Jiang, D. J. Nelson, K. C. Rogers, C. P. Decicco, and R. A. Copeland. 2002. Mutagenesis and mechanism-based inhibition of Streptococcus pyogenes Glu-tRNAGln amidotransferase implicate a serine-based glutaminase site. Biochemistry 41:6398-6407. [DOI] [PubMed] [Google Scholar]
- 10.Hausinger, R. P. 2004. Metabolic versatility of prokaryotes for urea decomposition. J. Bacteriol. 186:2520-2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernandez, V. P., L. Higgins, and A. M. Fallon. 2003. Characterization and cDNA cloning of an immune-induced lysozyme from cultured Aedes albopictus mosquito cells. Dev. Comp. Immunol. 27:11-20. [DOI] [PubMed] [Google Scholar]
- 12.Kanamori, T., N. Kanou, H. Atomi, and T. Imanaka. 2004. Enzymatic characterization of a prokaryotic urea carboxylase. J. Bacteriol. 186:2532-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapphahn, R. J., C. M. Ethen, E. A. Peters, L. Higgins, and D. A. Ferrington. 2003. Modified αA crystalline in the retina: altered expression and truncation with aging. Biochemistry 24:15310-15325. [DOI] [PubMed] [Google Scholar]
- 14.Karp, P. 2004. BioCyc Database, World Wide Web. http://biocyc.org/BSUB/new-image?type=ENZYME&object=BG11231-MONOMER.
- 15.Kim, Y. S., and S. W. Kang. 1988. Assays for malonyl-coenzyme a synthase. Anal. Biochem. 170:45-49. [DOI] [PubMed] [Google Scholar]
- 16.Koo, H. M., S. O. Choi, H. M. Kim, and Y. S. Kim. 2000. Identification of active-site residues in Bradyrhizobium japonicum molonamidase E2. Biochem. J. 349:501-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labahn, J., S. Neumann, G. Buldt, M. R. Kula, and J. Granzin. 2002. An alternative mechanism for amidase signature enzymes. J. Mol. Biol. 322:1053-1064. [DOI] [PubMed] [Google Scholar]
- 18.Martinez, B. 2001. Characterization, complete nucleotide sequence and organization of atrazine catabolic plasmid, pADP-1 from Pseudomonas sp. strain ADP. Ph.D. thesis. University of Minnesota, Saint Paul, Minn. [DOI] [PMC free article] [PubMed]
- 19.Martinez, B., J. Tomkins, L. P. Wackett, R. Wing, and M. J. Sadowsky. 2001. Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J. Bacteriol. 183:5684-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patricelli, M. P., and B. F. Cravatt. 2000. Clarifying the catalytic roles of conserved residues in the amidase signature family. J. Biol. Chem. 275:19177-19184. [DOI] [PubMed] [Google Scholar]
- 21.Radosevich, M., S. J. Traina, H. Yue-Li, and O. H. Tuovinen. 1995. Degradation and mineralization of atrazine by a soil bacterial isolate. Appl. Environ. Microbiol. 61:297-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Segel, I. H. 1975. Other methods of plotting enzyme kinetics data, p. 208-224. In I. H. Segal (ed.), Enzyme kinetics: behavior and analysis of rapid equilibrium and steady-state enzyme systems. Wiley-Interscience, New York, N.Y.
- 24.Shin, S., T. H. Lee, C. N. Ha, H. M. Koo, S. Y. Kim, H. S. Lee, Y. S. Kim, and B. H. Oh. 2002. Structure of malonamidase E2 reveals a novel Ser-cisSer-Lys catalytic triad in a new serine hydrolase fold that is prevalent in nature. EMBO J. 21:2509-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin, S., Y. S. Yun, H. M. Koo, Y. S. Kim, K. Y. Choi, and B. H. Oh. 2003. Characterization of a novel Ser-cisSer-Lys catalytic triad in comparison with the classical Ser-His-Asp triad. J. Biol. Chem. 278:24937-24943. [DOI] [PubMed] [Google Scholar]
- 26.Struthers, J. K., K. Jayachandran, and T. B. Moorman. 1998. Biodegradation of atrazine by Agrobacterium radiobacter J14a and use of this strain in bioremediation of contaminated soil. Appl. Environ. Microbiol. 64:3368-3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sumrada, R. A., and T. G. Cooper. 1982. Urea carboxylase and allophanate hydrolase are components of a multifunctional protein in yeast. J. Biol. Chem. 257:9119-9127. [PubMed] [Google Scholar]
- 28.Topp, E., H. Zhu, S. M. Nour, S. Houot, M. Lewis, and D. Cuppels. 2000. Characterization of an atrazine-degrading Pseudaminobacter sp. isolated from Canadian and French agricultural soils. Appl. Environ. Microbiol. 66:2773-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Topp, E., W. M. Mulbry, H. Zhu, S. M. Nour, and D. Cuppels. 2000. Characterization of s-triazine herbicide metabolism by a Norcardiodes sp. isolated from agricultural soils. Appl. Environ. Microbiol. 66:3134-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wackett, L. P. 2004. Evolution of enzymes for the metabolism of new chemical inputs into the environment. J. Biol. Chem. 279:41259-41262. [DOI] [PubMed] [Google Scholar]
- 31.Wackett, L. P., M. J. Sadowsky, B. Martinez, and N. Shapir. 2002. Biodegradation of atrazine and related triazine compounds: from enzymes to field studies. Appl. Microbiol. Biotechnol. 58:39-45. [DOI] [PubMed] [Google Scholar]
- 32.Walsh, C. T. 1979. Enzymatic reaction mechanisms. W. H. Freeman and Company, San Francisco, Calif.
- 33.Weatherburn, M. W. 1967. Phenol-hypochlorite reaction for determination of ammonia. Anal. Chem. 39:791-794. [Google Scholar]
- 34.Whitney, P. A., and T. G. Cooper. 1972. Urea carboxylase and allophanate hydrolase. Two components of adenosine triphosphate:urea amido-lyase in Saccharomyces cerevisiae. J. Biol. Chem. 247:1349-1353. [PubMed] [Google Scholar]
- 35.Yan, J. X., N. H. Packer, A. A. Gooley, and K. L. Williams. 1998. Protein phosphorylation: technologies for the identification of phosphoamino acids. J. Chromatogr. A 808:23-41. [DOI] [PubMed] [Google Scholar]