Abstract
RNA interference represents an exciting new technology that could have therapeutic applications for the treatment of viral infections. Hepatitis C virus (HCV) is a major cause of chronic liver disease and affects over 270 million individuals worldwide. The HCV genome is a single-stranded RNA that functions as both an mRNA and a replication template, making it an attractive target for therapeutic approaches using short interfering RNA (siRNA). We have shown previously that double-stranded siRNA molecules designed to target the HCV genome block gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. However, we now show that this block is not complete. After several treatments with a highly effective siRNA, we have shown growth of replicon RNAs that are resistant to subsequent treatment with the same siRNA. However, these replicon RNAs were not resistant to siRNA targeting another part of the genome. Sequence analysis of the siRNA-resistant replicons showed the generation of point mutations within the siRNA target sequence. In addition, the use of a combination of two siRNAs together severely limited escape mutant evolution. This suggests that RNA interference activity could be used as a treatment to reduce the devastating effects of HCV replication on the liver and the use of multiple siRNAs could prevent the emergence of resistant viruses.
RNA interference (RNAi) is a biological process in which double-stranded RNA within the cell induces specific degradation of mRNA with homologous sequences. Protein components of the RNA interference machinery first cleave the long double-stranded RNA into 19- to 21-base pair short interfering RNA (siRNA) and then use the short RNA molecules as guides to target homologous RNA species (reviewed in references 7 and 30). The introduction or expression of siRNA in mammalian cells to target disease-causing genes or virus-specific sequences for degradation represents a potential new therapeutic strategy. Previous reports have shown that RNAi induction has promising antiviral activity against positive- and negative-stranded RNA viruses and DNA viruses in model systems (12, 23). A major concern regarding the use of RNA interference activity against virus infections, particularly RNA viruses, is the development of resistance. Many RNA viruses encode polymerase enzymes that lack proofreading abilities and as a result have high rates of mutation. Thus, there is a high probability that viruses with resistance to RNA interference induced by a particular siRNA will evolve during virus replication through the incorporation of nucleotide mutations within the target sequence of the siRNA. Previous reports have described the selection of human immunodeficiency virus and poliovirus escape mutants in response to prolonged RNA interference activity (2, 4).
We have analyzed the potential of hepatitis C virus (HCV) to escape siRNA treatments. HCV infections can lead to the development of liver cirrhosis and hepatocellular carcinoma. Currently, the only treatment available for HCV infections is combined interferon-ribavirin therapy, a regimen that fails to cure the infection in 30 to 50% of cases. This is particularly true for HCV genotypes 1 and 2. Clearly, other treatments are required. We and others have reported that RNA interference represents a promising treatment option for HCV infections (14, 21, 24, 29). Since HCV cannot currently be grown in culture, RNA interference activity against HCV was analyzed using the HCV subgenomic replicon, a self-replicating HCV RNA that propagates in cell culture but does not yield infectious virus particles (1, 18). RNA interference activity directed against multiple target sequences of the HCV genome has been found to effectively block the synthesis of replicon RNA (reviewed in reference 22).
We now report that HCV replicons are able to escape RNA interference activity through the accumulation of nucleotide point mutations within the siRNA target sequence. Importantly, multiple point mutations were generated within the siRNA target sequence following several sequential treatments with the same siRNA, suggesting that a single base change in the target sequence of this siRNA is not sufficient to confer resistance to siRNA antiviral activity. We also show that resistant replicons are susceptible to RNA interference induced by an siRNA targeting another region of the replicon. Thus, unlike the situation involving resistance to chemical antiviral agents, which necessitates the design and development of new drugs, further treatments with siRNA would simply require the use of a nucleotide directed against an alternative target sequence. In addition, we show that through the use of two siRNAs in combination, the evolution of resistant replicons was severely limited.
MATERIALS AND METHODS
Cell culture.
The cell line Huh-7 (20) was kindly provided by Stanley M. Lemon (The University of Texas Medical Branch at Galveston, Galveston, TX) and was routinely grown in Dulbecco's minimal essential medium (DMEM) supplemented with nonessential amino acids, 100 U/ml of penicillin, 100 μg/ml of streptomycin, and 10% fetal calf serum (Wisent Inc., Montreal, Canada). HCV replicon cell lines AB12-A2 (29) and BB7 (1) were grown in medium containing 800 μg/ml of G418 active ingredient (geneticin; Gibco/Invitrogen, Carlsbad, CA).
Short interfering RNA sequences and in vitro transcription of replicon RNA.
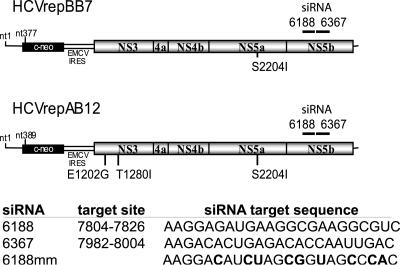
HCV-specific siRNAs 6367 and 6188, the nonsilencing control siRNA 6188 mm, and the HCV replicon plasmid HCVrepAB12 have been described previously (29). The target nucleotide sequences of each siRNA and the approximate location of the target in the HCV replicon genome are shown numerically in Fig. 1. The siRNA 6367 and siRNA 6188 numerical designations refer to the initial nucleotide numbers and their locations in the BB7 replicon transcript. Replicon RNA was transcribed from ScaI-linearized plasmid in vitro using the T7-Megascript in vitro transcription kit (Ambion, Austin, TX) according to the instructions of the manufacturer. After RNA synthesis, the DNA template was removed by three repeated digestions with 0.2 U/μl of DNase I enzyme at 37°C for 30 min.
FIG. 1.
(A) Schematic representation of the HCV replicon RNA and the approximate nucleotide locations of the siRNA target sequences. (B) The sequence of each siRNA and nucleotide base location of the target sequence in the HCV con1 genotype genome sequence are indicated. The siRNA 6367 and 6188 numerical designations refer to their locations in the BB7 replicon transcript. Boldface characters indicate the locations of mismatch base pairs in the control siRNA.
Coelectroporation of HCV replicon RNA and siRNA and selection with G418.
Cells were electroporated using the protocol described by Lohmann et al. (17). For serial coelectroporation of replicon RNA and siRNA, 10 ng of HCVrepAB12neo RNA and 40 pmol of siRNA were coelectroporated into naïve Huh-7 cells. To assay for colony formation, coelectroporated cells were transferred to 8 ml DMEM and seeded into one 10-cm-diameter tissue culture dish. Twenty-four hours later and every 3 to 4 days during selection, the medium was replaced with fresh DMEM supplemented with 800 μg G418 until colonies were visible. G418-resistant colonies were pooled and expanded, and total cellular RNA was purified from the replicon cell pool using Trizol. Three subsequent serial siRNA treatments were done by coelectroporation of 10 μg of total replicon cellular RNA from surviving G418-resistant colonies and 40 pmol of the indicated siRNA. For coelectroporation experiments designed to screen for siRNA-resistant replicon RNA, siRNA and total cellular RNA were coelectroporated into Huh-7 cells plated onto one 10-cm dish and selected with G418 as described above. The G418-resistant colonies were fixed and stained with 0.1% gentian violet prior to enumeration.
Treatment of stable replicon cells with siRNA.
To treat replicon cells with siRNA, either AB12-A2 or BB7 cells were prepared as described previously (29) and electroporated with the indicated concentrations of siRNA. Cells from each electroporation were seeded onto one 100-cm tissue culture dish and grown for 3 weeks in medium containing 800 μg G418. Cells that survived G418 selection were harvested, expanded, and re-treated with siRNA by electroporation. This was repeated three to five times for each replicon cell line.
RNA purification.
Total RNA was isolated from Huh-7 cells using Trizol reagent (Life Technologies, Invitrogen, Carlsbad, CA).
sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis of HCV protein levels.
Replicon cell lines were electroporated with 50 nM of the indicated siRNA using the method described above. Following electroporation, each sample was added to 8 ml of DMEM and 1 ml of this suspension was plated onto a 35-mm tissue culture dish. The cells were harvested at 72 h postelectroporation and were lysed in sodium dodecyl sulfate sample buffer. Protein was subjected to electrophoresis on polyacrylamide gels (Novex Invitrogen, Carlsbad, CA) and transferred to a Hybond-C Extra supported nitrocellulose membrane (Amersham Pharmacia, Piscataway, NJ). The blots were probed with monoclonal anti-NS3, anti-NS5B, and anti-actin using standard methods. Proteins were visualized using enhanced chemiluminescence (Amersham Pharmacia, Piscataway, NJ).
Reverse transcription (RT)-PCR, cloning, and sequencing of DNA fragments.
To analyze the development of escape mutations within the 6367 siRNA target sequence, the corresponding region of the HCV replicon was reverse transcribed, PCR amplified, and sequenced. Total cellular RNA from siRNA-treated or untreated replicon cells was reverse transcribed using the First-Strand cDNA Synthesis Kit and a random hexamer primer according to the manufacturer's recommended protocol (Amersham Biosciences, Little Chalfont, England). The 6367 siRNA target region of the HCV replicon was amplified by PCR using oligonucleotides NS5b 6100s TGCACTGAGCAACTCTTTGC and NS5b 6977as CAAGGTCGTCTCCGCATACG and Taq polymerase (Invitrogen, Carlsbad, CA) and cloned into the plasmid pCR2.1-TOPO using the TOPO-TA cloning kit and the recommended protocol (Invitrogen, Carlsbad, CA). Plasmids were sequenced using the M13 reverse and T7 promoter primers. Six individual escape mutant 6367 target sequences were cloned into the replicon cDNA construct by first transferring an HpaI-to-Sfil fragment for each mutant, an XhoI-to-EcoRV replicon subclone, and then inserting the XholI-to-EcoRV fragments into the AB12 snfBB7 replicon cDNA constructs. All constructs were confirmed by sequencing.
RESULTS
Attempts to select escape mutants that were resistant to RNA interference through serial coelectroporation of siRNA and HCV replicon RNA from surviving colonies.
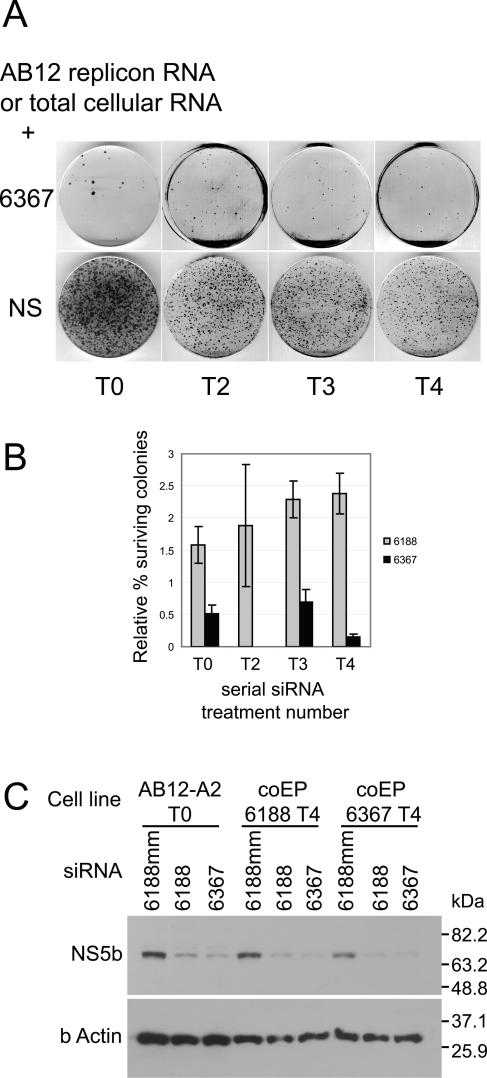
In our previous report, we showed that coelectroporation of HCV AB12 replicon (Fig. 1) RNA and siRNA caused a very potent RNA interference effect which was characterized by a 99% inhibition of colony formation compared to samples coelectroporated with a nonsilencing control siRNA (29). We wondered whether the resulting 1% of the surviving cell colonies contained replicons that were resistant to RNA interference in general or to the specific siRNA being used. To investigate these possibilities, we pooled and harvested the colonies that grew after each siRNA treatment and expanded them into new replicon cell lines. We then harvested total cellular RNA from these cell lines (which consisted of cellular RNA and HCV replicon RNA) and coelectroporated it with the same siRNA that was used during previous treatments. We repeated this process four successive times with either of two HCV-specific siRNAs, 6188 and 6367, and analyzed the numbers of colonies that formed after each siRNA treatment relative to coelectroporation with a nonsilencing control siRNA, 6188 mm. If the few colonies that grew after each successive siRNA treatment contained RNA interference-resistant replicons, we would expect the numbers of colonies that grew after each subsequent siRNA treatment to increase dramatically. Interestingly, we did not see any significant change in the colony growth after any of the four treatments with siRNA 6188 or 6367 (Fig. 2A and B). Thus, the colonies that grew after coelectroporation of replicon RNA with the HCV-specific siRNAs do not contain RNAi-resistant HCV replicon RNA. Instead, these colonies may have been derived from cells that contained replicon RNA but had insufficient amounts of siRNA to induce RNA interference. Alternatively, the few colonies that grew may have been derived from cells that are defective in RNA interference activity. However, cell lines from colonies that grew following four rounds of coelectroporation with replicon RNA and siRNA 6188 or 6367 were still susceptible to HCV-specific RNA interference activity. These cells exhibited decreased NS5b protein levels following electroporation with siRNAs 6188 and 6367 (Fig. 2C). Thus, it appears that that over the course of four serial coelectroporation treatments, the replicon is incapable of generating siRNA escape mutants. This is probably due to an almost complete elimination of replicon RNA by RNA interference before significant replication can occur and produce mutants. These experiments were repeated using total RNA derived from established BB7 replicon cells, which may contain greater sequence diversity and thus a greater chance of containing an escape mutant. However, no resistant replicons were detected (data not shown).
FIG. 2.
(A) Representative colony growth assay after coelectroporation of HCV replicon RNA with nonsilencing (NS) and replicon-specific siRNAs. (B) Histogram of relative percent HCV replicon colony growth after coelectroporation of HCV replicon RNA (T0) or total cellular RNA (T2, T3, T4) with either siRNA 6188 or 6367 versus coelectroporation with nonsilencing control siRNA. The sample numbers represent the serial siRNA treatment numbers. Error bars represent the standard error of three independent experiments. (C) RNA interference activity in T4 HCV replicon cell lines by immunoblot analysis of NS3 and NS5b protein levels after electroporation of siRNA.
Development of RNA interference escape mutants by successive electroporations of siRNA into established HCV replicon cells.
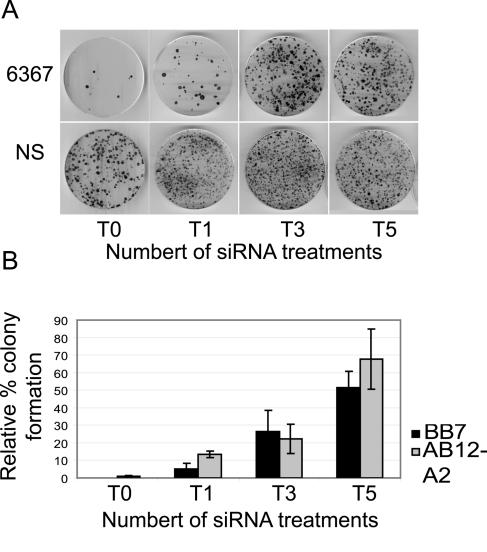
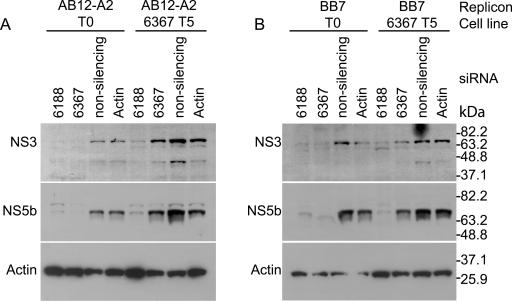
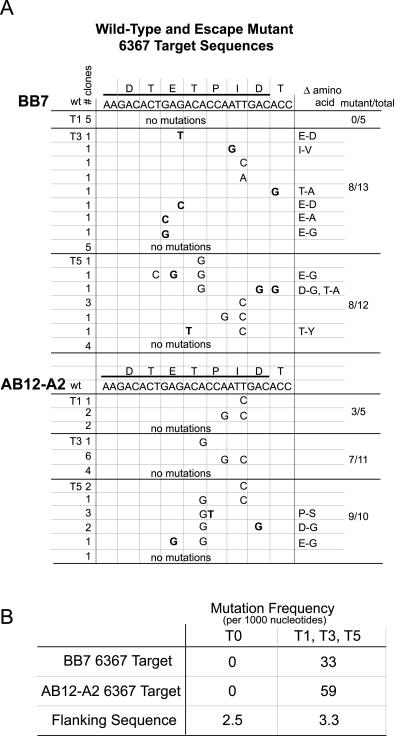
Repeated coelectroporation of siRNA with HCV replicon RNA in Huh-7 cells did not select replicon escape mutants. However, using a different method, repeated electroporation of siRNA into cell lines that harbored replicating HCV replicons, we successfully generated RNA interference escape mutants. We reported previously that electroporation of replicon cells with siRNA 6367 caused an 87.1% reduction in the levels of HCV replicon RNA and near-complete clearance of HCV replicon-specific proteins NS3 and NS5b from the cells (29). We speculated that some of the remaining replicon RNA might have developed resistance to the siRNA. To test this hypothesis, we challenged replicon cells AB12-A2 and BB7 five times with siRNA 6367. We first electroporated each replicon cell line with 100 pmol of siRNA 6367. Cells still harboring replicon RNA were selected by growth in G418 for 3 or 4 weeks. In subsequent treatments, the cells were electroporated with 20 pmol or 100 pmol of siRNA 6367 in AB12-A2 and BB7 cells, respectively. Each electroporation was followed by selection with G418. The development of siRNA-resistant replicons was assessed by coelectroporation of total RNA purified from the G418-resistant HCV replicon cell lines and siRNA 6367 into fresh Huh-7 cells following each round of electroporation and selection. Since we have established that coelectroporation of siRNA 6367 and replicon RNA is about 99% effective in blocking colony formation, we could assess the development of resistant replicons by analyzing the relative numbers of colonies that grew after coelectroporation with siRNA 6367 compared to coelectroporation with a nonsilencing control siRNA 6188 mm. Figure 3A shows an example of the HCV replicon colony formation ability from total RNA isolated from AB12-A2 replicon cells after each consecutive treatment and analyzed by coelectroporation into Huh-7 cells with siRNA 6367 (top row) or nonsilencing control siRNA 6188 mm (bottom row). The relative levels of resistant replicons increased during all five passages and reached a maximum level of 68% after five treatments with siRNA (Fig. 3A and B). We did not see evidence for the selection of cell lines that were deficient in RNA interference activity, since subsequent to five treatments with siRNA 6367, cell lines electroporated with siRNA 6188 still showed a significant decrease in the levels of HCV nonstructural proteins NS3 and NS5b by Western blot analysis (Fig. 4A, AB12 6367 T5, siRNA 6188, and Fig. 4B, BB7 6367 T5, siRNA 6188). Thus, the observed resistance to siRNA 6367 was a feature of the replicon RNA and not a general feature of the cell lines. RT-PCR amplification and sequencing of the target region of siRNA 6367 confirmed the development and selection of HCV escape mutant replicons (Fig. 5A). We cloned and sequenced five cDNAs from cells following treatment 1 and between 10 and 13 cDNA clones from cells following siRNA treatments 3 and 5, using both AB12-A2 and BB7 replicon cells. Replicon RNAs with point mutations within the siRNA 6367 target sequence were first seen after a single siRNA treatment in the case of AB12-A2 cells. A variety of point mutations subsequently accumulated within the siRNA 6367 target sequence, some being silent and some leading to amino acid changes (Fig. 5A). Few mutations occurred in the 300 nucleotides flanking the target sequence or in replicon negative controls that were selected in the absence of siRNA. When enumerated, the prevalence of mutations within the 6367 target sequence was at least 10-fold greater than that seen in the flanking sequence in T1, T3, and T5 replicon RNA samples (Fig. 5B). However, the number of mutations in the flanking sequence did not change significantly between the untreated and siRNA-treated RNA samples. After five treatments, some cDNA clones contained up to three point mutations (Fig. 5A), suggesting that one and even two point mutations within the 6367 siRNA target sequence do not completely block RNA interference activity.
FIG. 3.
(A) Colony growth assay of total cellular RNA from AB12-A2 replicon cell lines following the indicated consecutive electroporation treatment (T0 to T5). NS, nonsilencing. (B) Relative percent colony formation of replicon RNA (T0) or total RNA (T1, T3, T5) purified from replicon cell lines AB12-A2 or BB7 following consecutive electroporation treatments with siRNA 6367. Samples are labeled based on the numbers of serial siRNA treatments. Error bars show the standard error of three independent experiments.
FIG. 4.
RNA interference activities in HCV replicon cell lines (A) AB12-A2 and (B) BB7 before and after five serial treatments with siRNA 6367 (AB12 6367 T5 and BB7 6367 T5). The cell lines were electroporated with the indicated siRNAs, and HCV NS3 and NS5b protein levels were analyzed by Western blotting. The nonsilencing control siRNA is 6367 mm. Immunoblot analyses showing levels of β-actin protein are included as protein loading controls.
FIG. 5.
(A) Sequence analysis of HCV replicon escape mutants from AB12-A2 and BB7 replicon cell lines after siRNA treatments T1, T3, and T5. The target sequence of siRNA 6367 was determined for several cDNA clones amplified from RNA interference-resistant replicon cell lines. The nucleotide sequence and corresponding amino acid sequence of the wild-type (wt) 6367 target sequence are shown. Nucleotide point mutations observed in the replicon cDNA clones are shown below the wild-type sequence. Boldface nucleotides are point mutations that led to amino acid changes. All other mutations are silent. The amino acid changes caused by the point mutations shown in boldface are listed in the column labeled Δ amino acid. The “# clones” column shows the number of sequenced clones that contained that point mutation. The cDNAs sequenced from each RNA sample were derived from at least three independent RT-PCRs. (B) Mutation frequency within the 6367 target sequence and of BB7 and AB12-A2 replicon RNA and from flanking sequences was calculated by counting the number of nucleotide changes within at least 10 different sequences and expressing that number relative to the total number of nucleotides examined. The mutation frequency was calculated for sequences from untreated replicon RNA clones and for sequences isolated from siRNA-treated replicon RNA clones.
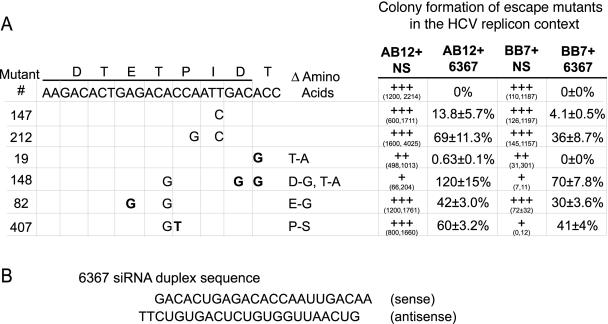
Six of the adaptive mutations were inserted into the AB12 and BB7 replicon cDNA constructs in order to analyze their abilities to confer siRNA resistance on the replicon. In vitro-transcribed mutant replicon RNA was coelecroporated with nonsilencing siRNA or with 6367 into Huh-7 cells, and after selection, colony growth was enumerated. The results are shown in Fig. 6. The ability to resist siRNA 6367 was expressed as the relative percent colony formation following coelectroporation with 6367 versus the colony growth following coelectroporation with the nonsilencing siRNA, 6188 mm. The results confirm a resistance phenotype for most of the isolated replicon mutants. The presence of a single base change in mutation 147 led to recovery of 4 to 14% of the colony formation activity in the presence of coelectroporated siRNA 6367, and the presence of combinations of two and three mutations led to higher levels of resistance, between 30 and 100% (Fig. 6A). The variable levels of resistance shown by the escape mutant replicons suggest that the coelectroporation method used to enumerate the levels of resistance within an RNA population may underestimate the actual numbers of replicons carrying point mutations. We also saw some alterations in the colony formation abilities of some of the mutants. Colony growth was enumerated from the two independent experiments and presented separately (Fig. 6A) due to variability in HCV replicon colony growth in different passages of Huh-7 cells (16). A single mutation present in mutant 19 caused a T-to-A amino acid change that appeared to affect replication fitness. Replicons that contained this mutation, those containing escape mutant sequences 19 and 148, showed 90% fewer colonies than wild-type replicon RNA when coelectroporated with the nonsilencing control siRNA. The presence of this mutation on two separate escape mutant clones is also somewhat curious, since it lies in a region outside of the siRNA target sequence and provides no significant resistance to coelectroporation with siRNA 6367. However, this nucleotide is located within the sequence that would be annealed by the siRNA sense strand on the negative strand of the replicon RNA (Fig. 6B). Selection for this mutation suggests the possibility that the minus strand may be susceptible to RNAi. It remains to be determined if this mutation affords resistance to siRNA 6367 during RNA replication. Mutation 407 also caused a severely reduced growth phenotype (100-fold fewer colonies), but only in the context of the BB7 replicon. It conferred normal colony-forming abilities when cloned into the AB12 replicon construct. The reason for this difference is unknown. The AB12 replicon was derived from BB7 by the addition of 12 nucleotides of the HCV internal ribosome entry sequence-core coding sequence upstream of the neomycin resistance gene and the introduction of two additional adaptive mutations within the NS3 coding region (Fig. 1). The poor growth of a BB7 replicon with mutation 407 suggests that the proline-to-serine mutation may not be tolerated well by BB7. Perhaps the conformational change of NS5b caused by the loss of a proline, or putative phosphorylation of the introduced serine residue, may alter an interaction between NS5b and a host or viral binding partner and somehow the additional presence of adaptive mutations in AB12 NS3 makes the NS5b interaction less sensitive to the alteration.
FIG. 6.
Colony-forming abilities and 6367 resistance of AB12 and BB7 replicon RNAs carrying escape mutations. Escape mutations that were cloned into AB12 and BB7 replicon cDNAs are shown on the left. Boldface characters indicate the nucleotide mutations that led to amino acid changes. Replicon RNA was coelecroporated into Huh-7 cells with nonsilencing siRNA 6188 mm (NS) or with siRNA 6367 to assess colony growth and 6367 resistance phenotypes. The growth phenotypes for each mutant replicon after coelectroporation with the nonsilencing siRNA 6188 mm are shown in columns AB12+NS and BB7+NS. The growth phenotype for each replicon is expressed as +++ for the wild-type replicon RNA and for mutant replicons that were able to produce colony numbers within 2 times the wild-type levels. Replicon growth was graded as ++ or + if colony counts were less than 2 times or less than 10 times wild-type levels, respectively. The colony numbers for two independent experiments are given in parentheses. The data from the two independent experiments are presented separately due to variability in HCV replicon colony growth in different passages of Huh-7 cells. The relative percent resistance of colony growth following coelectroporation with siRNA 6367 was calculated for each experiment, and the average and standard error for each mutant replicon are shown in the columns labeled AB12 + 6367 and BB7 + 6367. (B) The sequence of the 6367 siRNA duplex is shown.
Serial electroporation with two siRNAs together significantly reduces the propensity for HCV to escape silencing.
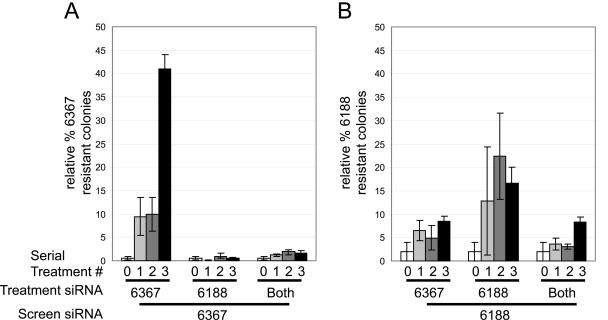
We observed a rapid evolution of siRNA escape mutant replicon RNA following treatments with a single siRNA. We speculated that the use of multiple siRNAs in combinations would limit the chances of developing viral escape mutants. We have tested this hypothesis by repeating the serial siRNA electroporation experiments on HCV replicon cells using two siRNAs, 6367 and 6188, in combination or using either siRNA alone. When treatments were done using two siRNAs together, we observed a dramatic decrease in the emergence of resistant replicons (Fig. 7). We electroporated AB12-A2 HCV replicon cells three times with either 100 nM siRNA 6367, 100 nM siRNA 6188, or a combination of 50 nM 6367 and 50 nM siRNA 6188. Each electroporation was followed by 3 weeks of selection in media containing G418 to remove cells in which the replicon RNA had been cleared. Total-RNA samples were purified from the G418-resistant cell lines following each siRNA electroporation and screened for the presence of replicons resistant to either siRNA by enumerating the colony formation ability following coelectroporation with siRNAs 6367 or 6188 relative to the total colony-forming ability when coelectroporated with a nonsilencing control siRNA, 6188 mm. The results are shown in Fig. 7. As expected, there was a clear increase in the presence of 6367-resistant replicons following repeated treatments with siRNA 6367 (Fig. 7A, treatment 6367). In addition, we saw evidence for the emergence of 6188-resistant replicons following serial treatment with siRNA 6188 alone (Fig. 7B, treatment 6188). However, when both siRNAs 6188 and 6367 were used together, there was little or no evidence for the development of resistance to siRNA 6367 or 6188. The relative colony formation levels in screens of RNAs from cells treated with both siRNAs (Fig. 7A, treatment Both, and B, treatment Both) were similar to those seen in negative control screens, in which the siRNA used to determine resistance was not the one used in the serial treatments (Fig. 7A, treatment 6188, and B, treatment 6367). In the coelectroporation screens using siRNA 6188, the background levels of colony growth are higher than those seen in coelectroporation screens using 6367 because 6188 is slightly less effective at blocking replicon colony formation than 6367 (29). For both siRNAs, treatment with a combination of both siRNAs led to significantly less, or no, detectible development of resistant replicons. Thus, the use of multiple highly effective siRNAs in combination to treat HCV-infected patients would increase the chances of success.
FIG. 7.
The use of two siRNA duplexes to treat HCV replicon cell lines dramatically inhibits the development of resistance. Histograms A and B show the relative percentages of siRNA-resistant replicon RNA in total-RNA samples from replicon cell lines treated with either siRNA 6367, 6188, or both. AB12-A2 cell lines were electroporated with the indicated “treatment siRNA” three consecutive times. The number directly beneath each bar represents the treatment number after which the total-RNA sample was harvested. Each bar shows the percent colony formation for each total-RNA sample after coelectroporation with the screen siRNA relative to the total colony numbers when coelectroporated with the nonsilencing control siRNA, 6188 mm. Histogram A shows the results of coelecroporation of treatment T0, T1, T2, and T3 RNAs with screen siRNA 6367 and shows the relative percentage of 6367-resistant replicon in each total-RNA sample. Histogram B shows the relative percentages of 6188-resistant replicons in the same RNA samples as those analyzed in histogram A. Error bars represent the standard error of at least three independent coelectroporation experiments.
DISCUSSION
After only one treatment with an siRNA targeting the NS5b coding region of the HCV genome, we were able to generate HCV replicon escape mutants containing sequence point mutations within the siRNA target sequence. This result is not unexpected, since HCV possesses an error-prone RNA-dependent RNA polymerase. Importantly, after five consecutive siRNA treatments, several of the HCV replicon escape mutants continued to accumulate point mutations within the siRNA target sequence. A similar situation was previously reported with human immunodeficiency virus RNA interference escape mutants (4). These results suggested that a single point mutation in the target sequence does not block RNA interference activity completely and that there is further selective pressure to accumulate more point mutations. In fact, a replicon containing a single point mutation was only 4 or 13% resistant to the target siRNA, while the addition of a second point mutation increased the resistance to as high as 70 to 80%. The activity of siRNA 6367 may have been as a micro-RNA (miRNA) on replicons having one or more nucleotide mismatches, since miRNAs are identical in structure to siRNAs but have sequence mismatches with their target mRNA and act to block mRNA translation rather than to induce RNA degradation (27). A survey of the escape sequences after five consecutive siRNA treatments showed a high level of diversity. This was somewhat surprising to us, since one would expect that the most fit and resistant replicon would eventually dominate the population. We speculate that the diverse HCV sequence population may occur because the differences in the levels of resistance between the escape mutant replicons is not great enough for any of the mutants to become dominant within the time frame of the experiments.
After repeated treatments with siRNA, we did not see evidence of any general resistance to RNA interference activity, since replicons that resisted one siRNA were fully susceptible to another siRNA directed against another target sequence. However, it is still possible that stably replicating HCV replicons may have an inherent resistance to RNA interference. Established replicon RNA did not succumb to RNA interference activity immediately and replicated in the presence of siRNA to subsequently develop escape mutations. On the other hand, HCV replicon RNA that was coelectroporated with siRNA failed to establish escape mutants immediately after transfection, likely due to the high susceptibility of the RNA to interference activity in the Huh-7 cells. This suggests that HCV replicon RNA may be more resistant to RNA interference during the actual process of RNA replication but is sensitive immediately following electroporation, prior to replication. This hypothesis is supported by previous experiments in which coelectroporation of siRNA, along with replicon RNA, inhibited the growth of replicon colonies by 97 to 99% (27). In contrast, established HCV replicon RNA was less susceptible to siRNA treatment, and electroporation of replicon cells with the same HCV-specific siRNA caused only a 90% reduction in replicon RNA levels (27). It is possible that the replicative form of replicon RNA may be resistant to RNA interference activity due to its localization in membranous compartments (9, 19). However, the presence of mutation 19 at a nucleotide which would only be targeted by siRNA on the HCV replicon minus strand suggests that the negative strand might be susceptible. Another possibility is that HCV encodes a gene that inhibits RNA interference activity. Previous reports have shown no difference in RNAi susceptibility between the genomic and subgenomic replicons (15, 21) and suggest that the structural proteins and NS2 do not influence RNAi susceptibility. Other viral genes, such as those encoding influenza virus NS1 (3, 5) and vaccinia virus E3L proteins (6), can inhibit RNA interference activity when expressed in plant or insect cell lines. Both NS1 and E3L were first characterized for their activities against innate cellular immunity induced by interferon, and both are viral virulence factors. It remains to be determined whether genes expressed by the subgenomic replicon, perhaps NS5a (10) or NS3 (8), which have previously been reported to inhibit host interferon-related activities, also affect RNA interference activity.
There is great interest in the use of RNA interference for antiviral therapy, and HCV represents an attractive target for treatment using siRNA. The observation that HCV replicon RNA is highly susceptible to RNA interference activity immediately after electroporation suggests that the HCV genomic RNA may also be highly susceptible following virion entry into the host cell. The HCV genome may be protected from RNA interference activity due to encapsidation by core protein, but after uncoating it is possible that the viral RNA is highly susceptible to RNA interference activity during primary translation. If this is the case, we suggest that RNA interference may be particularly useful as a prophylactic treatment. Tissues that are pretreated with siRNA, or stably expressing siRNA from an expression vector, may be resistant to HCV infection. A potential application of this strategy could be to protect a donor liver prior to transplantation into an HCV-infected patient.
An obvious challenge in the use of siRNA as a therapeutic agent is the development of suitable delivery methods. There have been recent advances in delivery of siRNA using peptide and polymeric vehicles (13, 25) and in vivo application through injection of a large volume of liquid siRNA solutions into the tail veins of mice. The latter treatment could induce RNA interference in the mouse liver and prevent Fas-mediated apoptosis (26). We are investigating the use of different peptide delivery systems for the induction of RNA interference in cell culture and in mice, which could potentially be useful in treating HCV infections.
The likelihood of HCV developing escape mutations illustrates the importance of careful siRNA target sequence selection during the development of treatment strategies. The target sequence for siRNA 6367 is within the NS5b coding region. Since the sequence was able to tolerate several different combinations of mutations, this suggests that the sequence of this region of the virus is conducive to moderate change and is required primarily to encode a functional NS5b RNA-dependent RNA polymerase. The ability of the virus to tolerate such mutations is partly due to the degeneracy of the genetic code. In addition, several escape mutations were predicted to cause amino acid changes within NS5b, some of which did not appear to affect the function of the polymerase. This highlights the importance of selecting target regions of HCV whose functions are critical for viral transcription and replication.
Several conserved regions within the HCV 5′ untranslated region (5′ UTR) have been found to be viable targets for RNA interference (22) and have potential in therapeutic RNA interference strategies. For example, the 5′ UTR acts as an internal ribosome entry sequence, and its activity is determined by RNA structural characteristics (11, 28). As such, the sequence does not tolerate nucleotide changes and is highly conserved between different HCV genotypes. Therefore, siRNA target sequences based on the 5′ UTR region offer promise for siRNA-based treatments (15). We and others have predicted that one strategy to prevent the development of resistance is through the use of multiple siRNA targets in combination (15). We have now shown conclusively that by using two highly active siRNA targets we can dramatically decrease the ability of the replicon to develop resistance to either siRNA. Since the design of different siRNA targets relies simply on the knowledge of nucleotide sequence information, the number and variability of siRNA target combinations that can be designed and tested are vast. Future treatment strategies will rely on the identification of several highly effective target sequences that can be used in combination.
Acknowledgments
We thank Charles M. Rice for sending us the plasmid pHCVrep1bBB7. Huh-7 cells were kindly provided by Stanley M. Lemon.
This work was supported by Canadian Institutes of Health Research Grant EOP-38155 and CANVAC (Canadian Network of Centres of Excellence).
REFERENCES
- 1.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 2.Boden, D., O. Pusch, F. Lee, L. Tucker, and B. Ramratnam. 2003. Human immunodeficiency virus type 1 escape from RNA interference. J. Virol. 77:11531-11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bucher, E., H. Hemmes, P. de Haan, R. Goldbach, and M. Prins. 2004. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J. Gen. Virol. 85:983-991. [DOI] [PubMed] [Google Scholar]
- 4.Das, A. T., T. R. Brummelkamp, E. M. Westerhout, M. Vink, M. Madiredjo, R. Bernards, and B. Berkhout. 2004. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J. Virol. 78:2601-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delgadillo, M. O., P. Saenz, B. Salvador, J. A. Garcia, and C. Simon-Mateo. 2004. Human influenza virus NS1 protein enhances viral pathogenicity and acts as an RNA silencing suppressor in plants. J. Gen. Virol. 85:993-999. [DOI] [PubMed] [Google Scholar]
- 6.Ding, S. W., H. Li, R. Lu, F. Li, and W. X. Li. 2004. RNA silencing: a conserved antiviral immunity of plants and animals. Virus Res. 102:109-115. [DOI] [PubMed] [Google Scholar]
- 7.Dykxhoorn, D. M., C. D. Novina, and P. A. Sharp. 2003. Killing the messenger: short RNAs that silence gene expression. Nat. Rev. Mol. Cell. Biol. 4:457-467. [DOI] [PubMed] [Google Scholar]
- 8.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 9.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He, Y., and M. G. Katze. 2002. To interfere and to anti-interfere: the interplay between hepatitis C virus and interferon. Viral Immunol. 15:95-119. [DOI] [PubMed] [Google Scholar]
- 11.Honda, M., E. A. Brown, and S. M. Lemon. 1996. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA 2:955-968. [PMC free article] [PubMed] [Google Scholar]
- 12.Joost Haasnoot, P. C., D. Cupac, and B. Berkhout. 2003. Inhibition of virus replication by RNA interference. J. Biomed. Sci. 10:607-616. [DOI] [PubMed] [Google Scholar]
- 13.Kakizawa, Y., S. Furukawa, and K. Kataoka. 2004. Block copolymer-coated calcium phosphate nanoparticles sensing intracellular environment for oligodeoxynucleotide and siRNA delivery. J. Control Release 97:345-356. [DOI] [PubMed] [Google Scholar]
- 14.Kapadia, S. B., A. Brideau-Andersen, and F. V. Chisari. 2003. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc. Natl. Acad. Sci. USA 100:2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kronke, J., R. Kittler, F. Buchholz, M. P. Windisch, T. Pietschmann, R. Bartenschlager, and M. Frese. 2004. Alternative approaches for efficient inhibition of hepatitis C virus RNA replication by small interfering RNAs. J. Virol. 78:3436-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 19.Mottola, G., G. Cardinali, A. Ceccacci, C. Trozzi, L. Bartholomew, M. R. Torrisi, E. Pedrazzini, S. Bonatti, and G. Migliaccio. 2002. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology 293:31-43. [DOI] [PubMed] [Google Scholar]
- 20.Nakabayashi, H., K. Taketa, K. Miyano, T. Yamane, and J. Sato. 1982. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 42:3858-3863. [PubMed] [Google Scholar]
- 21.Randall, G., A. Grakoui, and C. M. Rice. 2003. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc. Natl. Acad. Sci. USA 100:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Randall, G., and C. M. Rice. 2004. Interfering with hepatitis C virus RNA replication. Virus Res. 102:19-25. [DOI] [PubMed] [Google Scholar]
- 23.Saleh, M. C., R. P. Van Rij, and R. Andino. 2004. RNA silencing in viral infections: insights from poliovirus. Virus Res. 102:11-17. [DOI] [PubMed] [Google Scholar]
- 24.Seo, M. Y., S. Abrignani, M. Houghton, and J. H. Han. 2003. Small interfering RNA-mediated inhibition of hepatitis C virus replication in the human hepatoma cell line Huh-7. J. Virol. 77:810-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simeoni, F., M. C. Morris, F. Heitz, and G. Divita. 2003. Insight into the mechanism of the peptide-based gene delivery system MPG: implications for delivery of siRNA into mammalian cells. Nucleic Acids Res. 31:2717-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song, E., S. K. Lee, J. Wang, N. Ince, N. Ouyang, J. Min, J. Chen, P. Shankar, and J. Lieberman. 2003. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat. Med. 9:347-351. [DOI] [PubMed] [Google Scholar]
- 27.Tijsterman, M., and R. H. Plasterk. 2004. Dicers at RISC; the mechanism of RNAi. Cell 117:1-3. [DOI] [PubMed] [Google Scholar]
- 28.Wang, C., S. Y. Le, N. Ali, and A. Siddiqui. 1995. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA 1:526-537. [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson, J. A., S. Jayasena, A. Khvorova, S. Sabatinos, I. G. Rodrigue-Gervais, S. Arya, F. Sarangi, M. Harris-Brandts, S. Beaulieu, and C. D. Richardson. 2003. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc. Natl. Acad. Sci. USA 100:2783-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson, J. A., and C. D. Richardson. 2003. Induction of RNA interference using short interfering RNA expression vectors in cell culture and animal systems. Curr. Opin. Mol. Ther. 5:389-396. [PubMed] [Google Scholar]