Abstract
The early transcriptional region 4 (E4) of adenovirus type 5 (Ad5) encodes gene products that modulate splicing, apoptosis, transcription, DNA replication, and repair pathways. Viruses lacking both E4orf3 and E4orf6 have a severe replication defect, partially characterized by the formation of genome concatemers. Concatemer formation is dependent upon the cellular Mre11 complex and is prevented by both the E4orf3 and E4orf6 proteins. The Mre11/Rad50/Nbs1 proteins are targeted for proteasome-mediated degradation by the Ad5 viral E1b55K/E4orf6 complex. The expression of Ad5 E4orf3 causes a redistribution of Mre11 complex members and results in their exclusion from viral replication centers. For this study, we further analyzed the interactions of E4 proteins from different adenovirus serotypes with the Mre11 complex. Analyses of infections with serotypes Ad4 and Ad12 demonstrated that the degradation of Mre11/Rad50/Nbs1 proteins is a conserved feature of the E1b55K/E4orf6 complex. Surprisingly, Nbs1 and Rad50 were localized to the replication centers of both Ad4 and Ad12 viruses prior to Mre11 complex degradation. The transfection of expression vectors for the E4orf3 proteins of Ad4 and Ad12 did not alter the localization of Mre11 complex members. The E4orf3 proteins of Ad4 and Ad12 also failed to complement defects in both concatemer formation and late protein production of a virus with a deletion of E4. These results reveal surprising differences among the highly conserved E4orf3 proteins from different serotypes in the ability to disrupt the Mre11 complex.
Viruses utilize multiple approaches to modify the host cell environment to promote efficient viral replication. The early region 4 (E4) of adenovirus type 5 (Ad5) is essential for efficient virus production and encodes six gene products, some of which are not required for growth in cultured cells (reviewed in references 22 and 36). Deletions of the E4 region result in a number of severe phenotypes, including defects in viral mRNA accumulation, transcription, splicing, late protein synthesis, host cell shutoff, and viral DNA replication. The defect in virus production is due in part to the production of genome concatemers (38). This imprecise repair mechanism results in the covalent joining of viral genomes into molecules that exceed the packaging capacity of the capsid. Concatemerization of viral genomes is mediated by the host cell nonhomologous end-joining pathway and also requires the Mre11/Rad50/Nbs1 complex (3, 35).
The products of E4 open reading frames 3 and 6 (E4orf3 and E4orf6) have been shown to serve redundant functions in complementing concatemer formation as well as the other defects of an E4 deletion (5, 14, 17, 35, 38). Concatemerization is prevented by either E1b55K/E4orf6 or E4orf3, at least in part by targeting the cellular Mre11/Rad50/Nbs1 complex (35). The expression of E4orf6 and E1b55K results in the proteasome-mediated degradation of Mre11 complex members (6, 35). The E4orf3 protein can redistribute Mre11, Rad50, and Nbs1 from their normal diffuse nuclear localization into large nuclear and cytoplasmic accumulations during infections and transfections (35). The gene products of E4orf3 and E4orf6 both physically interact with the viral E1b55K protein and alter its cellular localization (4, 20, 23, 32).
The E4orf3 protein has been reported to be localized mainly in the nucleus, where it is tightly associated with the nuclear matrix, but cytoplasmic accumulations have also been observed during infection (20, 33, 34). Previously, E4orf3 has been shown to reorganize nuclear structures known as promyelocytic leukemia (PML) oncogenic domains (PODs/ND10) (7, 10). These distinct nuclear structures contain a growing list of proteins (reviewed in references 2 and 27) and have been implicated in multiple functions, including genomic stability and DNA repair (42). Many viruses express proteins that associate with PODs/ND10 and may affect their functions (reviewed in references 12 and 26). The predominant component of PODs/ND10 is the PML protein, and upon E4orf3 expression, PML is redistributed into nuclear track-like structures (7, 10). A subset of the Mre11 complex localizes to PODs/ND10 (24, 25, 39), and the Mre11/Rad50/Nbs1 proteins are also redistributed into track structures upon expression of the Ad5 E4orf3 protein (35). Other DNA repair proteins have been reported to be associated with PODs, including the BLM helicase that is mutated in Bloom's syndrome (1, 18, 42) and TopBP1 (40), but the effect of E4orf3 on these proteins is unknown.
The mechanism by which E4orf3 accomplishes the dramatic changes in nuclear architecture and the consequences of mislocalization of cellular proteins remain unclear. It has been suggested that E4orf3 may have functions that are important for Ad DNA replication which are genetically separable from the inhibition of concatemer formation (11, 34). In order to further characterize the effects of adenoviral E4 proteins on POD/ND10 structures and the cellular Mre11 complex, we utilized the naturally occurring diversity between different Ad serotypes. POD/ND10 proteins which have previously been examined for altered localization during Ad5 infection include PML, Sp100, and the Mre11 complex (10, 35). Immunofluorescence revealed that the rearrangement of proteins associated with PODs/ND10 is selective. Analyses of serotypes Ad4 and Ad12 showed that while Mre11 degradation by E1b55K/E4orf6 is conserved, some of this cellular repair complex is localized to viral replication centers. When the Ad5 E4orf3 protein was expressed via transfection prior to infection with a virus with an E4 deletion, the rearrangement of the Mre11 complex prevented its recruitment to virus replication centers and the production of genomic concatemers. In contrast, the Ad4 E4orf3 and Ad12 E4orf3 proteins failed to redistribute the Mre11 complex and could not prevent the accumulation of the Mre11 complex at viral replication centers and concatemer formation. Our observations reveal unexpected differences in the functions of the conserved E4orf3 proteins between Ad serotypes and demonstrate that reorganization of the Mre11 complex is independent of the disruption of other POD/ND10 components.
MATERIALS AND METHODS
Cell lines.
HeLa and 293 cells were maintained in Dulbecco modified Eagle's medium supplemented with 10% fetal bovine serum and penicillin-streptomycin. Cells that stably express the Ad5 E1b55K protein have been previously described (6). All cells were maintained as monolayers in Dulbecco modified Eagle's medium supplemented with 10 or 20% fetal bovine serum at 37°C in a humidified atmosphere containing 5% CO2. Cells were transfected by the use of calcium phosphate or Effectene according to standard protocols.
Plasmids.
The cDNA of Ad5 E4orf3 was a gift from Ron Evans. The E4orf3 cDNAs of Ad4 and Ad12 were PCR amplified from DNAs extracted from infected A549 cells and then cloned into the pL2 and pL2-FLAG vectors (gifts from M. Tini) for expression from the cytomegalovirus promoter. The pRK5.Ad5-E4orf6 expression vector has been previously described (8). The N82A mutant of Ad5 E4orf3 has been previously described (35). The E4orf6 cDNAs of Ad4 and Ad12 were PCR amplified from DNAs extracted from infected cells and then cloned into the pRK5 vector (a gift from T. Hunter) for expression from the cytomegalovirus promoter.
Antibodies.
Antibodies to the following proteins were purchased from commercial sources and used at the indicated dilutions: Rad50 (1:300), Mre11 (1:300), Nbs1 (1:500), and BLM (1:100) (Novus Biologicals); PML (1:200), SUMO1 (1:100), γ-tubulin (1:100), and DAXX (1:100) (Santa Cruz); TopBP1 (1:100) (BD Biosciences); FLAG (1:20,000) (Sigma); and p53 Ab6 (1:500) (Oncogene). An antibody to RPA32 was a gift from T. Melendy. Antibodies to viral proteins recognized the Ad5 E1b55K protein (1:200; a gift from A. Levine), the Ad DBP (1:200; a gift from A. Levine) the Ad5 E4orf3 protein (1:2,000; a gift from G. Ketner), and the Ad5 fiber protein (1:10,000; Neomarkers). Secondary antibodies for immunofluorescence and immunoblotting were obtained from Sigma or Jackson Laboratories.
Viruses and infections.
Wild-type adenovirus serotypes Ad4, Ad5, and Ad12 were obtained from the American Type Culture Collection and propagated on 293 cells. The E4 mutant viruses dl1004 and dl1017 were gifts from G. Ketner and were propagated on W162 and 293 cells, respectively. All viruses were purified by two sequential rounds of ultracentrifugation in CsCl gradients and stored in 40% glycerol at −20°C.
Immunofluorescence.
Cells were grown on glass coverslips in 24-well dishes and infected with virus or transfected with vectors. Cells were washed with phosphate-buffered saline (PBS) and fixed for 10 min at −20°C in ice-cold methanol:acetone (1:1). Cells were rehydrated in PBS and blocked with PBS-B (5% bovine serum albumin in PBS) for 30 min. Primary antibodies were incubated for 1 h at room temperature in PBS-B and then washed four times with PBS. Secondary antibodies were incubated with the cells for 1 h at room temperature in PBS-B. Cells were washed four times with PBS and mounted in Fluoromount-G. In all cases, control staining experiments showed no cross-reactions between the fluorophores, and images obtained by staining with individual antibodies were the same as those obtained by double labeling. Nuclear DNA was stained with 4′,6′-diamidino-2-phenylindole (DAPI), and coverslips were mounted by the use of Fluoromount-G (Southern Biotechnology Associates). Immunoreactivity was visualized by use of a Nikon microscope in conjunction with a charge-coupled-device camera (Cooke Sensicam). Images were obtained in double or triple excitation mode and processed with SlideBook and Adobe Photoshop.
Immunoblotting.
Cells were lysed in PBS containing 1% NP-40 and 0.1% sodium dodecyl sulfate and then were cleared by centrifugation. Protein concentrations were determined with the Lowry assay (Bio-Rad), and equal concentrations were loaded for analysis in polyacrylamide gels. Proteins were separated by electrophoresis and transferred to Hybond ECL membranes. Membranes were blocked overnight in PBS with 5% dry milk. Primary antibodies were incubated with the membranes for 1 h at room temperature in PBS with 3% bovine serum albumin. Proteins were visualized by enhanced chemiluminescence after incubation with secondary antibodies coupled to horseradish peroxidase for 1 h at room temperature.
PFGE and RT-PCR.
For analysis of the complementation of a virus with an E4 deletion, HeLa cells in 60-mm dishes were transfected by the use of Lipofectamine 2000 with an empty vector or expression plasmids (7.5 μg) for E4orf3 proteins, and after 24 h, the cells were infected with dl1004 (5) at a multiplicity of infection (MOI) of 25. After a further 48 h of infection, DNA, RNA, and protein lysates were analyzed. Pulsed-field gel electrophoresis (PFGE) was performed as previously described (38). RNAs were isolated from half of the cells used for the concatemer assay by use of an RNeasy mini kit (QIAGEN). Reverse transcription-PCR (RT-PCR) was performed with a One-Step RT-PCR kit (QIAGEN) and 100 ng of RNA. The reaction conditions for each serotype were similar except for the primers and the annealing temperature used. For Ad5 E4orf3, the forward primer was 5′-TGATTCGCTGCTTGAGGCTG-3′, the reverse primer was 5′-GCTTCCAAAAGGCAAACGGC-3′, and the annealing temperature was 46.8°C. For Ad4 E4orf3, the forward primer was 5′-GCTTTCGCTGTCCTCTTGTTTCTG-3′, the reverse primer was 5′-GTCTCGGAGCACTTCAAAATGC-3′, and the annealing temperature was 56.9°C. For Ad12 E4orf3, the forward primer was 5′-GAATGGTTCCAGTCAGGCAGTTTC-3′, the reverse primer was 5′-TAAATCTCGCAGGTGGCAGC-3′, and the annealing temperature was 53.3°C. Standards consisted of stepwise dilutions, from 0.0001 ng to 1 ng, of the respective plasmid.
Molecular phylogeny.
Predicted amino acid sequences for the E4orf1, E4orf2, E4orf3, E4orf4, E4orf6, and E1b55K proteins were obtained from sequences available in GenBank. The sequences were aligned with the CLUSTAL algorithm (16) implemented in ClustalX version 1.83 (37). The Phylogeny Inference Package (PHYLIP) suite of software tools, version 3.63 (distributed by J. Felsenstein, Department of Genome Sciences, University of Washington, Seattle), was used to perform phylogenic comparisons as follows. A sequence alignment with gaps was sampled by bootstrapping to yield 2,000 replicates. The replicates were evaluated by the maximum parsimony algorithm in PROTPARS, and a predominant, unrooted tree was identified by the majority rule algorithm. The resulting consensus trees were in excellent agreement with those derived by Kovacs and associates based on the E2B, pIIIa, and pVI proteins of primate adenoviruses (21). The topology of the consensus tree was used as the basis for determining sequence divergence by the maximum likelihood method using the Jones-Taylor-Thornton model of amino acid changes (19). The uprooted distance trees are presented with the Ad12 species A protein selected as the outgroup, or as an unrooted graph.
RESULTS
Differential rearrangement of POD components by E4orf3.
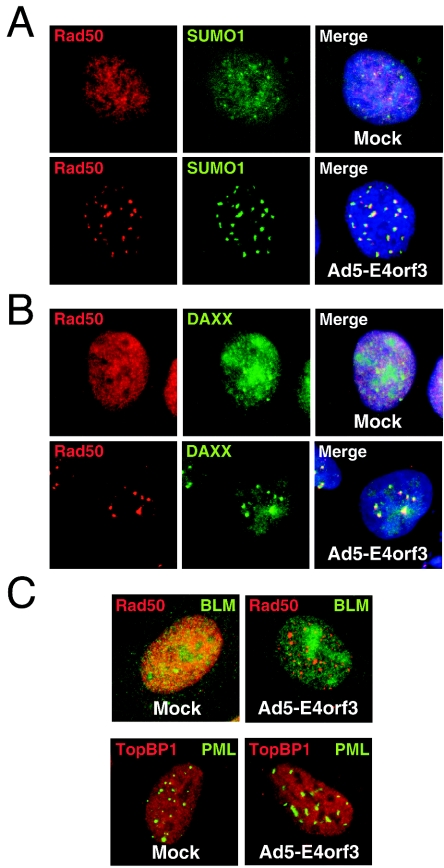
We previously described the effect of the E4orf3 protein from Ad5 on nuclear PML structures (10) and the cellular Mre11 complex (35). We observed that PML and the Mre11 complex members were reorganized by Ad5 E4orf3 expression into nuclear speckles. Both PML and the Mre11 complex are components of the POD/ND10 structures, and therefore we investigated whether reorganization by E4orf3 was a feature shared by all proteins reported to be associated with PODs/ND10. We examined the localization of a number of POD components by immunofluorescence during infection with wild-type Ad5, a mutant that lacks expression of E1b55K and E4orf6 (dl1017), or after transfection of a plasmid vector expressing Ad5 E4orf3. We found that some of these proteins were relocalized into track-like structures that partially colocalized with PML or members of the Mre11 complex. These included SUMO-1 and DAXX (Fig. 1A and B). However, other components of the PODs/ND10 were unaffected, including the repair factors BLM and TopBP1 (Fig. 1C). The results obtained from infections (data not shown) were similar to those obtained from transfections of E4orf3 alone. These results show that a disruption of PML localization does not lead to a redistribution of all other components of the PODs/ND10, and they suggest that some proteins are specifically targeted by the Ad5 E4orf3 protein.
FIG. 1.
Effect of E4orf3 on localization of components of PODs/ND10. HeLa cells were transfected with an expression vector for Ad5 E4orf3, and the localization of proteins was assessed by immunofluorescence. E4orf3 induced the reorganization of SUMO1 (A) and DAXX (B) into nuclear track-like structures that partially colocalized with Rad50. (C) Neither BLM nor TopBP1 was recruited to the structures induced for Rad50 and PML by E4orf3 expression.
The Mre11 complex is differentially affected by Ad serotypes 4, 5, and 12.
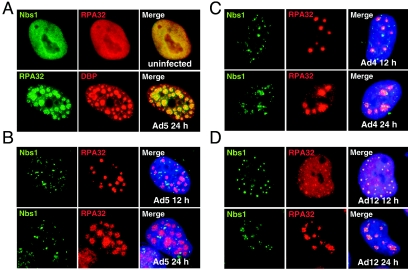
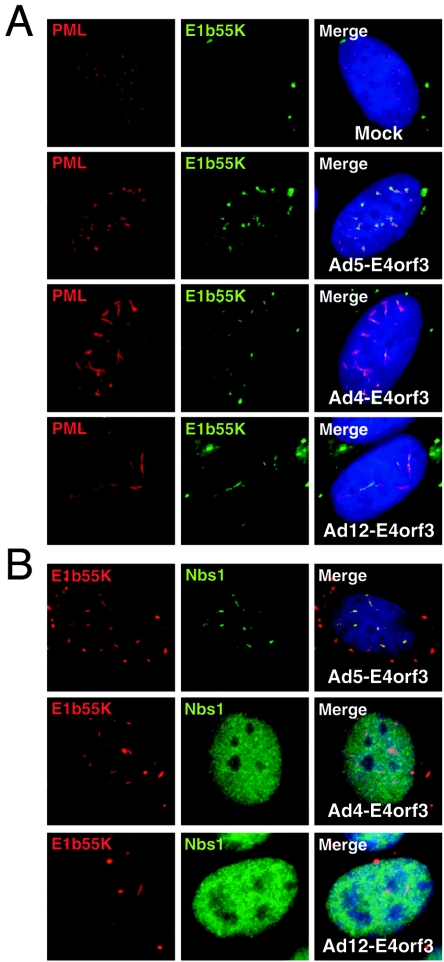
To further investigate the effects of adenoviral E4 proteins on the cellular Mre11 complex, we examined infections with different Ad serotypes. We analyzed infections with Ad5 from subgroup C, Ad12 from subgroup A, and Ad4 from subgroup E. Immunoblotting of lysates from HeLa cells infected with the various serotypes showed that the Mre11 and Rad50 proteins were down-regulated by Ad4 and Ad12 as well as Ad5 infection (data not shown). We also analyzed components of the Mre11 complex by immunofluorescence during infection with different Ad serotypes (Fig. 2). Antibodies directed against the viral DNA binding protein (DBP) of Ad5 could not be used to identify replication centers in cells infected with Ad4 and Ad12 due to a lack of cross-reactivity across serotypes (data not shown). Therefore, in order to identify infected cells and viral replication centers, we used an antibody against the 32-kDa subunit of replication protein A (RPA32). In uninfected cells, both Nbs1 and RPA32 were found diffusely throughout the nucleus (Fig. 2A), but in cells infected with wild-type Ad5, the RPA32 protein was relocalized in a pattern overlapping with the DBP-positive viral replication centers (Fig. 2A). This relocalization was observed throughout the infection and was seen with all serotypes tested, suggesting that it can therefore be used a surrogate marker of viral replication centers. For all serotypes tested, decreased signal intensities were observed for the Mre11 complex proteins in cells that displayed mature viral replication centers, consistent with the targeted degradation described above. Despite the degradation and lower signal intensity, sufficient Nbs1 protein remained at this time point for us to assess its location during different virus infections. As previously reported (35), during infection with Ad5 the Mre11 complex members were observed to be excluded from viral replication domains and were present in nuclear and cytoplasmic aggregates (Fig. 2B). In contrast, during infection with Ad4 (Fig. 2C) and Ad12 (Fig. 2D), there was an absence of Nbs1 in nuclear tracks, and instead the protein was localized to viral replication centers. The Nbs1 protein accumulated in foci around the viral centers, in a pattern that resembled that seen during infection with a virus with an E4 deletion (dl1004) derived from Ad5 (35). These results suggest that degradation of the Mre11 complex is a conserved feature but that reorganization of the complex is specific to certain adenovirus serotypes.
FIG. 2.
Localization of Mre11 complex during infection with different Ad serotypes. (A) In uninfected HeLa cells, the Nbs1 and RPA32 proteins were localized diffusely throughout the cellular nucleoplasm. In Ad-infected cells, the RPA32 protein was found to be relocalized in a pattern that overlapped with that of viral replication centers detected by staining with an antibody to the viral DBP. This shows that RPA32 relocalization can be used as a surrogate marker for viral replication centers. Localization of the Mre11 complex was determined by staining for Nbs1 during infections of HeLa cells with Ad5 (B), Ad4 (C), and Ad12 (D). Cells infected with Ad5 (MOI of 25), Ad 4 (MOI of 100), and Ad12 (MOI of 25) were fixed and analyzed by immunofluorescence at 12 and 24 h postinfection. Staining with RPA32 was used to localize the replication centers because the Ad5 DBP monoclonal antibody does not cross-react with the Ad4 DBP and Ad12 DBP proteins. (B) Nbs1 was excluded from Ad5 replication centers after 12 h and 24 h of infection. Nbs1 localized to sites of viral replication during Ad4 (C) and Ad12 (D) infections.
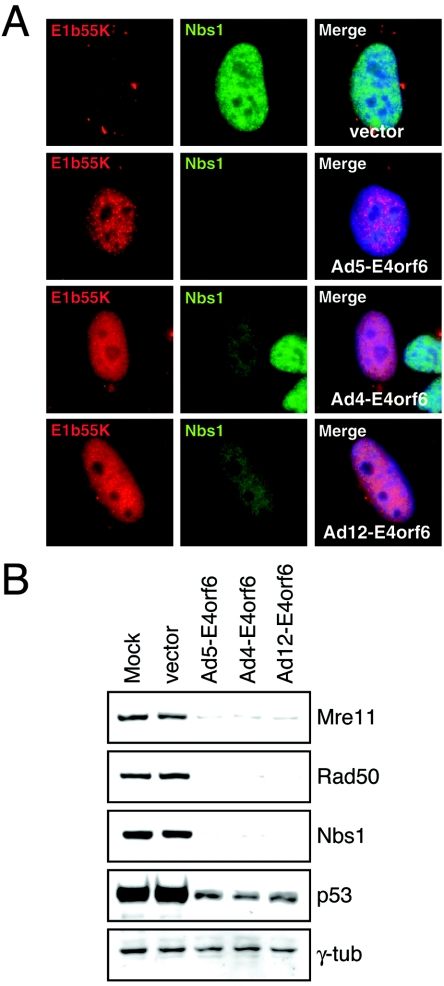
Our previous work with Ad5 has demonstrated that the E1b55K/E4orf6 complex is responsible for the degradation of Mre11/Rad50/Nbs1 (6, 35), possibly through the recruitment of cellular proteins to form a ubiquitin ligase (15, 31). Therefore, to study this degradation further, we PCR amplified and cloned the cDNAs for the E4orf6 proteins from lysates of cells infected with Ad4 and Ad12. In stable cell lines expressing Ad5 E1b55K, the viral protein accumulates in cytoplasmic speckles (6), but the transfection of E4orf6 plasmids resulted in relocalization to the nucleus for the proteins from serotypes Ad4, Ad5, and Ad12 (Fig. 3A). This suggests that the E1b55K interaction domain in E4orf6 is conserved among the serotypes. This was also accompanied by degradation of the Nbs1 protein, demonstrating that the complex of Ad5 E1b55K with the various E4orf6 proteins was functional for targeting the Mre11 complex for degradation. This was further supported by the observation that the transfection of 293 cells that contain the Ad5 E1b55K gene with E4orf6-expressing plasmids for Ad4, Ad5, and Ad12 also led to the degradation of Mre11, Rad50, and Nbs1 (Fig. 3B). These results indicate that degradation of the Mre11 complex members by E4orf6 and E1b55K is conserved among serotypes Ad4, Ad5, and Ad12.
FIG. 3.
Degradation of the Mre11 complex is conserved among Ad serotypes. (A) Transfection of HeLa cells that stably express the Ad5 E1b55K protein (6) with plasmids encoding the E4orf6 proteins of Ad5, Ad4, and Ad12. All E4or6 proteins resulted in the nuclear accumulation of Ad5 E1b55K and the degradation of Nbs1. (B) Immunoblotting of 293 cells (expressing Ad5 E1b55K) transfected with cDNA expression vectors (4 μg) for E4orf6 proteins of Ad5, Ad4, and Ad12. Negative controls included untransfected cells (mock) and cells transfected with the empty vector construct. The expression of all three E4orf6 proteins led to the down-regulation of Mre11, Rad50, and Nbs1. The p53 protein was also degraded with each E4orf6 protein, and γ-tubulin served as a loading control.
The E4orf3 proteins of Ad4 and Ad12 mislocalize PML but not the Mre11 complex.
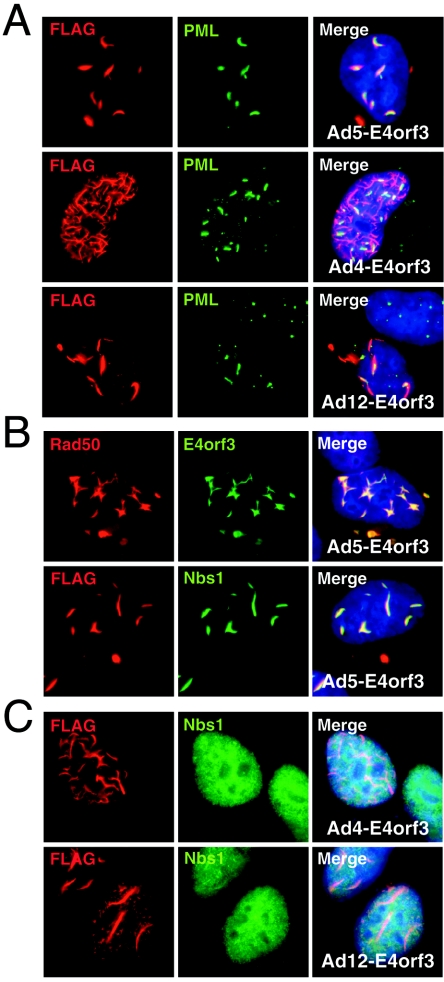
The E4orf3 protein of Ad5 is clearly sufficient to redistribute both the PML protein and the Mre11 complex (35). To determine if the E4orf3 proteins of Ad4 and Ad12 disrupt the Mre11 complex in the absence of viral infection, we cloned the cDNAs from these serotypes and expressed them by transfection with or without an N-terminal FLAG epitope tag. Transfections of HeLa cells with FLAG-tagged E4orf3 proteins of Ad5, Ad4, and Ad12 followed by anti-FLAG immunofluorescence revealed track-like structures in each case (Fig. 4A). Each of the E4orf3 proteins was also sufficient for the disruption of PML bodies (Fig. 4A). The tagged Ad5 E4orf3 protein efficiently targeted the Mre11 complex and also partially colocalized with Rad50 and Nbs1 in nuclear tracks and cytoplasmic accumulations (Fig. 4B). Untagged constructs were also generated, and a similar PML disruption was observed upon transfection, but unfortunately the antibody raised against the C terminus of Ad5 E4orf3 did not recognize the Ad4 E4orf3 and Ad12 E4orf3 proteins (data not shown). Interestingly, the FLAG-tagged E4orf3 proteins of Ad4 and Ad12 failed to disrupt Mre11, Rad50, or Nbs1 in transfections (Fig. 4C and data not shown). These results suggest that disruptions of the normal localization of PML and the Mre11 complex are separable events and that Mre11 reorganization is a function specific to Ad5 E4orf3 and is not shared by all Ad serotypes.
FIG. 4.
Redistribution of the Mre11 complex is a specific activity of E4orf3 from serotype Ad5. HeLa cells were transfected with plasmids carrying FLAG-tagged cDNAs encoding the E4orf3 proteins of Ad5, Ad4, and Ad12. After 24 h, the localization of E4orf3 and cellular proteins was analyzed by immunofluorescence. (A) E4orf3 proteins from all three serotypes disrupted PML bodies and generated track-like structures. The expression of FLAG-tagged E4orf3 from Ad5 (top), Ad4 (middle), and Ad12 (bottom) caused a dispersion of the usually punctate PML staining. (B) Expression of FLAG-tagged Ad5 E4orf3 leads to disruption of the Mre11 complex, and E4orf3 colocalizes with Rad50 and Nbs1. (C) Nbs1 localization was unaffected by the expression of FLAG-tagged E4orf3 proteins from Ad4 and Ad12.
E4orf3 also physically interacts with the adenoviral E1b55K protein (23). When expressed alone by transfection or in stable cell lines, the E1b55K protein is predominantly found in cytoplasmic bodies (6, 23, 41), but it is imported into the nucleus by the expression of E4orf3 (20). During infection, the E1b55K protein is predominantly nuclear and initially associates with PODs/ND10 and the track-like PML structures (10, 23). We therefore assessed the effect of the E4orf3 proteins on E1b55K localization (Fig. 5A). Expression constructs for the E4orf3 proteins were transfected into a HeLa-derived cell line that expresses Ad5 E1b55K from a retrovirus vector (6). Immunofluorescence revealed that the E4orf3 proteins of Ad4, Ad5, and Ad12 were all capable of recruiting E1b55K into nuclear tracks that partially colocalized with PML. When E1b55K localization was examined in conjunction with Nbs1 staining, the two proteins colocalized in tracks in the presence of Ad5 E4orf3 (Fig. 5B). However, although the E1b55K protein was present in nuclear tracks induced by Ad4 E4orf3 and Ad12 E4orf3, the Nbs1 protein remained diffusely nuclear. These results indicate that there is a selective recruitment of proteins into E4orf3-induced nuclear tracks.
FIG. 5.
E1b55K is recruited into nuclear PML track-like structures by E4orf3 proteins. HeLa cells expressing the E1b55K viral protein were transfected with E4orf3 expression vectors. (A) In mock-transfected cells, E1b55K was located predominantly in cytoplasmic speckles and PML was localized to PODs/ND10. The expression of E4orf3 from Ad4, Ad5, and Ad12 resulted in the import of E1b55K into the nucleus, where it partially colocalized with PML in track-like structures. (B) Nuclear tracks of E1b55K colocalized with Nbs1 when induced by Ad5 E4orf3 but not when induced by the Ad4 E4orf3 and Ad12 E4orf3 proteins.
The Ad4 E4orf3 and Ad12 E4orf3 proteins fail to complement defects of a virus with an E4 deletion.
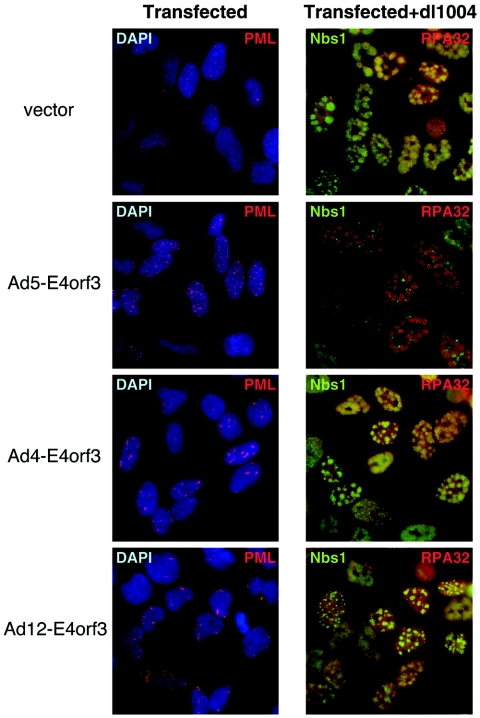
Infections of cells with an Ad5-derived virus with an E4 deletion lead to an accumulation of the Mre11 complex at foci around viral replication centers and are accompanied by concatemerization of viral genomes (35). Neither foci nor concatemers are observed for viruses that express Ad5 E4orf3, and therefore we assessed the ability of the different E4orf3 proteins to complement these functions during infection with an adenovirus with an E4 deletion. Cells were transfected with expression vectors for the E4orf3 proteins and then infected with the virus dl1004, which has an E4 deletion (Fig. 6). Immunofluorescence analysis for PML confirmed E4orf3 expression and the induction of track-like structures. Viral replication centers were located by RPA32 staining, and localization of the Mre11 complex was determined with an Nbs1 antibody. In cells transfected with an empty vector, there were foci of Nbs1 at viral centers, but in the presence of Ad5 E4orf3, the Nbs1 protein was found in tracks and was excluded from viral centers. Although the Ad4 E4orf3 and Ad12 E4orf3 proteins caused a redistribution of PML, they did not prevent accumulation of the Mre11 complex at viral replication centers.
FIG. 6.
Expression of Ad5 E4orf3 prevents accumulation of the Mre11 complex at virus replication centers during infection with a virus with an E4 deletion. HeLa cells were transfected with expression vectors for E4orf3 proteins or with an empty vector (0.8 μg). The left column shows images of transfected cells stained for PML localization after 24 h, demonstrating track-like structures in cells expressing all of the E4orf3 proteins. The images on the right are from transfected cells that were also infected with the virus dl1004, which has an E4 deletion, for a further 24 h and then stained for Nbs1 and RPA32 (marks viral replication centers). The Nbs1 protein was excluded from virus centers by Ad5 E4orf3 but not by the E4orf3 proteins of Ad4 and Ad12.
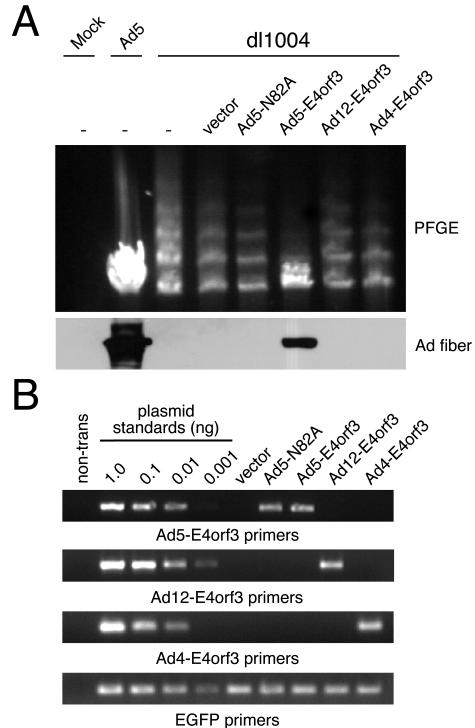
We used PFGE to analyze DNA from infected cells for the formation of concatemers (Fig. 7A). The viral genome was detected predominantly as a linear monomer during infection with wild-type Ad5, but concatemers were observed for the virus with the E4 deletion. Concatemerization was greatly reduced by the expression of Ad5 E4orf3 but not by a mutant (Ad5 N82A) that did not reorganize the Mre11 complex (35). In contrast, neither of the E4orf3 proteins from Ad4 and Ad12 prevented concatemers. RT-PCR analysis of RNAs from transfections using primers specific to each serotype gene demonstrated comparable levels of E4orf3 transcripts (Fig. 7B). We also looked for the production of viral late proteins by immunoblotting with an antifiber antibody (Fig. 7A). No fiber protein was produced by infection with the virus with the E4 deletion. Only wild-type Ad5 E4orf3 complemented the virus for late protein production. Taken together, these results demonstrate that the Ad4 and Ad12 E4orf3 proteins, which do not mislocalize the Mre11 complex, also fail to prevent concatemer formation and to promote the production of late proteins.
FIG. 7.
Ability of E4orf3 proteins to complement a virus with an E4 deletion. (A) HeLa cells were transfected with expression vectors for the E4orf3 proteins of Ad5, Ad4, and Ad12. After 24 h, the cells were infected with the virus dl1004, containing an E4 deletion, and DNAs were harvested for PFGE analysis after a further 48 h. Concatemerization was abrogated only by wild-type Ad5 E4orf3. An empty vector and the N82A mutant of Ad5 E4orf3 served as negative controls. The production of late proteins was assessed by immunoblotting with an antifiber antibody. Infection with Ad5 served as a positive control for this experiment. (B) Transcription of the different E4orf3 constructs was confirmed by RT-PCR with lysates from transfected cells, using serotype-specific primer sets.
Molecular phylogeny of adenovirus E4 proteins.
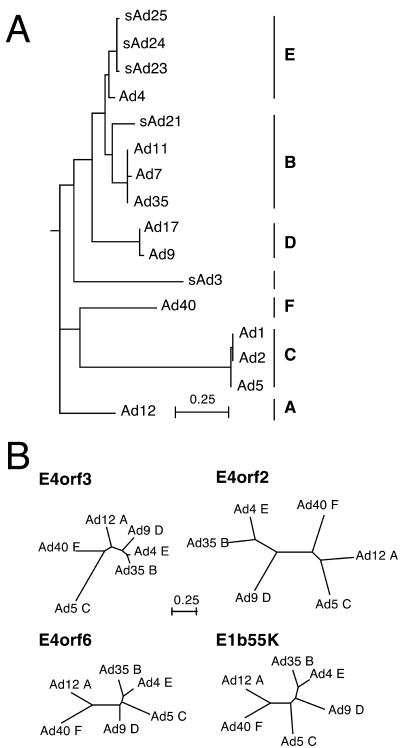
The E4orf3 proteins of Ad12 (subgroup A) and Ad4 (subgroup E) differed from that of Ad5 (subgroup C) by their failure to mislocalize the Mre11 complex and prevent viral DNA concatemerization. Sequence comparisons among these proteins were uninformative because they share approximately 50% identity and 60% similarity at the amino acid level (data not shown). However, molecular phylogeny suggests that the E4orf3 proteins of subgroup C have diverged substantially from those of other human adenovirus species (Fig. 8A). Representative proteins from each subgroup of human adenoviruses were analyzed further by the unrooted branching graphs shown in Fig. 8B. In contrast to the proximity of the E4orf3 proteins of subgroup A, D, E, and B adenoviruses, the E4orf3 protein of Ad5 (subgroup C) lies at a substantial distance from its nearest neighbor (Fig. 8B). The E4orf3 protein of Ad40 (subgroup F) shows the next largest divergence, although it appears to be more closely related to the cluster of other virus subgroups. The apparent divergence of the E4orf3 protein from subgroup C adenoviruses is not shared by other E4 proteins or by the E1b55K protein. For example, no similar cluster was observed in the graphs for the E4orf1, E4orf2, E4orf4, and E4orf6 proteins (Fig. 8B and data not shown). It is curious that the E4orf2 protein, which has no known function, appears to have experienced the most divergence between subgroups.
FIG. 8.
The E4orf3 protein of Ad5 exhibits divergence from the E4orf3 proteins of other adenovirus species. (A) The relationships among E4orf3 proteins of the indicated human and monkey adenoviruses were calculated by application of the PHYLIP suite of software as described in Materials and Methods. The uprooted tree obtained by this method was presented with the Ad12 subgroup A protein as the outgroup. The length of each branch corresponds to the expected number of amino substitutions, with the scale bar corresponding to a rate of change of 0.25 substitutions per residue. Simian viruses are identified as “sAd,” and human viruses are identified as “Ad.” The vertical bar to the right of the virus names identifies viruses of the indicated subgroup, as suggested by the analysis of Kovacs and associates (21). (B) The divergence of E4orf3, E4orf6, E4orf2, and E1b55K proteins from representative human adenoviruses is presented as branching graphs in which the length of each branch corresponds to the expected number of amino substitutions at each position. All graphs are presented on the same scale. The specific viruses represented in this analysis are indicated, as are their subgroup designations (A to F). Bar, rate of change of 0.25 substitutions per residue.
DISCUSSION
We previously reported that the E4orf3 protein of Ad5 reorganizes the cellular distribution of the Mre11 complex (35). The formation of viral genomic DNA concatemers requires the Mre11 complex and is prevented by the Ad5 E4orf3 protein (35, 38). We therefore further analyzed the reorganization of the Mre11 complex by E4orf3 proteins in order to explore the correlation between mislocalization and inhibition of concatemerization. The Ad5 E4orf3 protein was observed in a pattern of nuclear track-like structures and cytoplasmic accumulations which colocalized with the rearranged Mre11/Rad50/Nbs1 complex. The partial overlap of the patterns for PML and the Mre11 complex that were induced by E4orf3 suggests that they may form through a related process. One possibility is that E4orf3 reorganization of the Mre11 complex occurs through its association with PML and the PODs/ND10. The lack of reorganization of other proteins associated with PODs/ND10, together with an analysis of E4orf3 proteins from different serotypes, makes this possibility seem unlikely.
Many viruses encode proteins that interact with the PODs/ND10 or disrupt these structures during infection (12, 26). It has therefore been suggested that POD/ND10 reorganization may be a prerequisite for efficient viral DNA replication. At least in the case of adenoviruses, the disruption of PODs may not be sufficient to initiate viral replication, but rather there may be a specific pattern of reorganization of POD components as viral replication centers are formed (11). Our data from an analysis of different Ad serotypes clearly demonstrate that the mislocalization of PML and that of the Mre11 complex are separable events. Ad4 and Ad12 both encode E4orf3 proteins that efficiently target PML but do not affect Mre11 localization. It is possible that the reorganization of the Mre11 complex by Ad5 E4orf3 is mediated by a direct interaction that is lacking in the other serotypes, despite the high level of sequence conservation between proteins. The mechanism by which Ad5 E4orf3 causes redistribution of the Mre11 complex remains enigmatic. Further mutational analysis of the Ad5 E4orf3 protein may identify distinct residues of the protein which are involved in its specificity and may aid in both functional analysis and interaction studies.
An analysis of the functions of the Mre11 complex in the presence of E4orf3 may be valuable in addressing the extent of its inactivation after redistribution. While mislocalization of the Mre11 complex may not be completely sufficient to prevent viral concatemerization (11), our results suggest that it is a contributing factor. The complementation experiments with the Ad4 E4orf3 and Ad12 E4orf3 proteins strengthen the links between the reorganization of Mre11 complex localization and the prevention of viral genome concatemerization. In addition to a role in joining viral genomes, the Mre11 complex is also important for signaling in response to DNA damage and in the repair process (6, 9, 30). It will be interesting to determine which of these functions are affected by the relocalization of Mre11 induced by Ad5 E4orf3. In addition, in conjunction with the E1 proteins, the expression of E4orf3 potentiates the transformation of rodent cells, primarily in a “hit-and-run” fashion (28, 29). Understanding the mechanism of E4orf3 function could provide insight into functions of the Mre11 complex and elucidate its possible role in E4orf3-induced transformation.
Adenovirus mutants that do not express either E4orf3 or E4orf6 are phenotypically indistinguishable from wild-type viruses, suggesting that these two viral proteins have complementary functions (5, 14, 17). Each of these two proteins can overcome the defects of a complete E4 deletion, and they have multiple functions that include effects on the initiation of viral DNA replication, splicing, RNA processing, and late protein expression. The actions of these two proteins are likely to be distinct, and their apparent redundancy may reflect different requirements that may depend upon the cell type or cell cycle status of the infected host cell (13, 34). One of the common functions of E4orf3 and E4orf6 is their ability to prevent concatemerization of the adenovirus genome (35, 38). Since the joining event is dependent on the Mre11 complex (35), it would seem that both proteins perform this function by targeting components of this repair complex. Degradation of the Mre11/Rad50/Nbs1 proteins by the E1b55K/E4orf6 complex is conserved among the adenovirus serotypes that we have studied, whereas only Ad5 E4orf3 can alter their cellular location and prevent their accumulation at viral replication centers. This may reflect variations in the target cell type or mode of replication of the different serotypes.
Molecular phylogenic comparisons confirm that the E4orf3 protein is highly conserved among adenoviruses (reviewed in reference 36) but also suggest that the subgroup C adenovirus E4orf3 protein has diverged more sharply from those of the other serotypes. The findings reported here demonstrate that a representative subgroup C protein from Ad5 differs at the functional level by its ability to mislocalize the Mre11 complex and prevent concatemers. Molecular comparisons between the E4orf3 proteins of different Ad serotypes may suggest other naturally occurring variants that are worth studying. For example, it will be interesting to determine which properties are shared by the E4orf3 protein of the enteric Ad40 from subgroup F, which has diverged to a lesser extent from the subgroup A, B, D, and E proteins (Fig. 8B). Perhaps the need to express redundant activities that target DNA repair mechanisms is related to the natural history of the virus in the human host. Most of the mutations generated in the E4 region have been done in the context of human Ad5 or Ad2, and therefore it is not known whether the redundancy between E4orf3 and E4orf6 is maintained in other serotypes.
In addition to its effect on concatemer formation, the E4orf3 protein has several other functions that are important for replication and its effects on viral growth (11, 34). The naturally occurring differences we have uncovered for serotypes Ad4 and Ad12 provide valuable reagents to facilitate a functional dissection of the multifunctional E4orf3 protein. The variable regions between different serotypes may also serve to guide future mutagenesis studies aimed at understanding the functions of E4orf3. A mutational analysis of Ad5 E4orf3 was initiated with point mutants that were generated and expressed under the endogenous E4 promoter when recombined into a virus lacking E4orf6 (11). It will be interesting to determine which defects of a virus with an E4 deletion will be complemented by the E4orf3 proteins derived from the various serotypes and whether the multiple functions of E4orf3 are connected to each other through common cellular targets.
Acknowledgments
We are grateful to R. Evans, T. Hunter, G. Ketner, A. Levine, T. Melendy, T. Sternsdorf, M. Tini, and P. van der Vliet for their generous gifts of reagents. We thank members of the Weitzman lab for discussions and critical readings of the manuscript. We acknowledge the James B. Pendleton Charitable Trust for providing the Pendleton Microscopy Facility.
This work was supported in part by NIH Public Health Service grants AI43341 (M.D.W.), CA97093 (M.D.W.), and CA77342 (D.A.O.) and by gifts from the Joe W. and Dorothy Dorsett Brown Foundation and the Lebensfeld Foundation. T.H.S. was supported by an NIH graduate training grant to UCSD and by fellowships from the H. A. and Mary K. Chapman Charitable Trust, the Legler Benbough Foundation, and the Salk Institute. C.T.C. was supported by the Timken-Sturgis Foundation and by an NIH training grant to the Salk Institute.
REFERENCES
- 1.Bischof, O., S. H. Kim, J. Irving, S. Beresten, N. A. Ellis, and J. Campisi. 2001. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J. Cell Biol. 153:367-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borden, K. L. 2002. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 22:5259-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer, J., K. Rohleder, and G. Ketner. 1999. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology 263:307-312. [DOI] [PubMed] [Google Scholar]
- 4.Bridge, E., and G. Ketner. 1990. Interaction of adenoviral E4 and E1b products in late gene expression. Virology 174:345-353. [DOI] [PubMed] [Google Scholar]
- 5.Bridge, E., and G. Ketner. 1989. Redundant control of adenovirus late gene expression by early region 4. J. Virol. 63:631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carson, C. T., R. A. Schwartz, T. H. Stracker, C. E. Lilley, D. V. Lee, and M. D. Weitzman. 2003. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 22:6610-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carvalho, T., J.-S. Seeler, K. Öhman, P. Jordan, U. Pettersson, G. Akusjärvi, M. Carmo-Fonseca, and A. Dejean. 1995. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J. Cell Biol. 131:45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cathomen, T., and M. D. Weitzman. 2000. A functional complex of adenovirus proteins E1B-55kDa and E4orf6 is necessary to modulate the expression level of p53 but not its transcriptional activity. J. Virol. 74:11407-11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Amours, D., and S. P. Jackson. 2002. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 3:317-327. [DOI] [PubMed] [Google Scholar]
- 10.Doucas, V., A. M. Ishov, A. Romo, H. Juguilon, M. D. Weitzman, R. M. Evans, and G. G. Maul. 1996. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10:196-207. [DOI] [PubMed] [Google Scholar]
- 11.Evans, J. D., and P. Hearing. 2003. Distinct roles of the adenovirus E4 ORF3 protein in viral DNA replication and inhibition of genome concatenation. J. Virol. 77:5295-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett, R. D. 2001. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 20:7266-7273. [DOI] [PubMed] [Google Scholar]
- 13.Goodrum, F. D., and D. A. Ornelles. 1999. Roles for the E4 orf6, orf3, and E1B 55-kilodalton proteins in cell cycle-independent adenovirus replication. J. Virol. 73:7474-7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halbert, D. N., J. R. Cutt, and T. Shenk. 1985. Adenoviral early region 4 encodes functions required for efficient DNA replication, late expression, and host cell shutoff. J. Virol. 56:250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harada, J. N., A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 76:9194-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higgins, D. G., and P. M. Sharp. 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73:237-244. [DOI] [PubMed] [Google Scholar]
- 17.Huang, M.-H., and P. Hearing. 1989. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J. Virol. 63:2605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss III, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 20.Konig, C., J. Roth, and M. Dobbelstein. 1999. Adenovirus type 5 E4orf3 protein relieves p53 inhibition by E1B-55-kilodalton protein. J. Virol. 73:2253-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovacs, G. M., A. J. Davison, A. N. Zakhartchouk, and B. Harrach. 2004. Analysis of the first complete genome sequence of an Old World monkey adenovirus reveals a lineage distinct from the six human adenovirus species. J. Gen. Virol. 85:2799-2807. [DOI] [PubMed] [Google Scholar]
- 22.Leppard, K. N. 1997. E4 gene function in adenovirus, adenovirus vector and adeno-associated virus infections. J. Gen. Virol. 78:2131-2138. [DOI] [PubMed] [Google Scholar]
- 23.Leppard, K. N., and R. D. Everett. 1999. The adenovirus type 5 E1b 55K and E4 Orf3 proteins associate in infected cells and affect ND10 components. J. Gen. Virol. 80:997-1008. [DOI] [PubMed] [Google Scholar]
- 24.Lombard, D. B., and L. Guarente. 2000. Nijmegen breakage syndrome disease protein and MRE11 at PML nuclear bodies and meiotic telomeres. Cancer Res. 60:2331-2334. [PubMed] [Google Scholar]
- 25.Maser, R. S., O. K. Mirzoeva, J. Wells, H. Olivares, B. R. Williams, R. A. Zinkel, P. J. Farnham, and J. H. Petrini. 2001. Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol. Cell. Biol. 21:6006-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moller, A., and M. L. Schmitz. 2003. Viruses as hijackers of PML nuclear bodies. Arch. Immunol. Ther. Exp. (Warsaw) 51:295-300. [PubMed] [Google Scholar]
- 27.Negorev, D., and G. G. Maul. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20:7234-7242. [DOI] [PubMed] [Google Scholar]
- 28.Nevels, M., B. Tauber, E. Kremmer, T. Spruss, H. Wolf, and T. Dobner. 1999. Transforming potential of the adenovirus type 5 E4orf3 protein. J. Virol. 73:1591-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nevels, M., B. Tauber, T. Spruss, H. Wolf, and T. Dobner. 2001. “Hit-and-run” transformation by adenovirus oncogenes. J. Virol. 75:3089-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petrini, J. H. 1999. The mammalian Mre11-Rad50-Nbs1 protein complex: integration of functions in the cellular DNA-damage response. Am. J. Hum. Genet. 64:1264-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarnow, P., P. Hearing, C. W. Anderson, D. N. Halbert, T. Shenk, and A. J. Levine. 1984. Adenovirus early region 1B 58,000-dalton tumor antigen is physically associated with an early region 4 25,000-dalton protein in productively infected cells. J. Virol. 49:692-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarnow, P., P. Hearing, C. W. Anderson, N. Reich, and A. J. Levine. 1982. Identification and characterization of an immunologically conserved adenovirus early region 11,000 Mr protein and its association with the nuclear matrix. J. Mol. Biol. 162:565-583. [DOI] [PubMed] [Google Scholar]
- 34.Shepard, R. N., and D. A. Ornelles. 2004. Diverse roles for E4orf3 at late times of infection revealed in an E1B 55-kilodalton protein mutant background. J. Virol. 78:9924-9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348-352. [DOI] [PubMed] [Google Scholar]
- 36.Tauber, B., and T. Dobner. 2001. Molecular regulation and biological function of adenovirus early genes: the E4 ORFs. Gene 278:1-23. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiden, M. D., and H. S. Ginsberg. 1994. Deletion of the E4 region of the genome produces adenovirus DNA concatemers. Proc. Natl. Acad. Sci. USA 91:153-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, G., W. H. Lee, and P. L. Chen. 2000. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. Implication of NBS1 in alternative lengthening of telomeres. J. Biol. Chem. 275:30618-30622. [DOI] [PubMed] [Google Scholar]
- 40.Xu, Z. X., A. Timanova-Atanasova, R. X. Zhao, and K. S. Chang. 2003. PML colocalizes with and stabilizes the DNA damage response protein TopBP1. Mol. Cell. Biol. 23:4247-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zantema, A., J. A. Fransen, A. Davis-Olivier, F. C. Ramaekers, G. P. Vooijs, B. DeLeys, and A. J. Van der Eb. 1985. Localization of the E1B proteins of adenovirus 5 in transformed cells, as revealed by interaction with monoclonal antibodies. Virology 142:44-58. [DOI] [PubMed] [Google Scholar]
- 42.Zhong, S., P. Hu, T. Z. Ye, R. Stan, N. A. Ellis, and P. P. Pandolfi. 1999. A role for PML and the nuclear body in genomic stability. Oncogene 18:7941-7947. [DOI] [PubMed] [Google Scholar]