Abstract
Our objective was to develop data-based algorithms for definition of immunologic response to AIDS therapies in pediatric patients, taking account of T-cell subset measurement errors. The study design involved cross-protocol analysis of 2,148 enrollees in six completed Pediatric AIDS Clinical Trials Group trials. We used standard quantitation of T-cell subsets; linear modeling with mean-dependent measurement error variance was used to develop 95% tolerance limits for change in CD4%. For individuals with a CD4% of approximately 25%, the measurement error-based 95% tolerance interval ranges from 15% to 35%, whereas for individuals with a CD4% of approximately 5%, the tolerance interval ranges from 3% to 7%. When pairs of CD4% measures taken within a time interval of less than 30 days are averaged to estimate steady-state CD4%, tolerance interval width decreases by approximately 30%. A simple graphical tool that provides a data-based criterion for immunologic response over and above variation ascribable to T-cell measurement error is provided. Variability in CD4% due to measurement error is substantial, increases with level of CD4%, and complicates assessment of immunologic response to therapy. Replicates of CD4% measures could be used to improve precision of interpretation of CD4% measures.
Methods of managing human immunodeficiency virus (HIV) disease are evolving rapidly. The most mature and well-established markers of disease progression are CD4 T-helper cell count and RNA copy number (2, 3, 5, 7). While these parameters are straightforwardly used for cross-sectional classification of patients with respect to clinical state, much uncertainty exists regarding evaluation and interpretation of changes in these markers over time (4). Recovery of CD4 T cells is an important criterion for immune reconstitution in patients who are given antiretroviral therapy. This report focuses on the short-term variability of repeated measures of CD4% in HIV-infected children enrolled in the Pediatric AIDS Clinical Trials Group (PACTG) treatment trials. Data from six clinical trials conducted in the past decade are combined for approximate estimation of measurement error variability. The impact of using replicated CD4% measures (i.e., averaging two measures obtained simultaneously or separated by a very short time intervals) to reduce effects of measurement error is illustrated.
MATERIALS AND METHODS
Participants.
Data were collected from six clinical trials conducted in the PACTG: protocols 152, 190, 300, 338, 377, and 382. Of these, protocols 152, 190, and 300 were the earlier treatment trials, consisting of one- or two-nucleoside analog reverse transcriptase inhibitor therapies, with protocols 152 and 190 completing by 1996 and 300 completing in 2001. Protocols 338, 377, and 382 were instituted later (ending by 2002) and evaluated triple-antiretroviral therapies, including protease inhibitors and/or nonnucleoside analogues.
Measurement protocols.
The six trials enumerated above used the same protocol for T-cell subset measurement. Immunophenotyping for T-cell subsets was performed by two-color standard flow cytometry according to the PACTG consensus protocol (http://pactg.s-3.com/immeth.html). Laboratories that performed these assays were certified by the PACTG immunology quality assurance program. All trials used “pre-entry” and “entry” timed T-cell subset measurements to characterize patient baseline state. These pairs of T-cell subset measurements usually occurred over a period of about one week, during which the patient is reasonably assumed to be clinically stable and on a stable treatment regimen preparatory to starting a new trial.
Statistical methods.
Standard descriptive statistics and scatterplots were derived using the R statistical computing environment (www.r-project.org). Variance component analysis was conducted using the linear mixed effects models software package for R by Pinheiro and Bates (6).
The basic model for response variable y (e.g., CD4%) measured on subjects from study s is
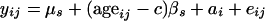 |
where i indexes patients in study s, j = 1, 2 indexes the pre-entry and entry measures, μs is the overall mean value of response for a subject aged c years, βs is the study-specific slope of mean response on age at time of measurement, ai is a subject-specific random effect with distribution 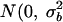 ), and eij is a residual error term.
), and eij is a residual error term.
The residuals eij are of central interest in this study. Let mi denote the true mean CD4% for subject i. If mi is very small (say 5% or less), then the measurement error variability for measures on subject i tends also to be small, in part because negative values of CD4% are not possible. Among subjects for whom mi is larger, measurement error variability tends to be larger as well. We therefore adopt a heteroskedastic measurement error model. Conditionally on the value of ai, eij has distribution 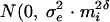 ). Note that as mi tends to zero, so does the measurement error variance. As mi increases, measurement error variability increases proportionally to
). Note that as mi tends to zero, so does the measurement error variance. As mi increases, measurement error variability increases proportionally to 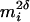 . The parameter δ can be used to control the growth of measurement error variability with magnitude of mean CD4%.
. The parameter δ can be used to control the growth of measurement error variability with magnitude of mean CD4%.
Given the generally short time elapsed between repeated measures and the likely clinical and therapeutic stability of the patient in the pre-entry to entry interval, the eij are reasonably regarded as data on irreducible measurement error in the subset measurement process. For a specified value of m, “intraclass correlation coefficient”
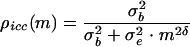 |
varies between 0 and 1 and is a dimensionless measure of repeatability, high values reflecting good repeatability.
RESULTS
Demographics and baseline characteristics.
Table 1 (columns 2 to 6) presents basic descriptive statistics regarding the patients in the various protocols. This table includes information on all patients who supplied valid data on pre-entry and entry CD4 counts, regardless of treatment assignment or reappearance of patients in multiple protocols. Individuals whose pre-entry and entry measures were separated in time by more than 30 days were excluded.
TABLE 1.
Descriptive statistics and selected results of variance component modeling stratified by protocol and overalla
| Study no. | n | Age (yr) | LRNA | CD4P | CD4/CD8 ratio | 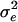 |
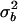 |
δ | ρicc(15) | ρicc(25) |
|---|---|---|---|---|---|---|---|---|---|---|
| 152 | 738 | 2.27 | 5.15 | 24 | 0.63 | 0.24 | 105.94 | 0.69 | 0.91 | 0.84 |
| 190 | 227 | 5.12 | N/A | 24 | 0.53 | 0.16 | 108.36 | 0.69 | 0.94 | 0.89 |
| 300 | 614 | 2.72 | N/A | 23 | 0.59 | 0.28 | 127.41 | 0.61 | 0.94 | 0.90 |
| 338 | 289 | 7.11 | 4.19 | 28 | 0.61 | 0.07 | 61.39 | 0.77 | 0.93 | 0.86 |
| 377 | 190 | 5.64 | 4.44 | 28 | 0.72 | 0.20 | 74.64 | 0.59 | 0.94 | 0.89 |
| 382 | 90 | 4.09 | 4.89 | 29 | 0.79 | 0.11 | 112.63 | 0.74 | 0.95 | 0.89 |
| All | 2148 | 3.41 | 4.83 | 25 | 0.61 | 0.27 | 104.29 | 0.63 | 0.93 | 0.87 |
Abbreviations and symbols: n, number of patients in each study; LRNA, median log10 HIV RNA copy number; CD4P, mean study-specific CD4%; CD4/CD8, median ratio of CD4 count to CD8 count; 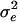 and
and 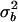 , the intra- and interpatient variability; δ, relationship between error variability and magnitude of CD4% measurement; ρicc(15) and ρicc(25), repeatability of results at mean CD4% of 15% and 25%, respectively; N/A, not applicable.
, the intra- and interpatient variability; δ, relationship between error variability and magnitude of CD4% measurement; ρicc(15) and ρicc(25), repeatability of results at mean CD4% of 15% and 25%, respectively; N/A, not applicable.
Median baseline patient age varied from 2.2 to 7.1 years. Protocols 152, 300, and 382 provided data on patients as young as six months at baseline; minimum ages on other protocols ranged from 9 months on 377 to 15 months on 338. The overall median age was 3.35 years.
In general, HIV RNA loads were uncontrolled, with medians well above 10,000 copies in all protocols for which viral loads were measured.
Immunosuppression as measured by median CD4% varied among the cohorts, with a median of 23% for protocol 300 and a median of 29% for protocol 382. Median CD8 counts were above 1,000 for all protocols (data not shown), and CD4/CD8 ratios ranged from 0.53 (protocol 190) to 0.79 (protocol 382). Forty-nine percent of patients were Centers for Disease Control and Prevention (CDC) category I (CD4% of >25), 32% were category II (15 < CD4% < 24), and 19% were category III (CD4% of <15%). A total of 116 patients (5.4%) had a CD4% of <5%.
After excluding individuals whose time elapsed from pre-entry to entry T-cell counts exceeded 30 days, median lag between pre-entry and entry measurements was generally considerably less than two weeks.
Graphical depiction of CD4% repeatability.
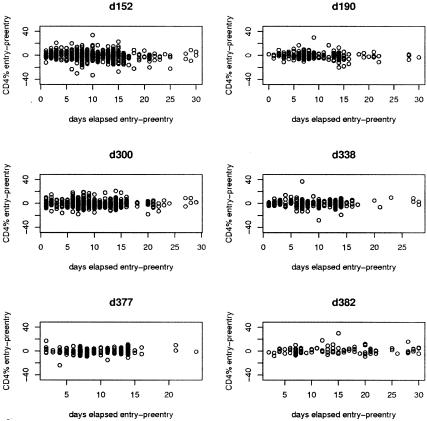
Figure 1 depicts on a study-specific basis the dispersion in entry-pre-entry CD4% measures. If CD4% measurements were perfect, we would expect these scatterplots of individual-level changes to be tightly concentrated around the line y = 0, as in general there is no biological basis for variation in CD4% in this pretreatment interval. For very short lags (up to three days), variability is very modest, but it appears that short-term variability is substantial at five days and that longer lags are not associated with greater variability. The appearance of these figures, in conjunction with the numerical analyses to be discussed, led us to conclude that amalgamation of the data across these protocols was a reasonable step in learning about CD4% repeatability in pediatric clinical trials.
FIG. 1.
Scatterplots of CD4% at entry minus CD4% at pre-entry visit, versus time elapsed between the two visits. Data show considerable variability between CD4% measurements. The variability was similar across studies.
Modeling of CD4% repeatability.
Table 1 (columns 7 to 11) presents results of fitting the variance component model of the “Participants” section in two basic forms: to data stratified by study and to the amalgamated complete data set. The data show that study-specific mean CD4% varied from 24.5% to 30.4%. Despite the fact that intra- and interpatient measures varied considerably among studies, the parameter δ, which describes the association between CD4% measurement error and CD4% mean, varied remarkably less, between 0.59 and 0.77. Furthermore the repeatability of CD4% measurements at means of 15% and 25% was remarkably stable across studies, with values of 0.91 to 0.95 and 0.84 to 0.90, respectively.
Tolerance limits for CD4% change.
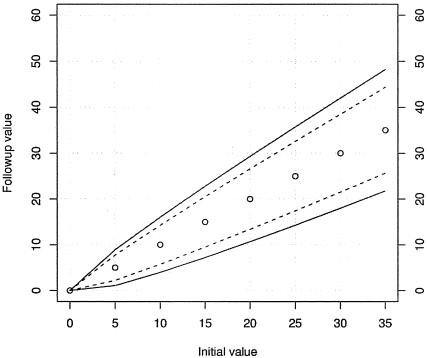
Figure 2 depicts 95% tolerance limits for change in true mean value when comparing two CD4% measures. To use the figure, find the baseline or other reference value on the x axis and draw a vertical line from that point. Note where the vertical line intersects the solid lines and project to the y axis. The interval thus defined on the y axis is the tolerance limit. This identifies the central 95% of the distribution of CD4% measures consistent with fluctuation due to measurement error alone, with no change in underlying mean between baseline and follow-up. Follow-up measures that lie outside the interval are highly unlikely under a hypothesis of no change in underlying mean CD4%. Note that the choice of 95% limits is conventional. Other choices of threshold can be used.
FIG. 2.
Ninety-five percent tolerance limits for change in CD4%. The current value is located on the x axis, the boundaries of tolerance limit for single CD4% measurements are projected to the y axis using solid curves, and the boundaries for replicated CD4% measurements are projected to the y axis using dashed curves. In the construction of the dashed curves, it is assumed that both initial and follow-up values are obtained by averaging a pair of measures at each time point.
Figure 2 also provides information on the effect of reducing measurement error variability by averaging pairs of CD4% measures. The error decreases by approximately 30% when the CD4% result is derived from the average of two measurements performed within a time interval of ≤30 days.
DISCUSSION
Loss of CD4 T cells is a hallmark of HIV infection. This trend is reversed in patients given antiretroviral therapy that successfully achieve virologic suppression, but it is now well recognized that gains in CD4 counts may also occur in the absence of durable virologic suppression. Clear criteria for designating an individual as an immunologic responder are lacking, and many studies have relied upon arbitrary measures of CD4 increase from baseline values. The goal of this study was to establish how to interpret a change in CD4 from an initial value. By estimating measurement error variability using repeated CD4% measures taken over a short time interval, we provide a basis for identifying changes that are likely to represent true immunologic response or deterioration.
Based on the variance analysis, we constructed a model depicting 95% tolerance limits for change in true mean value of CD4%. As shown in Fig. 2, the model indicates that the variance is greater at higher levels of CD4% than at lower CD4%. Thus using a standard definition of gain in CD4 either in absolute number or in percentages could be misleading, failing to identify clinically meaningful CD4 changes at lower initial CD4 percentages or overinterpreting changes at higher initial CD4 percentages. The data support the idea of computing averages of two measurements performed within a time interval of <30 days.
This study is the first model-based analysis of pediatric patients for assessing CD4 measurement variability. While we assembled records on over 2,000 participants in PACTG trials conducted over the past ten years, we acknowledge that our cohort does not represent a systematic or random sample of HIV-positive children. The protocols selected for this study included infants and young children <8 years of age with various degrees of immunosuppression, and our basic model included an adjustment for an age effect. The size and diversity of the cohort on which our model is constructed contribute to, but do not guarantee, the face validity of our inferences on measurement variability. It was noted by a referee that some of the observations shown in Fig. 1 appear to be outliers. A formal test for outliers in the marginal distribution of all CD4% measures used in the model did not reject the null hypothesis of no outliers. However, a test applied to the marginal distribution of all entry-pre-entry differences did identify 15 putative outliers, with magnitude of the difference exceeding 21 percentage units. When the model was refit, excluding these observations, the estimate of δ was 0.56, that of 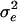 was 0.35, and the impact on the analog of Fig. 2 was slight, with a slight narrowing of the bands. In the absence of frank evidence of data error, we prefer to employ the entire data set to fit the model of interest. Robustness to data anomalies is a concern with any application of sophisticated statistical modeling. A related analysis with HIV-1 RNA measures was conducted by Brambilla and colleagues (1), who obtained robust estimates of assay standard deviation by rescaling empirical quantiles.
was 0.35, and the impact on the analog of Fig. 2 was slight, with a slight narrowing of the bands. In the absence of frank evidence of data error, we prefer to employ the entire data set to fit the model of interest. Robustness to data anomalies is a concern with any application of sophisticated statistical modeling. A related analysis with HIV-1 RNA measures was conducted by Brambilla and colleagues (1), who obtained robust estimates of assay standard deviation by rescaling empirical quantiles.
The construct validity of the tolerance limits presented in Fig. 2 can be assessed by evaluating the association between declared “genuine immunologic response” (changes beyond the bounds of the tolerance limit for given initial value) and other clinical, immunologic, or virologic events. Such analyses will be important steps toward formation of an evidence-based criterion for immunologic response to antiretroviral therapy.
Acknowledgments
This work was supported by NIH/NIAID contract/grant number 2 U01 AI41110-06.
REFERENCES
- 1.Brambilla, D., P. S. Reichelderfer, J. W. Bremer, D. E. Shapiro, R. C. Hershow, D. A. Katzenstein, S. M. Hammer, B. Jackson, A. C. Collier, R. S. Sperling, M. G. Fowler, R. W. Coombs, et al. 1999. The contribution of assay variation and biological variation to the total variability of plasma HIV-1 RNA measurements. AIDS 13:2269-2279. [DOI] [PubMed] [Google Scholar]
- 2.Demeter, L. M., M. D. Hughes, R. W. Coombs, J. B. Jackson, J. M. Grimes, R. J. Bosch, S. A. Fiscus, S. A. Spector, K. E. Squires, M. A. Fischl, and S. M. Hammer. 2001. Predictors of virologic and clinical outcomes in HIV-1-infected patients receiving concurrent treatment with indinavir, zidovudine, and lamivudine. AIDS Clinical Trials Group Protocol 320. Ann. Intern. Med. 135:954-964. [DOI] [PubMed] [Google Scholar]
- 3.HIV-Surrogate-Marker-Collaborative-Group. 2000. Human immunodeficiency virus type 1 RNA level and CD4 count as prognostic markers and surrogate end points: a meta-analysis. AIDS Res. Hum. Retrovir. 16:1123-1133. [DOI] [PubMed] [Google Scholar]
- 4.Mildvan, D., R. J. Bosch, R. S. Kim, J. Spritzler, D. W. Haas, D. Kuritzkes, J. Kagan, M. Nokta, V. DeGruttola, M. Moreno, and A. Landay. 2004. Immunophenotypic markers and antiretroviral therapy (IMART): T cell activation and maturation help predict treatment response. J. Infect. Dis. 189:1811-1820. [DOI] [PubMed] [Google Scholar]
- 5.Mildvan, D., A. Landay, V. De Gruttola, S. G. Machado, and J. Kagan. 1997. An approach to the validation of markers for use in AIDS clinical trials. Clin. Infect. Dis. 24:764-774. [DOI] [PubMed] [Google Scholar]
- 6.Pinheiro, J. C., and D. M. Bates. 2000. Mixed effects models in S and S-plus. Springer Verlag, New York, N.Y.
- 7.Schmid, A., H. H. Hirsch, et al. 2003. Highly active antiretroviral therapy of HIV patients. Internist (Berlin). 44:1597-1600. (In German.) [DOI] [PubMed] [Google Scholar]