Abstract
Genetically engineered mouse models (GEMMs) are used extensively to study the effects of a range of cancer-associated mutations in vivo, but current models only capture a small fraction of genetic lesions observed in human cancer. GEMMs based on CRISPR-Cas9 have enabled rapid assessment of additional mutations, primarily loss-of-function alleles that arise from insertions or deletions. However, the utility of these models for engineering precise mutations is limited by their reliance on error-prone DNA repair mechanisms. Here, we describe the development of a facile system for performing in vivo prime editing in murine tissues by encoding a Cre-inducible prime editor enzyme in the mouse germline. We show this model allows rapid and precise engineering of a wide range of mutations in cell lines and organoids derived from multiple primary tissues, including a clinically relevant Kras mutation associated with resistance to targeted therapies and Trp53 hotspot mutations commonly observed in pancreatic cancer patients. We also demonstrate somatic prime editing in vivo using lentiviruses and lipid nanoparticles, and further illustrate the utility of these mice for modeling lung and pancreatic cancer in vivo through viral delivery of prime editing guide RNAs or syngeneic orthotopic transplantation of prime edited organoids. We anticipate that prime editing GEMMs will accelerate preclinical functional studies of cancer-associated alleles and complex genetic combinations that are challenging to model by traditional approaches.
Cancer is driven by somatic mutations that accumulate throughout progression and often display extensive intertumoral heterogeneity, occurring in thousands of different combinations across human cancer1,2. The precise nature of driver mutations and their combinations can profoundly influence how cancers initiate, progress, and respond to therapy, establishing tumor genotype as a critical determinant of disease outcome3,4. Emerging precision oncology treatment paradigms aim to match specific therapies with tumor genotypes, and this strategy has shown promise for several driver mutations5,6. To expand the promise of precision oncology to more patients, it is critical to develop tools to systematically interrogate the effects of distinct genetic lesions and combinations thereof on the overall tumor phenotype, particularly in vivo.
GEMMs have proven invaluable for elucidating the mechanisms by which cancer drivers promote tumor development and progression in vivo7,8. However, generating new GEMMs using traditional approaches is an expensive, laborious, and time-consuming process. Established GEMMs can also take months for investigators to acquire and often require laborious breeding programs to combine multiple alleles of interest and to establish a colony of sufficient size for experimental cohorts. These factors impede studies aimed at developing precision oncology treatments for tumors driven by specific genetic variants, which continue to be identified on a regular basis9.
Genome editing technologies like CRISPR-Cas9 can be used to rapidly engineer somatic mutations when delivered exogenously or when installed as germline alleles10–14. While these models have accelerated the study of putative cancer driver genes, they are most frequently used to induce DNA double-stranded breaks (DSBs), leading to inactivation of tumor suppressor genes via error-prone repair and frameshifting insertion/deletion (indel) formation. Although CRISPR-based homology-directed repair (HDR) has been used to model precise single nucleotide variants (SNVs) in Cas9-knockin mice, this method requires an exogenous DNA donor template and is limited by low efficiency and high rates of indel byproducts15. Furthermore, the requirement for DSBs to induce frameshifts or HDR-based precise edits can lead to confounding genotoxic effects, including on-target chromothripsis events and artificial fitness costs incurred through continued disruption of edited oncogenes16,17.
Precision genome editing technologies like base editing18 can be used to model cancer in mice by installing specific transition mutations with high efficiency and negligible indel byproducts11. Though precise and highly efficient, base editors also have limitations, including the requirement for different base editor enzymes depending on the mutation being studied (e.g., cytosine or adenine base editors), and their propensity for bystander editing, which can prohibit introducing desired amino acid substitutions. While the recent development of C:G and A:Y transversion base editors will expand the scope of cancer modeling19–22, current base editing technology is not amenable to modeling the full spectrum of small somatic mutations.
In contrast to base editing and standard CRISPR-Cas9, prime editing enables engineering the full spectrum of single nucleotide substitutions and indels with high product purity23,24. Prime editors employ a Cas9 nickase coupled with a reverse transcriptase that complexes with prime editing guide RNAs (pegRNAs). pegRNAs encode mutations of interest within a reverse transcriptase template23,24, enabling highly precise and programmable editing. Prime editing thus offers a versatile approach to study the full spectrum of cancer driver mutations, their combinations, and the growing catalog of secondary mutations that confer resistance to targeted therapies25–28.
Beyond editing versatility, prime editing also avoids the formation of indel byproducts associated with DSBs. This is particularly important for studying SNVs with putative neomorphic qualities in tumor suppressor genes, as HDR-directed mutations would be diluted by the higher rate of naturally selected indels. Prime editing also exhibits lower rates of unintended activity at off-target loci, reducing the risk of confounding off-target effects24,29. These advantages, combined with broad editing capacity, provide an unprecedented opportunity to generate faithful models of human cancer.
With these considerations in mind, we developed both conditional and tissue-restricted prime editing GEMMs (PE GEMMs) that eliminate the need for exogenous delivery of prime editors, which can be challenging given their significant size30,31. Encoding the prime editing machinery within the mouse germline also minimizes confounding acute or chronic anti-tumor immune responses that could be induced by exogenous delivery of a Cas9-based fusion protein32–34. In conjunction with the development of PE GEMMs, we also developed a range of DNA vectors and engineered pegRNAs (epegRNAs) that promote efficient prime editing in a variety of cell lines and organoids derived from these mice. With this toolset, we established new organoid models harboring Trp53 mutations frequently found in pancreatic cancer patients but not modeled by current GEMMs of the disease, as well as a clinically relevant Kras mutation associated with resistance to KRASG12C inhibitors. We further show that PE GEMMs enable efficient prime editing in vivo via viral or nonviral delivery of pegRNAs to a variety of tissues. Extending these studies, we harnessed PE GEMMs to model cancer in vivo through somatic initiation of autochthonous lung and pancreatic adenocarcinoma, and by orthotopic transplantation of prime edited pancreatic organoids. We also investigated the oncogenic potential of a variety of primary Kras mutations in the lung, including the poorly understood KrasG12A mutation present in more than ten percent of lung adenocarcinoma patients. We expect PE GEMMs to both expand the landscape of achievable cancer-associated mutations and accelerate techniques required to study their function and associated therapeutic vulnerabilities.
Results:
Quantification of human cancer-associated mutations amenable to modeling by prime editing in mice.
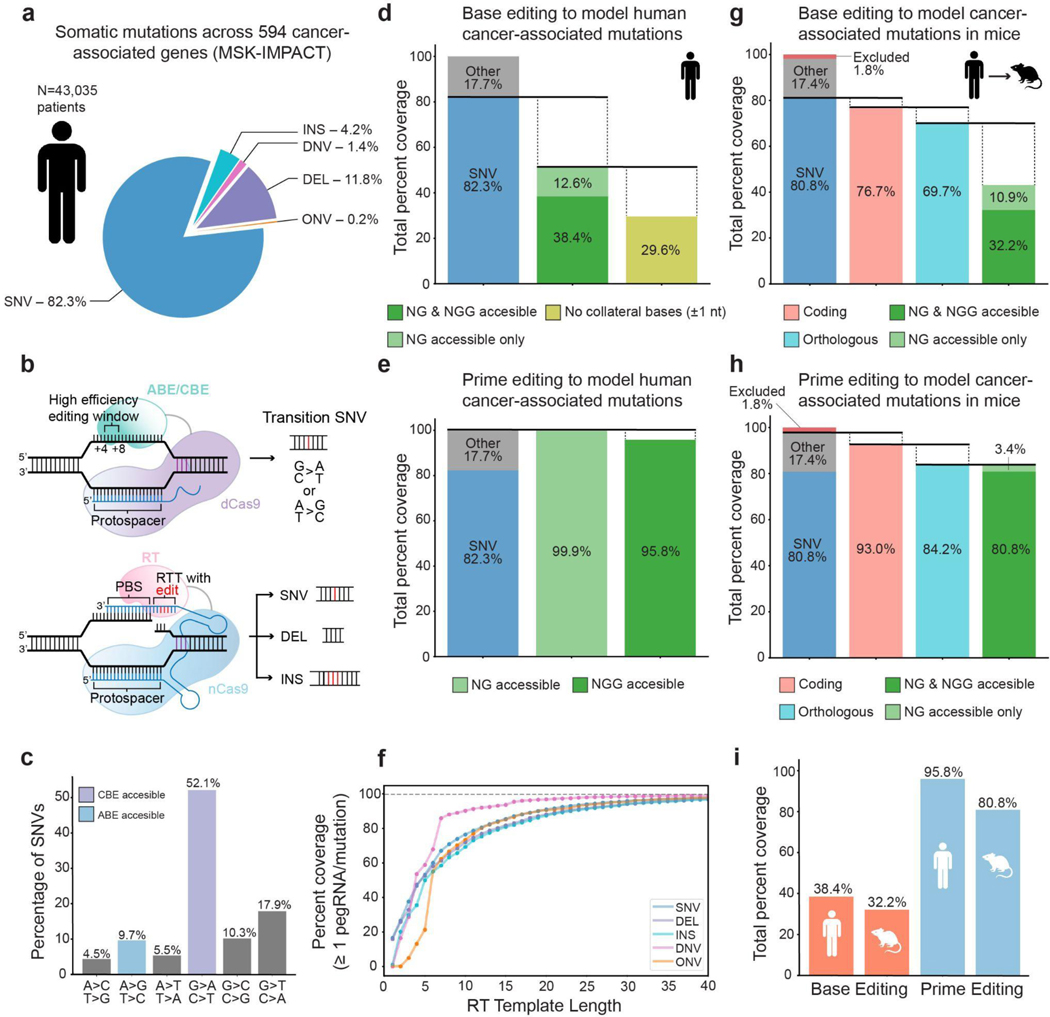
Recent work has shown that base editing can be used to elucidate the function of specific cancer-associated genetic variants35 and to systematically probe a large fraction of all possible alleles for genes and proteins of interest36. Base editors are primarily capable of engineering transition SNVs23 (A•T>G•C or G•C>A•T), though the base editor architecture has recently been adapted to produce C•G>G•C transversions with variable efficiency19,20,37–39. In contrast, prime editors are capable of engineering all transition and transversion SNVs24, as well as defined indel alleles40,41, expanding the potential for rapid modeling of genetic variants even further. To define the expanded editing capacity afforded by prime editing, we quantified the abilities of both base and prime editing to install specific somatic mutations identified from a cohort of 43,035 genetically-profiled cancer patients from the Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) dataset (Fig. 1a,b and Supplementary Fig. 1)9,35. Out of 422,822 mutations identified from targeted exon sequencing of 594 cancer-associated genes, 82.3% are SNVs, while the remaining 17.7% are deletions (DEL), insertions (INS), and di/oligo-nucleotide variants (DNVs, ONVs), in descending order of frequency (Fig. 1a).
Figure 1.
Quantification of human cancer-associated mutations amenable to modeling by base editing or prime editing in humans and mice.
a. Distribution of somatic variant types in a cohort of 43,035 patients with 422,822 mutations observed in 594 cancer-associated genes. Single nucleotide variants = SNV, deletions = DEL, insertions = INS, di-nucleotide variants = DNV, oligo-nucleotide variants = ONV.
b. Schematic of the modeling capabilities of base editing (top) and prime editing (bottom).
c. Quantification of somatic SNVs by type, illustrating enrichment for transition SNVs. Transition SNVs amenable to modeling by cytosine base editors (CBE) are shown in purple, while transition SNVs amenable to modeling by adenine base editors (ABE) are shown in blue. Transversions are shown in gray.
d. Quantification of mutations amenable to modeling with cytosine or adenine base editors that use an NG or NGG PAM. All percentages are given as a percentage of all mutations in the dataset. 38.4% of all mutations are amenable to base editing and fall within the protospacer of an NGG PAM (dark green), while an additional 12.6% of all mutations are amenable to base editing and fall within an NG PAM protospacer (light green). Of this subset of mutations that fall within either an NG or NGG PAM protospacer, only ~60%, or 29.6% of all mutations, lack matching collateral bases within one nucleotide (nt) of the mutation site.
e. 95.8% of all mutations in the dataset are potentially amenable to modeling by a prime editor using an NGG PAM (dark green) coupled with a pegRNA with a reverse transcription (RT) template length of 30 nucleotides. 99.9% of all mutations can be modeled by a prime editor using an NG PAM with the same pegRNA specifications.
f. Percentage of mutations with at least one suitable pegRNA as a function of the RT template length of the pegRNA, excluding the additional length of a homologous region in the RT template. Calculations assume the prime editor recognizes an NGG PAM.
g. Quantification of orthologous coding mutations potentially amenable to modeling by base editing in mice. Mutations are defined as orthologous if they derive from a wild-type amino acid conserved in the murine ortholog, as determined by pairwise protein alignment between human and mouse protein sequences. The rightmost bar indicates the fraction of orthologous coding mutations that can be modeled by base editors that recognize NG or NGG PAMs. “Excluded mutations” refers to mutations that fall in a gene lacking an ortholog. All percentages are given as a percentage of all mutations in the dataset.
h. Quantification of orthologous coding mutations potentially amenable to modeling by prime editing. Orthologous mutations are defined as in Fig. 1g. The rightmost bar indicates the ability of an NG or NGG prime editor to model these orthologous mutations, assuming an RT template greater than 30 nt. Excluded mutations are defined as in Fig. 1g.
i. Summary of the cancer mutation modeling capabilities of base and prime editing assuming an NGG PAM.
To estimate what fraction of common cancer-associated mutations are captured in currently available transgenic mouse models, we analyzed a dataset curated from the Mouse Genome Informatics database (see Methods)42,43. We found that 65 of the 100 most frequent SNVs in MSK-IMPACT, including 50 of 84 missense SNVs, are not represented by published mouse cancer models (Supplementary Tables 1 and 2). Notably, the majority of these SNVs are transitions, which comprise 61.8% of all SNVs in the overall MSK-IMPACT dataset and are theoretically compatible with engineering using base editors (Fig. 1c). In general, 38.4% of all mutations in the dataset are amenable to base editing using a canonical NGG PAM sequence23,35 (Fig. 1d). The total mutation coverage with base editing increases to 51% when accounting for base editors that utilize more abundant NG PAM sequences.
With base editors, adjacent identical nucleotides can be collaterally edited and result in undesired editing outcomes. When considering only mutations without identical bases present within one adjacent nucleotide, the total mutation coverage drops to 29.6% (Fig. 1d). This analysis does not account for the location of a desired edit within the protospacer, which can influence base editing efficiency and the total fraction of amenable mutations (Supplementary Fig. 2).
We used a similar approach to quantify the modeling capabilities of prime editors that use an NGG or NG PAM coupled with variable reverse transcription template (RTT) lengths encoded within pegRNAs (Supplementary Fig. 1). Using an NGG PAM and RTT length of 30 base pairs (bp), excluding the additional length of a homologous region in the RTT, prime editing theoretically reaches 95.8% coverage of all mutations in this dataset (Fig. 1e). This value increases to 99.9% for prime editors that could theoretically use an NG PAM (Fig. 1e). Moreover, analysis of the relationship between RTT length and modeling capabilities reveals that ~85% of mutations in this dataset can be modeled by placing the mutation within the first 15 bp of the RTT (Fig. 1f). These parameters are well within the recommended guidelines for pegRNA RTT length, even with the additional size required for a region of homology23. Collectively, this analysis suggests that both base editing and prime editing can serve as versatile technologies for modeling cancer-associated mutations.
We also sought to determine the fraction of cancer-associated mutations that derive from protein sequences conserved in murine orthologs. We reasoned that only this subset of conserved sequences, when mutated in mouse systems, could be expected to mimic effects seen in human cancer. To quantify the ability of base and prime editors to model cancer-associated mutations in mice, we performed pairwise alignment on orthologous mouse and human proteins to define whether mutations derive from a conserved wild-type amino acid and reside in a region of homology (Supplementary Fig. 1). Of the SNVs that occur in coding sequences, 90.9% derive from codons that encode conserved amino acids between mouse and human. Of these conserved, cancer-associated SNVs, 61.8% are amenable to base editing (NG or NGG PAM), which translates to 43.1% of all mutations in the dataset (Fig. 1g). In contrast, NG or NGG prime editors are capable of modeling 100% of coding mutations that occur at conserved amino acid residues in mice (84.2% of all mutations in the dataset) (Fig. 1h). In total, 80.8% of human cancer-associated mutations observed in this dataset could be modeled in mice with prime editors using a traditional NGG PAM (Fig. 1f,i). This same pattern holds when filtering the dataset to only mutations that occur in multiple patients, and when considering various stringencies of homology in the regions flanking the mutations of interest (Supplementary Figs. 2 and 3). In total, these results demonstrate that prime editing could significantly broaden both the diversity and number of human cancer-associated mutations that can be rapidly modeled in murine orthologs.
Design and construction of a Cre-inducible prime editor allele.
We sought to develop a transgenic system capable of precisely engineering the majority of cancer-associated mutations without requiring exogenous delivery of a prime editor enzyme. To accomplish this, we targeted a transgene expression cassette encoding the PE2 enzyme and the mNeonGreen (mNG)44 fluorescent reporter, separated by the P2A ribosome skipping sequence, into the Rosa26 locus10,45 (Fig. 2a). Like previous Cre-inducible Rosa26 alleles10,46,47, transgene expression is driven by the CAGG promoter and is induced by Cre-mediated excision of a loxP-stop-loxP (LSL) cassette. A neomycin resistance gene was included to enable selection of cells containing the targeted allele. We also incorporated FRT/FRT3 sequences flanking the central construct to enable Flp recombinase-mediated replacement of the Rosa26PE2 allele with future generations of prime editor enzymes or other desirable editors29,48. This vector was targeted to Trp53flox/flox C57BL/6J embryonic stem cells, where Trp53 can be deleted upon expression of Cre recombinase (Supplementary Fig. 4). Chimeric mice were then crossed to wild-type C57BL/6J mice to generate pure strain heterozygous Trp53flox/+; Rosa26PE2/+ mice. These mice were subsequently crossed with Trp53+/+ and Trp53flox/flox mice to generate Rosa26PE2/+ mice on wild-type and Trp53flox/flox backgrounds.
Figure 2.
Design and functional validation of the Rosa26PE2 prime editor allele.
a. Schematic depicting the design of the Cre-inducible Rosa26PE2 allele.
b. Schematic depicting the formation of hU6-pegRNA-EF-1α-Cre (UPEC) and hU6-pegRNA-EFS-mScarlet (UPEmS) vectors from templates encoding a red fluorescent protein (RFP) by Golden Gate assembly.
c. Bright-field images of pancreatic organoids derived from chimeric prime editor mice and wild-type mice. With and without treatment with neomycin.
d. Bright-field and fluorescent images showing PE2-P2A-mNG expression only after exposure to Cre encoded by a UPEC vector.
e. Schematic depicting the derivation of multiple organoids and a fibroblast cell line from Rosa26PE2/+ prime editor mice.
f. Editing efficiency of a trinucleotide (+GGG) insertion located eight base pairs downstream of the start codon in Dnmt1 in pancreatic organoids, lung organoids, and tail tip-derived fibroblasts. Unintended indel byproducts in all conditions were present in <1% of sequencing reads. Data and error bars indicate the mean and standard deviation of three independent transductions.
g. Editing efficiency and indel byproduct frequency of Dnmt1+GGG in liver tissue one week after tail vein injection with LNPs harboring either Cre mRNA and pegRNA (left) or pegRNA alone (right). Data and error bars indicate the mean and standard deviation of three to five independent mice.
h. Bright-field and fluorescent images of pancreases derived from Rosa26PE2/+ (left) or Pdx-1 Cre;Rosa26PE2/+ mice (right).
i. Immunofluorescence imaging of intestinal tissue derived from Villin-CreERT2; Rosa26PE2/+ mice that were either untreated (left) or exposed to tamoxifen (right; 4-OHT). Tissue slides were stained with the DNA stain DAPI (4′,6-diamidino-2-phenylindole; top) or with an antibody specific to Cas9 (bottom). Scale bar indicates 100 μm.
Functional validation of the prime editor allele in organoids.
To confirm the functionality of the Rosa26PE2 allele, we developed two lentiviral vectors that co-express a pegRNA and either Cre recombinase (UPEC) or the red fluorescent protein, mScarlet49 (UPEmS) (Fig. 2b). We derived pancreatic organoids from chimeric Trp53flox/flox;Rosa26PE2/+ mice and developed a pure culture of transgene-containing cells via selection with neomycin (Fig. 2c; Supplementary Fig. 5). As expected, these pancreatic organoids displayed Cre-dependent mNG expression upon transduction with UPEC vectors (Fig. 2d; Supplementary Fig. 5). To test the prime editing functionality of this allele, we designed a Dnmt1-targeting pegRNA encoding a +1 CCC insertion, which templates a trinucleotide insertion of a GGG codon encoding glycine at residue 4 of Dnmt1. UPEC-transduced organoids were selected using nutlin-3a, a mouse double minute 2 homolog (MDM2) inhibitor that induces cell cycle arrest in Trp53-proficient (but not Trp53-deficient) cells50, enriching for those Trp53flox/flox cells that underwent Cre-mediated recombination following UPEC transduction. After selection, we detected up to 33.8% editing efficiency and minimal indel byproducts at Dnmt1 (Supplementary Fig. 5). These results validate the functionality of the Rosa26PE2 allele, including its ability to mediate prime editing of endogenous loci when using optimized pegRNAs.
Ex vivo prime editing in organoids and cell lines derived from the Rosa26PE2 model.
We next sought to evaluate prime editing across multiple tissues. To accomplish this, we derived lung organoids, pancreatic organoids, and tail tip-derived fibroblasts (TTFs) from multiple Rosa26PE2/+ mice (Fig. 2e). Consistent with results using chimera-derived organoids, we observed highly efficient Dnmt1 editing across all investigated tissues (Fig. 2f). Corroborating the well-established on-target fidelity of prime editing24,29,51, we did not detect off-target prime editing across multiple loci prioritized based on protospacer homology52 (Supplementary Fig. 6).
Prior work established a subset of DNA damage repair (DDR) genes as key factors influencing prime editing efficiency29. Given p53’s fundamental role in DDR, we examined whether Dnmt1+GGG editing levels differed substantially across Trp53+/+ and Trp53flox/flox conditions. In both TTFs and pancreatic organoids, we noticed a consistent 2–3-fold decrease in Dnmt1+GGG editing in Trp53+/+ relative to Trp53flox/flox tissues (Supplementary Fig. 6). This result suggests that Trp53 status may affect prime editing efficiency, although we still observe highly efficient editing across loci in Trp53-proficient tissues.
Prime editing in vivo with lipid nanoparticles.
To determine whether PE GEMMs enable prime editing in vivo, we co-formulated a synthetic pegRNA encoding the Dnmt1+GGG insertion and Cre mRNA within lipid nanoparticles (LNPs). We then treated Rosa26PE2/+ and Rosa26PE2/PE2 mice with one of two LNP formulations53 (see Methods) via tail vein injection. After one week, we observed Cre-induced fluorescence in the livers of mice that received pegRNA-bearing LNPs, but not in a control mouse that received phosphate-buffered saline (PBS) (Supplementary Fig. 7). Importantly, we also detected moderately efficient prime editing (up to 3.4%) at Dnmt1 as assessed by bulk liver analysis, and we did not detect significant editing in mice that received LNPs harboring only the Dnmt1 pegRNA (i.e., lacking Cre mRNA) (Fig. 2g). To our knowledge, these results represent the first published demonstration of LNP-mediated prime editing in vivo, and they confirm PE GEMMs are amenable to precision edits in vivo.
Generation of constitutive and inducible tissue-restricted PE GEMMs.
Prime editing in vivo could be more convenient if the need for Cre co-delivery was eliminated. To demonstrate compatibility of the conditional PE2 allele with tissue-restricted Cre drivers, we generated additional PE GEMMs through genetic crosses with mice harboring alleles that express Cre recombinase from endogenous loci. First, we crossed Rosa26PE2/PE2 mice to Pdx-1 Cre54, a pancreas-specific Cre driver allele, and Villin-CreERT2, an inducible, intestinal epithelial Cre driver allele55. As expected, Pdx-1 Cre;Rosa26PE2/+ animals showed bright and robust evidence of mNG expression in the pancreas (Fig. 2h), and Villin-CreERT2;Rosa26PE2/+ mice demonstrated PE2 expression in intestinal epithelial cells upon treatment with tamoxifen (Fig. 2i). Notably, histologic analysis of the pancreas and intestinal epithelia, respectively, revealed no gross or pathologic abnormalities, suggesting that constitutive or inducible expression of the PE2 enzyme does not lead to toxicity in these tissues (Supplementary Fig. 7).
Optimization of Kras-targeted pegRNAs and functional testing in pancreatic organoids.
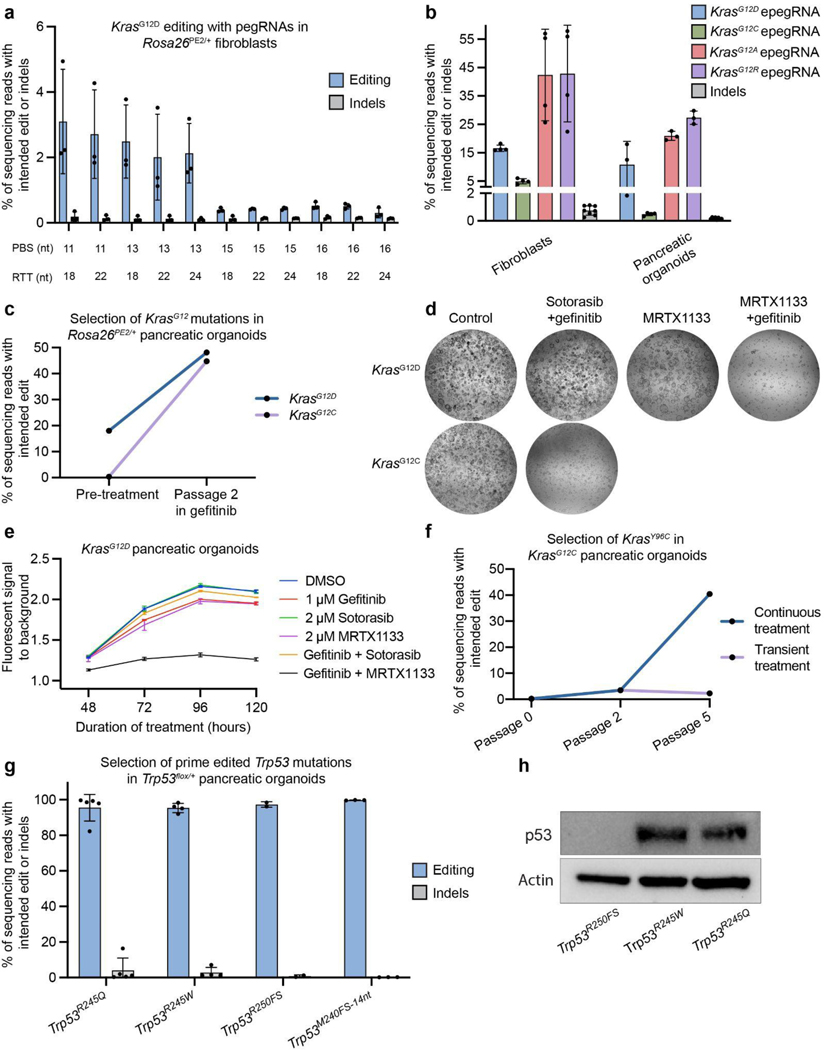
We next sought to empirically identify highly efficient pegRNAs that introduce the KrasG12D transition as a single nucleotide variant (GGT>GAT). Based on previous work56, we hypothesized that spacer sequences capable of producing the highest Cas9 indel efficiency in mouse N2A cells would serve as ideal scaffolds for high-efficiency pegRNA designs (Supplementary Fig. 8 and Supplementary Table 3). Using TTFs, we observed up to ~5% editing efficiency of KrasG12D with spacer-optimized pegRNAs (Fig. 3a and Supplementary Fig. 8). To further increase editing efficiency, we modified our best-performing pegRNA with a structured RNA pseudoknot motif, prequeosine1-1 riboswitch aptamer (tevopreQ1), recently shown to enhance prime editing efficiency by more than 3-fold in cell lines51. This resulted in up to ~18.4% editing efficiency of KrasG12D in pancreatic organoids and TTFs (Fig. 3b). We then modified this epegRNA to template the KrasG12C transversion and observed ~0.5% editing efficiency in pancreatic organoids and ~5% in TTFs. We also generated KrasG12A and KrasG12R epegRNAs and observed up to ~30% editing efficiency with both epegRNAs in TTFs (Fig. 3b).
Figure 3.
Ex vivo prime editing and functional testing of Kras and Trp53 mutations.
a. Editing efficiency and indel byproduct frequency of the KrasG12D transition mutation (G:C to A:T) templated by a cohort of pegRNAs based on a single Cas9 spacer (n = 3 for each pegRNA). pegRNAs are delineated by differences in the lengths of the primer binding site (PBS) and reverse transcriptase template (RTT). Data and error bars indicate the mean and standard deviation of three independent transductions.
b. Editing activity of four engineered pegRNAs (epegRNAs) templating either the KrasG12D transition or the KrasG12C, KrasG12A, or KrasG12R transversions in tail tip-derived fibroblasts or pancreatic organoids (KrasG12D and KrasG12C). Data and error bars indicate the mean and standard deviation of three independent transductions. epegRNAs were generated by appending the trimmed evopreQ1 motif after the primer binding site of the leftmost pegRNA depicted in Fig. 1a. Indel byproduct calculations were pooled from all conditions within each tissue.
c. Allele frequencies of KrasG12D or KrasG12C mutations in pancreatic organoids before and after two passages of treatment with gefitinib (1 μM) (n = 1). Gefitinib treatment selects for cells containing prime edited KrasG12D or KrasG12C mutations.
d. Bright-field images of prime edited KrasG12C or KrasG12D organoids treated for four days with either control DMSO, sotorasib (2 μM) and gefitinib (1 μM), MRTX1133 (5 μM), or MRTX1133 and gefitinib.
e. Viability of KrasG12D pancreatic organoids under various treatment conditions. Viability was quantified using the alamarBlue HS Cell Viability Reagent, which is metabolized into a fluorescent derivative in living cells. Bars represent the range across two independent replicates.
f. Allele frequency of KrasY96C in KrasG12C organoids during and after treatment with sotorasib (2 μM) and gefitinib (n = 1). After two passages, organoids were split into two groups, which included continued treatment (continuous treatment) in one group and removal of treatment in a second group (transient treatment).
g. Allele frequencies of three Trp53 mutations in Trp53flox/+ pancreatic organoids treated with nutlin-3a for three to five passages after transduction with UPEC vectors. Indel byproduct frequencies are also included. Note that the highest indel frequency depicted for Trp53R245W derives primarily from a scaffold insertion in a single replicate. Trp53R245Q and Trp53R245W are homologous to mutations commonly observed in pancreatic cancer patients (TP53R248Q and TP53R248W), as described in Supplementary Figure 8. Trp53R250FS denotes a dinucleotide deletion that induces a frameshift mutation. Trp53M240FS−14nt denotes a fourteen-nucleotide deletion. Data and error bars indicate the mean and standard deviation of two to five independent transductions.
h. Immunoblot indicating detectable levels of p53 protein in prime edited Trp53flox/R245Q and Trp53flox/R245W organoids and an absence of detectable protein in Trp53flox/R250FS organoids.
Both KrasG12A and KrasG12R epegRNAs template G·C-to-C·G substitutions, which proceed from C·C mismatch intermediates. These mismatches are not efficiently repaired by mismatch repair (MMR) and are thought to have higher basal prime editing rates as a consequence29. Work from Chen and colleagues has indicated that co-installation of silent or benign MMR-evasive edits can promote higher prime editing efficiency, consistent with the increased editing efficiency in producing KrasG12A and KrasG12R over KrasG12C. To further probe this phenomenon, we compared a variety of epegRNAs templating cancer-associated mutations across Kras, Trp53, and Egfr to counterparts modified with silent or inconsequential edits. In nearly every case, we found that installing MMR-evasive edits amplified prime editing efficiencies by >3-fold, often resulting in efficiencies greater than 20% (Supplementary Fig. 9). Collectively, these data demonstrate that the Rosa26PE2 allele enables efficient installation of SNVs, multinucleotide alterations, and insertions and deletions across a diverse array of cell lines and organoids.
To confirm the functional effects of these mutations, we installed either KrasG12D or KrasG12C mutations in Trp53flox/flox;Rosa26PE2/+ pancreatic organoids and selected transduced cells with nutlin-3a. We then treated prime edited organoids with the epidermal growth factor receptor (EGFR) inhibitor, gefitinib, to select for the oncogenic Kras mutation57 and evaluated the fraction of cells containing the intended edits before and after treatment. Consistent with receptor-independent signaling downstream of EGFR, only KrasG12D and KrasG12C prime-edited cells survived treatment with gefitinib, while control cells infected with the template UPEC lacking a pegRNA did not (Fig. 3c and Supplementary Fig. 10). We then tested whether cells transduced with KrasG12C epegRNAs were sensitive to sotorasib, a KRASG12C-specific inhibitor, alone or in combination with gefitinib. Consistent with previous work58, we found that KrasG12C pancreatic organoids were uniquely sensitive to the combination of sotorasib and gefitinib, while KrasG12D organoids were unaffected by these treatments (Fig. 3d and Supplementary Fig. 10). While KRASG12C inhibition has shown promising signs of clinical efficacy in pancreatic cancer5,59, current preclinical efforts focused on KRASG12D inhibition have the potential to benefit a broader fraction of patients with this disease (>38%)60,61. We therefore treated prime edited KrasG12D pancreatic organoids with MRTX113362, a KRASG12D inhibitor, alone or in combination with gefitinib. Consistent with results using sotorasib, we found that KrasG12D organoids were significantly more sensitive to the combination treatment compared with MRTX1133 alone (Fig. 3d,e and Supplementary Fig. 10), suggesting that concomitant EGFR inhibition may be a broadly effective strategy to augment the overall efficacy of KRAS mutant inhibitors in pancreatic cancer cells.
Prime editing enables rapid functional interrogation of putative resistance mutations.
While targeted therapies have revolutionized modern cancer treatment, therapy resistance is common and frequently arises through the acquisition of secondary missense mutations affecting the drugged driver28,63,64. Recent work by Awad and colleagues revealed a novel class of secondary KRAS mutations occurring in over 10% of non-small cell lung cancer and colorectal cancer patients with acquired resistance to adagrasib, a KRASG12C inhibitor63. Intriguingly, several mutations occur in codons 95–96, which occupy the switch II pocket targeted by adagrasib and sotorasib.
To test the utility of the Rosa26PE2 model to functionally interrogate novel mutations associated with resistance, we developed an epegRNA designed to introduce the KrasY96C transversion and tested its capacity to promote resistance in prime edited KrasG12C pancreatic organoids treated with gefitinib and sotorasib (Supplementary Fig. 10). All organoids were initially treated with both inhibitors for two passages, followed by continued treatment for three additional passages in one group (continuous treatment) and treatment removal in the second group (transient treatment). Consistent with patient data63, organoids transduced with the KrasY96C epegRNA were resistant to combined treatment with gefitinib and sotorasib and exhibited increased allele frequency of the KrasY96C mutation over time (Fig. 3f). Positive selection for composite KrasG12C;Y96C mutant organoids was not observed in organoids following the removal of gefitinib and sotorasib, confirming the requirement of the selective pressure exerted by the treatment. Though initially discovered in lung cancer patients treated with sotorasib monotherapy, these data indicate that secondary KRAS mutations can also confer therapy resistance in other tissue and combination treatment contexts. The above results demonstrate that the Rosa26PE2 allele can be harnessed for rapid preclinical evaluation of emerging mechanisms of resistance to targeted therapies in tissues of interest and, ultimately, for testing second-generation therapies designed to overcome resistance.
Rapid engineering of common p53 mutations using prime editing.
A key advantage of PE GEMMs is the ability to mediate nearly any codon substitution in accessible tissues, enabling tissue-specific functional studies of genetic variants with putative effects on tumor progression. TP53 is the most frequently mutated gene in human cancer and is often altered via missense mutations that can confer gain-of-function properties in certain contexts65. In an analysis of data from cBioPortal66,67, we found that some of the most frequent p53 amino acid substitutions observed in lung (TP53R158L and TP53R270L) and pancreatic adenocarcinoma (TP53R248W and TP53R248Q) have not been targeted to the endogenous Trp53 locus in murine models (Supplementary Fig. 11), despite having putative gain-of-function effects68–70. Notably, three of these mutations are transversions that cannot be modeled using base editing, and the human amino acid (p53R248), but not the human codon (CGG vs CGC), is conserved in murine Trp53. Therefore, engineering the Trp53R245W mutation in mice requires a dinucleotide substitution uniquely suitable to prime editing (Supplementary Fig. 11). We developed a suite of epegRNAs designed to introduce both Trp53R245W and Trp53R245Q and two truncating deletions, Trp53R250FS and Trp53M240FS−14nt, using a Trp53+/+ cell line derived from murine 3TZ cells (Supplementary Fig. 12). After selection with nutlin-3a, most Trp53flox/+;Rosa26PE2/+ pancreatic organoids transduced with each of these epegRNAs exhibited a prime edited allele frequency near 100% (Fig. 3g).
We also observed an average of >90% editing purity in these organoids (Fig. 3g; Supplementary Fig. 13). Western blots confirmed that Trp53R245Q and Trp53R245W cells retained p53 protein expression, while Trp53R250FS cells did not (Fig. 3h). While the ratio of prime edited reads to random indel-bearing reads was typically high, we did observe a variable unintended single nucleotide substitution (0.24% - 11.34% of reads) attributable to partial reverse transcription of the scaffold sequence when prime editing Trp53R245Q (Supplementary Fig. 11). In one instance, we also observed an insertion of the scaffold sequence when prime editing Trp53R245W (~7% of reads). Notably, we did not observe any of these unintended events with an epegRNA templating Trp53M240FS−14nt, which was designed to evade MMR and exhibited a high basal editing efficiency (Supplementary Fig. 13).
In all cases, we observed negligible off-target activity at computationally predicted loci, even after more than four weeks of culturing organoids with sustained pegRNA expression (Supplementary Fig. 13). This result is most striking for Trp53R245W, which is templated by a pegRNA bearing a protospacer that shares 100% sequence homology with an off-target locus on chromosome 17 (Supplementary Table 4). We detected an average of 0.002% editing at this locus, which was significantly greater than the sequencing error rate in control samples (Supplementary Fig. 13). No other loci displayed editing levels higher than those observed in controls. Collectively, these results establish the utility of our approach for high-fidelity installation of novel mutations using systems that can be rationally engineered and easily translated to an in vivo setting.
Modeling lung and pancreatic adenocarcinoma in vivo using prime editing.
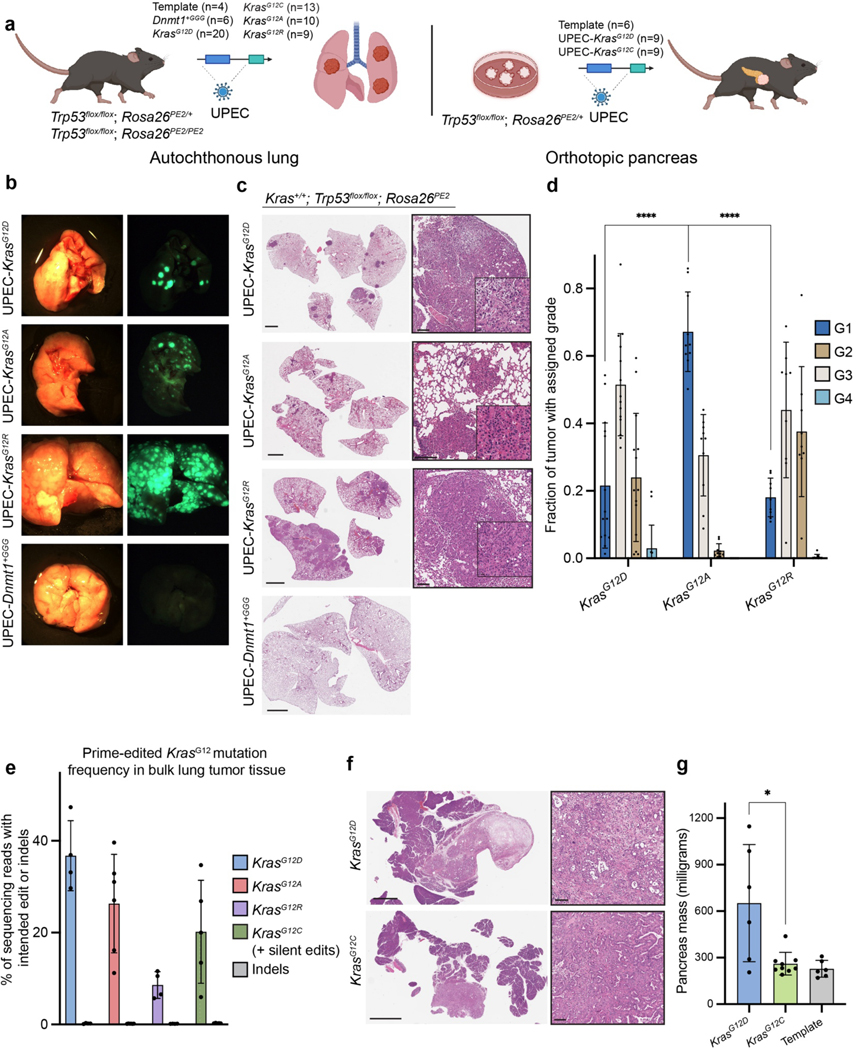
To benchmark the utility of PE GEMMs to model cancer in vivo, we initiated lung and pancreatic adenocarcinomas using autochthonous and orthotopic transplantation strategies (Fig. 4a). To model lung cancer, we intratracheally transduced the lungs of Trp53flox/flox;Rosa26PE2/+ and Trp53flox/flox;Rosa26PE2/PE2 mice with UPEC lentiviruses encoding the template vector (n=4) or pegRNAs for KrasG12D (n=20), KrasG12R (n=9), KrasG12A (n=10), KrasG12C (n=13), or the neutral Dnmt1+GGG (n=6). We also infected Trp53+/+;Rosa26PE2/+ mice with UPEC-KrasG12D to model low-grade lesions and assess in vivo prime editing in a Trp53-proficient setting.
Figure 4.
PE GEMMs enable autochthonous and orthotopic modeling of lung and pancreatic cancer.
a. Schematic depicting the design of in vivo experiments. Autochthonous lung tumors were initiated with lentivirus encoding UPEC vectors. Pancreatic tumors were initiated by orthotopic transplantation of prime edited pancreatic organoids. “Template” refers to the template UPEC vector lacking a pegRNA.
b. Representative bright-field and fluorescent images of lungs derived from mice infected with the UPEC vector encoding the neutral Dnmt1+GGG pegRNA, KrasG12D, KrasG12A, KrasG12R epegRNAs described in Fig. 3b.
c. Hematoxylin and eosin (H&E) staining of representative tissue from a control mouse infected with UPEC-Dnmt1+GGG (bottom), and tumor-bearing mice infected with UPEC-KrasG12D, UPEC-KrasG12A, and UPEC-KrasG12R (top). Callout boxes highlight histopathology consistent with lung adenoma and adenocarcinoma. Scale bars indicate 2 mm, 100 μm, and 20 μm respectively.
d. Bar charts indicating the distribution of grades across 16-week lesions from UPEC-KrasG12D (n = 14 mice), UPEC-KrasG12A (n = 10 mice), UPEC-KrasG12R (n = 9 mice). Lesion grades were called using the Aiforia algorithm as described in Methods. Data and error bars indicate the mean and standard deviation of all biological replicates in each condition. Statistical significance was calculated using unpaired, two-tailed t-tests comparing the fraction of Grade 1 lesions in KrasG12A-driven tumor tissue to KrasG12D-driven tumor tissue (P < 0.0001) or KrasG12R-driven tumor tissue (P < 0.0001).
e. Allele frequencies of KrasG12D, KrasG12A, KrasG12R (16 weeks) and KrasG12C + silent edits (12 weeks) in bulk lung tumors. Indel byproduct frequency was calculated as <1% in all cases.
f. H&E staining of representative pancreatic adenocarcinomas from a mouse transplanted with KrasG12D organoids (top) and a mouse transplanted with KrasG12C organoids (bottom). Scale bars indicate 2 mm, 25 μm, respectively.
g. Mass of pancreata of organoid transplant recipients measured in milligrams (n = 6–9 mice). Pancreata bearing KrasG12D-driven tumors were significantly larger than those bearing KrasG12C-driven tumors. Statistical significance was calculated using a two-tailed Mann-Whitney U test (P = 0.036).
In Trp53flox/flox recipients, tumors initiated by UPEC-KrasG12D were readily visible by μCT at 14 weeks post-injection (Supplementary Fig. 14). At 16 weeks, we observed multifocal fluorescent lesions in 16/20 (80%) UPEC-KrasG12D recipients and in none of the controls (Fig 4b). Histopathological analysis confirmed that lesions induced by prime editing recapitulated the full spectrum of lung cancer progression, from grade 1 atypical adenomatous hyperplasia through grade 4 adenocarcinoma. By immunohistochemistry, prime edited tumors recapitulated the cellular and molecular evolution seen in the classical KrasLSL-G12D/+;Trp53flox/flox (KP) GEMM model, demonstrating downregulation of lung lineage transcription factor Nkx2–1 and expression of chromatin regulator Hmga2 in poorly differentiated, advanced lesions71–73 (Fig. 4c–d, Supplementary Fig. 15).
We confirmed that tumors were initiated through on-target prime editing by sequencing genomic DNA derived from several bulk tumors (Fig. 4e). Prime editing in vivo did not require loss of p53, as 2/3 Trp53+/+;Rosa26PE2/+ mice developed fluorescent tumors upon infection with UPEC-KrasG12D, consistent with prior studies demonstrating that oncogenic Kras is sufficient to drive lung adenoma formation in vivo74 (Supplementary Fig. 16). These adenomas also harbored the intended KrasG12D mutation.
Similar to UPEC-KrasG12D recipients, UPEC-KrasG12A and UPEC-KrasG12R recipients consistently presented multifocal fluorescent lesions driven by on-target prime editing throughout the lung (Fig. 4c–e). However, both UPEC-KrasG12A and UPEC-KrasG12R recipients presented with greater tumor numbers than UPEC-KrasG12D recipients (Fig. 4d). While this is likely attributable in part to more efficient editing with the KrasG12A and KrasG12R epegRNAs (Fig. 3b), there were also discernible differences in the apparent oncogenic capacity of these mutations. In 8/9 UPEC-KrasG12R recipients, the overall tumor burden was significantly higher than the KrasG12A setting (Supplementary Fig. 16). Furthermore, histopathological analysis revealed that KrasG12R and KrasG12D tumors were of consistently higher grades relative to KrasG12A lesions (Fig. 4d). This is particularly striking given the relative rarity of KRASG12R in human lung cancer patients (<1% of KRAS mutations; see Discussion), although, of note, our data is consistent with prior work demonstrating that KrasG12R is highly oncogenic in murine models15. Taken together, these results highlight significant allele-specific differences in the oncogenic capacity of different Kras mutations and showcase the utility of PE GEMMs for rapidly discovering such phenotypes.
In contrast to other Kras mutations, only 4/13 (31%) UPEC-KrasG12C recipients presented tumors when harvested at 19 weeks, likely a consequence of the lower prime editing efficiency of the KrasG12C epegRNA. Furthermore, deep amplicon sequencing of these tumors occasionally revealed unintentional edits, including a KrasG12R mutation in one case and an additional silent substitution in codon 11 in another (data not shown). To address this shortcoming, we designed an improved KrasG12C epegRNA encoding MMR-evasive substitutions, which edits at a 3.2-fold higher efficiency (Supplementary Fig. 9). At 12 weeks, 8/9 Trp53flox/flox;Rosa26PE2/PE2 mice infected with this epegRNA developed multifocal tumor burden (Supplementary Fig. 16). Targeted sequencing confirmed the presence of the multinucleotide substitution encoding KrasG12C, without any unintended byproducts (Fig. 4e).
To further test the potential of PE GEMMs for cancer modeling in vivo, we transplanted prime edited KrasG12D/+;Trp53flox/flox;Rosa26PE2/+ and KrasG12C/+;Trp53floxflox;Rosa26PE2/+ pancreatic organoids into immunocompetent mice harboring the Rosa26PE2 allele (to ensure immunological tolerance75 to the prime editor enzyme). As controls, we transplanted Trp53flox/flox;Rosa26PE2/+ organoids infected with the template UPEC vector. Tumors were visible via ultrasound by five weeks (Supplementary Fig. 14), and fluorescent tumors that reflected the spectrum of pancreatic neoplasia were observed in 8/9 KrasG12D/+ recipients nine weeks post-transplantation (Fig. 4f; Supplementary Fig. 17). Notably, only 4/9 mice (44%) from the cohort of animals transplanted with KrasG12C/+ pancreatic organoids developed lesions. Of the remaining five, one developed a high-grade PanIN while the rest did not develop any lesions. Tumor burden in KrasG12C mice was significantly lower than in KrasG12D mice, as reflected in pancreatic weight measurements (Fig. 4g). These results are consistent with prior observations suggesting that KrasG12C may be less tumorigenic in the pancreas58. Metastases were only observed in KrasG12D recipients (Supplementary Fig. 17), indicative of a more aggressive phenotype of these tumors. We did not observe tumor formation in control recipients by ultrasound, microscopy, or histology, consistent with prior work showing that Trp53 knockout alone is insufficient for pancreatic tumorigenesis47,76.
To model autochthonous pancreatic adenocarcinoma, we adapted a strategy of retrograde pancreatic duct viral delivery47,77. We infected Trp53flox/flox;Rosa26PE2/+ mice with UPEC vectors encoding either KrasG12D or KrasY96C as a control. 3/4 KrasG12D infected animals developed pancreatic adenocarcinoma, while no tumors were detected in KrasY96C infected animals (Supplementary Fig. 17).
Discussion:
Advances in genome editing technologies have accelerated functional genetic studies, yet most approaches to model cancer mutations have relied on Cas9-mediated gene disruption via non-homologous end joining, failing to recapitulate many genetic lesions observed in human cancer. Emerging precision genome editing technologies like base editing and prime editing are poised to fill this gap by allowing the engineering of specific cancer-associated mutations. Nevertheless, the considerable size of base editors and prime editors makes delivery to most tissues and cell types challenging, posing significant limitations for in vivo studies. Prior studies have addressed this using split-prime editor systems that enable prime editing in vivo when delivered by dual adeno-associated virus (AAV) vectors. However, dual-AAV approaches remain hampered by delivery challenges to many tissues and, importantly, they can elicit an immune response against the prime editor enzyme34,78. The immunogenicity of genome editing reagents delivered exogenously significantly complicates cancer modeling experiments. With these challenges in mind, we developed a PE GEMM capable of rapidly installing a variety of genetic lesions with single nucleotide precision across in vitro, ex vivo, or in vivo contexts, as well as in an autochthonous, immunocompetent setting. By expressing the PE2 enzyme endogenously, we bypass the risk of a confounding immune response and significantly expand the capacity to deliver other functional cargo, such as Cre.
We used this model to install a variety of cancer-associated mutations, including transversions, transitions, multinucleotide substitutions, and deletions across Trp53, Egfr, and Kras. In the context of our pancreatic orthotopic transplant experiments, we observed that different Kras mutations exhibit variable in vivo tumor initiating potential, consistent with prior work comparing KrasG12C and KrasG12D autochthonous models in the pancreas58. In the lung, we found that KrasG12A, KrasG12D, and KrasG12R promote efficient but variable tumor formation. Tumor burden differences across genotypes are likely driven in part by variable pegRNA efficiencies, yet we also observed significant differences in the phenotype and grade of tumors when using rationally optimized pegRNAs. For example, KrasG12A-driven tumors exhibited a less advanced, more differentiated histopathology than KrasG12R and KrasG12D.
The significant tumor-initiating potential of KrasG12R is notable, given the rarity of KRASG12R in human non-small cell lung cancer patients61, but is consistent with prior results from Winters and colleagues15. Intriguingly, KRASG12R is known to have significantly impaired GTP hydrolysis relative to other KRASG12 mutants79. This property could enhance oncogenicity, yet KRASG12R is found at low frequency in most solid tumor types, except pancreatic cancer61. In pancreatic models, Zafra et al. (2020) previously found that KrasG12R mutations exhibit little to no PanIN formation potential in Trp53+/+ mice constitutively expressing KrasG12R in the pancreas, while KrasG12D promoted significant PanIN formation in most of the entire organ58. In contrast, transplanted KrasG12R;Trp53flox/flox organoids generated tumors with efficiency similar to KrasG12D;Trp53flox/flox organoids. These findings and our work suggest that mutation-specific properties may subject KRASG12R to especially potent tumor suppressive mechanisms that are lost in the context of concomitant Trp53 knockout specific to the murine experiments described. This warrants further investigation in the context of other genotypes (e.g., Trp53+/+) and experiments in which the sequence of mutations is temporally controlled.
We also observed Kras allele-specific responses to mutant-specific targeted therapies. For example, similar to prior studies of KRASG12C inhibitors58,80, we found that a KRASG12D inhibitor, MRTX1133, elicits a more powerful effect on prime edited KrasG12D pancreatic organoids when combined with the EGFR inhibitor, gefitinib. Several other clinical agents targeting a broader spectrum of oncogene mutations are undergoing clinical evaluation, and sotorasib and adagrasib, two KRASG12C inhibitors, have now been approved by the Food and Drug Administration60,62. PE GEMMs represent ideal systems for rapid interrogation of the effects of targeted therapies in the context of virtually any oncogenic mutation, including secondary resistance mutations like KRASY96C that are now being identified in patients. PE GEMMs also enable in vivo interrogation of these mutations in the context of syngeneic and immunocompetent mice. This broad utility for modeling Kras mutations in vivo is critical, as mutant KRAS inhibition has been shown to impact the tumor-immune microenvironment in models of colon cancer81,82 and may synergize with immune checkpoint blockade in other tissues not yet examined.
Beyond KRAS, we demonstrate in pancreatic organoids the precise installation and selection of two Trp53 dinucleotide substitutions encoding two mutant amino acid residues frequently observed at the same codon in human pancreatic cancer, as well as out-of-frame multinucleotide deletions at a nearby codon. Importantly, we observed over 90% editing purity after selection of all these mutations in vitro. Despite a high intended edit-to-unintended indel ratio, we also observed an unintended single nucleotide substitution at variable frequency when prime editing Trp53R245Q (Supplementary Fig. 8). We attribute this event to partial homology between the genomic region immediately following the reverse transcriptase template and the few nucleotides in the pegRNA scaffold that are commonly reverse-transcribed and excised during DNA repair, a prime editing intermediate noted by Nelson and colleagues51. Unintended edits such as this could be avoided by using an alternative pegRNA with a reverse transcriptase template ending a few nucleotides up or downstream to eliminate the homology, or could be reduced by introducing silent edits that prevent repeated editing of the same target site, as we demonstrated with the epegRNA encoding Trp53M240FS−14nt. This pegRNA is based on the same protospacer as Trp53R245Q, yet has a longer RTT and encodes a deletion that eliminates both the seed and PAM sequences. However, this phenomenon merits additional caution during pegRNA design and may be exacerbated in long-term prime editing experiments, such as when selecting cell lines over several passages with continuous expression of the prime editor and pegRNA.
The overall editing purity highlights the utility of prime editing for precise engineering of mutations with negligible indel byproducts. This is a key advantage over Cas9 HDR-based approaches, in which the high rate of indel byproducts could dilute intended point mutations in vitro and in vivo. Low editing purity could especially limit the study of specific point mutations in tumor suppressor genes, as unintended indels in these genes can produce frameshift mutations subject to positive selection. This limitation is especially important when considering that many genes, including TP53, often harbor point mutations that confer different properties relative to loss-of-function truncations, including gain-of-function effects68,83–85. For instance, Schulz-Heddergott and colleagues demonstrated that TP53R248Q exhibits a gain-of-function effect by hyperactivating the JAK2/STAT3 pathway, leading to more aggressive tumor progression in models of colon cancer68. These observations remain largely untested in models of pancreatic cancer in vivo due to a lack of suitable transgenic mouse models and human cell lines84. PE GEMMs are poised to fill critical gaps like this by allowing rapid and fine-tuned mutation control in a variety of tissue settings.
Though we did not explore them here, a variety of techniques are available to optimize prime editing efficiency, such as PE3 and PE3b editing strategies that combine nicking guides to bias DNA repair toward incorporation of prime edited nucleotides. Nevertheless, strategies based on single pegRNAs are more straightforward, have better multiplexing capacity because they rarely cause indels, and are better suited for high-throughput studies like genetic screens. In general, we found that spacer optimization and testing of up to 15 guides was sufficient to identify epegRNAs suitable for our experiments. We also found that silent or benign MMR-evasive edits close to the intended mutation reliably amplify prime editing efficiency by several fold, even for epegRNAs with optimized spacer sequences and PBS and RTT lengths. These techniques enabled us to identify epegRNAs that edit with greater than 20% efficiency across several cancer-associated genes. Future users should consider these and other strategies, including the co-delivery of an MLH1 dominant negative gene (PE4/5)29 or sensor-based pegRNA library approaches35, to maximize overall prime editing efficiencies, which may be especially helpful for in vivo applications.
We generally observed negligible off-target activity at computationally predicted loci, including one example with a protospacer identical to the intended target. This result corroborates the high on-target fidelity of prime editing. As established in prior studies24,29,51, additional homology required for repair using the reverse transcription product limits activity at off-target loci. While our results are consistent with prior literature, future studies could employ whole-genome sequencing to fully characterize off-target prime editing beyond a limited number of prioritized loci.
While we focused on installing somatic cancer driver mutations, we anticipate that PE GEMMs could be employed for broader applications. In principle, germline Rosa26PE2 alleles could be used to construct heritable mutations by modifying zygotes with pegRNAs encoding known drivers of inherited disease. We also envision sophisticated tumor modeling with the insertion of custom neoepitopes and other functional genetic sequences. These applications would enable investigators to address key questions in cancer genetics, immunology, and diverse genetic diseases while reducing the need to generate, genotype, and otherwise maintain traditional GEMMs. Finally, the combination of multiple epegRNAs in the context of a modified UPEC vector or LNP formulation should enable autochthonous generation of tumors defined by custom sets of multiple driver mutations in wild-type prime editor mice. This would enable increasingly complex studies of cooperating driver mutations. With these capabilities, PE GEMMs can provide a rapid preclinical avenue to complement both fundamental and clinical investigations aimed at treating cancer with novel precision treatment paradigms.
Methods:
Bioinformatic analysis of prime and base editor capabilities for modeling cancer-associated mutations.
We constructed a Python-based computational pipeline to compare the abilities of prime and base editors to model cancer-associated mutations. Data were retrieved from MSK-IMPACT datasets previously described35. The pipeline, all related scripts, and intermediate data needed to reproduce our results are available at https://github.com/samgould2/prime-vs-base-editing.
Analysis of recurrent cancer mutations incorporated in published transgenic mouse models.
To estimate the fraction of frequent cancer driver mutations captured by currently available transgenic mouse models, we used the MouseMine tool from the Mouse Genome Informatics database42,43 and obtained a comprehensive list of published transgenic alleles. We initiated our search using the mammalian phenotype code MP:0002006 (“neoplasm”) to retrieve all mouse models related to the study of cancer. We then modified the search by adding the following parameters: “Allele Type,” “Mutations (Name)”, “Alleles (Name and Molecular Note and Attribute string),” and “Subjects (synonyms → names).” We then filtered the results to retain only allele types annotated as “Targeted,” “Transgenic,” or “Endonuclease-mediated.”
After exporting these data (Supplementary Table 2), we then identified the 100 most frequent single nucleotide variants present in the MSK-IMPACT dataset. We then manually cross-referenced these two lists using standard Bash commands (e.g., grep to isolate relevant genes and codons) to identify available models representing specific mutations. In cases where models were absent in the MouseMine list, we performed a manual literature search to confirm an absence of models in the published literature. Using this approach, we designated for each mutation whether 1) any transgenic allele exists that can be used to model cancer in mice and 2) whether any existing models enable selective expression in a tissue of interest (e.g., through Cre recombinase-induced removal of a LoxP-STOP-LoxP cassette). The manually annotated results for the 100 most frequent mutations, as well as the entire MouseMine list are included as Supplementary Tables 1 and 2.
Design and cloning of the Cre-inducible prime editor allele.
The PE2-P2A-mNG Rosa26 targeting vector was generated with a backbone formed via BstBI and AscI restriction enzyme digestion of the SpCas9-NLS-P2A-EGFP Rosa26 targeting vector10,45. A fragment encoding the PE2 enzyme was generated by PCR amplification from the pCMV-PE2 plasmid obtained from Addgene24 (Addgene no. 132775), and a fragment containing the P2A-mNG sequence was amplified from a lentivirus plasmid encoding Cre-P2A-mNG. Two additional fragments containing WPRE-pA-PGK (Woodchuck Hepatitis Virus Posttranscriptional Regulatory Element-poly(A)-PGK promoter) and a neomycin resistance gene (NeoR-pA) were PCR-amplified from the SpCas9-NLS-P2A-EGFP vector. A FRT3 site was installed by incorporating overlapping portions of this motif into the PCR primers. All primers used are listed and described in Supplementary Table 5. A 5-part Gibson assembly reaction generated the final targeting vector using these components86. Successful assembly was confirmed with diagnostic restriction enzyme digestions and Sanger sequencing. The plasmid encoding the PE2-P2A-mNG Rosa26 targeting vector will be deposited at Addgene for distribution.
Embryonic stem cell targeting, validation, and chimera generation.
P4*, a C57BL/6J Trp53flox/flox (P) murine embryonic stem (ES) cell line, was generated by crossing a hormone primed C57Bl6J Trp53flox/flox female with a C57Bl6J KrasLSL-G12D;Trp53flox/flox male. At 3.5 days post coitum, blastocysts were flushed from the uterus, isolated, and cultured individually on a mouse embryonic fibroblast (MEF) feeder layer in ESCM+LIF+2i (Knockout DMEM (GIBCO), 15% FBS (Hyclone), 1% NEAA (Sigma), 2 mM Glutamine (GIBCO), 0.1 mM b-mercaptoethanol (SigmaAldrich) 50 IU Penicillin, 50 IU Streptomycin, 1000 U/ml LIF (Amsbio), 3 μM CHIR99021 (Abmole), and 1 μM PD0325901 (Abmole)). After 5–7 days in culture, the outgrown inner cell mass was isolated, trypsinized and replated on a fresh MEF layer. ES cell lines were genotyped for KrasLSL-G12D (P4* was wildtype +/+ for Kras), Trp53flox/flox, and Zfy (Y-chromosome specific). Primer sequences are available upon request. ES cell lines were tested for pluripotency by injection into host blastocysts from albino mice to generate chimeric mice.
Briefly, 36 μg of the prime editor targeting vector (R26–CAGG-LoxStopLox-Cas9(H840A)-MMLVRTP2A-mNeonGreen-WPRE-bHGpA; PGK-Neo-PGKpA) was linearized with PvuI, phenol/CHCl3 extracted, and then was ethanol precipitated. After resuspending the DNA in 150 μl of PBS it was mixed with 3 × 106 P4* ES cells in 650 μl of PBS in a 4 mm electroporation cuvette. The cell/DNA mixture was pulsed once in a Biorad Genepulsar 2 (00 V and 25 μF) followed by re-plating of the cells on irradiated mouse embryonic fibroblasts (MEFs). After 48 hours, the ES cell cultures were placed under selection with Geneticin (GIBCO) at 350 μg/mL. When colonies were large enough to pick, 45 colonies were manually picked using a stereomicroscope. Each clone was expanded and evaluated for correct integration by PCR with primers spanning the 5’ homology arm. Eleven PCR–positive clones were further evaluated using southern blot analysis. Briefly, genomic DNA was digested with EcoRV-HF (NEB) overnight. Digestions were electrophoresed on 0.7% agarose gels and blotted to Amersham Hybond XL nylon membranes (GE Healthcare). Samples were probed with 32P-labeled “Rosa26 3’ “external” and Cas9 “internal” probes applied in Church buffer (probe sequences available on request). Five clones (100A8, 100A10, 100C5, 100C7, and 100C8) displayed correct integration.
Correctly targeted clones verified by both PCR and southern blot analysis were injected into albino C57BL/6J blastocysts. High degree chimeras (visually assessed by coat color percentage) from the 100C7 and 100C8 ES cell clones successfully transmitted the prime editor allele through the germline.
Nucleofection of Neuro-2a cells and genomic DNA preparation
To evaluate spacers near the genetic locus encoding G12 in Kras, Neuro-2a cells were nucleofected using the SF Cell Line 4D-Nucleofector X Kit (Lonza) with 2 × 105 cells per sample (program DS-137). 800 ng of SpCas9-expressing plasmid and 200 ng of single guide RNA (sgRNA)-expressing plasmid were used according to the manufacturer’s protocol. Following nucleofection, samples were brought to 100 μL total volume with DMEM supplemented with GlutaMax (Thermo Fisher Scientific) and 10% (vol/vol) FBS (Gibco, qualified) at 37 °C. After incubation for 10 minutes at room temperature, the cells were seeded on 48-well poly-D-lysine coated plates (Corning). 3 days following nucleofection, the cells were washed with PBS after removing the media and then lysed by addition of 150 μL of freshly prepared lysis buffer (10 mM Tris-HCl, pH 8 at 23 °C; 0.05% SDS; 25 μg ml−1 of proteinase K (Qiagen)). Lysates were incubated at 37 °C for one hour and then heat-inactivated for 30 min at 80 °C. The Kras amplicon was amplified from the genomic DNA samples, sequenced on an Illumina MiSeq, and analyzed with CRISPResso287 for indel quantification as previously described37. Primers used for amplification of the Kras amplicon are listed in Supplementary Table 5.
pegRNA design and cloning.
pegRNAs were designed in part using the pegRNA design tool, Prime Design88. In some cases (e.g., editing at KrasG12D), CRISPR sgRNAs were tested prior to pegRNA design to select spacers that exhibited the highest level of Cas9 activity. For some designs (e.g., all Trp53-targeted pegRNAs), the trimmed evopreQ1 motif was included to form epegRNAs and optimize editing efficiency within a limited cohort of initial candidates51. pegRNAs and their sequences are provided in Supplementary Table 3. Generally, the editing efficiency of all Trp53-targeted pegRNAs was tested first in single replicates in split-PE2 Trp53+/+ 3TZ cells, while Kras-targeted and other pegRNAs were tested in Rosa26PE2/+ tail tip-derived fibroblasts. pegRNAs with the highest editing efficiencies were selected for later assays in organoids and mice.
All pegRNAs were tested within the context of hU6-pegRNA-EF-1α-Cre (UPEC) or hU6-RFP/pegRNA-EFS-mScarlet (UPEmS) vectors. All pegRNA-expressing vectors were assembled via Golden Gate Assembly89 using the uncut template plasmid and three annealed oligo pairs consisting of the spacer sequence, the scaffold, and the 3’ extension, all with compatible overhangs. Assembly was facilitated using the Golden Gate Assembly Kit (BsmBI-v2) from New England BioLabs. Plasmid DNA was extracted from transformed bacterial cultures using QIAprep Miniprep or Midiprep kits according to the manufacturer’s instructions.
The UPEmS template vector was generated via Gibson assembly of three insert fragments and a linearized backbone. Two fragments were formed by PCR amplification from the “pU6 pegRNA GG acceptor” plasmid (Addgene plasmid no.132777)24. Specifically, the hU6 promoter was amplified using primers modified to install a BsmBI recognition site and the pAF Gibson adapter sequence on either side of the promoter (pAF-hU6-BsmBI), and the red fluorescent protein (RFP) component was also amplified in part using a primer that installed another BsmBI recognition site (forming BsmBI-RFP-BsmBI-pAR/gBF). A third fragment, gAR/pBF-EFS-mScarlet-gBR, was amplified from a separate lentiviral plasmid containing U6-sgRNA-EFS-mScarlet. All fragments were designed to contain compatible overhangs for Gibson assembly. All vectors with detailed maps and sequences will be deposited into Addgene. All primer names and sequences are listed in Supplementary Table 5.
The UPEC template plasmid (hU6-RFP-EF-1ɑ-Cre; hU6-pegRNA-EF-1ɑ-Cre after cloning) was developed by Gibson assembly of two insert fragments and the same backbone used to clone pUPEmS. The pBF-EF-1alpha-Cre-gBR fragment was generated using pBF and gBR PCR primers targeting the pUSEC (U6-sgRNA-EF-1alpha-Cre) vector86,90. The pAF-U6-RFP-gAR fragment was amplified from the UPEmS vector. The final UPEC vector enables concomitant assembly of the pegRNA spacer, scaffold, and 3’ extension. Plasmids encoding the template UPEC and UPEmS vectors used for Golden Gate cloning will be deposited at Addgene.
Generation of tail tip-derived Rosa26PE2/+ fibroblasts.
To generate Rosa26PE2 cell lines for convenient testing of pegRNAs, a 2 cm piece was excised from the tail tip of an anesthetized, 3.5-week-old male. The sample was sprayed with ethanol and then dipped in PBS several times. A lengthwise incision was made, and outside skin and hair were removed. The sample was then incubated at 37 °C in digestion buffer comprised of 5 mL DMEM, 25 μL penicillin-streptomycin, 5 μL Amphotericin B, 10 μL DNase (40 U/mL −20 °C; 1:500), 50 μL collagenase (100 mg / mL; 1:100), and 50 μL CaCl2 (36 mM; 1:100). Samples were then washed twice with PBS, and dissociated chunks were added to a 6 cm dish. Additional media containing Amphotericin B was added the following day.
HEK293 and fibroblast cell culture conditions.
HEK293-FS, split-PE2 3TZ and tail tip-derived Rosa26PE2/+ fibroblast cells were cultured in standard media consisting of Dulbecco’s Modified Eagle’s Medium (DMEM) (Corning), penicillin-streptomycin, and 10% (volume/volume) fetal bovine serum (FBS). All cultured cells were incubated at 37 °C and 5% CO2.
Pancreatic ductal organoid isolation and culture.
Pancreata from mice of the desired genotype were dissected manually and minced with a razor blade. Pancreas tissue was then dissociated by 20 minutes of gentle agitation in pancreas digestion buffer [1X PBS (Corning), 125 U/ml collagenase IV (Worthington)] at 37 °C. Tissue suspensions were then strained through 70 μM filters, washed with 1X PBS, and pelleted with slow deceleration by centrifugation. Cells were resuspended in 100% Matrigel (Corning) and plated as 50 μL domes into 24-well plates (GenClone). Upon solidification of domes, cells were cultured in organoid complete media as previously described47, or alternatively, in complete medium as follows: AdDMEM/F12 medium supplemented with HEPES (1x, Invitrogen), GlutaMAX (1x, Invitrogen), penicillin/streptomycin (1x, VWR), B27 (1x Invitrogen), R-Spondin1-Conditioned Medium (10% v/v), A83–01 (0.5 μM, Tocris), murine Epidermal Growth Factor (mEGF, 0.05 μg/mL, PeproTech), Fibroblast Growth Factor 10 (FGF-10, 0.1μg/mL, PeproTech), Gastrin I (0.01 μM, Tocris), recombinant murine Noggin (0.1 μg/mL, PeproTech), N-acetyl-L-cysteine (1.25 mM, Sigma-Aldrich), Nicotinamide (10 mM, Sigma-Aldrich) and Y-27632 (10.5 μM, Cayman Chemical Company).
Organoids were passaged using TrypLE Express (Life Technologies) for Matrigel digestion for 15–30 minutes at 37 °C. Organoids were passaged for at least four passages prior to treatment with lentivirus. Organoids were infected at a high multiplicity of infection to ensure 100% recombination. Briefly, concentrated lentivirus (either diluted 1:9 or undiluted) was introduced to cell suspensions at the time of passage. Fluorescence microscopy confirmed Cre-mediated expression of mNG in organoids treated with UPEC, or mScarlet expression in organoids treated with UPEmS. For Trp53flox/flox lines, Nutlin-3a was added to organoid media (10 μM, Sigma-Aldrich) to ensure purification of recombined organoids. For prime edited organoids harboring KrasG12D or KrasG12C mutations, organoids were cultured in the presence of 1 μM Gefitinib in full organoid media (Cayman) to select for the intended edit. Sotorasib (Selleck) was added to media at 1, 2, and 5 μM for experiments as indicated in the main text. MRTX1133 (MedChem) was added to the media at 2 or 5 μM as indicated in the main text. Selection of prime edited mutations was confirmed by deep amplicon sequencing of organoids several days after the initial infection with lentivirus, and then again after several passages under treatment with drug. For selection of transgene-containing cells from chimera-derived pancreatic organoids, organoids were treated with 800 μg / mL of Geneticin (GIBCO). All drug stocks were dissolved in DMSO (Sigma-Aldrich). Pancreatic organoids were maintained in culture for <30 passages.
Organoid viability and proliferation were quantified using the alamarBlue HS Cell Viability Reagent (ThermoFisher Scientific). Viability reagent was directly added to organoid culture at 1/10 media volume. After 24 hours, 200 μl of reagent-containing media was removed and assayed in replicate in a Tecan Infinite Pro m200 using the manufacturer’s parameters. For time course experiments, media containing 10% viability reagent was replenished after 1 PBS wash.
Lung organoid culture.
Lung organoids were cultured as previously described91. Briefly, lungs were derived from 8–20-week-old mice and transferred into 500μL dispase and minced. 3–5 mL digestion buffer containing Advanced DMEM/F-12, Penicillin/Streptomycin, Amphotericin B, 1 mg/mL Collagenase (Sigma, C9407–500MG), 40 U/mL DNase I (Roche, 10104159001), 5 μM HEPES, and 0.36 mM CaCl2 was added for a 20–60 minute incubation at 37 °C in a rotating oven. The resulting suspension was incubated in 1 mL ACK Lysis Buffer (Thermo, A1049201) for 3–5 minutes at room temperature to lyse red blood cells. Samples were then washed two times with fluorescence-activated cell sorting (FACS) buffer (1X PBS with 1 mM EDTA and 0.1% BSA) and filtered through 40μm mesh to remove chunks. Samples were resuspended in 150 μL FACS buffer and CD45 cells were depleted using the EasySep™ Mouse CD45 Positive Selection kit (STEMCELL technologies, 18945) and preserving the flow through. Cells were then resuspended in FACS buffer containing 1x PBS, 0.1% BSA, and 2 mM EDTA and were stained with anti-mouse CD31-APC (1:500, Biolegend, 102507), CD45-APC (1:500, BD Biosciences, 559864), EpCAM-PE (1:500, Biolegend, 118206), MHCII-APC-eFluor-780 (1:500, Thermo, 47–5321-82) on ice for 30 minutes and then resuspended in FACS buffer containing DAPI (1μg/mL, Thermo, D1306). The suspensions were then sorted for DAPI-, CD31-, CD45-, EpCAM+, MHCII+ cells. Approximately 20,000 sorted AT2 cells were mixed with Growth Factor Reduced Matrigel (Corning) at a ratio of 1:9 and seeded onto multi-well plates as 20μL drops. The drops were incubated at 37 °C for 15 minutes to solidify, and then overlaid with F7NHCS medium supplemented with Y-27632 (Cayman). The cultures were maintained in a humidified 37 °C / 5% CO2 incubator at ambient O2 pressure. Media was replenished every 3–4 days using F7NHCS medium without Y-27632 and organoids were passaged 6–12 days after plating. For passaging, matrigel drops were dissolved in TrypLE Express (Sigma, 12604–013) and incubated at 37 °C for 7–15 minutes. The organoid suspensions were then dissociated into single cells by vigorous pipetting, washed twice, resuspended in 1x PBS, and plated as described above. We typically plated 10,000 cells per drop.
Generation of a 3TZ fibroblast cell line with the PE2 enzyme expressed from split inteins.
A cell line based on murine 3TZs cells was developed to test Trp53-targeted pegRNAs on a Trp53+/+ background. Two plasmids containing halves of the PE2 enzyme and distinct antibiotic resistance genes were generated via Gibson Assembly. The split intein-based constructs described by Anzalone et al. (2019) were utilized to enable post-translational splicing of the intein motifs and subsequent joining of the halves to form the full PE2 enzyme24,92. Specifically, the N-terminal half of PE2 (the first 573 amino acids of the Cas9 nickase joined to the Npu N-intein) was PCR-amplified from the U6-DNMT1-hSynapsin-PE2-Nterminal-P2A-EGFP-KASH-lenti plasmid (Addgene no. 135955) and then cloned into a puromycin resistance gene-containing backbone. A blasticidin resistance gene-containing backbone was assembled into a second vector with a PCR-amplified DNA fragment encoding the C-terminal half of PE2 (Npu C-intein joined to the remaining C-terminal half of PE2), amplified from the hSynapsin-PE2-Cterminal-lenti plasmid (Addgene no. 135956).
After Gibson assembly and subsequent plasmid validation by Sanger sequencing, the two constructs were incorporated into lentivirus, which was used to transduce murine 3TZ fibroblast cells. Transduced cells were then incubated for 48 hours. After 48 hours, both infected and control cells were plated into 12-well plates and treated with a range of dilutions of both puromycin and blasticidin, maximally delivered at 10 μg / mL and 20 μg / mL, respectively. After confirming lentivirus-mediated resistance to both antibiotics in infected cells, a polyclonal cell line was maintained generally with standard media containing 8 μg / mL of puromycin and 12 μg / mL of blasticidin. Prime editing in the resulting split-PE2 3TZ cells was confirmed using the Dnmt1 pegRNA described in Figs. 2–3 as a positive control (Supplementary Figure 12). This cell line was then used to test cohorts of pegRNAs targeted to Trp53.
Production of lentivirus and transduction.
Lentivirus was produced by transfection of the expression vector into 293FS* cells along with psPAX2 (psPAX2 was a gift from Didier Trono - Addgene plasmid # 12260 ; http://n2t.net/addgene:12260 ; RRID:Addgene_12260) and pMD2.G (pMD2.G was a gift from Didier Trono - Addgene plasmid # 12259 ; http://n2t.net/addgene:12259 ; RRID:Addgene_12259) packaging plasmids at a 4:3:1 ratio using polyethylenimine or Mirus transfection reagent. For small-scale transfections, 4 × 105 cells were seeded per well in a 6-well plate (Corning). 24 hours after seeding, cells were transfected using 4 μL PEI or Mirus (Mirus Bio) according to the manufacturer’s specifications. For large-scale transfections, 7.5 × 106 cells were seeded in 15 cm plates (Corning). 24 hours after seeding, cells were transfected using 80 μl Mirus or PEI. Media was replaced 24 hours after transfection. 48–72 hours post-transfection, viral supernatant was collected and filtered through 0.45 μm filters. Large-scale virus was concentrated by ultracentrifugation at 25,000 rpm at 4°C and then resuspended in Opti-MEM (Thermo-Fisher). 1 mL of small-scale viral supernatant was added directly to 1 × 10^5 cells at seeding in a 6-well plate (Corning) for transduction. Small-scale transductions were supplemented with polybrene (10 mg/mL, 1:1000, Sigma). Concentrated large-scale lentivirus and small-scale virus were stored at −80° C if not used immediately. Generally, cell lines were infected with small-scale virus while organoids were infected with large-scale virus.
Lentivirus titer assay.
Quantification of lentiviral titer was performed as previously described using a GFP Cre reporter 3TZ cell line14. Briefly, concentrated lentivirus was serially diluted and added to 10,000 cells. 48 hours post-infection, cells were trypsinized and analyzed to calculate the percentage of GFP positivity by flow cytometry.
Intratracheal delivery of lentivirus into the lung.
Intratracheal lentiviral infection has been previously described93. Mice were anesthetized in an isoflurane chamber. 6 × 104 TU and 1 X 105 TU of lentivirus containing UPEC vectors encoding pegRNAs and Cre recombinase were injected intratracheally into Trp53flox/flox;Rosa26PE2/+, Trp53+/+;Rosa26PE2/+, or Trp53flox/flox;Rosa26PE2/PE2 mice. Animals were monitored after injection to confirm recovery. Mice were at least six weeks old at the time of infection. Mice were sex and age matched within 4 weeks across experimental arms.
Orthotopic transplantation of pancreatic organoids.
Orthotopic transplants of pancreatic organoids were performed with minor modifications from previously described protocols47,94,95. Briefly, animals were anesthetized with Isoflurane, the left abdominal side was depilated with Nair, and the surgical region was disinfected with Chloraprep swabstick (BD). A small incision (~1.5cm) was made in the left subcostal area, and the spleen and pancreas were exteriorized with ring forceps. The organoid suspension (containing 1×105 organoid cells in 100μL of 50% PBS + 50% Matrigel) was injected using a 30-gauge needle into the pancreatic parenchyma parallel to the main pancreatic artery. Successful injection was verified by the appearance of a fluid bubble without signs of intraperitoneal leakage. The pancreas and spleen were gently internalized, and the peritoneal and skin layers were sutured independently using 4/0 PGA suture and 4/0 silk suture respectively (AD Surgical). All mice received pre-operative analgesia Buprenorphine Sustained-Release (Bup-SR, 0.5mg/kg) and were followed post-operatively for any signs of distress. Organoid/Matrigel mixtures were kept on ice throughout the whole procedure to avoid solidification. For orthotopic transplantation, syngeneic C57BL/6J Rosa26PE2 mice (aged 6–17 weeks) were used as recipients. Male pancreatic organoids were only transplanted into male recipients.
Autochthonous pancreatic tumor modeling.
Retrograde pancreatic duct infection with lentivirus was modified from previously reported techniques77. Briefly, the ventral abdomen was depilated (using Nair) 1 day prior to surgery. Animals were anesthetized with Isoflurane. The surgical area was disinfected with betadine/isopropanol and a 2–3 cm incision was made in the anterior abdomen. A subsequent vertical incision was made through the abdominal wall, securing the incision edges with a Colibri retractor. A Nikon stereomicroscope was used to visualize the pancreas, common bile duct and sphincter of Oddi. The common bile duct and cystic duct were gently separated from the portal vein and hepatic artery using blunt dissection with Moria forceps. A microclip was placed over the common bile duct to prevent influx of the viral particles into the liver or gallbladder. A 30-gauge needle was used to cannulate the common bile duct at the level of the sphincter of Oddi, and 150 μL virus was injected over 30s. After injection and removal of instruments, the peritoneum was closed using running 5–0 Vicryl sutures. The abdominal wall and fascia were closed using simple interrupted 5–0 Vicryl sutures. Animals were administered post-operative sustained-release Buprenorphine (Bup-SR) and were monitored post-operatively for signs of discomfort or distress. For retrograde pancreatic ductal installation, male mice (aged 8–20 weeks) and female mice (aged 8–20 weeks) were transduced with 500,000 TU (transducing units, see viral titering) in serum-free media (Opti-MEM; Gibco).
Lipid Nanoparticle (LNP) formulation and injection
Lipid nanoparticles were formulated as previously described53 with slight modifications. Dnmt1+GGG Synthetic pegRNA (1 mg) was ordered from Agilent Technologies. The first three and last three nucleotides were modified with 2’O-Methyl groups. The first three and last three nucleotide bonds were phosphorothioate-modified bonds. Both modifications were made to increase stability of the guide. Cre mRNA was obtained from Trilink. A weight ratio of 1:7.5 total mRNA ionizable lipid was used for lipid nanoparticle formulation, with a 1:2 ratio of Cre mRNA : pegRNA. The aqueous phase was prepared with 25 mM sodium acetate (pH 5.2), Cre mRNA and pegRNA solution. Two organic phase preparations were made by adding an ionizable lipid (Lipid 10 or 306-O12B) to cholesterol (Sigma-Aldrich), DOPC (Avanti), and DMG-PEG (Sunbright) stock solutions in 100% ethanol, at a 50:38.5:10:1.5 molar ratio. Nanoparticles were prepared by combining the organic and aqueous phases at a 1:3 ratio, and assembled using a NanoAssemblr (Precision Nanosystems). LNPs were dialyzed for 4 hours against PBS in ThermoScientific Slide-A- Lyzer dialysis Cassettes (3.5 K MWCO). LNPs were kept on ice prior to animal dosing. Mice were administered a maximal dose of 60 μg total RNA via tail vein injection, corresponding to roughly 200 μl per mouse.
Animal studies.
All mouse experiments described in this study were approved by the Massachusetts Institute of Technology Institutional Animal Care and Use Committee (IACUC) (institutional animal welfare assurance no. A-3125–01. KrasLSL-G12D and Trp53flox/flox mice have previously been described74,96. All mice were maintained on a pure C57Bl/6 background. For Vilin-CreERT2;Rosa26PE2/+ animals, Tamoxifen was administered in the diet (Envigo TD.130860) for 2 weeks prior to tissue collection. Mice with the appropriate genotypes and between 7–20 weeks were chosen for in vivo experiments. Mice of both sexes were used for autochthonous lung tumor initiation, and male mice were chosen for orthotopic pancreatic organoid experiments as the transplanted organoid line was male-derived. Mice were assessed for morbidity according to guidelines set by the MIT Division of Comparative Medicine and were humanely sacrificed prior to natural expiration.
Genotyping.
Mouse genotyping was performed on genomic DNA isolated from mouse ear tissue. Genotyping primers are listed in Supplementary Table 5.
Ultrasound imaging.
Observation of murine pancreatic tumors with high resolution ultrasound was performed as described47,97. Animals were anesthetized with Isoflurane and the left subcostal region of animals was depilated with Nair. Animals were imaged with a Vevo3100/LAZRX ultrasound and photoacoustic imaging system (Fujifilm-Visualsonics). Anesthetized animals were positioned supine and angled on an imaging platform for visualization of peritoneal organs. Landmark organs including kidney and spleen were first identified prior to imaging. A thin layer of ultrasound gel was applied over the depilated region of the abdomen. The transducer (VisualSonics 550S) was positioned above the abdomen and set at the scanning midpoint of the healthy pancreas or tumor. Approximately 1 cm of scanning area was used to capture the entirety of pancreas tumors, using a Z-slick thickness of 0.04 mm. Ultrasound scans were uploaded to Vevo Lab Software, from which representative images were exported.
Rodent μCT.
Control and tumor bearing mice were anesthetized by Isoflurane (3%, then maintained at 2.0–2.5% in oxygen – VetEquip, Livermore, CA) and scanned in a prone position using a Skyscan 1276 (Bruker Corp, Billerica, MA) with the following parameters: 100kVp source voltage, 200 uA current, 0.5mm Aluminum X-ray filter, 108ms exposure time, 0.65 degree rotational step size over 360 degrees in a continuous rotation. With 4×4 detector binning, the nominal pixel size after reconstruction (Bruker NRecon software) was 40.16 micron. Data were visualized using ImageJ.
Histology, immunohistochemistry, and immunofluorescence
Upon sacrifice, pancreata from control and tumor-bearing animals were manually dissected from the peritoneal cavity. A tissue dye (Davidson) was used to mark tumor regions for histology. Tumor-bearing lung was flushed with 1X PBS and separated into separate lobes. Tissue was fixed in Zinc Formalin overnight, transferred to 70% Ethanol, and then embedded in paraffin. Hematoxylin and eosin (H&E) staining was performed per a standard protocol by the Hope Babette Tang Histology Facility at the Koch Institute. Digitally scanned images of H&E slides were obtained with an Aperio ScanScope at 20X magnification. Digital slides were uploaded to the Aiforia™ image analysis software. Histologic quantification of tumor grade was performed by an automated deep neural network and in consultation with veterinary pathologist Dr. Roderick Bronson. Training of the Aiforia algorithm has been previously described73. For grade calling, the nsclc_v25 algorithm was used.
Lungs were removed and inflated with Zinc formalin, then left in approximately 5 mL Zinc formalin for 24 hours. Afterwards, samples were switched to 70% ethanol, and later processed for paraffin embedding. Sections were cut at 4 μm and dewaxed. Antigen retrieval was performed using citrate buffer (pH = 6.0).
For IHC, Endogenous peroxidase was blocked using DAKO Dual Endogenous Enzyme Block, and endogenous species protein was blocked using 2.5% Horse Serum (Vecta). Slides were incubated at 4 °C overnight with the following antibodies: anti-NKX2–1 (1:1000, abcam, Cat# ab76013; RRID:AB_1310784), anti-SFTPC (1:5000, Millipore Sigma, Cat# AB3786; RRID:AB_91588), and anti-HMGA2 (1:1000, Cell Signaling Technologies, Cat# 8179S; RRID:AB_11178942). ImPRESS anti-Rabbit horseradish peroxidase and DAB Peroxidase Substrate Kit (Vector) were used to develop slides. Tissues were counterstained with hematoxylin. Slides were digitally scanned and analyzed using QuPath98.
For IF, permeabilization was performed with 0.5% Triton X-100 in PBS. Endogenous species protein was blocked using 2.5% Horse Serum (Vecta). Slides were incubated at 4° C with anti-Cas9 (E7M1H, 1:100, CST Cat#19526). Horse anti-Rabbit secondary (AF488, 1:400) was used. All slides were counterstained with DAPI (1:20,000), and imaged using a Nikon 80 Eclipse 80i fluorescence microscope using 10x and 20x objectives and an attached Andor camera.
Isolation of genomic DNA.
Genomic DNA was extracted from organoids or tissue using either the KAPA Express Extract kit (Sigma) or the Qiagen Puregene Core Kit A. Briefly, for extraction with KAPA Express, dissociated cells were pelleted, washed with PBS, and pelleted again prior to resuspension (per condition) in 100 μL consisting of 88 μL PCR-grade water, 10 μL of 10X KAPA Express Extract Buffer, and 2 μL of KAPA Express Extract enzyme. This and subsequent steps were followed per the manufacturer’s instructions. For the Qiagen Puregene Core Kit A, briefly, minced tumors or dissociated cells were pelleted and resuspended in Qiagen Cell Lysis Solution and vortexed. Protein and ethanol precipitation were then performed to isolate pure genomic DNA, which was used for PCR amplification, all steps executed according to the manufacturer’s instructions. Genomic DNA was then stored at 4 °C or −20 °C until use.
DNA sequencing and analysis of genomic DNA samples.
Target loci were amplified from genomic DNA using PCR primers listed and described in Supplementary Table 5. Amplicons were then purified using either agarose gel extraction or using a QIAquick PCR purification kit (Qiagen). Purified amplicons were typically then submitted to the Massachusetts General Hospital Center for Computational and Integrative Biology’s DNA Core for next-generation sequencing (samples prepared according to guidelines provided for the CRISPR Sequencing service).
Amplicons prepared for evaluating prime editing efficiency of the initial Trp53245- and Kras96-targeted pegRNAs described in Supplementary Figs. 7 and 9 were sequenced as previously described18,99. Following amplification from genomic DNA, samples were given unique Illumina TruSeq barcodes for pooled sequencing. Barcoded PCR products were pooled and purified by electrophoresis with a 2% agarose gel using a Gel Extraction Kit (QIAGEN), eluting with 30 μL H2O. DNA concentration was quantified using a Qubit dsDNA High Sensitivity Assay Kit (ThermoFisher Scientific) and sequenced on an Illumina MiSeq instrument (single-end read, 250–300 cycles) according to the manufacturer’s protocols.
Sequencing reads stored in FASTQ files were aligned to reference amplicons and analyzed using the deep sequencing analysis program, CRISPResso287, V2.2.6, as a command-line tool in prime editing mode. General CRISPResso2 parameters employed for each target are described in Supplementary Note 1. The --split_interleaved_input parameter was included for paired-end data. Generally, prime editing efficiency was calculated as the percentage of reads aligning to the prime edited amplicon (excluding indels) relative to all reads aligning to both the prime edited and reference amplicons (including indels). Only reads with an average phred score ≥30 were considered44. Indel percentages were calculated in similar fashion using the total number of indel-bearing reads designated as “discarded” by CRISPResso2. In rare cases containing contaminant reads from off-target amplicons, contaminating reads were removed in silico by requiring every analyzed read to possess 60–90% homology to the reference amplicon. For experiments involving pegRNAs that alter multiple nucleotides, the allele frequency tables output by CRISPResso2 were consulted to confirm that the majority of prime edited reads contained all of the intended nucleotide alterations.
Sequencing and analysis of off-target loci.
Off-target loci were computationally identified using Cas-OFFinder52. For each of the four indicated protospacers (Supplementary Table 4), all off-target sites with three or fewer mismatches relative to the target protospacer were identified. Bulges were not permitted. These results were then filtered down to up to four off-target loci per protospacer. First, loci were prioritized by selecting candidates with the lowest number of protospacer mismatches. When loci contained the same number of protospacer mismatches, off-target sites were ranked by the lowest number of primer binding site mismatches, as this sequence would be critical to enable off-target prime editing. Finally, if loci contained the same number of both protospacer and primer binding site mismatches, the difference in DNA melting temperature between the mismatched target and the original primer binding site sequence was computed. This calculation was performed with the OligoAnalyzer Tool, version 3.1, from Integrated DNA Technologies100 with default parameters. Loci with the smallest difference in melting temperature compared to the non-mismatched strand were then prioritized. This was used to estimate the relative thermodynamic cost incurred among different mismatches. The off-target loci identified by this analysis are described in Supplementary Table 4.
For each off-target locus, amplicons were generated as described for on-target loci. Next-generation sequencing was also performed as described for on-target loci. Off-target prime editing was assessed by aligning sequencing reads to off-target amplicons using CRISPResso2 in batch mode87. The parameters “-w 20” and “-q 30” were used in all cases, along with the corresponding off-target protospacer as the guide RNA input sequence. Prime editing was then assessed using an approach described by Chen et al. (2021)29. For each sample, the “Nucleotide_percentage_summary” file output by CRISPResso2 was used to compare the DNA sequence immediately downstream of the nick site to the sequence encoded by the pegRNA. The first mismatched nucleotide site was then examined to quantify the percentage of reads bearing the allele encoded by the pegRNA. For each off-target locus, this percentage of prime edited reads was compared between samples cultured in the presence of the examined pegRNA versus samples cultured in the presence of an irrelevant pegRNA. All off-target samples were sequenced within the same MiSeq run to ensure a similar sequencing error rate between tested and control groups.
Immunoblotting.
Pancreas organoids were dissociated with TrypLE for 30 minutes at 37 °C, washed 6x with PBS, and then lysed in cell lysis buffer (RIPA with 100X HALT protease and phosphatase inhibitors). Protein concentration was quantified with the bicinchoninic acid (BCA) assay (Pierce) and boiled in 4x Laemmli Buffer + 355μM 2-mercaptoethanol for five minutes. 15 μg of reduced samples were loaded onto 4–12% Bis-Tris SDS-PAGE gels (Invitrogen) and run at 100V. Gels were transferred onto a PVDF membrane (60 V, 3 hours) and blocked with PBST (PBS + 0.5% Tween20) + 5% milk for one hour. Blots were incubated with primary antibody (p53 clone 1C12, Cell Signaling Technology (CST), β-Actin clone 13E5, CST, 1:5000) overnight at 4 °C, washed 3 times in PBST, incubated with HRP-conjugated secondary antibody in 5% milk for one hour at room temperature, and then washed 3 times in PBST. Blots were developed with Clarity or Clarity-Max ECL substrate (BioRad) and imaged on a ChemiDoc Gel Imaging System (BioRad).
Mutation frequency analysis using cBioPortal.
Somatic mutation frequencies of TP53 in human pancreatic cancer were retrieved from cBioPortal66,67 by querying this gene from four patient cohorts: CPTAC, TCGA (Firehose Legacy), QCMG, and ICGC. Overall mutation frequency was calculated as the fraction of patients possessing mutations at a specific codon divided by the total number of patients for which TP53 mutation status had been profiled. The same analysis was applied for human lung adenocarcinoma, with the following patient cohorts: Non-Small Cell Lung Cancer101 (TRACERx, NEJM and Nature 2017) and Pan-Lung Cancer102 (TCGA, Nat Genet 2016).
Supplementary Material
Acknowledgments:
We thank the entire Jacks Laboratory with particular thanks to Demi Sandel, Carla Concepcion, Megan Burger, William Hwang, Morgan Heileman, and Alex Jaeger for useful discussions. We also thank Karen Yee, Judy Teixeira, Katherine Anderson, and Margaret Magendantz for administrative support. We thank Steve Murray, Roderick Bronson, Grace Johnson, Jeffrey Kuhn, and Milton Cornwall-Brady for helpful advice. S.I.G. and F.J.S.R. thank Edward Kastenhuber for useful advice on human and mouse ortholog analyses, as well as David Solit, Niki Schultz, Michael Berger, and Benjamin Gross for access to MSK-IMPACT data. Z.A.E. acknowledges support from the Ludwig Center at MIT and the National Cancer Institute of the National Institutes of Health under Award Number F31CA268835. S.I.G. was supported by T32GM136540. F.J.S.R. is a HHMI Hanna Gray Fellow and was supported by the V Foundation for Cancer Research (V2022-028), NCI Cancer Center Support Grant P30-CA1405, the Ludwig Center at MIT (2036636), Koch Institute Frontier Awards (2036648 and 2036642), and the MIT Research Support Committee (3189800). G.A.N. was supported by a Helen Hay Whitney Postdoctoral Fellowship and K99 award HL163805. We also thank the Koch Institute Swanson Biotechnology Center for technical support, especially the Histology core facility. This work was also supported by the Howard Hughes Medical Institute, David H. Koch Graduate Fellowship Fund, NCI Cancer Center Support Grant P30-CA1405, NIH Pre-Doctoral Training Grant T32GM007287, NIH grants U01 AI142756, RM1 HG009490, R35 GM118062, and the Lustgarten Foundation for Pancreatic Cancer Research.
Footnotes
Declaration of Interests:
T.J. is a member of the Board of Directors of Amgen and Thermo Fisher Scientific, and a co-Founder of Dragonfly Therapeutics and T2 Biosystems. T.J. serves on the Scientific Advisory Board of Dragonfly Therapeutics, SQZ Biotech, and Skyhawk Therapeutics. T.J. is also the President of Break Through Cancer. None of these affiliations represent a conflict of interest with respect to the design or execution of this study or interpretation of data presented in this manuscript. His laboratory currently receives funding from the Johnson & Johnson Lung Cancer Initiative, but this funding did not support the research described in this manuscript. D.R.L. is a consultant for Prime Medicine, Beam Therapeutics, Pairwise Plants, Chroma Medicine, and Nvelop Therapeutics, companies that use or deliver genome editing or genome engineering agents and owns equity in these companies. K.H. and A.V.A. are currently employees of Prime Medicine.
Data availability:
Rosa26PE2 mice on wild-type (JAX Stock Number: JR037953) and Trp53flox/flox (JAX stock number: JR037954) backgrounds are available from the Jackson Laboratory. Plasmids will be made available through Addgene upon publication. All other materials and data are available from the corresponding author upon reasonable request.
References:
- 1.Vogelstein B. et al. Cancer Genome Landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garraway LA & Lander ES Lessons from the cancer genome. Cell 153, 17–37 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Hyman DM, Taylor BS & Baselga J. Implementing Genome-Driven Oncology. Cell 168, 584–599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang MT et al. Accelerating Discovery of Functional Mutant Alleles in Cancer. Cancer Discov 8, 174–183 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong DS et al. KRASG12C Inhibition with Sotorasib in Advanced Solid Tumors. New England Journal of Medicine 383, 1207–1217 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch TJ et al. Activating Mutations in the Epidermal Growth Factor Receptor Underlying Responsiveness of Non–Small-Cell Lung Cancer to Gefitinib. New England Journal of Medicine 350, 2129–2139 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Hill W, Caswell DR & Swanton C. Capturing cancer evolution using genetically engineered mouse models (GEMMs). Trends in Cell Biology 31, 1007–1018 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Kersten K, de Visser KE, van Miltenburg MH & Jonkers J. Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol Med 9, 137–153 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zehir A. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 23, 703–713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Platt RJ et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Annunziato S. et al. In situ CRISPR-Cas9 base editing for the development of genetically engineered mouse models of breast cancer. EMBO J 39, e102169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han T. et al. R-Spondin chromosome rearrangements drive Wnt-dependent tumour initiation and maintenance in the intestine. Nat Commun 8, 15945 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dow LE et al. Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol 33, 390–394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sánchez-Rivera FJ et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 516, 428–431 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winters IP et al. Multiplexed in vivo homology-directed repair and tumor barcoding enables parallel quantification of Kras variant oncogenicity. Nat Commun 8, 2053 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leibowitz ML et al. Chromothripsis as an on-target consequence of CRISPR–Cas9 genome editing. Nat Genet 53, 895–905 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blair LM et al. Oncogenic context shapes the fitness landscape of tumor suppression. http://biorxiv.org/lookup/doi/10.1101/2022.10.24.511787 doi: 10.1101/2022.10.24.511787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurt IC et al. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat Biotechnol 39, 41–46 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koblan LW et al. Efficient C•G-to-G•C base editors developed using CRISPRi screens, target-library analysis, and machine learning. Nat Biotechnol 39, 1414–1425 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lam DK et al. Improved cytosine base editors generated from TadA variants. Nat Biotechnol 1–12 (2023) doi: 10.1038/s41587-022-01611-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tong H. et al. Programmable A-to-Y base editing by fusing an adenine base editor with an N-methylpurine DNA glycosylase. Nat Biotechnol 1–5 (2023) doi: 10.1038/s41587-022-01595-6. [DOI] [PubMed] [Google Scholar]
- 23.Anzalone AV, Koblan LW & Liu DR Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol 38, 824–844 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Anzalone AV et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu P. et al. Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice. Nat Commun 12, 2121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gainor JF et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 6, 1118–1133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gainor JF et al. Patterns of Metastatic Spread and Mechanisms of Resistance to Crizotinib in ROS1-Positive Non-Small-Cell Lung Cancer. JCO Precis Oncol 2017, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi S. et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 352, 786–792 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Chen PJ et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 184, 5635–5652.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canté-Barrett K. et al. Lentiviral gene transfer into human and murine hematopoietic stem cells: size matters. BMC Res Notes 9, 312 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zinn E. & Vandenberghe LH Adeno-associated virus: fit to serve. Current Opinion in Virology 8, 90–97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang D. et al. Adenovirus-Mediated Somatic Genome Editing of Pten by CRISPR/Cas9 in Mouse Liver in Spite of Cas9-Specific Immune Responses. Hum Gene Ther 26, 432–442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Annunziato S. et al. Modeling invasive lobular breast carcinoma by CRISPR/Cas9-mediated somatic genome editing of the mammary gland. Genes Dev. 30, 1470–1480 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Böck D. et al. In vivo prime editing of a metabolic liver disease in mice. Science Translational Medicine 14, eabl9238 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sánchez-Rivera FJ et al. Base editing sensor libraries for high-throughput engineering and functional analysis of cancer-associated single nucleotide variants. Nat Biotechnol 1–12 (2022) doi: 10.1038/s41587-021-01172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erwood S. et al. Saturation variant interpretation using CRISPR prime editing. Nat Biotechnol 1–11 (2022) doi: 10.1038/s41587-021-01201-1. [DOI] [PubMed] [Google Scholar]
- 37.Chen L. et al. Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins. Nat Commun 12, 1384 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grünewald J. et al. A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing. Nat Biotechnol 38, 861–864 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan T. et al. Optimization of C-to-G base editors with sequence context preference predictable by machine learning methods. Nat Commun 12, 4902 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anzalone AV et al. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat Biotechnol 40, 731–740 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang T, Zhang X-O, Weng Z. & Xue W. Deletion and replacement of long genomic sequences using prime editing. Nat Biotechnol 40, 227–234 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bult CJ et al. Mouse Genome Database (MGD) 2019. Nucleic Acids Res 47, D801–D806 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MouseMine, Mouse Genome Informatics Web Site, The Jackson Laboratory, Bar Harbor, Maine. World Wide Web. (2022). [Google Scholar]
- 44.Shaner NC et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods 10, 407–409 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madisen L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ng SR et al. CRISPR-mediated modeling and functional validation of candidate tumor suppressor genes in small cell lung cancer. Proc Natl Acad Sci U S A 117, 513–521 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freed-Pastor WA et al. The CD155/TIGIT axis promotes and maintains immune evasion in neoantigen-expressing pancreatic cancer. Cancer Cell 39, 1342–1360.e14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schlake T. & Bode J. Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry 33, 12746–12751 (1994). [DOI] [PubMed] [Google Scholar]
- 49.Bindels DS et al. mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat Methods 14, 53–56 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Vassilev LT et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Nelson JW et al. Engineered pegRNAs improve prime editing efficiency. Nat Biotechnol 40, 402–410 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bae S, Park J. & Kim J-S Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qiu M. et al. Lipid nanoparticle-mediated codelivery of Cas9 mRNA and single-guide RNA achieves liver-specific in vivo genome editing of Angptl3. Proceedings of the National Academy of Sciences 118, e2020401118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hingorani SR et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4, 437–450 (2003). [DOI] [PubMed] [Google Scholar]
- 55.el Marjou F. et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Kim HK et al. Predicting the efficiency of prime editing guide RNAs in human cells. Nat Biotechnol 39, 198–206 (2021). [DOI] [PubMed] [Google Scholar]
- 57.Drost J. et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 521, 43–47 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Zafra MP et al. An In Vivo Kras Allelic Series Reveals Distinct Phenotypes of Common Oncogenic Variants. Cancer Discov 10, 1654–1671 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strickler JH et al. First data for sotorasib in patients with pancreatic cancer with KRAS p.G12C mutation: A phase I/II study evaluating efficacy and safety. JCO 40, 360490–360490 (2022). [Google Scholar]
- 60.Hofmann MH, Gerlach D, Misale S, Petronczki M. & Kraut N. Expanding the Reach of Precision Oncology by Drugging All KRAS Mutants. Cancer Discovery 12, 924–937 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waters AM & Der CJ KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb Perspect Med 8, a031435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang X. et al. Identification of MRTX1133, a Noncovalent, Potent, and Selective KRASG12D Inhibitor. J Med Chem 65, 3123–3133 (2022). [DOI] [PubMed] [Google Scholar]
- 63.Awad MM et al. Acquired Resistance to KRASG12C Inhibition in Cancer. N Engl J Med 384, 2382–2393 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pao W. et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2, e73 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Freed-Pastor WA & Prives C. Mutant p53: one name, many proteins. Genes Dev 26, 1268–1286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cerami E. et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discovery 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gao J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schulz-Heddergott R. et al. Therapeutic Ablation of Gain-of-Function Mutant p53 in Colorectal Cancer Inhibits Stat3-Mediated Tumor Growth and Invasion. Cancer Cell 34, 298–314.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barta JA, Pauley K, Kossenkov AV & McMahon SB The lung-enriched p53 mutants V157F and R158L/P regulate a gain of function transcriptome in lung cancer. Carcinogenesis 41, 67–77 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shakya R. et al. Mutant p53 upregulates alpha-1 antitrypsin expression and promotes invasion in lung cancer. Oncogene 36, 4469–4480 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Jackson EL et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res 65, 10280–10288 (2005). [DOI] [PubMed] [Google Scholar]
- 72.Winslow MM et al. Suppression of lung adenocarcinoma progression by Nkx2–1. Nature 473, 101–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.LaFave LM et al. Epigenomic State Transitions Characterize Tumor Progression in Mouse Lung Adenocarcinoma. Cancer Cell 38, 212–228.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jackson EL et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 15, 3243–3248 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aoyama N. et al. Transgenic mice that accept Luciferase- or GFP-expressing syngeneic tumor cells at high efficiencies. Genes Cells 23, 580–589 (2018). [DOI] [PubMed] [Google Scholar]
- 76.Hingorani SR et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 7, 469–483 (2005). [DOI] [PubMed] [Google Scholar]
- 77.Chiou S-H et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev 29, 1576–1585 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu B. et al. A split prime editor with untethered reverse transcriptase and circular RNA template. Nat Biotechnol (2022) doi: 10.1038/s41587-022-01255-9. [DOI] [PubMed] [Google Scholar]
- 79.Hunter JC et al. Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations. Molecular Cancer Research 13, 1325–1335 (2015). [DOI] [PubMed] [Google Scholar]
- 80.Amodio V. et al. EGFR Blockade Reverts Resistance to KRASG12C Inhibition in Colorectal Cancer. Cancer Discov 10, 1129–1139 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Canon J. et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575, 217–223 (2019). [DOI] [PubMed] [Google Scholar]
- 82.Kemp SB et al. Efficacy of a small molecule inhibitor of KrasG12D in immunocompetent models of pancreatic cancer. Cancer Discov CD-22–1066 (2022) doi: 10.1158/2159-8290.CD-22-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Escobar-Hoyos LF et al. Altered RNA Splicing by Mutant p53 Activates Oncogenic RAS Signaling in Pancreatic Cancer. Cancer Cell 38, 198–211.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klemke L. et al. The Gain-of-Function p53 R248W Mutant Promotes Migration by STAT3 Deregulation in Human Pancreatic Cancer Cells. Front Oncol 11, 642603 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schoenfeld AJ et al. The Genomic Landscape of SMARCA4 Alterations and Associations with Outcomes in Patients with Lung Cancer. Clin Cancer Res 26, 5701–5708 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gibson DG et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 87.Clement K. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol 37, 224–226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hsu JY et al. PrimeDesign software for rapid and simplified design of prime editing guide RNAs. Nat Commun 12, 1034 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Engler C, Kandzia R. & Marillonnet S. A One Pot, One Step, Precision Cloning Method with High Throughput Capability. PLOS ONE 3, e3647 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Akama-Garren EH et al. A Modular Assembly Platform for Rapid Generation of DNA Constructs. Sci Rep 6, 16836 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Naranjo S. et al. Modeling diverse genetic subtypes of lung adenocarcinoma with a next-generation alveolar type 2 organoid platform. Genes Dev 36, 936–949 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zettler J, Schütz V. & Mootz HD The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Lett 583, 909–914 (2009). [DOI] [PubMed] [Google Scholar]
- 93.DuPage M, Dooley AL & Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc 4, 1064–1072 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Boj SF et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim MP et al. Generation of orthotopic and heterotopic human pancreatic cancer xenografts in immunodeficient mice. Nat Protoc 4, 1670–1680 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Marino S, Vooijs M, van Der Gulden H, Jonkers J. & Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev 14, 994–1004 (2000). [PMC free article] [PubMed] [Google Scholar]
- 97.Sastra SA & Olive KP Quantification of murine pancreatic tumors by high-resolution ultrasound. Methods Mol Biol 980, 249–266 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bankhead P. et al. QuPath: Open source software for digital pathology image analysis. Sci Rep 7, 16878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gaudelli NM et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Owczarzy R. et al. IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Research 36, W163–W169 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jamal-Hanjani M. et al. Tracking the Evolution of Non–Small-Cell Lung Cancer. N Engl J Med 376, 2109–2121 (2017). [DOI] [PubMed] [Google Scholar]
- 102.Campbell JD et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 48, 607–616 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Rosa26PE2 mice on wild-type (JAX Stock Number: JR037953) and Trp53flox/flox (JAX stock number: JR037954) backgrounds are available from the Jackson Laboratory. Plasmids will be made available through Addgene upon publication. All other materials and data are available from the corresponding author upon reasonable request.