Abstract
Aichi virus is a member of the family Picornaviridae. It has already been shown that three stem-loop structures (SL-A, SL-B, and SL-C, from the 5′ end) formed at the 5′ end of the genome are critical elements for viral RNA replication. In this study, we further characterized the 5′-terminal cis-acting replication elements. We found that an additional structural element, a pseudoknot structure, is formed through base-pairing interaction between the loop segment of SL-B (nucleotides [nt] 57 to 60) and a sequence downstream of SL-C (nt 112 to 115) and showed that the formation of this pseudoknot is critical for viral RNA replication. Mapping of the 5′-terminal sequence of the Aichi virus genome required for RNA replication using a series of Aichi virus-encephalomyocarditis virus chimera replicons indicated that the 5′-end 115 nucleotides including the pseudoknot structure are the minimum requirement for RNA replication. Using the cell-free translation-replication system, we examined the abilities of viral RNAs with a lethal mutation in the 5′-terminal structural elements to synthesize negative- and positive-strand RNAs. The results showed that the formation of three stem-loops and the pseudoknot structure at the 5′ end of the genome is required for negative-strand RNA synthesis. In addition, specific nucleotide sequences in the stem of SL-A or its complementary sequences at the 3′ end of the negative-strand were shown to be critical for the initiation of positive-strand RNA synthesis but not for that of negative-strand synthesis. Thus, the 5′ end of the Aichi virus genome encodes elements important for not only negative-strand synthesis but also positive-strand synthesis.
Picornaviruses are small, nonenveloped, icosahedral viruses, and this virus family includes many human and animal pathogens, including poliovirus, rhinovirus, hepatitis A virus (HAV), and foot-and-mouth disease virus (38). Their genomes comprise single-stranded positive-sense RNAs of 7,200 to 8,500 nucleotides (nt) and consist of a 5′ untranslated region (UTR), a single coding region, a 3′ UTR, and a poly(A) tail from the 5′ end. After its release into the cytoplasm of infected cells, the genomic RNA serves as an mRNA for the synthesis of a long polyprotein, which is cleaved into functional proteins by virus-encoded proteinases. The positive-strand genomic RNA also acts as a template for the synthesis of negative-strand RNA, which is then transcribed into positive-strand RNA. The newly synthesized positive-strand RNA is packaged into capsid proteins to form virions.
The 5′ UTR of the picornavirus genome is very long (620 to 1,200 nucleotides in length) and contains several secondary and tertiary structural elements (38). This region contains sequences that control genome replication and translation (40). The 5′ ends of the genomes of enteroviruses and rhinoviruses fold into a cloverleaf-like structure (39, 50). In poliovirus, this structure functions as a cis-acting element for RNA replication and forms a ribonucleoprotein (RNP) complex by interacting with viral protease polymerase precursor polypeptides, 3CD, and a host protein, poly(rC)-binding protein (PCBP) (2, 3, 16, 36). In addition, this 5′-end RNP complex interacts with the 3′ poly(A) tail through binding to poly(A)-binding protein and the formation of this circular RNP complex is required for the initiation of negative-strand RNA synthesis (7, 20, 26). On the other hand, the 5′ ends of the genomes of cardio-, aphtho-, hepato-, and parechoviruses fold into stem-loop and pseudoknot structures (11, 13, 17, 24). The 5′ end of the genome of human parechovirus 1 consists of two stem-loops (SL-A and SL-B) and a pseudoknot structure (pk-C), and these structures are critical for RNA replication (34). Additionally, the pseudoknot structure formed at the 5′ end of the mengovirus genome plays an essential role in viral RNA synthesis (27). However, there is no evidence showing which step these structures are required for, i.e., positive- or negative-strand RNA synthesis during RNA replication.
Aichi virus, which is associated with acute gastroenteritis in humans (47), is a member of the genus Kobuvirus of the family Picornaviridae (37, 48). Computer-assisted prediction of its RNA secondary structure suggested the formation of three stem-loop structures (SL-A, SL-B, and SL-C) within the 120 nucleotides of the 5′ end of its genome. We have already investigated the functions of these structures and have shown that all three stem-loop structures are critical for viral RNA replication (33, 42). In addition, the nucleotide sequence of the stem segment in the middle part of SL-A has been shown to be important for viral RNA encapsidation (43).
In this study, we further characterized the 5′-terminal cis-acting replication elements of Aichi virus. We found an additional essential structural element, a pseudokot structure, which is formed through base-pairing interaction between the loop region of SL-B (nt 57 to 60) and a sequence downstream of SL-C (nt 112 to 115). Using a chimeric virus whose 5′ UTR consists of the Aichi virus 5′-terminal sequence and the encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES), the 5′-terminal region required for RNA replication was mapped. The results indicated that the 5′-end 115 nucleotides including the pseudoknot are the minimum requirement for RNA replication. In addition, using a cell-free translation-replication system, we examined which step the structural elements at the 5′ end of the genome are required for, i.e., negative-strand synthesis or positive-strand synthesis. The results showed that the formation of all three stem-loop structures is required for negative-strand RNA synthesis. Furthermore, a specific nucleotide sequence in the lower part of the stem of SL-A or its complementary sequences at the 3′ end of the negative-strand RNA was shown to be critical for the initiation of positive-strand RNA synthesis but not for that of negative-strand RNA synthesis. Thus, these results indicate that the 5′ end of the Aichi virus genome encodes RNA elements important for not only negative-strand synthesis but also positive-strand synthesis.
MATERIALS AND METHODS
Cell culture.
Vero cells were grown in Eagle's minimum essential medium containing 5% fetal calf serum (growth medium) at 37°C.
Aichi virus cDNA clones and replicons.
pAV-FL is an infectious cDNA clone of Aichi virus (42). pAV-FL-Luc is a replicon in which the capsid-coding region is replaced with a firefly luciferase gene (33).
Plasmid. (i) pAV-FL-Luc-3Dmut.
The PstI (nt 6771)-HindIII [just downstream of the poly(A) tract] fragment of pAV-FL was subcloned in pUC118, generating pUP-H. To change the GDD motif (23) of the 3D RNA-dependent RNA polymerase to Gly-Ala-Ala, PCR-based mutagenesis was performed with primers 7626D-A-P (5′ GCCGTCATCTACGCTACGGAA) and 7625D-A-M (5′ GGCACCGTACGCCAGGATGT) by using pUP-H as a template. The PCR product was self-ligated, and the nucleotide sequence between the PstI and HindIII sites of the derived plasmid was confirmed. The PstI-HindIII fragment with the GDD-GAA mutation was substituted for the corresponding fragment of pAV-FL-Luc to yield pAV-FL-Luc-3Dmut.
(ii) Hammerhead ribozyme-containing plasmids.
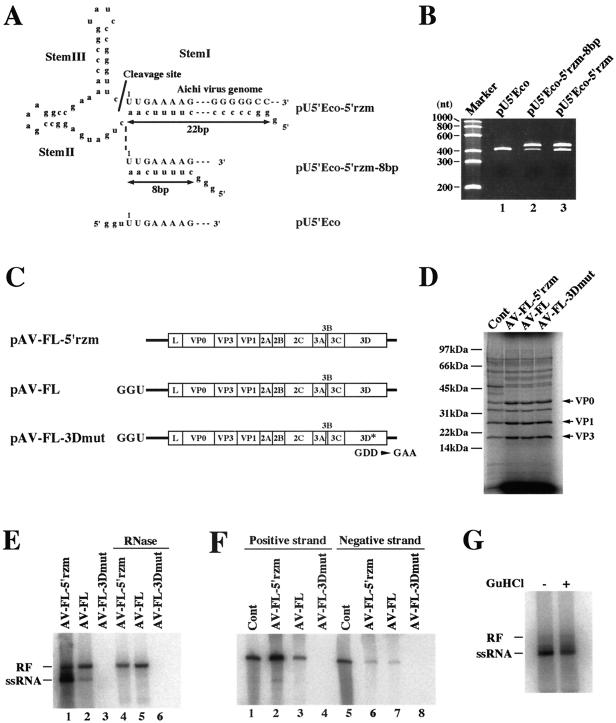
An Aichi virus-specific cis-active hammerhead ribozyme was designed based on the ribozyme used for the poliovirus cDNA clone (19). First, a hammerhead ribozyme in which stem I (8) comprised 8 bp was introduced to the 5′ end of the Aichi virus genome. Using pU5′Eco, which contains the T7 promoter and the 5′-end 391 nucleotides of the Aichi virus genome (42), as a template, PCR-based mutagenesis was performed with primers 5′rzm-P (5′ GGCCGAAAACCCGGTATCCCGGGTTCTTGAAAAGGGGGTGGGGGGGC) and 5′rzm-M (5′ TTTCGGCCTCATCAGTTGAAAAGCCCTATAGTGAGTCGTATTACAATTC). The PCR products were self-ligated, and the nucleotide sequence of the derived plasmid, termed pU5′Eco-5′rzm-8bp, was verified. To construct a hammerhead ribozyme with stem I comprising 22 bp, PCR was carried out with primers 5′rzmP-2 (5′ CCCCCTTTTCAACTGATGAGGCCGAAAG) and 5′rzmM-2 (5′ TGGGGGGGCCCTATAGTGAGTCGTATTACA) by using pU5′Eco-5′rzm-8bp as a template. The PCR products were self-ligated, and the derived plasmid was termed pU5′Eco-5′rzm. The EcoRI fragment of pU5′Eco-5′rzm was substituted for the corresponding fragment of pAV-FL, an Aichi virus full-length cDNA clone (42), generating pAV-FL-5′rzm.
We previously reported that the 5′-end sequence of the Aichi virus genome is UUUGAAAAG (42). However, only two, not three, uridine residues exist at the 5′ end of most picornavirus genomes. We designed the hammerhead ribozyme of pAV-FL-5′rzm so as to obtain in vitro transcripts with two uridine residues at the 5′ end. As described in the Results section, transcripts derived from pAV-FL-5′rzm generated viable viruses in the cell-free replication system. In addition, we constructed a full-length cDNA clone with a hammerhead ribozyme to produce transcripts with three uridine residues at the 5′ end. The in vitro transcript with three uridine residues produced positive-strand RNAs in the cell-free replication system, but the amount was less than that produced by the transcript with two uridine residues at the 5′ end (data not shown). This suggests that the accurate 5′-end sequence of the Aichi virus genome is UUGAAAAG. Since we determined the 5′-end sequence of the Aichi virus genome by sequencing plasmids obtained through 5′ rapid amplification of cDNA ends in the previous study (42), the 5′-terminal nucleotide of the three uridine residues would be added independent of the template by using reverse transcriptase (12). In this study, we used a full-length cDNA clone harboring a hammerhead ribozyme, which produces transcripts with two uridine residues at the 5′ end (see Fig. 6A). On the other hand, the in vitro transcript synthesized from pAV-FL, which has no hammerhead ribozyme, has three uridine residues at the 5′ end of the genome, following two guanine residues; that is, the transcripts have three nonviral nucleotides, GGU, at the 5′ end (see Fig. 6A and C). The nucleotide numbers given in this study are for the sequence with two uridine residues at the 5′ end.
FIG. 6.
(A) The 5′ end sequences of in vitro transcripts synthesized from pU5′Eco-5′rzm, pU5′Eco-5′rzm-8bp, and pU5′Eco. pU5′Eco have the T7 promoter followed by the 5′-end 391 nucleotides of the Aichi virus sequence. pU5′Eco-5′rzm and pU5′Eco-5′rzm-8bp were constructed by introducing the cis-active hammerhead ribozyme sequence between the T7 promoter and the virus sequence. In pU5′Eco-5′rzm and pU5′Eco-5′rzm-8bp, stem I segments of the ribozyme were 22 bp and 8 bp in length, respectively. The Aichi virus sequences and nonviral nucleotides are represented by uppercase letters and lowercase letters, respectively. (B) Cleavage by the hammerhead ribozyme. In vitro transcripts synthesized from pU5′Eco-5′rzm, pU5′Eco-5′rzm-8bp, and pU5′Eco were electrophoresed on a 3.5% polyacrylamide-7 M urea gel, and then the gel was stained with ethidium bromide. (C) Schematic diagrams of in vitro transcripts derived from full-length cDNA clones. The in vitro transcripts synthesized from plasmids containing the hammerhead ribozyme have a precise 5′ end, while those from plasmids without the hammerhead ribozyme have three nonviral nucleotides, GGU, at the 5′ end. In pAV-FL-3Dmut, the GDD motif within the 3D RNA polymerase-coding sequence was changed to Gly-Ala-Ala to inactivate the RNA polymerase. (D) Protein synthesis in a cell-free reaction programmed with AV-FL-5′rzm RNA, AV-FL RNA, or AV-FL-3Dmut RNA. Reactions were performed in the presence of [35S]methionine-cysteine for 4 h, and then proteins were analyzed by SDS-polyacrylamide gel electrophoresis. In lane represented as Cont, viral proteins that were metabolically labeled with [35S]methionine-cysteine in transfected cells were loaded. The positions of capsid proteins are indicated. (E) RNA products in a cell-freereaction programmed with AV-FL-5′rzm RNA, AV-FL RNA, or AV-FL-3Dmut RNA. RNA was labeled with [α-32P]CTP at 3 to 5 h after the start of incubation, and then total RNA was extracted. The extracted RNA was analyzed by nondenaturing agarose gel electrophoresis after being either treated with RNase A and RNase T1 (lanes 4 to 6) or untreated (lanes 1 to 3). The positions of double-stranded RF and ssRNA are indicated. (F) RPA. Labeled viral RNA products of a cell-free reaction were subjected to RPA using an unlabeled probe that hybridizes with the Aichi virus positive-strand RNA or negative-strand RNA. To detect the negative-strand RNA, two-cycle RPA was carried out. As a positive control (Cont), in vitro-labeled full-length positive-strand or negative-strand RNA was used. Protected RNAs were analyzed by 3.5% polyacrylamide-7 M urea gel electrophoresis. (G) Effect of guanidine HCl on viral RNA replication in cell extracts. AV-FL-Luc-5′rzm RNA was used for a cell-free reaction with or without 2 mM guanidine HCl. Labeled RNA products were analyzed by nondenaturing agarose gel electrophoresis. The positions of RF and ssRNA are indicated.
Site-directed mutagenesis.
An EcoRI fragment of pAV-FL was subcloned into pUC118. The resultant plasmid contains the T7 promoter sequence and the 5′-end 391 nucleotides of the genome. Single mutations were introduced into this plasmid by inverse PCR as described previously (42). The primer pairs used for construction of the mutants and their sequences were as follows: for mut112-118, primers A (5′ CCATACTCCCCCCACCCCCCTTTTGT; plus sense) and B (5′ cacgagcTTTAATTCCTCCGAACAGGTTCC; minus sense); for mut157-163, primers C (5′ cacgggcTGATCTTGACTCCCACGGAAC; plus sense) and D (5′ ACATACTTAGTTACAAAAGG; minus sense); for mut120-124, primers E (5′ TCCCCCCACCCCCCTTTTGTAACTA; plus sense) and F (5′ catacGGTGCCCGTTTAATTCCTCCGAACA; minus sense); and for mut151-155, primers G (5′ catacTGTGCTCGTGATCTTGACTCC; plus sense) and H (5′ TTAGTTACAAAAGGGGGGTGGGGGGAGTA; minus sense). Double mutations were introduced by inverse PCR into the plasmid that had a mutation at nt 112 to 118. The sequences of the primers were as follows: for mut112-118/157-163, primers C and D; for mut55-60/112-118, primers I (5′ cagagcGACCACCGTTACTCCATTCAGCT; plus sense) and J (5′ CAGACCACCGGAAAAGAGGGTGA; minus sense); for mut55-58/112-118, primers K (5′ cagaCGGACCACCGTTACTCCATTCAGCT; plus sense) and J; and for mut57-60/112-118, primers L (5′ gagcGACCACCGTTACTCCATTCAGCTTC; plus sense) and M (5′ ACCAGACCACCGGAAAAGAGG; minus sense). In these primer sequences, the mutated nucleotides are indicated by lowercase letters. The nucleotide sequences of the derived plasmids were confirmed, and then the EcoRI fragments were ligated into pAV-FL and pAV-FL-Luc from which the EcoRI fragment had been removed, yielding various mutant full-length cDNA clones and replicons. In addition, the EcoRI fragments of mut112-118, mut112-118/157-163, and mut55-58/-112-118 were ligated into pAV-FL-Luc-3Dmut from which the EcoRI fragment had been removed, yielding mut112-118-3Dmut, mut112-118/157-163-3Dmut, and mut55-58/112-118-3Dmut, respectively.
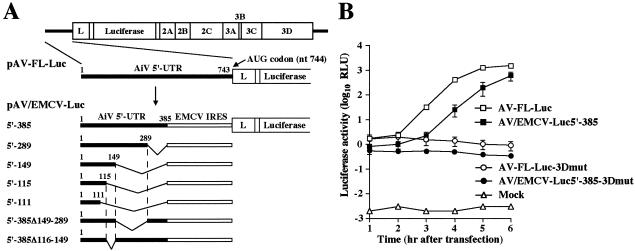
Aichi virus-EMCV chimera replicons containing a luciferase gene. (i) pAV/EMCV-Luc5′-385.
pAV-FL has been linearized with HindIII for in vitro transcription; however, a HindIII site exists in the EMCV IRES. To linearize chimeric replicons, a unique MluI site was introduced downstream of the poly(A) tail as follows. The Aichi virus sequence spanning from nt 6735 to the HindII site downstream of the poly(A) tail was amplified by PCR with primers 6735P (5′ AAACAACCCGCTCCCCTCAAG; plus-strand sequence from nt 6735 to 6755) and Hind-Mlu-poly(A)M (5′ AAAGCTTACGCGTT39GTAAGAACAGT; HindIII and MluI sites underlined, poly(A) tract and minus-strand sequence from nt 8269 to 8279) by using pAV-FL-Luc as a template. The PCR product was digested with PstI (nt 6771) and HindIII and then cloned into the PstI-HindIII sites of pUC118, and the nucleotide sequence of the derived clone was confirmed. The PstI-HindIII fragment of this clone was substituted for the PstI-HindIII fragment of pAV-FL-Luc, generating pAV-FL-Luc-3′Mlu.
Next, the EMCV IRES was introduced into pAV-FL-Luc-3′Mlu as described below. pAV-FLΔP1 (33), which is a full-length cDNA clone with deletion of the capsid-coding region (nt 1275 to 3770), was digested with SacI and HindIII, blunt-ended, and then self-ligated to remove the SacI (upstream of nt 3771)-HindIII fragment [downstream of the poly(A) tail]. Furthermore, the region upstream of the translation initiation site (nt 386 to 742) was deleted from the derived plasmid by PCR using the Spe-385 M primer (5′ TTACTAGTACACGCCGGGGCTAGCCTTAA 3′; minus-strand sequence from nt 365 to 385, with the SpeI site underlined) and the Nco-748P primer (5′ AACCATGGCTGCAACACGGGTTTCACG 3′; plus-strand sequence from nt 748 to 766, with the NcoI site underlined). The PCR product was self-ligated, and the nucleotide sequence between the EcoRI (upstream of the T7 promoter) and XbaI (nt 1275) sites of the derived clone, termed pAV-FLΔP1-HindΔIRES, was confirmed. The EMCV IRES with SpeI and NcoI recognition sites at the 5′ and 3′ ends, respectively, was amplified by PCR with the Spe-EMCV IRES-5′P primer (5′ TACTAGTTTGGAATAAGGCCGGTGTGCGT 3′; SpeI site is underlined) and Nco-EMCV IRES-3′M primer (5′ TCCCATGGTATCATCGTGTTTTTCAAAGGA 3′; NcoI site is underlined) by using pIRES2-EGFP (Clontech) as a template. The PCR product was digested with SpeI and NcoI and then ligated into the SpeI-NcoI sites of pAV-FLΔP1-HindΔIRES, generating pAV-FLΔP1-Hind/EMCV-IRES. The sequence validity of the PCR-generated region of this plasmid was confirmed. Finally, the EcoRI-XbaI fragment of pAV-FLΔP1-Hind/EMCV-IRES, which contains the T7 promoter, the Aichi virus sequence (nt 1 to 385), the EMCV IRES, and the leader protein coding region, was ligated into the EcoRI-XbaI sites of pAV-FL-Luc-3′Mlu, yielding pAV/EMCV-Luc5′-385.
(ii) pAV/EMCV-Luc5′-385-3Dmut.
The Aichi virus sequence spanning from nt 6735 to the HindIII site downstream of the poly(A) tail containing the 3D mutations was amplified by PCR with primers 6735P and Hind-Mlu-poly(A)M by using pAV-FL-Luc-3Dmut. The PCR product was digested with PstI (nt 6771) and HindIII and then cloned into the PstI-HindIII sites of pUC118, and the nucleotide sequence and MluI site of the derived clone were confirmed. The PstI-HindIII fragment of this clone was substituted for the PstI-HindIII fragment of pAV-FL-Luc, generating pAV-FL-Luc-3′Mlu-3Dmut. The EcoRI-XbaI (nt 1275) fragment of pAV/EMCV-Luc5′-385 was ligated into pAV-FL-Luc-3′Mlu-3Dmut, from which the EcoRI-XbaI fragment had been removed, yielding pAV/EMCV-Luc5′-385-3Dmut.
(iii) Deletion mutants of the chimera replicon.
Various lengths of the Aichi virus 5′-end sequence with EcoRI and SpeI sites at the 5′ and 3′ ends, respectively, were amplified by PCR. The sequences of the primers were as follows: for pAV-EMCV-Luc5′-289, primer M13-RV, which anneals to the vector sequence upstream of the T7 promoter sequence in pAV-FL, and primer Spe-289 M (5′ TTACTAGTCAGCCTGTGATACGTCAGGCT; minus sense, from nt 269 to 289, with the SpeI site underlined); for pAV-EMCV-Luc5′-149, primers M13-RV and Spe-149 M (5′ TTACTAGTTAGTTACAAAAGGGGGGTGGG; minus sense, from nt 129 to149, with the SpeI site underlined); for pAV-EMCV-Luc5′-115, primers M13-RV and Spe-115 M (5′ TACTAGTCCCGTTTAATTCCTCCGAACA; minus sense, from nt 95 to 115, with the SpeI site underlined); and for pAV-EMCV-Luc5′-111, primers M13-RV and Spe-111 M (5′ TACTAGTTTTAATTCCTCCGAACAGGTT; minus sense, from nt 91 to 111, with the SpeI site underlined). The PCR fragments were cloned into the pCRII-TOPO vector (Invitrogen), and then the nucleotide sequences of the PCR-derived fragments were confirmed. A deletion mutation, from nt 149 to 289 or nt 116 to 149, was introduced by inverse PCR into pAV-FLΔP1-HindΔIRES. The sequences of the primers were as follows: for pAV-EMCV-Luc5′-385Δ149-289, primers 290P (5′ TGTGAAGCCCCCGCGAAAGCT; plus sense, from nt 290 to 310) and 149 M (5′ TAGTTACAAAAGGGGGGTGGG; minus sense, from nt 129 to 149); and for pAV-EMCV-Luc5′-385Δ116-149, primers 150P (5′ AGTATGTGTGCTCGTGATCTT; plus sense, nt 150 to 170) and 115 M (5′ CCCGTTTAATTCCTCCGAACA; minus sense, from nt 95 to 115). The PCR products were self-ligated, and then the nucleotide sequences of the resultant plasmids were confirmed. The EcoRI-SpeI fragments of the derived clones were substituted for the EcoRI-SpeI fragment of pAV/EMCV-Luc5′-385, yielding various mutant chimera replicons. In addition, the EcoRI-XbaI fragment of pAV/EMCV-Luc5′-111 was ligated into pAV-FL-Luc-3′Mlu-3Dmut, from which the EcoRI-XbaI fragment had been removed, yielding pAV/EMCV-Luc5′111-3Dmut.
In vitro transcription.
pAV-FL and its mutants were linearized by digestion with HindIII or MluI, and RNA transcripts were synthesized with T7 RNA polymerase using a T7 RiboMAX Express large-scale RNA production system (Promega). The integrity of the synthesized RNAs was confirmed by agarose gel electrophoresis.
Titration of viable viruses generated from transcripts.
Vero cell monolayers in 35-mm dishes were transfected with 1 μg of in vitro transcripts using Lipofectin reagent (Life Technologies) according to the manufacturer's recommendations. After incubation at 37°C for 6 h, the cells were washed and then 2.5 ml of growth medium was added to each dish. After incubation for 72 h, the cells were lysed by three consecutive freeze-thaw cycles and then the lysates were subjected to the plaque assay. The numbers and sizes of plaques were determined at 96 h after infection.
Luciferase assay.
RNA transcripts (20 μg) containing a luciferase gene were electroporated into Vero cells as described previously (42), and then the cells were cultured in six 35-mm dishes. At various times after electroporation, cell lysates were prepared and then luciferase activity was measured as described previously (33).
Vero S10 preparation.
A Vero cell S10 extract was prepared according to the method of Svitkin and Sonenberg (46) with modifications. Vero cells grown in tissue culture dishes were trypsinized and then harvested. The cells were washed twice with phosphate-buffered saline and then resuspended in methionine-free Dulbecco's minimal essential medium (5 × 106 to 10 × 106 cells/ml), followed by incubation at 37°C for 2 h with rotation. After incubation, the cells were washed three times with ice-cold isotonic buffer (35 mM HEPES-KOH [pH 7.3], 146 mM NaCl, 11 mM glucose) (46) and then pelleted by centrifugation at 750 × g at 4°C. The pelleted cells were resuspended in 1.5 volumes of hypotonic buffer (25 mM HEPES-KOH [pH 7.3], 50 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol) (46) and then incubated on ice for 20 min. Then, the cells were broken with 30 strokes in a tightly fitting Dounce homogenizer (Wheaton) and centrifuged at 10,000 × g for 20 min at 4°C. The resultant supernatant (40 to 45 U/ml at A260) was treated with micrococcal nuclease (150 U/ml) in the presence of 0.75 mM CaCl2 for 15 min at 20°C, and then EGTA was added to a concentration of 2 mM. The Vero S10 cell extract prepared was divided into aliquots and stored at −80°C.
Virus production, RNA translation, and RNA replication in the cell-free system.
The standard reaction was carried out according to the method of Svitkin and Sonenberg (46) with the exception that the salt solution used was comprised of 1.25 M KCH3COO, 7.5 mM MgCl2, and 2.5 mM spermidine. A 25-μl standard reaction mixture contained 12.5 μl of S10 extracts, 5 μl of the master mix (25 mM HEPES-KOH [pH 7.3]; 10 mM ATP; 2 mM concentrations each of GTP, CTP, and UTP; 100 mM creatine phosphate; 1 mg/ml creatine kinase; 0.2 mM each of amino acids), 5 μl of the salt solution, and 250 ng of viral RNA. To examine the virus yield, the reaction mixture was incubated at 32°C for 15 h. After incubation, the reaction mixture was treated with RNase T1 (400 U/ml) and RNase A (140 μg/ml) at room temperature for 1 h and then subjected to plaque assay. To label translation products, 5 μCi of [35S]methionine-cysteine (Amersham) was added to 25 μl of reaction mixture. Five μl of the reaction mixture was subjected to sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis, and then the gel was dried and exposed to a FujiFilm imaging plate. Radioactive signals were detected with BAS1000 or BAS2000 (FujiFilm). For the standard reaction for RNA labeling, the reaction mixture (25 μl) was incubated without the addition of CTP for 3 h and then 1 μl of 30 μM unlabeled CTP and 5 μCi of [α-32P]CTP (10 mCi/ml, 3,000 Ci/mmol; Amersham) were added. After incubation for 2 h, RNA was extracted as described by Barton et al. (5). When indicated, the extracted RNA was treated with 25 U of RNase T1 and 3.5 μg of RNase A at 37°C for 15 min. The product RNA was analyzed by nondenaturing agarose gel electrophoresis. The gel was dried, and radioactive signals were detected as described above.
RNA stability.
To investigate RNA stability in cell extracts, a 10-μl reaction mixture containing the 32P-labeled RNA transcripts (approximately 10,000 cpm) was incubated at 32°C. At various times after incubation, RNA was extracted, denatured with glyoxal, and then analyzed by 1% agarose gel electrophoresis as described by Sambrook and Russell (41). The gel was dried, and radioactive signals were detected as described above. Signal intensities were measured using a Fujix BAS 2000 phosphorimager.
RNase protection assay.
To detect 32P-labeled positive- or negative-strand RNA synthesized in the cell-free reaction, the RNase protection assay (RPA) was performed with unlabeled probes. To synthesize probes, a BamHI fragment comprising nt 4789 to 5252 of pAV-FL was subcloned into pGEM-3Zf, yielding pGB4789. To synthesize a negative-strand RNA probe, pGB4789 was linearized by digestion with EcoRI and then the RNA was transcribed with SP6 RNA polymerase. For positive-strand RNA probe synthesis, pGB4789 was digested with HindIII and then a transcript was synthesized with T7 RNA polymerase. Hybridization, digestion with RNase T1 and RNase A, and preparation of protected RNAs were carried out under the conditions described by Sambrook and Russell (41). 32P-labeled full-length Aichi virus positive- and negative-strand RNAs were synthesized from pAV-FL and pAV-FL-neg, respectively, using a T7 Maxiscript kit (Ambion) and [α-32P]CTP, and approximately 20,000 cpm of the transcribed RNAs was used as positive controls in RPA.
To detect the positive-strand RNA synthesized in the cell-free reaction, total RNA extracted from a 25-μl cell-free reaction mixture was hybridized with 500 ng of the unlabeled negative-strand probe. For detection of the negative-strand RNA synthesized in the cell-free reaction, total RNA extracted from a 25-μl reaction mixture was subjected to two-cycle RPA (35), 50 ng of the unlabeled positive-strand probe being used in the second hybridization step. After digestion with RNase T1 and RNase A, the protected RNAs were analyzed by 3.5% polyacrylamide-7 M urea gel electrophoresis.
RESULTS
Site-directed mutagenesis of a predicted stem-loop structure downstream of SL-C.
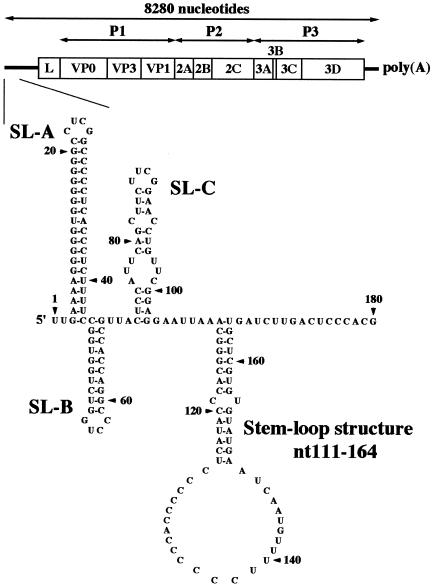
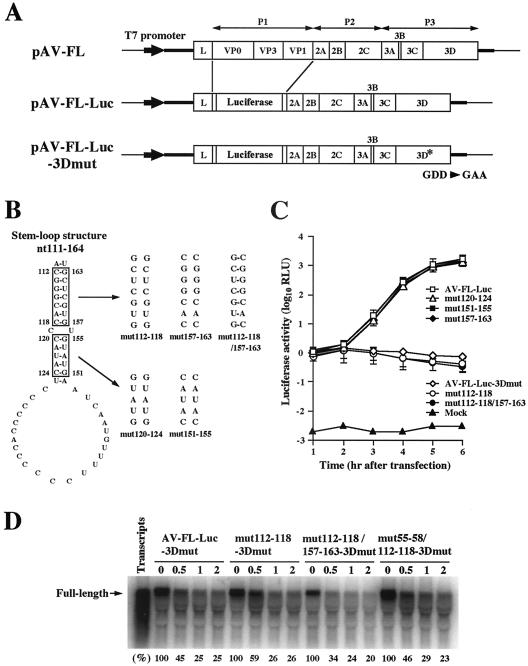
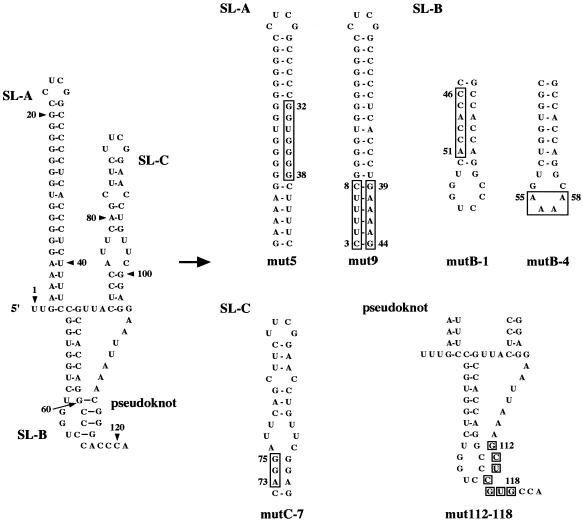
It has already been shown that three stem-loop structures formed within the 120 nucleotides of the 5′ end of the Aichi virus genome are critical for viral RNA replication (33, 42). Here, we analyzed the 180 nucleotides of the 5′ end using the MFOLD program (28) and another stem-loop structure (nt 111 to 164) downstream of SL-C was predicted (Fig. 1). To determine whether this stem-loop structure is required for virus replication, we carried out site-directed mutational analysis using pAV-FL, an infectious cDNA clone of Aichi virus, and pAV-FL-Luc, an Aichi virus replicon harboring a luciferase gene (Fig. 2A). We introduced nucleotide changes on either side (nt 112 to 118, nt 157 to 163, nt 120 to 124, or nt 151 to 155) of the two putative helical segments to disrupt the base-pairings, yielding single mutants mut112-118, mut157-163, mut120-124, and mut151-155 (Fig. 2B). In addition, we introduced nucleotide changes on both sides of the upper stem (nt 112 to 118 and nt 157 to 163) to restore the base-pairings, generating a double mutant, mut112-118/157-163. To examine the effects of these mutations on the translation efficiency, in vitro transcription/translation analysis was carried out in a rabbit reticulocyte lysate. No significant differences in translation efficiency were observed among the mutants (data not shown).
FIG. 1.
Schematic diagram of the Aichi virus genome and the predicted secondary structure of the 5′-end 180 nucleotides of the genome. The thick lines and open box indicate the untranslated regions and coding region, respectively. Vertical lines within the box represent putative cleavage sites for viral proteinases. The three stem-loop structures were designated SL-A, SL-B, and SL-C, and another stem-loop structure (nt 111 to 164) downstream of SL-C was predicted.
FIG. 2.
(A) Organization of the Aichi virus infectious cDNA clone, pAV-FL, and the Aichi virus replicon harboring a luciferase gene, pAV-FL-Luc, and pAV-FL-Luc-3Dmut. The thick lines and open boxes show the untranslated regions and coding regions, respectively. The thin lines indicate the vector sequences. The Aichi virus sequence was cloned just downstream of the T7 promoter sequence. (B) Site-directed mutations introduced into the stem-loop structure (nt 111 to 164) predicted downstream of SL-C. (C) Luciferase activity in cells transfected with AV-FL-Luc and mutants as to the predicted stem-loop structure. Vero cells were electroporated with the RNAs, and then at the indicated times after electroporation, cell lysates were prepared and analyzed for luciferase activity. The experiment was repeated at least three times. Standard deviation bars are shown. (D) RNA stability in Vero cell extracts. 32P-labeled RNA transcripts were incubated at 32°C. At the indicated times after incubation, RNA was extracted, treated with glyoxal, and analyzed by 1% agarose gel electrophoresis. The gel was dried, and radioactive signals were detected with a phosphorimager. Signal intensities of viral RNAs were quantitated, and the percentages of intensities relative to that obtained at 0 h after incubation are shown. Labeled AV-FL-Luc-3Dmut RNA was loaded in the lane represented as input RNA to show its electrophoretic mobility.
RNA transcripts of pAV-FL and its mutants were transfected into Vero cells by lipofection, and the virus titers at 72 h after transfection were determined by plaque assay. On the other hand, RNA transcripts of pAV-FL-Luc and its mutants were transfected into Vero cells by electroporation and luciferase activity in the transfected cells was measured to determine the efficiency of RNA replication. Of the four single mutants, mut157-163, mut120-124, and mut151-155 exhibited efficient RNA replication and virus production in the luciferase and plaque assays, respectively (Fig. 2C and Table 1). In contrast, no increase in luciferase activity was observed in the cells transfected with mut112-118 RNA and the mutant had lost its plaque-forming ability. Additionally, mut112-118/157-163, a double mutant, also lacked the abilities to replicate and to produce viruses, in spite of maintenance of the secondary structure (Fig. 2C and Table 1). To examine RNA stability of these mutants, radiolabeled transcripts, in which the GDD motif of the 3D RNA-dependent RNA polymerase was changed to eliminate RNA replication, were incubated in Vero cell S10 extracts over a timecourse. No significant differences in RNA stability were observed for mut112-118-3Dmut and mut112-118/157-163-3Dmut compared with that for AV-FL-Luc-3Dmut (Fig. 2D). These results suggest that folding of the stem-loop structure (nt 111 to 164) is not important but that the nucleotide sequence of nt 112 to 118 is critical for viral RNA replication.
TABLE 1.
Ability of mutant RNAs to produce viruses
| RNA | Virus titera (PFU/ml) | Plaque sizeb (mm) |
|---|---|---|
| AV-FL | 1.4 × 106 | 2.1 ± 0.1 |
| mut112-118 | 0 | |
| mut157-163 | 1.0 × 106 | 2.1 ± 0.1 |
| mut112-118/157-163 | 0 | |
| mut120-124 | 1.1 × 106 | 1.7 ± 0.2 |
| mut151-155 | 1.3 × 106 | 1.8 ± 0.1 |
| mut55-58/112-118 | 0 | |
| mut55-60/112-118 | 3.8 × 104 | 0.9 ± 0.1 |
| mut57-60/112-118 | 1.1 × 104 | 0.9 ± 0.1 |
Titers of infectious viruses in the cells collected at 72 h after transfection.
Values are the average diameters of 20 plaques ± standard deviations. The sizes were determined at 96 h after transfection.
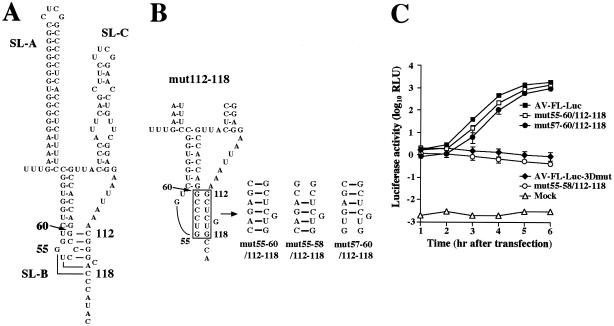
Folding of a pseudoknot structure is critical for viral RNA replication.
The previous study showed that nucleotide changes in the loop segment of SL-B (mutB-4) abolish the replication ability (33). This study suggested the importance of the nucleotide sequence of bases 112 to 118 for viral RNA replication. We compared the two sequences and found that the sequence from nt 55 to 60 of SL-B, GUCCCG, is complementary to that from nt 112 to 118, excluding the cytosine at nt 116 (lowercase), CGGGcAC. This suggests that these sequences potentially interact with each other, resulting in the formation of a pseudoknot structure (Fig. 3A). In mut112-118, which cannot replicate, the pseudoknot structure is disrupted (Fig. 3B). To determine whether folding of the pseudoknot structure is required for RNA replication, we introduced 6-nucleotide mutations into the loop region (nt 55 to 60) of SL-B of mut112-118 to restore the base-pair interaction with nt 112 to 118, excluding the guanosine at nt 116 (mut55-60/112-118) (Fig. 3B). As shown by the luciferase assay (Fig. 3C) and the plaque assay (Table 1), the RNA replication and plaque-forming abilities of this mutant were recovered, albeit there were a slight decrease in the RNA replication efficiency and a 36-fold decrease in the virus titer compared to the wild type. This indicated that a pseudoknot structure formed through interaction between nt 55 to 60 and 112 to 118 is important for RNA replication. To identify further the region forming base-pairings required for RNA replication, we introduced two kinds of 4-nucleotide compensatory mutations into the loop segment of SL-B to generate mut55-58/112-118 and mut57-60/112-118 (Fig. 3B). mut55-58/112-118 involves a 4-base-pair interaction between nt 55 to 58 and nt 114, 115, 117, and 118, while mut57-60/112-118 involves a 4-base-pair interaction between nt 57 to 60 and nt 112 to 115. In the luciferase and plaque assays, RNA replication and virus production were not observed for cells transfected with mut55-58/112-118 RNA (Fig. 3C and Table 1). No significant differences in RNA stability were observed between mut55-58/112-118-3Dmut and AV-FL-Luc-3Dmut (Fig. 2D). On the other hand, mut57-60/112-118 RNA replicated and generated viable viruses, the efficiency being similar to that in the case of mut55-60/112-118 RNA. These results indicate that the pseudoknot structure formed through interaction between nt 57 to 60 (CCCG) in the loop segment of SL-B and nt 112 to 115 (CGGG) is critical for viral RNA replication.
FIG. 3.
(A) The predicted pseudoknot structure. Nt 55 to 60 in the loop segment of SL-B can interact with nt 112 to 118, excluding the cytosine at nt 116. (B) Site-directed mutations introduced into mut112-118 to examine the base-pairing interactions. (C) Luciferase activity in cells transfected with AV-FL-Luc and mutants as to the predicted pseudoknot structure. Vero cells were electroporated with RNAs, and then at the indicated times after electroporation, cell lysates were prepared and analyzed for luciferase activity. The experiment was repeated at least three times. Standard deviation bars are shown.
Construction of AiV-EMCV chimera replicons.
The IRES elements of picornavirus genomes or the hepatitis C virus (HCV) subgenomic replicon can be replaced by the IRES elements of other viruses, and the resulting chimeric viruses have been used to analyze the IRES function and to map the sequences responsible for viral RNA replication without any influence on translation (1, 10, 14, 22, 25, 40). We tried to determine the Aichi virus 5′-end sequence required for RNA replication by using chimera replicons whose 5′ UTRs consist of various lengths of the Aichi virus 5′-terminal sequence and the EMCV IRES (Fig. 4A). We first constructed pAV/EMCV-Luc5′-385. In this construct, the approximately 350 nucleotides (nt 386 to 742) upstream of the translation initiation codon were replaced with the EMCV IRES (Fig. 4A). The IRES element of Aichi virus has not been mapped yet. However, since almost all picornavirus IRESs consist of approximately 450 nucleotides, we thought that the 385 nucleotides of the 5′ end of the Aichi virus genome would include the 5′-terminal part of an IRES, i.e., that the 5′-terminal 385 nucleotides would contain the region required for RNA replication. As a negative control, we constructed pAV/EMCV-Luc5′-385-3Dmut, in which the GDD motif of the 3D RNA-dependent RNA polymerase was changed. Upon in vitro transcription/translation analysis with rabbit reticulocyte lysate, no apparent decrease in the translation efficiency was observed for pAV/EMCV-Luc5′-385 and pAV/EMCV-Luc5′-385-3Dmut compared with that for pAV-FL-Luc (data not shown).
FIG. 4.
(A) Schematic diagram of the 5′ UTR of Aichi virus and chimera replicons. The thick lines indicate the Aichi virus sequences in the 5′ UTR. Each plasmid name shows the number of Aichi virus nucleotides retained at the 5′ end. The open boxes indicate EMCV IRES. (B) Luciferase activity in the transfected cells with AV-FL-Luc, AV/EMCV-Luc5′-385, AV-FL-Luc-3Dmut, and AV/EMCV-Luc5′-385-3Dmut. Vero cells were electroporated with the RNAs, and then at the indicated times after electroporation, cell lysates were prepared and analyzed for luciferase activity. The experiment was repeated at least three times. Standard deviation bars are shown.
To examine the translation and RNA replication abilities in transfected cells, in vitro transcripts of pAV/EMCV-Luc5′-385 and pAV/EMCV-Luc5′-385-3Dmut were electroporated into Vero cells and then the luciferase activities in the transfected cells were measured (Fig. 4B). At 1 h after transfection, the luciferase activities of AV-FL-Luc and AV-FL-Luc-3Dmut were at almost the same level with each other. This indicates that, at this time, replication of the replicons had hardly occurred and that the luciferase activity in transfected cells represents translation from input RNAs. At 1 h after transfection, AV/EMCV-Luc5′-385 RNA and AV/EMCV-Luc5′-385-3Dmut RNA were slightly lower than those of AV-FL-Luc and AV-FL-Luc-3Dmut in cells transfected with AV/EMCV-Luc5′-385 RNA and AV/EMCV-Luc5′-385-3Dmut RNA, respectively. This indicates that the chimeric replicons had only a slight defect in translation in transfected cells compared to wild-type replicons. At 3 h after transfection, the luciferase activity in the AV/EMCV-Luc5′-385 RNA-transfected cells began to increase. Although the increase in the luciferase activity in the AV/EMCV-Luc5′-385 RNA-transfected cells was slower than that in the AV-FL-Luc RNA-transfected cells, the level at 6 h was similar to that in the case of AV-FL-Luc (Fig. 4B). These results show that AV/EMCV-Luc5′-385 RNA replicates efficiently in transfected cells. We considered that we could map the 5′-terminal sequence of the Aichi virus genome required for RNA replication using this chimera replicon without any influence on translation.
Mapping of the essential element for viral RNA replication.
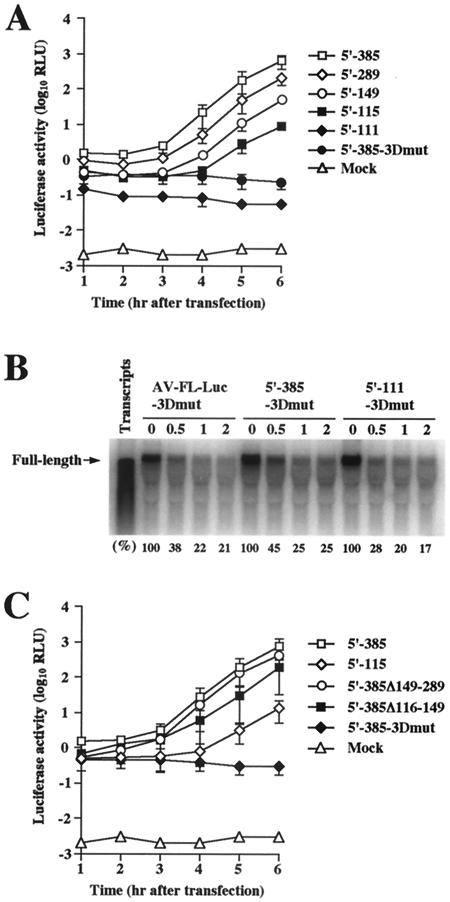
We investigated the 5′-end sequence required for RNA replication by using AV/EMCV-Luc5′-385 and a series of deletion mutants (AV/EMCV-Luc5′-289, AV/EMCV-Luc5′-149, AV/EMCV-Luc5′-115, and AV/EMCV-Luc5′-111) (Fig. 4A).
The replicon with the 5′-end 111 nucleotides of Aichi virus at the 5′ end (AV/EMCV-Luc5′-111), which does not include the pseudoknot structure, could not replicate (Fig. 5A). This replicon appeared to have a slight defect in the translation activity, as indicated by the luciferase activity at 1 h after transfection; however, its activity was significantly higher than that in mock-transfected cells. No significant decrease in RNA stability was observed for AV/EMCV-Luc5′-111-3Dmut compared with that for AV-FL-Luc-3Dmut and AV/EMCV-Luc5′-385-3Dmut (Fig. 5B). The inability of AV/EMCV-Luc5′-111 to replicate would be due mainly to disruption of the pseudoknot structure. In contrast, replication of the replicon carrying the 5′-end 115 nucleotides of Aichi virus at the 5′ end (AV/EMCV-Luc5′-115), in which the pseudoknot structure could be formed, was observed. The mutants with the longer Aichi virus 5′-terminal sequences at the 5′ end (AV/EMCV-Luc5′-149 and AV/EMCV-Luc5′-289) replicated more efficiently (Fig. 5A). These results indicate that the 5′-end 115 nucleotides of the genome including the pseudoknot structure are the minimum requirement for RNA replication. In addition, these results also imply that a sequence downstream of the pseudoknot structure participates in enhancement of the RNA replication efficiency.
FIG. 5.
(A and C) Luciferase activity in cells transfected with AV/EMCV-Luc5′-385 and a series of deletion mutants as to the 5′ UTR. Vero cells were electroporated with the RNAs, and then at the indicated times after electroporation, cell lysates were prepared and analyzed for luciferase activity. The experiment was repeated at least three times. Standard deviation bars are shown. (B) RNA stability of AV-FL-Luc-3Dmut RNA, AV/EMCV-Luc5′-385-3Dmut RNA, and AV/EMCV-Luc5′-111-3Dmut RNA in Vero cell extracts. RNA stability was analyzed as described in the legend to Fig. 2D. Signal intensities of viral RNAs were quantitated, and the percentages of intensities relative to that obtained at 0 h after incubation are shown.
Then, to determine whether any element enhancing the replication efficiency was present in the sequence downstream from nt 115, deletions of nt 116 to 149 and nt 149 to 289 were introduced into pAV/EMCV-Luc5′-385, yielding pAV/EMCV-Luc5′-385Δ116-149 and pAV/ENCV-Luc5′-385Δ149-289, respectively (Fig. 4A). The luciferase assay showed that the deletion of nt 149 to 289 had only a modest effect on the RNA replication efficiency (Fig. 5C). The deletion of nt 116 to 149 also did not severely affect the efficiency of replication. These results, together with the finding that the deletion of nt 289 to 385 had a modest effect on replication (AV/EMCV-Luc5′-289) (Fig. 5A), suggest that the presence of the sequence downstream of the pseudoknot structure enhances the RNA replication efficiency but that a region absolutely required for enhancement might not exist.
Cell-free replication system for Aichi virus.
The cell-free translation/replication system was first developed for poliovirus by Molla et al. (29), and since then, this system has been frequently used as a powerful tool for analyzing the mechanism of poliovirus replication. In this study, we tried to develop a cell-free translation/replication system for Aichi virus.
(i) A cis-active hammerhead ribozyme.
It has been reported that efficient replication of poliovirus RNA in HeLa cell extracts requires a precise 5′ end and that a cis-active hammerhead ribozyme is used to remove the nonviral nucleotides present at the 5′ ends of T7 RNA polymerase-synthesized transcripts derived from a poliovirus full-length cDNA clone (19). We performed mutagenesis to introduce a cis-active hammerhead ribozyme at the 5′ end of the Aichi virus cDNA sequence using a plasmid, pU5′Eco (42), which contained the T7 promoter and the 5′-end 391 nucleotides of the Aichi virus genome. First, we constructed a hammerhead ribozyme whose stem I comprised 8 base pairs (pU5′Eco-5′rzm-8bp) (Fig. 6A ). pU5′Eco linearized by digestion with HindIII would give rise to RNA 444 nt in length upon in vitro transcription with T7 RNA polymerase, and, on the other hand, transcripts derived from pU5′Eco-5′rzm-8bp would be 505 nt in length before cleavage. When transcripts synthesized from pU5′Eco-5′rzm-8bp were electrophoresed on a denaturing polyacrylamide gel, two products were detected, the shorter one being the same size as the transcripts synthesized from pU5′Eco (Fig. 6B, lanes 1 and 2). This shorter product, which is thought to be the cleaved form, comprised only a small portion of the transcripts. We presumed that the formation of a stable stem-loop structure (SL-A) at the 5′ end of the genome (42) may inhibit the formation of stem I of the hammerhead ribozyme, resulting in inefficient cleavage. Next, we constructed a hammerhead ribozyme with stem I comprising 22 base pairs (pU5′Eco-5′rzm) (Fig. 6A). Transcripts derived from pU5′Eco-5′rzm also contained the uncleaved form (Fig. 6B, lane 3), but the hammerhead ribozyme functioned more efficiently than that in the transcripts derived from pU5′Eco-5′rzm-8bp. We used this hammerhead ribozyme with stem I comprising 22 base pairs in the following experiments.
(ii) Virus production in Vero cell S10 extracts.
For the cell-free Aichi virus replication system, we used Vero cell S10 extracts. An Aichi virus cDNA clone with the 5′ hammerhead ribozyme sequence, pAV-FL-5′rzm, was constructed (Fig. 6C). To optimize the concentrations of K+ and Mg2+ in cell-free reactions, transcripts synthesized from pAV-FL-5′rzm were incubated in the cell-free reaction mixtures containing various concentrations of K+ and Mg2+ at 32°C for 15 h and then the virus yield was determined by plaque assay. As a result, optimal concentrations of K+ and Mg2+ were determined to be 150 mM and 1.5 mM, respectively, and the virus yield was 3 × 104 PFU/ml. Under these conditions, the reaction programmed with the virion RNA produced viable viruses at the titer of 3 × 105 PFU/ml. This titer is 10-fold higher than that for the reaction programmed with AV-FL-5′rzm RNA. The decreased virus production observed for the reaction programmed with AV-FL-5′rzm RNA was probably due mainly to the incomplete cleavage by the 5′ hammerhead ribozyme. There was no morphological difference between the plaques formed by viruses derived from virion RNA and AV-FL-5′rzm RNA (data not shown). A 25-μl reaction mixture programmed with AV-FL RNA produced no viable viruses.
(iii) Translation and replication of Aichi virus RNA in cell extracts.
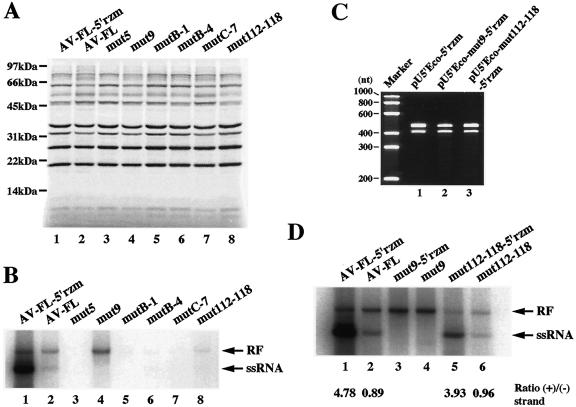
Reactions were programmed with AV-FL-5′rzm RNA, AV-FL RNA, or AV-FL-3Dmut RNA. AV-FL RNA and AV-FL-3Dmut RNA have three nonviral nucleotides, GGU, at their 5′ ends (Fig. 6C). Translation products of the three RNAs were labeled by adding [35S]methionine-cysteine into the cell-free reaction mixture. Translation of these three RNAs occurred with similar efficiency (Fig. 6D). For detection of RNA synthesized in cell-free reactions, [32P]CTP was added to the reaction mixture at 3 h after the start of the reaction and then the mixture was incubated for an additional 2 h. Product RNAs with or without RNase A and RNase T1 treatment were analyzed by nondenaturing agarose gel electrophoresis (Fig. 6E). The reaction with AV-FL-5′rzm RNA produced both RNase A/T1-resistant double-stranded replicative form (RF) and RNase A/T1-sensitive single-stranded RNA (ssRNA) (lanes 1 and 4). The labeled RF consists of unlabeled inputted positive-strand RNA and labeled negative-strand RNA, while the labeled ssRNA consists of newly synthesized positive-strand RNA (19, 20). When AV-FL RNA was used for a reaction, RF and ssRNA were also detected (lanes 2 and 5); however, the amount of ssRNA was much less than that in the reaction with AV-FL-5′rzm RNA whereas the amounts of RF were similar. This indicates that the extra 3 nucleotides at the 5′ end of the transcript do not affect negative-strand RNA synthesis but significantly impair positive-strand synthesis.
It has been reported that a cell-free reaction programmed with poliovirus RNA with two nonviral guanosine residues at the 5′ end does not produce a detectable level of positive-strand RNA (6, 19). In contrast, in our cell-free system, extra nucleotides at the 5′ end of the transcript appeared not to abolish positive-strand synthesis completely. To further confirm that the reaction programmed with AV-FL RNA produces a detectable amount of positive-strand RNA, RPA was performed with unlabeled probes. For the detection of negative-strand RNA, two-cycle RPA (35) was carried out. AV-FL RNA and AV-FL-5′rzm RNA produced similar amounts of negative-strand RNA (Fig. 6F, lanes 6 and 7). In addition, consistent with the results described above, positive-strand RNA was detected in a much lower amount in the reaction programmed with AV-FL RNA than in that with AV-FL-5′rzm (lanes 2 and 3). Thus, the extra 3 nucleotides at the 5′ ends of Aichi virus RNA transcripts remarkably impaired positive-strand RNA synthesis in our cell-free replication system but synthesized positive-strand RNA was detectable.
(iv) Effect of guanidine hydrochloride (HCl) on Aichi virus RNA replication.
It is known that guanidine HCl is an inhibitor of poliovirus RNA replication. Indeed, we confirmed that Vero cells infected with poliovirus exhibited no cytopathic effect in the presence of 2 mM of guanidine HCl (data not shown). In contrast, addition of 2 mM of guanidine HCl to the cell culture medium did not inhibit Aichi virus replication. AV-FL-Luc RNA replicated in transfected cells in the presence of 2 mM of guanidine HCl as efficiently as in the absence of guanidine HCl (data not shown). In addition, in cell extracts, the presence of 2 mM of guanidine HCl did not affect the replication of AV-FL-5′rzm RNA (Fig. 6G). Thus, guanidine HCl is not an inhibitor of Aichi virus RNA replication.
In most studies on poliovirus using the cell-free system, preinitiation replication complexes isolated from the cell-free reaction containing 2 mM guanidine HCl (4) are used for analyzing RNA replication. In the following experiments of this study, however, analyses of RNA synthesis in cell extracts were performed without isolating preinitiation replication complexes, due to the insensitivity of Aichi virus to guanidine HCl.
In summary, a cell-free translation/replication system for Aichi virus was established and it became possible to analyze the replication of genetically engineered Aichi virus RNAs in cell extracts by using transcripts synthesized from a cis-active hammerhead ribozyme-containing full-length cDNA clone.
The 5′ end of the Aichi virus genome is important for both positive- and negative-strand RNA synthesis.
We have already carried out site-directed mutational analysis of the 5′ end of the genome and obtained many mutants that could not replicate in transfected cells (33, 42). In this study, using cell extracts that can support the translation, replication, and encapsidation of Aichi virus RNA, we investigated the abilities of these mutants to synthesize positive- and negative-strand RNAs. mut5, mut9, mutB-1, mutB-4, mutC-7, and mut112-118 were used for this experiment (Fig. 7). mut5 contains a 7-nucleotide mutation (nt 32 to 38) in the middle part of the stem of SL-A to disrupt the base-pairings of the stem, and mut9 was constructed by exchanging 6-nucleotide stretches (nt 3 to 8 and 39 to 44) in the lower part of the stem of SL-A with each other to maintain the base-pairings of the stem of SL-A (42). mutB-1 and mutC-7 contain mutations that disrupt the base-pairings of the stem of SL-B and the lower stem of SL-C, respectively (33). In mutB-4, which has nucleotide changes in the loop region of SL-B (33), and mut112-118, the formation of the pseudoknot structure was prevented (Fig. 3B). Upon dot blot hybridization or luciferase assay, replication of these mutants was not observed in transfected cells (33, 42) (Fig. 2C).
FIG. 7.
Site-directed mutations introduced into the three stem-loop and pseudoknot structures. The mutated regions and nucleotides are boxed.
First, translation of mutant RNAs in Vero cell extracts was examined. In this system, newly synthesized proteins can be detected independent of RNA replication. No significant differences in the translation efficiency among these mutants were observed (Fig. 8A).
FIG. 8.
(A) Translation of mutant RNAs in Vero cell extracts. RNA transcripts were subjected to in vitro translation reaction in the presence of [35S]methionine-cysteine for 4 h at 32°C. The translation products were electrophoresed on an SDS-polyacrylamide gel, and then the gel was dried. Radioactive signals were detected with a phosphorimager. (B) Positive- and negative-strand RNA synthesis in the cell-free reactions. The product RNA was analyzed by native agarose gel electrophoresis. Radioactive signals were detected with a phosphorimager. The positions of RF and ssRNA are indicated. (C) Cleavage by the hammerhead ribozyme. In vitro transcripts synthesized from pU5′Eco-5′rzm, pU5′Eco-mut9-5′rzm, and pU5′Eco-112-118-5′rzm were electrophoresed on a 3.5% polyacrylamide-7 M urea gel, and then the gel was stained with ethidium bromide. (D) Positive- and negative-strand RNA synthesis in cell-free reactions of mut9 and mut112-118 harboring the 5′ hammerhead ribozyme. The ratios of positive-strand to negative-strand RNA synthesis are indicated.
Next, RNA synthesis of the mutants in a cell-free reaction was investigated. In the analysis using RNAs that have three nonviral nucleotides (GGU) at the 5′ end (Fig. 6C), no RF band was detected for the reaction programmed with mut5 (Fig. 8B, lane 3). mutB-1, mutB-4, and mutC-7 produced only faint bands (lanes 5 to 7). mut112-118 synthesized RF and positive-strand RNA at significantly lower levels than those for AV-FL, although the ratio of positive-strand RNA to RF was similar for AV-FL and mut112-118. These results indicate that mut5, mutB-1, mutB-4, mutC-7, and mut112-118 have a defect in negative-strand synthesis. In contrast, mut9 generated only an RF band, i.e., ssRNA was not detected (Fig. 8B, lane 4), indicating that mut9 has a defect in positive-strand synthesis but not in negative-strand synthesis.
The abilities of mut112-118 and mut9 to produce positive-strand RNA were further investigated using mut112-118-5′rzm RNA and mut9-5′rzm RNA. The hammerhead ribozymes introduced into these two mutants exhibited a similar level of the cleavage efficiency compared to that introduced into AV-FL-5′rzm (Fig. 8C). As expected, mut112-118-5′rzm generated positive-strand RNA more efficiently than mut112-118 (Fig. 8D, compare lanes 5 and 6). mut112-118-5′rzm and mut112-118 produced smaller amounts of positive-strand RNA and RF than AV-FL-5′rzm and AV-FL, respectively. Quantitative analysis showed that the amount of the RF synthesized by mut112-118 was sevenfold less than that synthesized by AV-FL and that the amount of ssRNA produced by mut112-118 to 5′rzm was sixfold less than that in AV-FL-5′rzm (Fig. 8D). However, the ratio of positive-strand RNA to RF was similar between the two RNAs (compare lanes 1 and 5). This further indicates that mut112-118 has a defect in negative-strand synthesis, not in positive-strand synthesis. In contrast, mut9-5′rzm synthesized no detectable level of ssRNA, indicating a defect of this mutant in positive-strand RNA synthesis.
Overall, the results of analysis of RNA synthesis by the mutants using the cell-free replication system indicate that the formation of the proper secondary and tertiary structures at the 5′ end of the genome is required for negative-strand RNA synthesis. In addition, it was shown that mutations of the primary sequence in the lower part of the stem of SL-A abolishes positive-strand RNA synthesis.
DISCUSSION
Computer-assisted secondary structure prediction suggested the formation of a stem-loop structure at nt 111 to 164, besides the previously defined three stem-loop structures SL-A, SL-B, and SL-C (Fig. 1). In this study, using various site-directed mutants, we examined whether the predicted stem-loop structure downstream of SL-C is important for virus replication. As a result, folding of this stem-loop structure (nt 111 to 164) was found to not be important for RNA replication (Fig. 2), but a novel structural element, a pseudoknot structure, was detected. This pseudoknot structure is formed through interaction between nt 57 to 60 in the loop segment of SL-B and nt 112 to 115, and its proper folding is critical for RNA replication (Fig. 3). The formation of a pseudoknot structure is predicted at the 5′ ends of the genomes of other picornaviruses including cardio-, parecho-, and hepatoviruses (11, 13, 17, 24), and, for mengovirus and human parechovirus 1, experimental data showing the importance of pseudoknot structures for RNA replication have been presented (27, 34). Recently, a virus, the U-1 strain, related to Aichi virus was discovered and its complete genome sequence was determined (49). The 5′-terminal region of the genome of strain U-1 is predicted to fold into three stem-loop structures similar to those in Aichi virus (49). We found that the 5′-terminal region of strain U-1 has the potential to form a pseudoknot structure similar to that found in Aichi virus (data not shown).
This study also indicated that the 5′-end 115 nucleotides including the pseudoknot are the minimum requirement for RNA replication. The chimera replicon harboring the 5′-terminal 111 nucleotides of the genome (AV/EMCV-Luc5′-111), which does not include the pseudoknot structure, did not replicate (Fig. 5A). In contrast, replication of AV/EMCV-Luc5′-115, in which the pseudoknot structure is formed, was observed. However, the chimera replicons harboring the longer 5′-terminal sequences of the Aichi virus genome replicated more efficiently (Fig. 4A and 5A). It has been reported that stem-loop II within the poliovirus IRES is involved in RNA replication (21, 45). In HAV, the 5′-end 94 nucleotides of the genome fold into stem-loop structures with the potential to form pseudoknot structures (11, 44); however, an experiment involving chimera replicons showed that the 5′-end 237 nucleotides are required for RNA replication (22). Since the 5′ border of the HAV IRES is mapped between nt 151 and 257 (10), the 5′-end 237 nucleotides may contain the 5′ part of the IRES. In addition, it has been shown in HAV that the nucleotide sequence of bases 140 to 144 is essential for efficient replication in cultured cells (44). In the case of Aichi virus, deletion of either nt 116 to 149, 149 to 289, or 289 to 385 did not severely decrease the RNA replication efficiency compared to that in the case of AV/EMCV-Luc5′-385 (Fig. 5A and C). Thus, there appears to be no region absolutely necessary for enhancement of the replication efficiency.
It is possible that the nucleotide sequence downstream of nt 115 serves to spatially separate the two elements, i.e., the 5′-terminal cis-acting replication element and the IRES, or to allow the two elements to fold into the proper structures. In the HCV replicon harboring the poliovirus IRES, when the HCV 5′ UTR is directly fused to the poliovirus IRES, the poliovirus IRES activity is severely impaired (14). The introduction of a spacer element between the HCV 5′ UTR and the poliovirus IRES of the replicon improves the translation activity. Also, in this study, the translation activity of AV/EMCV-Luc5′-111 was found to be reduced (Fig. 5A). Although a significant reduction in the translation activity of the other chimera replicons with reduced RNA replication ability was not observed, it is possible that the proper folding of the 5′-end cis-element or -binding of proteins required for RNA replication to the 5′-end cis-element is prevented. Very recently, it was shown that the poliovirus RNA with the complete IRES deletion replicates in cell-free translation-replication reactions (32). A similar approach will clarify further the role of the nucleotide sequence downstream of nt 115 in RNA replication. In addition, this approach will also help to exclude the possibility that the EMCV IRES introduced into chimera replicons contains sequences that affect Aichi virus RNA replication.
In this study, we established a cell-free replication system for Aichi virus. Vero S10 extracts programmed with in vitro transcripts synthesized from a full-length cDNA clone containing a cis-active hammerhead ribozyme generated viable viruses. Thus, it became possible to analyze Aichi virus replication in cell extracts using genetically engineered viral RNA.
The cell-free replication system has been used as a powerful tool to analyze the mechanism of poliovirus RNA replication, because using this system, it is possible to distinguish between mutant RNAs defective in the synthesis of negative- and positive-strand RNA. Our system also made this possible. By electrophoresis of the product RNAs of cell-free reactions on a nondenaturing agarose gel, labeled double-stranded RF and ssRNA were detected. Labeled RF and ssRNA are indications of the synthesis of negative-strand RNA and positive-strand RNA, respectively (19, 20).
As reported for poliovirus (19), removal of extra nucleotides at the 5′ end of the genome using a hammerhead ribozyme resulted in a significant increase of positive-strand synthesis in a cell-free reaction (Fig. 8D, compare lanes 1 and 2). However, the cis-active hammerhead ribozyme that we designed did not function completely (Fig. 6B, lane 3). A subset of AV-FL-5′rzm RNA, which still has the uncleaved ribozyme sequence at the 5′ end, would function as a template for the negative-strand RNA. But the negative-strand RNAs complementary to the sequence of the uncleaved ribozyme sequence at the 3′ end would not serve as templates for positive-strand synthesis. This would be a cause of the relatively low value of the ratio of ssRNA to RF (5:1) observed in a cell-free reaction of AV-FL-5′rzm RNA (Fig. 8D) compared to the ratio reported in a cell-free reaction of poliovirus (20:1) (19). It is necessary to enhance the cleavage efficiency of the ribozyme for a detailed quantitative analysis of positive-strand synthesis. On the other hand, for a quantitative analysis of negative-strand synthesis, RNA with extra nucleotides at the 5′ end would be more suitable than RNA synthesized from the plasmid harboring the 5′ ribozyme because the RF synthesized by RNA with the accurate 5′ end is used efficiently as a template for positive-strand synthesis, resulting in the conversion of the RF to replicative intermediate RNA.
Using cell extracts, we investigated the replication properties of mutants harboring a lethal mutation in the 5′-terminal region. Mutations disrupting structural elements abolished or impaired negative-strand RNA synthesis remarkably. mut112-118, in which formation of the pseudoknot structure is prevented, produced very small amounts of positive- and negative-strand RNAs in the cell-free reaction. Interestingly, the ratio of positive-strand RNA to RF was similar for AV-FL-5′rzm and mut112-118-5′rzm (Fig. 8D, lanes 1 and 5). Since the ribozymes in the two RNAs exhibited a cleavage efficiency similar to each other (Fig. 8C, lanes 1 and 3), this result implies that the two RNAs synthesize positive-strand RNA with an equivalent efficiency. That is, this suggests that disruption of the pseudoknot structure affects negative-strand RNA synthesis but not positive-strand RNA synthesis. On the other hand, nucleotide changes in the lower part (nt 3 to 8/39 to 44) of the stem of SL-A to maintain the base-pairings of the stem did not affect negative-strand synthesis but abolished positive-strand RNA synthesis (Fig. 8D, mut9).
In poliovirus, the 5′ cloverleaf structure binds to 3CD and PCBP (2, 3, 16, 36) and 3CD and PCBP interact with poly(A)-binding protein that binds to the poly(A) tract at the 3′ end of the genome (7, 20, 26). Such genome circularization is required for negative-strand RNA synthesis. It is likely that the 5′-terminal structural elements of the Aichi virus genome function in a similar manner in negative-strand synthesis. Disruption of the 5′-terminal structural elements would probably prevent interaction with proteins required for genome circularization. We intend to identify viral or host proteins interacting with the 5′-terminal structural elements of the Aichi virus genome.
Also, the involvement of the poliovirus 5′ cloverleaf structure in positive-strand synthesis has previously been reported (2). Mutation of stem-loop b or d, which affects formation of the RNP complex, reduced the ratio of positive- to negative-strand RNA accumulated in infected cells (2). However, it has also been shown that the same mutation introduced into stem-loop d inhibits negative-strand RNA synthesis severely in the cell-free reaction (7). In this study, we demonstrated clearly that a mutation in the 5′-end nucleotide sequence (mut9) abolishes positive-strand RNA synthesis without affecting negative-strand synthesis using the cell-free translation-replication system (Fig. 8D, lanes 3 and 4). For Sindbis virus, it has been reported that the 5′ end of the genome contains cis-acting elements that regulate minus- and plus-strand RNA synthesis. A notable finding is that deletion of the very 5′-end nucleotides (nt 2 to 4 or nt 5) had no or only a moderate effect on minus-strand RNA synthesis but almost completely abolished plus-strand synthesis (15).
It is possible that the 5′-end sequence functions in the complementary sequence, i.e., in the 3′ end of the negative-strand RNA, for positive-strand synthesis. It should be noted that, in mut9, the nucleotide change was introduced into the sequence that serves as the template for the addition of nucleotides directly to VPgpUpU, which is the primer for positive-strand RNA synthesis (18, 30, 31). The specific 3′-end nucleotide sequence of the negative-strand RNA may be required for direct binding with 3D polymerase or for promotion of the assembly of a complex required for the initiation of positive-strand RNA synthesis. Recently, it has been shown that the 3′ UTR of the poliovirus genome is involved in positive-strand RNA synthesis (9). This suggests that the 5′-end region of the negative-strand RNA, which is the complementary sequence to the 3′ UTR, interacts with the 3′ end of the negative-strand RNA to initiate positive-strand RNA synthesis. The 3′-end nucleotide sequence of the negative-strand RNA may interact with the viral or cellular proteins required to circularize the negative-strand RNA. Further experiments are required to determine the role of the 5′-end sequence in positive-strand RNA synthesis.
In conclusion, this study has indicated that the 5′-end 115 nucleotides of the Aichi virus genome are the minimum requirement for viral RNA replication and that they encode elements required for positive- and negative-strand RNA synthesis. It has been previously shown that the most 5′-end stem-loop structure, SL-A, is involved in RNA encapsidation. Thus, the 5′ terminus of the Aichi virus genome is a multifunctional region.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Alexander, L., H. H. Lu, and E. Wimmer. 1994. Polioviruses containing picornavirus type 1 and/or type 2 internal ribosomal entry site elements: genetic hybrids and the expression of a foreign gene. Proc. Natl. Acad. Sci. USA 91:1406-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andino, R., G. E. Rieckhof, and D. Baltimore. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63:369-380. [DOI] [PubMed] [Google Scholar]
- 3.Andino, R., G. E. Rieckhof, P. L. Achacoso, and D. Baltimore. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 12:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton, D. J., E. P. Black, and J. B. Flanegan. 1995. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J. Virol. 69:5516-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1996. Assays for poliovirus polymerase, 3DPol, and authentic RNA replication in HeLa S10 extracts. Methods Enzymol. 275:35-57. [DOI] [PubMed] [Google Scholar]
- 6.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1999. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 73:10104-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birikh, K. R., P. A. Heaton, and F. Eckstein. 1997. The structure, function and application of the hammerhead ribozyme. Eur. J. Biochem. 245:1-16. [DOI] [PubMed] [Google Scholar]
- 9.Brown, D. M., S. E. Kauder, C. T. Cornell, G. M. Jang, V. R. Racaniello, and B. L. Semler. 2004. Cell-dependent role for the poliovirus 3′ noncoding region in positive-strand RNA synthesis. J. Virol. 78:1344-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, E., A. J. Zajac, and S. M. Lemon. 1994. In vitro characterization of an internal ribosomal entry site (IRES) present within the 5′ nontranslated region of hepatitis A virus RNA: comparison with the IRES of encephalomyocarditis virus. J. Virol. 68:1066-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, E. A., S. P. Day, R. W. Jansen, and S. M. Lemon. 1991. The 5′ nontranslated region of hepatitis A virus RNA: secondary structure and elements required for translation in vitro. J. Virol. 65:5828-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark, J. M. 1988. Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res. 16:9677-9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duke, G. M., M. A. Hoffman, and A. C. Palmenberg. 1992. Sequence and structural elements that contribute to efficient encephalomyocarditis virus RNA translation. J. Virol. 66:1602-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frolov, I., R. Hardy, and C. M. Rice. 2001. Cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA 7:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamarnik, A. V., and R. Andino. 1997. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA 3:882-892. [PMC free article] [PubMed] [Google Scholar]
- 17.Ghazi, F., P. J. Hughes, T. Hyypiä, and G. Stanway. 1998. Molecular analysis of human parechovirus type 2 (formerly echovirus 23). J. Gen. Virol. 79:2641-2650. [DOI] [PubMed] [Google Scholar]
- 18.Goodfellow, I. G., C. Polacek, R. Andino, and D. J. Evans. 2003. The poliovirus 2C cis-acting replication element-mediated uridylylation of VPg is not required for synthesis of negative-sense genomes. J. Gen. Virol. 84:2359-2363. [DOI] [PubMed] [Google Scholar]
- 19.Herold, J., and R. Andino. 2000. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 74:6394-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishii, T., K. Shiroki, A. Iwai, and A. Nomoto. 1999. Identification of a new element for RNA replication within the internal ribosome entry site of poliovirus RNA. J. Gen. Virol. 80:917-920. [DOI] [PubMed] [Google Scholar]
- 22.Jia, X.-Y., M. Tesar, D. F. Summers, and E. Ehrenfeld. 1996. Replication of hepatitis A viruses with chimeric 5′ nontranslated regions. J. Virol. 70:2861-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koonin, E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72:2197-2206. [DOI] [PubMed] [Google Scholar]
- 24.Le, S.-Y., J.-H. Chen, N. Sonenberg, and J. V. Maizel, Jr. 1993. Conserved tertiary structural elements in the 5′ nontranslated region of cardiovirus, aphthovirus and hepatitis A virus RNAs. Nucleic Acids Res. 21:2445-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu, H.-H., and E. Wimmer. 1996. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosomal entry site of hepatitis C virus. Proc. Natl. Acad. Sci. USA 93:1412-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyons, T., K. E. Murray, A. W. Roberts, and D. J. Barton. 2001. Poliovirus 5′-terminal cloverleaf RNA is required in cis for VPg uridylylation and the initiation of negative-strand RNA synthesis. J. Virol. 75:10696-10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin, L. R., and A. C. Palmenberg. 1996. Tandem mengovirus 5′ pseudoknots are linked to viral RNA synthesis, not poly(C)-mediated virulence. J. Virol. 70:8182-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 29.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free, de novo synthesis of poliovirus. Science 254:1647-1651. [DOI] [PubMed] [Google Scholar]
- 30.Morasco, B. J., N. Sharma, J. Parilla, and J. B. Flanegan. 2003. Poliovirus cre(2C)-dependent synthesis of VPgpUpU is required for positive- but not negative-strand RNA synthesis. J. Virol. 77:5136-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murray, K. E., and D. J. Barton. 2003. Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J. Virol. 77:4739-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray, K. E., B. P. Steil, A. W. Roberts, and D. J. Barton. 2004. Replication of poliovirus RNA with complete internal ribosome entry site deletions. J. Virol. 78:1393-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagashima, S., J. Sasaki, and K. Taniguchi. 2003. Functional analysis of the stem-loop structures at the 5′ end of the Aichi virus genome. Virology 313:56-65. [DOI] [PubMed] [Google Scholar]
- 34.Nateri, A. S., P. J. Hughes, and G. Stanway. 2002. Terminal RNA replication elements in human parechovirus 1. J. Virol. 76:13116-13122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novak, J. E., and K. Kirkegaard. 1991. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J. Virol. 65:3384-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parsley, T. B., J. S. Towner, L. B. Blyn, E. Ehrenfeld, and B. L. Semler. 1997. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124-1134. [PMC free article] [PubMed] [Google Scholar]
- 37.Pringle, C. R. 1999. Virus taxonomy at the XIth International Congress of Virology, Sydney, Australia, 1999. Arch. Virol. 144:2065-2070. [DOI] [PubMed] [Google Scholar]
- 38.Racaniello, V. R. 2001. Picornaviridae: the viruses and their replication, p. 685-722. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott-Williams & Wilkins Co., Philadelphia, Pa.
- 39.Rivera, V. M., J. D. Welsh, and J. V. Maizel, Jr. 1988. Comparative sequence analysis of the 5′ noncoding region of the enteroviruses and rhinoviruses. Virology 165:42-50. [DOI] [PubMed] [Google Scholar]
- 40.Rohll, J. B., N. Percy, R. Ley, D. J. Evans, J. W. Almond, and W. S. Barclay. 1994. The 5′-untranslated regions of picornavirus RNAs contain independent functional domains essential for RNA replication and translation. J. Virol. 68:4384-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Sasaki, J., Y. Kusuhara, Y. Maeno, N. Kobayashi, T. Yamashita, K. Sakae, N. Takeda, and K. Taniguchi. 2001. Construction of an infectious cDNA clone of Aichi virus (a new member of the family Picornaviridae) and mutational analysis of a stem-loop structure at the 5′ end of the genome. J. Virol. 75:8021-8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sasaki, J., and K. Taniguchi. 2003. The 5′-end sequence of the genome of Aichi virus, a picornavirus, contains an element critical for viral RNA encapsidation. J. Virol. 77:3542-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaffer, D. R., E. A. Brown, and S. M. Lemon. 1994. Large deletion mutations involving the first pyrimidine-rich tract of the 5′ nontranslated RNA of human hepatitis A virus define two adjacent domains associated with distinct replication phenotypes. J. Virol. 68:5568-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shiroki, K., T. Ishii, T. Aoki, M. Kogashi, S. Ohka, and A. Nomoto. 1995. A new cis-acting element for RNA replication within the 5′ noncoding region of poliovirus type 1 RNA. J. Virol. 69:6825-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Svitkin, Y. V., and N. Sonenberg. 2003. Cell-free synthesis of encephalomyocarditis virus. J. Virol. 77:6551-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamashita, T., S. Kobayashi, K. Sakae, S. Nakata, S. Chiba, Y. Ishihara, and S. Isomura. 1991. Isolation of cytopathic small round viruses with BS-C-1 cells from patients with gastroenteritis. J. Infect. Dis. 164:954-957. [DOI] [PubMed] [Google Scholar]
- 48.Yamashita, T., K. Sakae, H. Tsuzuki, Y. Suzuki, N. Ishikawa, N. Takeda, T. Miyamura, and S. Yamazaki. 1998. Complete nucleotide sequence and genetic organization of Aichi virus, a distinct member of the Picornaviridae associated with acute gastroenteritis in humans. J. Virol. 72:8408-8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamashita, T., M. Ito, Y. Kabashima, H. Tsuzuki, A. Fujiura, and K. Sakae. 2003. Isolation and characterization of a new species of kobuvirus associated with cattle. J. Gen. Virol. 84:3069-3077. [DOI] [PubMed] [Google Scholar]
- 50.Zell, R., K. Sidigi, A. Henke, J. Schmidt-Brauns, E. Hoey, S. Martin, and A. Stelzner. 1999. Functional features of the bovine enterovirus 5′-non-translated region. J. Gen. Virol. 80:2299-2309. [DOI] [PubMed] [Google Scholar]