Abstract
This study utilized 16S rRNA high-throughput sequencing technology to analyze the community structure and function of endophytic bacteria within the roots of three plant species in the vanadium–titanium–magnetite (VTM) mining area. The findings indicated that mining activities of VTM led to a notable decrease in both the biodiversity and abundance of endophytic bacteria within the root systems of Eleusine indica and Carex (p < 0.05). Significant reductions were observed in the populations of Nocardioides, concurrently with substantial increments in the populations of Pseudomonas (p < 0.05), indicating that Pseudomonas has a strong adaptability to this environmental stress. In addition, β diversity analysis revealed divergence in the endophytic bacterial communities within the roots of E. indica and Carex from the VTM mining area, which had diverged to adapt to the environmental stress caused by mining activity. Functional enrichment analysis revealed that VTM mining led to an increase in polymyxin resistance, nicotinate degradation I, and glucose degradation (oxidative) (p < 0.05). Interestingly, we found that VTM mining did not notably alter the endophytic bacterial communities or functions in the root systems of Dodonaea viscosa, indicating that this plant can adapt well to environmental stress. This study represents the primary investigation into the influence of VTM mining activities on endophytic bacterial communities and the functions of nearby plant roots, providing further insight into the impact of VTM mining activities on the ecological environment.
Keywords: mining activity, soil pollution, bioremediation, microecology
1. Introduction
Vanadium–titanium–magnetite (VTM) is a common polymetallic ore that includes a range of metals, including iron (Fe), vanadium (V), titanium (Ti), cobalt (Co), and chromium (Cr), with Fe, V, and Ti being the predominant elements present in VTM [1]. VTM is a significant source of Fe, as well as a primary source of V and Ti. Therefore, it has significant value for comprehensive utilization [2]. The world’s VTM resources are primarily found in China, Canada, Australia, Russia, New Zealand, and South Africa, totaling a global reserve of almost 48 billion tons [3,4,5,6]. China has reserves of 9.38 billion tons of VTM resources, primarily located in the Panxi region, with the most substantial reserves of 3.6 billion tons found in the Hongge reserves in the Panzhihua–Xichang region [7,8,9,10]. V and Ti mainly exist in VTM as associated ores. V and Ti are crucial rare metals that find extensive applications in steel, metallurgy, chemicals, batteries, aerospace, and various other industries [11,12,13,14]. In the iron and steel industry, V is a commonly utilized component. The addition of V to steel can improve the wear resistance, strength, hardness, ductility, and other properties of steel [11]. Ti and its alloys have excellent properties, such as corrosion resistance, high strength, good strength at high and low temperatures, non-magnetic properties, good adaptability to humans, shape memory, and superconductivity [15]. Due to its lightweight and high-strength characteristics, it has been widely used in aerospace and other fields. In recent years, its application has gradually expanded to include shipbuilding, petrochemical equipment, offshore platforms, electric power equipment, medical treatment, and high-end consumer goods [15,16]. With the increasing demand for V and Ti, the mining scale of VTM is also increasing. Nevertheless, during mineral mining and ore processing processes, significant quantities of VTM tailings are produced, which are frequently disposed of in dams, resulting in various environmental consequences, including the discharge of harmful contaminants into the nearby surroundings. These conditions pose a danger to the local ecosystems [17,18,19]. However, the impact of VTM mining on local plants and microorganisms has not been determined.
Endophytic bacteria, which are present in healthy plant tissues, are essential microbial-plant symbionts and valuable microbial resources that do not pose a significant threat to the host [20,21,22,23]. Multiple studies have proven that a high quantity of endophytic bacteria can live within plant tissues, developing a variety of mutually beneficial symbiotic partnerships [24,25,26]. While colonizing plant tissues, these microbes may also enhance plant adaptability in contaminated soil [27,28]. Moreover, endophytic bacteria can promote seed germination and growth through various mechanisms [29,30,31,32,33,34]. Some Bacillus, Pseudomonas, and Pantoea species can synthesize secondary metabolites with antibacterial properties, exhibiting strong antagonistic effects against plant pathogenic bacteria [35,36,37]. Furthermore, endophytic bacteria are essential components of plant microbial communities and possess substantial potential for use in protecting plants [38]. Endophytic bacteria, like beneficial microbes present in different ecosystems, can function as transporters for emerging bioactive substances, thereby holding importance in medicine, agriculture, and industry [22,25,39]. The diversity of plant endophytes and the establishment of community structures are closely related to factors such as plant variety, growth environment, and geographic location [40]. Endophytic bacteria, which are associated with plant health and productivity, have become a focal point of interest in scientific and commercial realms worldwide in recent years [22,41,42]. However, the effects of VTM mining on the diversity and function of endophytic bacteria in the root systems of nearby plants, as well as the response strategies of root endophytes, have not been determined.
In this study, three local plant species (Dodonaea viscosa, Carex L., and Eleusine indica) were used as the research objects, and the influence of these three plant species on the diversity and function of the endophytic bacterial communities in the roots was investigated through 16S rRNA high-throughput sequencing. Meanwhile, the response strategies of these three plant root endophytic bacteria to VTM mining were analyzed. This study addresses the gap in understanding the ecological effects of VTM mining and provides insights into the ecological remediation of pollution caused by VTM mining.
2. Materials and Methods
2.1. Collection of Plant Roots and Extraction of Endophytic Bacterial DNA from the Roots
The sampling site was located in the VTM mining area near Hongge Village (101°56′ E, 26°32′ N), Yanbian County, Panzhihua City, Sichuan Province, China. The site has an average annual rainfall of 1065.6 mm, an average annual temperature of 19.2 °C, an average annual sunshine of 2307.2 h, and a relative humidity of 66.6%. We chose three different species of plants with the best local growing conditions for our study: D. viscosa, Carex, and E. indica. To collect the plants, we used the five-point sampling method. First, the midpoint of the diagonal line was determined as the center sampling point. Next, four points were selected on the diagonal line with a distance of 20 m from the center sampling point as sample points. Three plants of each species were randomly collected and a total of 45 plants were collected. In addition, we used the same sampling method to collect these three plants as control samples from the soil of a non-VTM mining site located 3 km away from the mine site. Extracted from the soil, the roots of these three plants had the adhering soil shaken off to obtain the root systems. The plants were subsequently placed in sterile sample bags filled with ice packs, shipped to the laboratory, refrigerated at 4 °C, and treated within 24 h. The residual soil on the surface of the plant roots was washed with sterile distilled water, soaked in 70% ethanol for surface disinfection, and finally rinsed repeatedly with sterile distilled water. All plants were treated in this manner, and each plant had three biological replicates. About 50× g of plant roots were weighed from each sample into a 15 mL sterile centrifuge tube for bacterial diversity analysis. Following this, 18 samples were transferred to the laboratory in a foam box with ice packs for DNA extraction and 16S rRNA sequencing. Genomic DNA was extracted from plant root samples by utilizing the Omega Bio-Tek Plant DNA Kit (Norcross, GA, USA). The DNA was then separated on a 1% (w/v) agarose gel for the purpose of evaluating the quality of the extracted DNA. The samples of D. viscosa, Carex, and E. indica from the VTM mining area were represented as VT-Dvi, VT-Car, and VT-Ein, respectively. The control samples of these three plant species were represented as CK-Dvi, CK-Car, and CK-Ein, respectively.
2.2. PCR Amplification and Detection
The concentration of DNA was quantified by Nanodrop One. The extracted genomic DNA was diluted to 1 ng/µL with sterile water, and the 16S rRNA V3-V4 region of the sample was subsequently carried out with primers that included a barcode (341F: 5′-cctacgggaggcagcagg-3′; 806R: 5′-GGACTACNVGGGTWTCTAAT-3′). PCR was performed using 15 μL of Phusion® High-Fidelity PCR Master Mix from New England Biolabs, 2 μL of forward and reverse primers, and 10 ng of template DNA. During thermal cycling, denaturation at 98 °C for 1 min was repeated 30 times, including denaturation for 10 s, annealing at 50 °C for 30 s, extension at 72 °C for 30 s, and storage at 72 °C for 5 min. After completion of PCR, the products were first mixed with an equal volume of loading buffer containing SYBR Green, after which the PCR products were detected via electrophoresis on a 2% (w/v) agarose gel. When amplification was complete, the PCR products were purified using the Qiagen Gel Extraction Kit (Qiagen, Hilden, Germany).
2.3. Library Preparation, Sequencing, and Raw Data Processing
As per the manufacturer’s guidelines, we proceeded to generate sequencing libraries using the TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, San Diego, CA, USA), which included index codes. For assessing the library’s quality, we used both a Qubit@2.0 Fluorometer (Thermo Scientific, Waltham, MA USA) and an Agilent Bioanalyzer 2100 system. Subsequently, the library underwent sequencing with the Illumina NovaSeq platform to yield a 250 bp paired-end sequence. The reads were identified according to their individual barcodes. Following this, the barcodes and primer sequences were eliminated. The merging of paired-end reads was carried out using FLASH V1.2.7 [43]. To obtain high-quality labels, the original labels underwent processing following the quality control procedure of QIIME V1.9.1 [44]. After comparing the labels to the Silva reference database, the dataset was cleansed of the identified chimeric sequences [45].
2.4. OTU Cluster and Species Annotation
By utilizing Uparse v7.0.1001, sequences exhibiting a similarity of 97% or higher were categorized into operational taxonomic units (OTUs) [46]. For the purpose of annotation facilitation, a specific sequence was chosen to represent each OTU. The representative sequences were assigned the classification information from the Silva database utilizing the Mothur algorithm [45]. To analyze the phylogenetic relatedness of OTUs and the variation in dominant species among different samples or taxa, we utilized MUSCLE v3.8.3 for multiple sequence alignment [47]. Normalization of OTU abundance data was carried out by aligning it with a benchmark sample that had the fewest sequences. Utilizing the normalized data, further analyses were conducted on α and β diversity.
2.5. α and β Diversity Analyses
We used seven indicators, Pielou evenness, Simpson, Chao1, observed species, Shannon, Good’s coverage, and Faith_pd, to analyze species diversity in each sample. For the calculation of these indicators, QIIME 1.7.0 software was used, and the results were displayed using R v2.15.3 [44]. For community richness evaluation, we employed three indicators: Pielou evenness, Chao1, and observed species. For community diversity assessment, we utilized the Shannon and Simpson indices. β diversity analysis was employed to examine the changes in species complexity among the samples. Lastly, the R vegan software was used to conduct Non-metric Multidimensional Scaling (NMDS) analysis and Principal Coordinates Analysis (PCoA).
2.6. Functional Prediction of Endophytic Bacteria
PICRUSt2 can elucidate the functions of endophytic bacteria within the root system [48,49], which utilizes the Gene Ontology (GO), KO, and MetaCyc databases [50,51]. Before the cluster analysis, we employed the FactoMineR and ggplot2 software packages in R v2.15.3 to conduct PCoA in order to decrease the dimensionality of the original variables.
2.7. Statistical Analysis
Statistical analysis was employed to identify the significance of differences among the samples. The t-test was applied for the direct comparison of two groups of samples, while Tukey’s test was used to analyze multiple sample groups. A p-value below 0.05 signifies a statistically significant difference between the groups.
3. Results
3.1. High-Throughput Sequencing Statistics of Endophytic Bacterial Communities
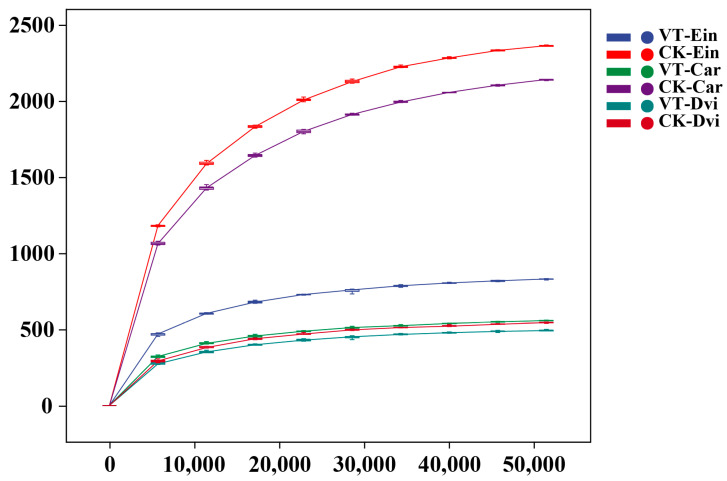
Utilizing 16S rRNA high-throughput sequencing technology, we examined the endophytic bacteria within the root systems of D. viscosa, Carex, and E. indica samples obtained from both the VTM mining region and the non-VTM mining region in this study. In Figure A1, the curve for the sparse OTUs is displayed. The incremental increase in the observed species was positively associated with the growing quantity of sequencing reads. When the number of sequencing reads reached over 20,000, the sparse curve gradually stabilized, indicating that the sequencing results were close to saturation. There were enough reads to effectively capture the comprehensive profile of endophytic bacteria residing in the root system of the sample. Therefore, the large amount of sequencing data can provide insight into the bacterial diversity present in the sample. After removing chimeras, poor-quality reads, and short reads, an average of 97,932 clean reads were successfully acquired per sample. These clean reads were then allocated to OTUs using a 97% similarity threshold. The number of OTUs per sample varied across all samples, ranging from a minimum of 380 to a maximum of 1283, with the average number across all samples being 723 OTUs.
3.2. α Diversity Indices
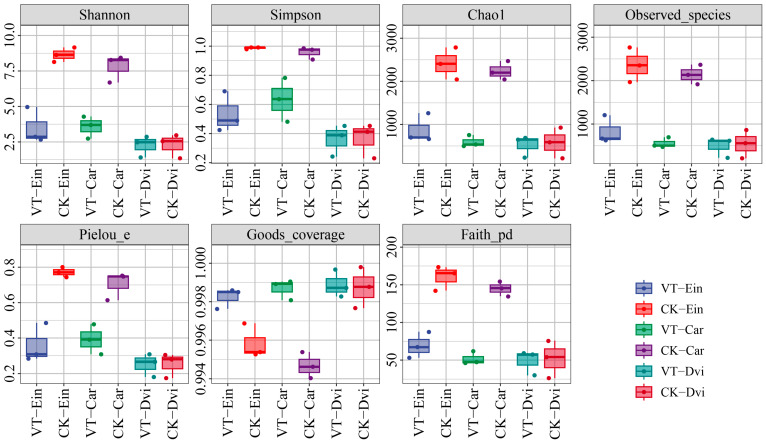
The diversity and richness of the samples were evaluated using seven indicators (Shannon, Pielou evenness, Simpson, Chao1, observed species, Good’s coverage, and Faith_pd), as illustrated in Figure 1. A higher α diversity index indicates a greater level of community diversity among the endophytic bacteria present in the root system of the sample. The Shannon and Simpson indices showed that the CK-Ein and CK-Car samples had the highest levels of community diversity, while the VT-Dvi and CK-Dvi samples had the lowest levels of community diversity. The VTM mining activities significantly reduced the Shannon and Simpson indices of the VT-Ein and VT-Car samples compared to those of the CK-Ein and CK-Car samples (p < 0.05). In contrast to the CK-Dvi sample, the mining activities conducted by VTM did not have a notable impact on the community diversity of the VT-Dvi sample. In addition, an analysis of community richness, including observed species, Chao1, and Pielou evenness, showed that the CK-Ein sample exhibited the greatest diversity, while the VT-Car sample showed the least. The Chao1, observed species, and Pielou evenness indices of endophytic bacteria in the root system of E. indica and Carex within the VTM mining area exhibited a significant decrease compared to those in the non-VTM mining area (p < 0.05). However, VTM mining did not significantly impact the diversity of endophytic bacteria within the root system of D. viscosa. In comparison to the CK-Ein and CK-Car samples, the Faith_pd index was significantly lower in the VT-Ein and VT-Car samples (p < 0.05), while the Good’s coverage index was notably higher (p < 0.05). Nevertheless, when compared to the samples of VT-Dvi and CK-Dvi, there were no significant changes in any of the indices.
Figure 1.
Box diagram of the differences in the α diversity indices of endogenous root bacteria between different samples. Colored dots represent values that are significantly different from other data in this group. VT-Dvi, Dodonaea viscosa in VTM mining area; VT-Car, Carex in VTM mining area; VT-Ein, Eleusine indica in VTM mining area; CK-Dvi, Dodonaea viscosa in non-VTM mining area; CK-Car, Carex in non-VTM mining area; CK-Ein, Eleusine indica in non-VTM mining area.
3.3. Taxonomic Analyses of the Endophytic Bacterial Communities
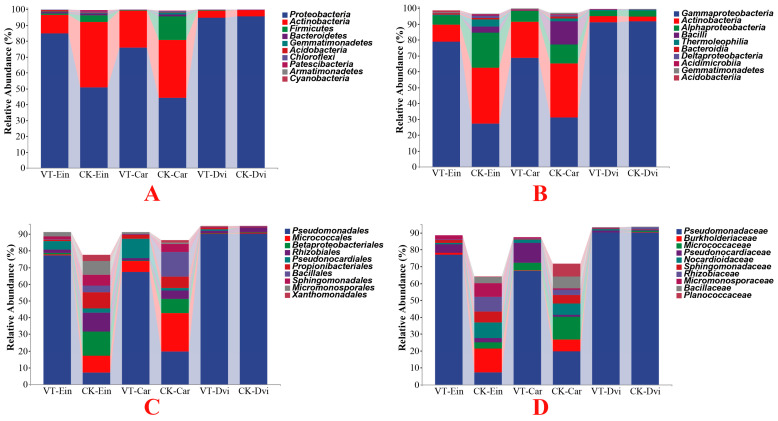
By examining the abundance of the top 10 phyla, differences among the samples were assessed (Figure 2A). Proteobacteria, the dominant phylum in the samples, was found to make up an average of 74.36% of the total bacteria, followed by Actinobacteria (20.19%), Firmicutes (3.44%), and Bacteroidetes (0.63%). Among all the samples, Proteobacteria (95.61% on average) was the predominant phylum in the CK-Dvi sample, Actinobacteria (41.23% on average) dominated the CK-Ein sample, while Firmicutes (14.83% on average) and Bacteroidetes (1.35% on average) were the most abundant in the CK-Car sample. Compared to the CK-Ein sample, there was a notable boost in the abundance of Proteobacteria within the VT-Ein samples (p < 0.05), whilst a considerable decrease in the quantity of Actinobacteria was observed (p < 0.05). Besides, the abundances of Firmicutes and Bacteroidetes decreased, but not significantly. A notable change was observed in the abundance of Proteobacteria in the VT-Car sample, which demonstrated a significant rise compared to the CK-Car sample (p < 0.05). Furthermore, the abundance of Bacteroidetes in the VT-Car sample was significantly reduced compared to the CK-Car sample (p < 0.05). Besides, the abundances of Actinobacteria and Firmicutes were decreased in the VT-Car sample, but not significantly. While a drop in the abundance of Proteobacteria was observed in the VT-Dvi sample compared to the CK-Dvi sample, this difference was not found to be statistically significant. Additionally, the abundances of Actinobacteria, Firmicutes, and Bacteroidetes were increased, but not significantly.
Figure 2.
Cumulative bar charts of the relative abundance of taxa (top 10) at the phylum (A), class (B), order (C), and family (D) levels. VT-Dvi, D. viscosa in the VTM mining area; VT-Car, Carex in the VTM mining area; VT-Ein, E. indica in the VTM mining area; CK-Dvi, D. viscosa in the non-VTM mining area; CK-Car, Carex in the non-VTM mining area; CK-Ein, E. indica in the non-VTM mining area.
At the class level, Gammaproteobacteria predominated in all samples, comprising an average of 64.82%, with Actinobacteria following at 18.23%, Alphaproteobacteria at 8.87%, and Bacilli at 3.24% (Figure 2B). Compared to the CK-Ein sample, the VT-Ein samples exhibited a significantly increased abundance of Gammaproteobacteria (p < 0.05), a notably decreased abundance of Actinobacteria and Alphaproteobacteria (p < 0.05), and a reduced abundance of Bacilli, but not significantly. In the VT-Car sample, it was noted that the relative abundance of Gammaproteobacteria significantly increased compared to the CK-Car sample (p < 0.05), while the abundances of Actinobacteria, Alphaproteobacteria, and Bacilli all showed decreases, although not statistically significant. In the VT-Dvi sample, the abundance of Actinobacteria was increased compared to the CK-Dvi sample, but not significantly. Additionally, the abundances of Gammaproteobacteria, Alphaproteobacteria, and Bacilli were decreased, but not significantly.
At the order level, members of the Pseudomonadales were found to be the most prevalent with an average abundance of 58.62%, followed by Micrococcales (6.77%), Betaproteobacteriales (4.17%), and Rhizobiales (3.98%) (Figure 2C). In comparison to the CK-Ein sample, the VT-Ein sample showed a notably higher abundance of Pseudomonadales (p < 0.05). Additionally, the abundances of Betaproteobacteriales and Rhizobiales exhibited a significant decrease (p < 0.05), while the abundance of Micrococcales also decreased, but not significantly. In the VT-Car sample, the abundance of Pseudomonadales showed a significant increase compared to the CK-Car sample (p < 0.05). A significant decrease in the abundance of Betaproteobacteriales was observed (p < 0.05), with the abundances of Micrococcales and Rhizobiales also showing decreases, although not statistically significant. Compared to the CK-Dvi sample, the abundances of Pseudomonadales, Betaproteobacteriales, Micrococcales, and Rhizobiales in the VT-Dvi sample were all reduced, but not significantly.
At the family level, Pseudomonadaceae had the highest proportion, representing an average of 58.62% of the total population in the samples, with Burkholderiaceae (3.81%), Micrococcaceae (3.74%), and Pseudomonocardiaceae (3.63%) following in abundance (Figure 2D). In comparison to the CK-Ein sample, the VT-Ein sample exhibited a significantly higher abundance of Pseudomonadaceae (p < 0.05). Furthermore, the abundance of Burkholderiaceae was notably lower (p < 0.05), while the abundance of Micrococcaceae was also lower, but not significantly. Additionally, the abundance of Pseudomonocardiaceae was greater, although not statistically significant. In comparison to the CK-Car sample, the VT-Car sample exhibited a significantly greater abundance of Pseudomonadales (p < 0.05). The overall abundance of Burkholderiaceae decreased notably (p < 0.05), whereas the decrease in Micrococcaceae was not statistically significant. Moreover, the prevalence of Pseudomonocardiaceae was higher, although not statistically significant. Compared to the CK-Dvi sample, the abundances of Pseudomonadaceae, Burkholderiaceae, and Micrococcaceae in the VT-Dvi sample were all decreased, but not significantly, while the abundance of Pseudonocardiaceae was increased, but not significantly.
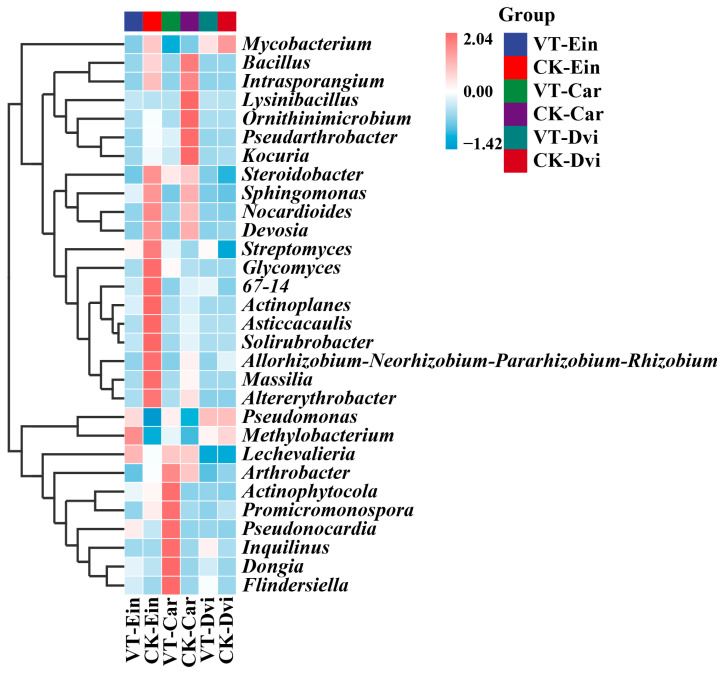
At the genus level, Pseudomonas emerged as the most prevalent, representing an average of 58.60% of the total bacteria in the samples, followed by Nocardioides (2.90%), Pseudonocardia (2.44%), and Pseudarthrobacter (1.94%) (Figure 3). Compared to the CK-Ein sample, the VT-Ein sample showed a significantly increased abundance of Pseudomonas (p < 0.05) and a significantly reduced abundance of Nocardioides (p < 0.05). The abundance of Pseudonocardia was also increased, but not significantly. Additionally, the abundance of Pseudarthrobacter was decreased, but not significantly. In comparison to the CK-Car samples, there was a significant increase in the abundance of Pseudomonas in the VT-Car sample (p < 0.05). Moreover, there was a notable decrease in the amount of Nocardioides (p < 0.05), while the abundance of Pseudonocardia increased, but not significantly. The abundance of Pseudarthrobacter exhibited a decrease, although it was not statistically significant. Compared to the CK-Dvi sample, the abundances of Pseudomonas and Pseudarthrobacter were both reduced, but not significantly, while the abundances of Nocardioides and Pseudonocardia were increased, but not significantly.
Figure 3.
Heatmap analysis of the 30 genes with the highest abundance of different taxa samples. The relative abundances of different genera are represented by different colors. Blocks, with colors ranging from blue to red indicate an increase in relative abundance. VT-Dvi, D. viscosa in the VTM mining area; VT-Car, Carex in the VTM mining area; VT-Ein, E. indica in the VTM mining area; CK-Dvi, D. viscosa in the non-VTM mining area; CK-Car, Carex in the non-VTM mining area; CK-Ein, E. indica in the non-VTM mining area.
3.4. Structural Differentiation of Endophytic Bacterial Communities
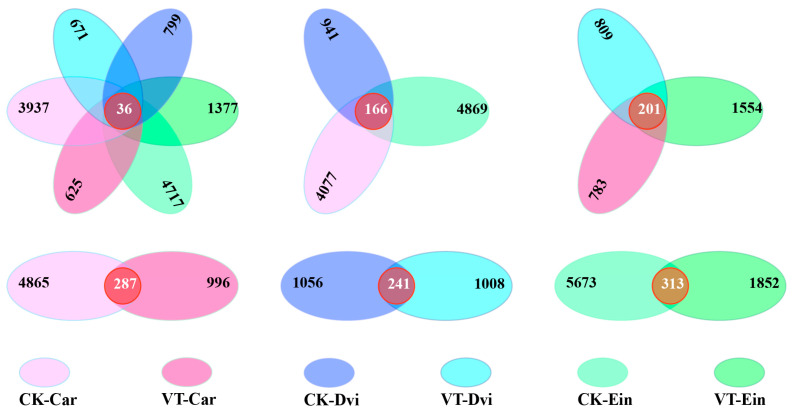
An examination of the distinct OTUs and the OTUs shared among the samples indicated the presence of 36 core OTUs in all samples. Additionally, there were 4717, 1377, 3937, 625, 799, and 671 OTUs specific to CK-Ein, VT-Ein, CK-Car, VT-Car, CK-Dvi, and VT-Dvi samples, respectively (Figure 4). Among the three types of plant root systems in the VTM mining area, the VT-Ein, VT-Car, and VT-Dvi samples had 1554, 783, and 809 specific OTUs and 201 shared OTUs, respectively. Among the three plant root systems in the non-VTM mining area, the CK-Ein, CK-Car, and CK-Dvi samples had 4869, 4077, and 941 specific OTUs and 166 shared OTUs, respectively. Between the CK-Ein and VT-Ein samples, the CK-Ein sample contained 5673 specific OTUs, while the VT-Ein sample contained 1852 specific OTUs. There were 313 common OTUs between the two samples. Among the CK-Car and VT-Car samples, the CK-Car sample contained 4865 specific OTUs, while the VT-Car sample contained 996 specific OTUs. There were 287 common OTUs between the two samples. Between the CK-Dvi and VT-Dvi samples, the CK-Dvi sample contained 1056 specific OTUs, while the VT-Dvi sample contained 1008 specific OTUs. There were 241 common OTUs between the two samples.
Figure 4.
Differential microbial comparison between different samples. VT-Dvi, D. viscosa in the VTM mining area; VT-Car, Carex in the VTM mining area; VT-Ein, E. indica in the VTM mining area; CK-Dvi, D. viscosa in the non-VTM mining area; CK-Car, Carex in the non-VTM mining area; CK-Ein, E. indica in the non-VTM mining area.
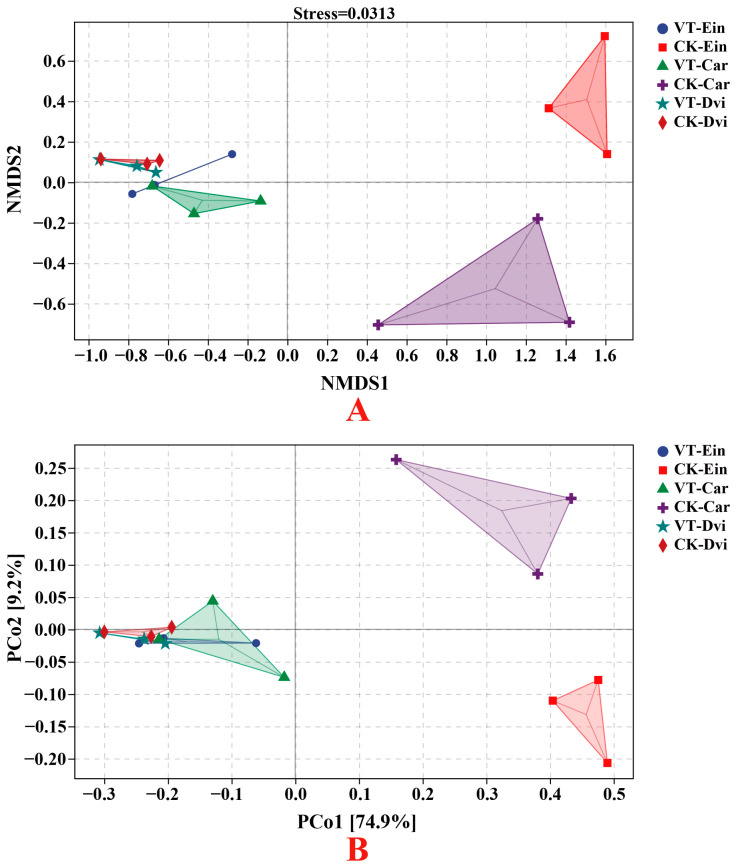
We employed NMDS and PCoA to assess variations in endophytic bacterial communities within the root systems across different samples (Figure 5). The results indicated that mining VTM resulted in variations in the composition of the endophytic bacterial community within the root systems of Carex and E. indica when compared to areas without VTM mining. In summary, the NMDS and PCoA analyses conducted on the bacterial communities present in the roots of Carex and E. indica demonstrated that the VTM mining activities significantly altered the overall structure and composition of the endophytic bacterial community. However, there was no significant difference in the community composition of endophytic bacteria in the root systems of D. viscosa between VTM mining and non-VTM mining areas. This suggests that, for endophytic bacteria in the root system of D. viscosa, internal environmental factors were more influential than external environmental factors.
Figure 5.
β diversity analysis of the root endophytic bacterial community in different samples based on NMDS (A) and PCoA (B). VT-Dvi, D. viscosa in the VTM mining area; VT-Car, Carex in the VTM mining area; VT-Ein, E. indica in the VTM mining area; CK-Dvi, D. viscosa in the non-VTM mining area; CK-Car, Carex in the non-VTM mining area; CK-Ein, E. indica in the non-VTM mining area.
3.5. Function Prediction of the Endophytic Bacterial Communities
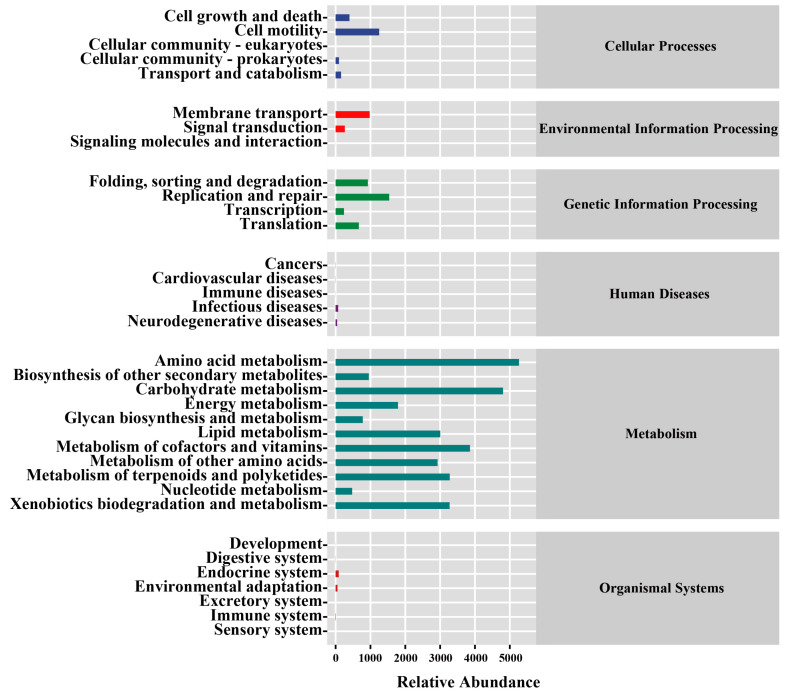
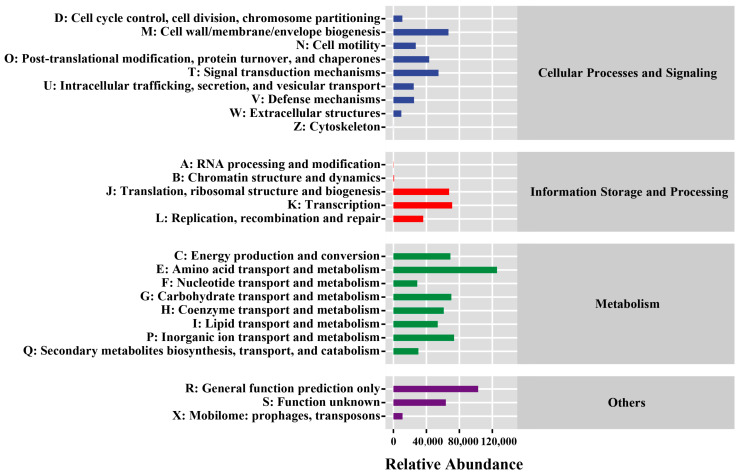
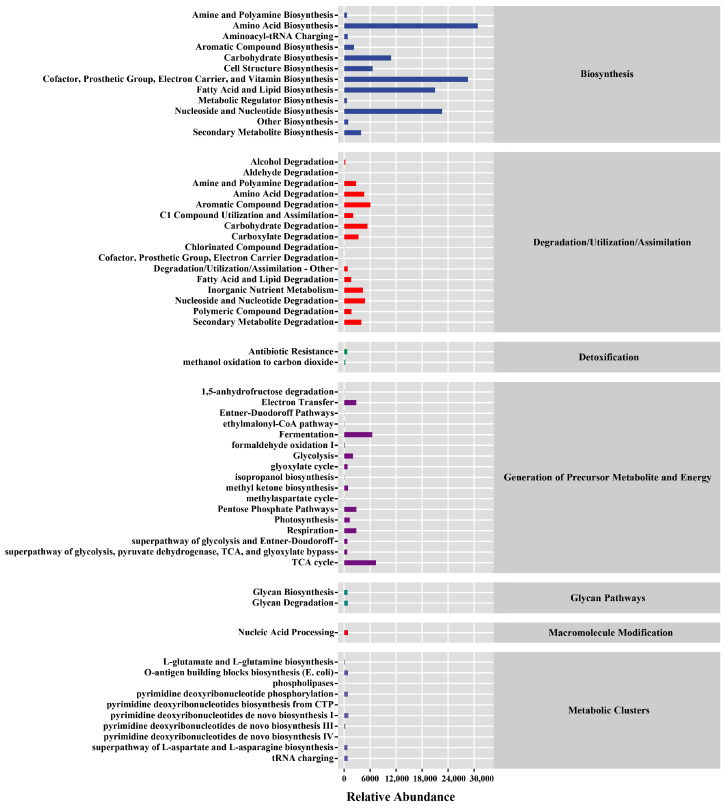
PICRUSt2 was employed to gain a thorough understanding of the specific functions of the root endophytic bacteria present in the samples. According to the data in the KEGG database, the various genes found in bacterial cells have been organized into six broad categories (Figure A2). The largest group of these genes can be categorized as metabolism genes, which were found to represent 81.78% of all the genes in the database. The second largest group, representing 9.06% of the total genes, can be categorized as genetic information processing genes, followed by cellular processes genes, which represent 5.10% of the total. At the second level, amino acid metabolism, carbohydrate metabolism, metabolism of cofactor and vitamins, xenobiotics biodegradation, and metabolism of terpenoids and polyketides were the most abundant and accounted for 14.16%, 12.93%, 10.37%, 8.82%, and 8.80%, respectively. Based on the COG database, approximately 84.28% of bacterial genes were categorized into three groups (Figure A3). Metabolism stood out as the most enriched function, encompassing 45.43% of the genes, while cellular process and signaling accounted for 23.23%, and information storage and processing for 18.52%. Second-level COG metabolism showed that genes involved in amino acid transport and metabolism, transcription, inorganic ion transport and metabolism, and carbohydrate transport and metabolism were the most abundant, accounting for 11.14%, 6.53%, 6.32%, and 6.23%, respectively. According to the MetaCyc database, bacterial genes can be divided into seven categories (Figure A4). In the observed data, it can be seen that biosynthesis is the most prevalent category among the samples, representing a staggering 62.15% of the total. The second most common category is degradation/utilization/assimilation, accounting for 20.04%, while the third is the generation of precursor metabolites and energy, making up 13.91% of all samples. In the second level, the largest proportion of genes was found in amino acid biosynthesis (14.79%), followed by cofactors, prosthetic groups, electron carrier, and vitamin biosynthesis (13.71%), nucleoside and nucleotide biosynthesis (10.85%), fatty acid and lipid biosynthesis (10.07%), and carbohydrate biosynthesis (5.19%).
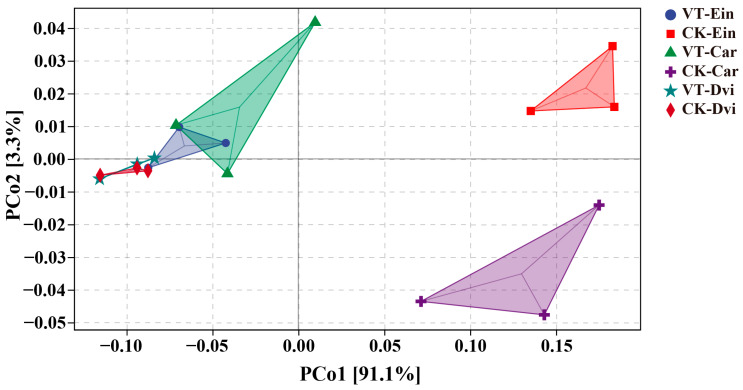
We employed PCoA to evaluate variations in endophytic bacterial function across different samples, as illustrated in Figure 6. The findings indicated that endophytic bacteria in the root systems of Carex and E. indica exhibited variations when subjected to VTM mining activities, as opposed to those from non-VTM mining areas. Moreover, the mineral mining operations conducted by VTM resulted in a convergence of the functionalities of endophytic bacteria within the root structures of these three plant species.
Figure 6.
PCoA of the functions of the root endophytic bacteria predicted by PICRUSt2. VT-Dvi, D. viscosa in the VTM mining area; VT-Car, Carex in the VTM mining area; VT-Ein, E. indica in the VTM mining area; CK-Dvi, D. viscosa in the non-VTM mining area; CK-Car, Carex in the non-VTM mining area; CK-Ein, E. indica in the non-VTM mining area.
3.6. Functional Enrichment of Endophytic Bacteria
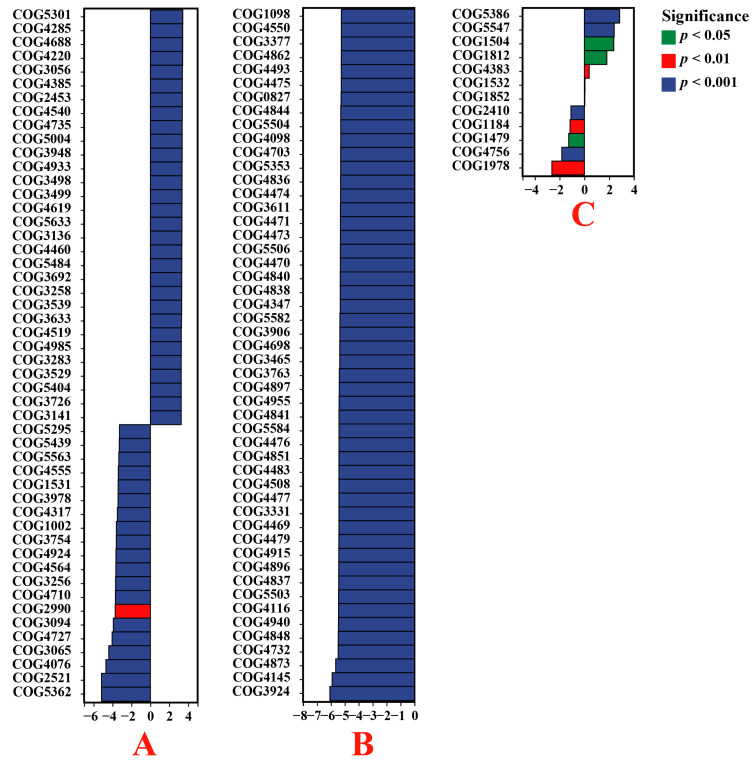
The analysis of the COG database revealed that the endophytic bacteria present in the roots of E. indica from the VTM mining area contained significantly increased levels of phage-related tail fiber protein (COG5301), uncharacterized protein (COG4285), uncharacterized protein (COG4688), ohage DNA packaging protein (COG4220), and uncharacterized protein (COG3056) compared to those from the non-VTM mining area (p < 0.05). In contrast, the levels of the phage terminase large subunit (COG5362), predicted archaeal methyltransferase (COG2521), predicted RNA methylase (COG4076), starvation-inducible outer membrane lipoprotein (COG3065), and uncharacterized protein (COG4727) were notably reduced (p < 0.05) (Figure 7). The levels of uncharacterized protein (COG3924), Na+/panthothenate symporter (COG4145), uncharacterized protein (COG4873), predicted membrane protein (COG4732), and uncharacterized protein (COG4848) in the endophytic bacteria of the Carex root system from the VTM mining area were significantly lower than those in the non-VTM mining area (p < 0.05). Compared to D. viscosa from the non-VTM area, the endophytic bacteria in the root systems of D. viscosa from the VTM mining area had higher levels of the heme-binding NEAT domain (COG5386), uncharacterized protein (COG5547), uncharacterized protein (COG1504), archaeal S-adenosylmethionine synthetase (COG1812), and Mu-like prophage protein gp29 (COG4383), which were significantly reduced (p < 0.05). In contrast, the contents of predicted RNase H-related nuclease YkuK, DUF458 family (COG1978), predicted cation transporter (COG4756), uncharacterized protein (COG1479), translation initiation factor 2B subunit, eIF-2B α/β/delta family (COG1184), and predicted nuclease (RNAse H fold) (COG2410) were significantly reduced (p < 0.05).
Figure 7.
Based on the COG database, the functions of endophytic bacteria in plant roots were significantly different between VTM mining areas and non-VTM mining areas. The vertical axis represents the ID of the COG, the horizontal axis represents the log2-fold change, and different colors indicate significant differences between samples at different levels. (A) VT-Ein vs. CK-Ein; (B) VT-Car vs. CK-Car; (C) VT-Dvi vs. CK-Dvi.
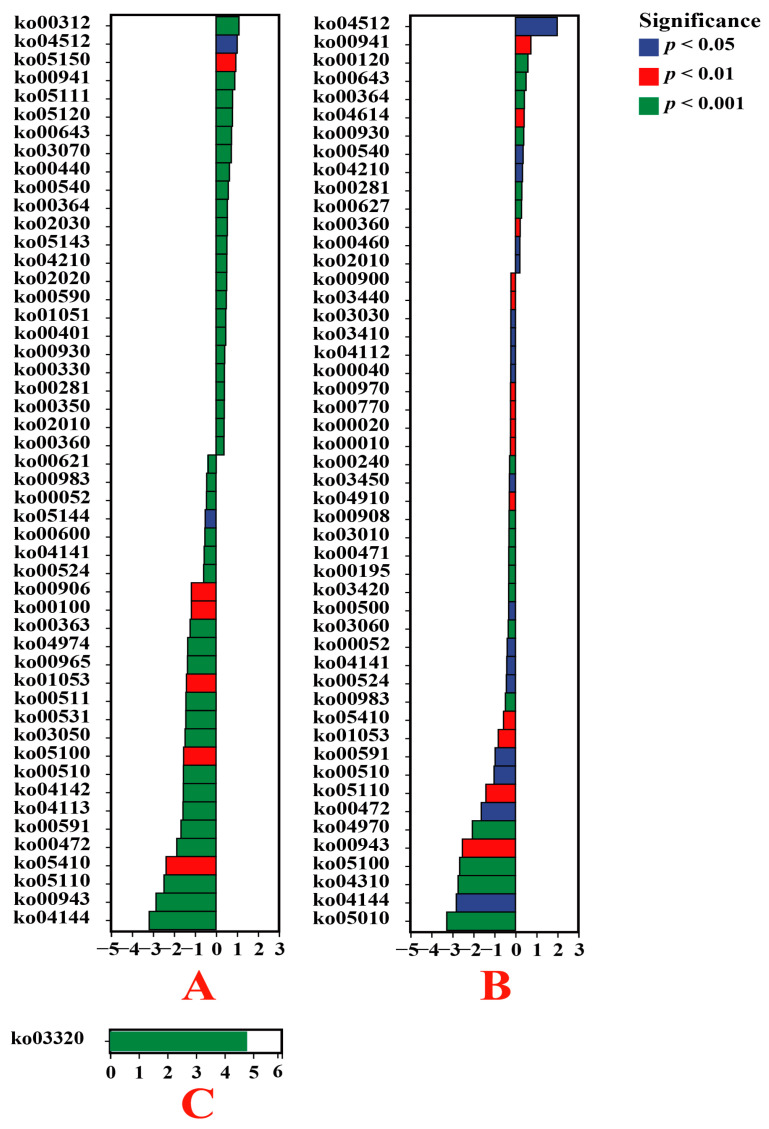
Owing to the impact of VTM extraction operations, the endophytic bacteria in the root systems of E. indica from the mining area included β-Lactam resistance (ko00312), ECM-receptor interaction (ko04512), Staphylococcus aureus infection (ko05150), flavonoid biosynthesis (ko00941), and Vibrio cholerae pathogenic cycle (ko05111), which significantly increased (p < 0.05). Meanwhile the content of endocytosis (ko04144), isoflavonoid biosynthesis (ko00943), Vibrio cholerae infection (ko05110), hypertrophic cardiomyopathy (ko05410), and D-arginine and D-ornithine metabolism (ko00472) significantly decreased (p < 0.05) (Figure 8). The endophytic bacteria of the Carex root system from the VTM mining area had significantly higher amounts of ECM-receptor interaction (ko04512), flavonoid biosynthesis (ko00941), primary bile acid biosynthesis (ko00120), styrene degradation (ko00643), and fluorobenzoate degradation (ko00364) than those from the non-VTM mining region (p < 0.05). Meanwhile, the abundances of genes involved in Alzheimer’s disease (ko05010), endocytosis (ko04144), Wnt signaling pathway (ko04310), bacterial invasion of epithelial cells (ko05100), and isoflavonoid biosynthesis (ko00943) were notably lower (p < 0.05). Compared to D. viscosa from the non-VTM mining area, only the content of the PPAR signaling pathway (ko03320) was significantly increased among endophytic bacteria in the root system of D. viscosa from the VTM mining area (p < 0.05). Conversely, the degree of ECM-receptor interaction (ko04512) and flavonoid biosynthesis (ko00941) was visibly augmented in root-dwelling endophytic bacteria of E. indica and Carex from the vicinity of VTM mining operations when contrasted with their counterparts unassociated with mining activities (p < 0.05). However, the levels of endocytosis (ko04144), isoflavonoid biosynthesis (ko00943), and Alzheimer’s disease (ko05010) in root endophytic bacteria of E. indica and Carex from the VTM mining compared to non-VTM mines were significantly reduced (p < 0.05).
Figure 8.
Based on the KO database, the functions of endophytic bacteria in plant roots were significantly different between VTM mining areas and non-VTM mining areas. The vertical axis represents the ID of the KO, the horizontal value is prelog2 (fold change), and different colors indicate significant differences between samples at different levels. (A) VT-Ein vs. CK-Ein; (B) VT-Car vs. CK-Car; (C) VT-Dvi vs. CK-Dvi.
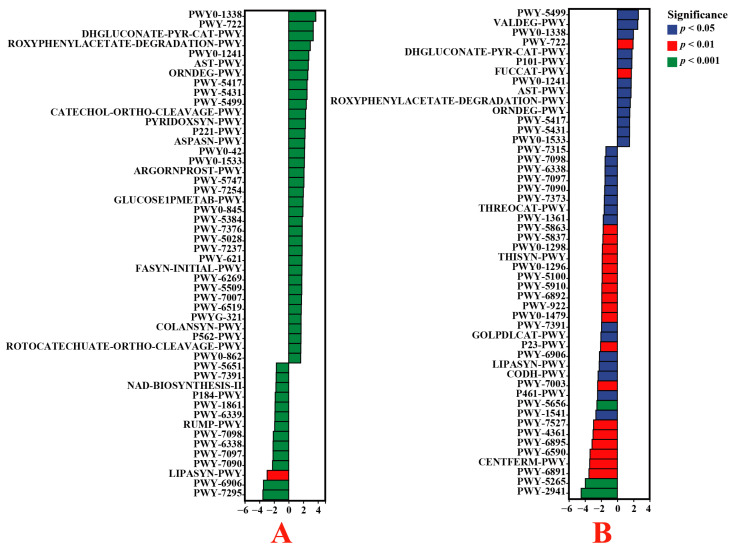
According to MetaCyc analysis, it was indicated that the functions of polymyxin resistance (PWY0-1338), nicotinate degradation I (PWY-722), glucose degradation (oxidative) (DHGLUCONATE-PYR-CAT-PWY), ADP-L-glycero-β-D-manno-heptose biosynthesis (PWY0-1241), and 4-hydroxyphenylacetate degradation (3-HYDROXYPHENYLACETATE-DEGRADATION-PWY) were significantly enhanced in the root bacteria of endophytic E. indica from the VTM mining area compared to those from the non-VTM mining region (p < 0.05). Additionally, the levels of L-arabinose degradation IV (PWY-7295), chitin-derivative degradation (PWY-6906), phospholipases (LIPASYN-PWY), UDP-2,3-diacetamido-2,3-dideoxy-α-D-mannuronate biosynthesis (PWY-7090), and vanillin and vanillate degradation I (PWY-7097) were significantly lower (p < 0.05) (Figure 9). In comparison with the non-VTM mining area, the endophytic bacterial population within Carex roots within the confines of the VTM mining area contained significantly increased levels of vitamin B6 degradation (PWY-5499), L-valine degradation I (VALDEG-PWY), polymyxin resistance (PWY0-1338), nicotinate degradation I (PWY-722), and glucose degradation (oxidative) (DHGLUCONATE-PYR-CAT-PWY) (p < 0.05). On the other hand, L-lysine biosynthesis II (PWY-2941), peptidoglycan biosynthesis II (staphylococci) (PWY-5265), thiazole biosynthesis II (Bacillus) (PWY-6891), the superpathway of Clostridium acetobutylicum acidogenic fermentation (PWY-6590), and pyruvate fermentation to butanoate (CENTFERM-PWY) were significantly reduced (p < 0.05). Nevertheless, the metabolic pathways associated with endophytic bacteria in the roots of D. viscosa exhibited no discernible disparity between the VTM mining area and the non-VTM mining area. In addition, compared to the non-VTM mining area, polymyxin resistance (PWY0-1338), nicotinate degradation I (PWY-722), and glucose degradation (oxidative) (DHGLUCONATE-PYR-CAT-PWY) were significantly higher in the endophytic bacteria in the roots of E. indica and Carex from the VTM mining area.
Figure 9.
Based on the MetaCyc database, the functions of endophytic bacteria in plant roots were significantly different between VTM mining areas and non-VTM mining areas. The vertical axis represents the ID of the pathway, the value of pre-axis log2 (fold change), and different colors indicate significant differences between samples at different levels. (A) VT-Ein vs. CK-Ein; (B) VT-Car vs. CK-Car.
3.7. Correlation Analysis of the Endophytic Bacteria Communities
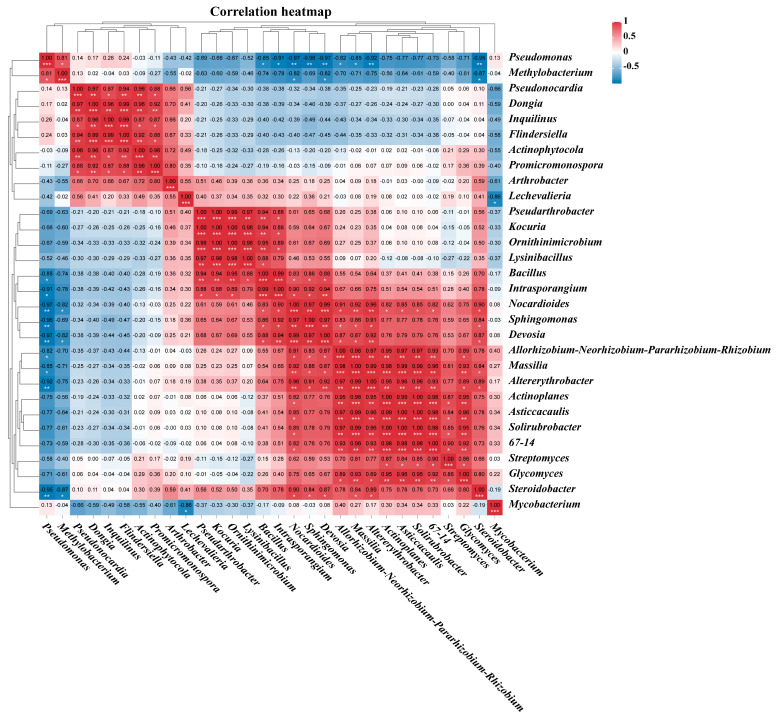
Pearson correlation analysis revealed significant interactions among the 30 most abundant genera of plant root endophytic bacteria from the VTM mining area (Figure 10). Positive correlations were observed between Altererythrobacter, Actinoplanes, Asticcacaulis, Solirubrobacter, 67–14, and Nocardioides (p < 0.05). Moreover, Dongia, Inquilinus, Flindersiella, Actinophytocola, Promicromonospora, and Pseudonocardia were also found to be positively correlated (p < 0.05). On the other hand, negative correlations were observed between Bacillus, Intrasporangium, Nocardioides, Sphingomonas, Devosia, Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium, Massilia, Altererythrobacter, and Pseudomonas. Additionally, Nocardioides, Devosia, Steroidobacter, and Methylobacterium were also negatively correlated (p < 0.05). The study illustrated a significant interplay evident among the primary cohorts of endophytic bacterial communities active within plant root systems.
Figure 10.
Pearson correlation analysis between the 30 genes with the highest abundance in different samples. The Pearson correlation index is marked in a square color block, with a blue color block indicating a negative correlation between two bacterial genera and a red color block indicating correlation. One asterisk indicates a significant correlation between two bacterial genera at the p < 0.05 level, two asterisks indicate a significant correlation between two bacterial genera at the p < 0.01 level, and three asterisks indicate a significantly greater correlation at the p < 0.001 level.
4. Discussion
VTM is an important mineral resource, and with increasing market demand, the mining scale of VTM is also constantly expanding. Nevertheless, significant VTM tailings are created when mining VTM. Tailings are frequently disposed of in dams, leading to the potential release of toxic pollutants into the surrounding environment, thereby endangering human health [17,18,19]. In China, Panzhihua City is a major mining area for VTM [52]. Therefore, we collected three types of plants near the mining area to study the ecological effects of mining activities. Although existing studies have analyzed the characteristics of microbial communities in the soil of the VTM mining area [53], no study has demonstrated a relationship between VTM mining operations and the diversification or population dynamics of endophytic bacterial communities within the plant roots adjacent to such operations. Endophytes are crucial for enhancing plant growth, development, stress resilience, and defense against pests and diseases, and some endophytes can also protect plants from heavy metals [54,55,56]. However, reductions in the richness of endophytes can elicit diminished plant attributes such as quality and resilience under stress [57]. Consequently, the potential impact of VTM mining on the biodiversity of endophytic bacteria within nearby plants necessitates careful evaluation. In this investigation, specific focus has been placed on three indigenous plants in close proximity to the VTM mining area, revealing the ramifications of VTM mining on the endophytic bacterial populations and functional activities within the root systems of these three plant species. These findings significantly augment our comprehension of the ecological implications of VTM mining activities.
4.1. Microbial Community Responses
Jose et al. demonstrated that increased levels of heavy metals result in alterations to the diversity of soil bacterial communities, which is consistent with our findings [58]. Our study showed that VTM mining activities directly led to a significant decrease in endophytic bacterial diversity and community richness indicators in the root systems of E. indica and Carex in the mining area. This could be attributed to the loss of soil nutrients caused by mining activities, resulting in a decline in the quality of living conditions for microbes and ultimately leading to a decrease in microbial diversity [53]. This result indicates that the negative impact of VTM mining on the endophytic bacteria in the roots of nearby plants may be due to the diffusion of harmful components, such as heavy metals, and alterations in soil physicochemical characteristics induced by VTM mining activities. Further study revealed that VTM mining activities resulted in the differentiation of endophytic bacterial communities within the roots of E. indica and Carex in the mining area. Therefore, the ecological impact of VTM mining deserves widespread attention. Furthermore, we observed lower abundances of Nocardioides and Pseudarthrobacter in the roots of E. indica and Carex from the vicinity of the VTM mining site compared to those present in the non-VTM mining region. The findings uncovered the susceptibility of these two strains to environmental alterations instigated by VTM mining. We detected Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes in our samples, which is similar to the findings of Zhang et al. and Tang et al. that these bacteria are widespread in contaminated environments [59,60]. Although environmental contamination from mining activities can severely damage the microecology of native plants, there are microorganisms that have been able to adapt to these adverse conditions under prolonged heavy metal stress [61]. These microorganisms have developed a variety of anti-stress strategies to increase their tolerance and ultimately mitigate the effects of toxicity [62,63]. Remarkably, we observed an enrichment of Pseudomonas within the root structures of E. indica and Carex growing in the vicinity of the VTM mining area. This is similar to the findings of Tang et al., Xiao et al., and Zhang et al., where the phylum Pseudomonas consistently constitutes the most dominant phylum in environments contaminated with heavy metals [60,64,65]. In addition, the impact of mining activities led to a significant increase in the relative abundance of Pseudomonas in the root systems of E. indica and Carex. This suggests that Pseudomonas is well adapted to environments contaminated with heavy metals, and its extreme adaptability to heavy metals can be explained by its efficient utilization of resources and metabolic activity [66,67]. It has been shown that Pseudomonas can reduce highly toxic pentavalent V to less toxic tetravalent V under certain conditions, thereby reducing its toxicity [67]. At the same time, some studies have shown that Pseudomonas has a certain resistance to heavy metals and is widely used in heavy metal remediation [68,69,70,71]. Additionally, Actinobacteria and Firmicutes were dominant among the root endophytes in these samples, which could be attributed to their high tolerance to heavy metals. According to Pan et al., Actinobacteria were found to be effectively enriched in heavy metal-contaminated soils, dominating all of the microbial communities [72]. Meanwhile, the study by Wang et al. found that Firmicutes could also play a role in reducing V to decrease exposure to toxicity. In addition, other dominant microbiota may also be involved in the reduction of V, since V is also a metal that is easily reduced by oxidation [73]. Therefore, these bacteria can be further developed and utilized for bioremediation in VTM mining areas. The abundance of two bacteria, Proteobacteria and Actinobacteria, accounted for a significant portion of the sum of all microbial communities in the samples, which may be due to their unique ability to bind heavy metals, allowing them to adapt to harsh heavy metal-contaminated environments [74,75,76].
Hierarchical taxonomic descriptions at the genus level allow for further analysis of the more complex details of bacterial community structure. At this level of genus, Pseudomonas, Nocardioides, Pseudonocardia, and Pseudarthrobacter are the dominant microbial communities. It has been shown that these bacteria have good resistance to heavy metals, that they coexist well with heavy metals, and that these bacteria are often isolated in a variety of environments heavily contaminated with heavy metals [72,77,78]. In addition, other studies have shown that Pseudarthrobacter is not only enriched in heavy metal-contaminated environments, but also plays a key role in promoting plant growth, and therefore, this bacterium also has great potential for application in bioremediation at VTM mining sites [79,80]. The strong resistance of these bacteria and their high tolerance to heavy metals have led to their complete dominance among the endophytic bacteria in the root systems of plants in VTM mining areas.
Furthermore, our study also revealed that the levels of enrichment of endophytic bacteria within plant roots in the VTM mining area were different, indicating that plants have certain screening and shaping abilities for endophytic bacterial communities. Throughout the extended coexistence and integration of endophytic bacteria within the plant root system, only competitive and adaptive bacteria are able to successfully colonize and form an advantageous mutualistic association with the host organism [81].
Overall, our study revealed that VTM mining significantly influenced the distribution of endophytic bacterial communities in the root systems of E. indica and Carex within the mining region. Under contaminated environmental conditions, the microbial communities in E. indica and Carex roots in VTM mining areas consist of heavy metal-resistant bacteria, which is the result of long-term selection by plants. Studying changes in microbial communities due to mining activities can help us identify bacteria with good environmental adaptations to aid in the bioremediation of mining sites.
4.2. Functional Enrichment Analysis
We also used PICRUSt2 to infer the function of the endophytic bacterial community in plant roots through three databases. We found that VTM mining also caused the development of distinct functions in the endophytic bacterial community in plant roots. Plants in VTM mining areas have different enriched microbial community functions compared to plants in non-VTM mining areas. The VTM mining has a profound impact on the functionality of endophytic bacteria within plant root systems prevalent in VTM mining regions. For example, the abundance of the phage-related tail fiber protein (COG5301), β-Lactam resistance (ko00312), and ECM-receptor interaction (ko04512) functions significantly augmented (p < 0.05). These functional changes revealed the local response mechanisms of endophytic bacteria in plant roots to VTM mining and environmental stress.
Jacquiod et al., found a high percentage of phage-related mRNA sequences in mRNAs in soils chronically contaminated with Cu, which is similar to the results we found [82]. We suggest that in the root systems of plants in the VTM mining area, their endophytic communities may maintain the stability of each function of the community by increasing the expression of the phage-related tail fiber protein (COG5301) function. The up-regulation of this function may potentially play a role in the survival of these microbial communities. Some studies have shown that β-Lactam is a widely used antibiotic, which is commonly used in the treatment of bacterial infections [83]. The functions of heavy metals and β-Lactam have some similarities, and they are both effective in inhibiting bacterial growth [84]. In VTM mining areas, mining activities can lead to elevated levels of heavy metals in nearby soil, posing a great challenge for the survival of local plants and microorganisms. Therefore, in the function of plant rhizosphere endophytes in the VTM mining area, microbial communities may reduce the repressive function caused by heavy metals by increasing the expression of β-Lactam resistance (ko00312) function, which enables them to survive in the heavy metal-exposed environment. Meng et al. found a significant enrichment of the ECM-receptor interaction (ko04512) function in the digestive glands of Cu-exposed Japanese scallops [85]. Similarly, Yao et al., also found a significant enrichment of this function in the livers of antimony-exposed zebrafish [86]. Our results were similar to theirs; the expression of the ECM-receptor interaction (ko04512) function was also significantly elevated as a function of the endophytic bacterial community in plant roots in the VTM mining area. ECM plays an important role in maintaining the structure and function of the cell [87]. Therefore, endophytic bacteria in the root systems of plants in the VTM mining area may ensure the function of the microbial community by enhancing the expression of the ECM-receptor interaction (ko04512) function. In conclusion, our study found that endophytic bacteria in the root systems of plants in the VTM mining area adapt to environmental changes by modulating various functions.
Furthermore, it was determined for the first time that the community structure and functional role of the endophytic bacteria within the roots of D. viscosa remained relatively unaffected between the VTM mining area and the non-VTM mining area. This indicates that D. viscosa itself has a strong adaptability to environmental changes. Therefore, it is also possible to consider using D. viscosa for the ecological restoration of VTM mining regions.
5. Conclusions
This study examined the impact of VTM mining on the diversity, community composition, and metabolic function of endophytic bacteria in the roots of E. indica, Carex, and D. viscosa. The study findings indicate that VTM mining negatively impacted the biodiversity of endophytic bacteria in the root systems of E. indica and Carex in the mining area. The Chao1, observed species, Shannon, Simpson, and Pielou evenness indices were all notably lower (p < 0.05). Furthermore, VTM mining has induced alterations in the community structure of endophytic bacteria within the root systems of E. indica and Carex, as opposed to the non-VTM mining area. Due to VTM mining, the abundance of Nocardioides in both E. indica and Carex notably reduced (p < 0.05). Interestingly, the abundance of Pseudomonas in the roots of both plants significantly increased (p < 0.05), indicating that this bacterium has good environmental adaptability. Through PICRUSt2 prediction, our study reveals that the endophytic bacteria in the root systems of E. indica and Carex in the VTM mining area responded to the environmental stress caused by mining activities through a series of methods. These include the enhancement of β-Lactam resistance (ko00312), flavonoid biosynthesis (ko00941), polymyxin resistance (PWY0-1338), and nicotinate degradation I (PWY-722). Conversely, no substantive alterations were observed in the diversity, community composition, and metabolic functionality of endophytic bacteria within the root tissue of D. viscosa following VTM mining activities. This shows that D. viscosa has good environmental adaptability. This paper, for the first time, studied the effect of VTM mining activities on endophytic bacteria in nearby plant roots. It emphasizes the ecological impact of VTM mining activities on endophytic bacteria in plant roots and provides a basis for soil ecological remediation in VTM mining areas.
Appendix A
Figure A1.
Rarefaction curves of root endophytic bacteria OTUs in different samples. VT-Dvi, D. viscosa in the VTM mining area; VT-Car, Carex in the VTM mining area; VT-Ein, E. indica in the VTM mining area; CK-Dvi, D. viscosa in the non-VTM mining area; CK-Car, Carex in the non-VTM mining area; CK-Ein, Eleusine indica in the non-VTM mining area.
Figure A2.
Functional enrichment analysis based on the KEGG predicted by PICRUSt2.
Figure A3.
Functional enrichment analysis based on the COG predicted by PICRUSt2.
Figure A4.
Functional enrichment analysis based on the MetaCyc database predicted by PICRUSt2.
Author Contributions
Literature retrieval and information collection: Q.L., Y.Z., X.C. and Z.X. Analysis of data: A.S., W.X., Y.L. and L.P. Writing and reviewing the paper: Q.L. and Z.X. Project management: L.P. and L.Z. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Funding Statement
This work is supported by Sichuan Natural Science Foundation Project (2023NSFSC1229) and Open Foundation of Hebei Key Laboratory of Wetland Ecology and Conservation (No. hklk202203).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Lai L., Zhou H., Lai B. Heterogeneous degradation of bisphenol A by peroxymonosulfate activated with vanadium-titanium magnetite: Performance, transformation pathways and mechanism. Chem. Eng. J. 2018;349:633–645. doi: 10.1016/j.cej.2018.05.134. [DOI] [Google Scholar]
- 2.Chen S.Y., Fu X.J., Chu M.S., Li X.Z., Liu Z.G., Tang J. Carbothermic Reduction Mechanism of Vanadium-titanium Magnetite. J. Iron Steel Res. Int. 2016;23:409–414. doi: 10.1016/S1006-706X(16)30065-6. [DOI] [Google Scholar]
- 3.Li W., Fu G.Q., Chu M.S., Zhu M.Y. Non-isothermal Reduction Behavior and Mechanism of Hongge Vanadium Titanomagnetite Pellet with Simulated Shaft Furnace Gases. Isij Int. 2018;58:415–421. doi: 10.2355/isijinternational.ISIJINT-2017-516. [DOI] [Google Scholar]
- 4.Feng C., Chu M.-S., Tang J., Liu Z.-G. Effects of smelting parameters on the slag/metal separation behaviors of Hongge vanadium-bearing titanomagnetite metallized pellets obtained from the gas-based direct reduction process. Int. J. Miner. Met. Mater. 2018;25:609–622. doi: 10.1007/s12613-018-1608-5. [DOI] [Google Scholar]
- 5.Li W., Wang N., Fu G.Q., Chu M.S., Zhu M.Y. Influence of roasting characteristics on gas-based direct reduction behavior of Hongge vanadium titanomagnetite pellet with simulated shaft furnace gases. Powder Technol. 2017;310:343–350. doi: 10.1016/j.powtec.2017.01.062. [DOI] [Google Scholar]
- 6.Zhou M., Jiang T., Yang S.T., Xue X.X. Vanadium-titanium magnetite ore blend optimization for sinter strength based on iron ore basic sintering characteristics. Int. J. Miner. Process. 2015;142:125–133. doi: 10.1016/j.minpro.2015.04.019. [DOI] [Google Scholar]
- 7.Wang S., Chen M., Guo Y., Jiang T., Zhao B. Reduction and Smelting of Vanadium Titanomagnetite Metallized Pellets. JOM. 2019;71:1144–1149. doi: 10.1007/s11837-018-2863-7. [DOI] [Google Scholar]
- 8.Li W., Wang N., Fu G., Chu M., Zhu M. Effects of Preheating Temperature and Time of Hongge Vanadium Titanomagnetite Pellet on Its Gas-Based Direct Reduction Behavior with Simulated Shaft Furnace Gases. ISIJ Int. 2018;58:594–603. doi: 10.2355/isijinternational.ISIJINT-2017-486. [DOI] [Google Scholar]
- 9.Yang J., Jiang T., Ma S.-H., Yang S.-T., Zhou M. Kinetics and mechanism of coal-based direct reduction of high-chromium vanadium–titanium magnetite. J. Iron Steel Res. Int. 2022;29:1723–1733. doi: 10.1007/s42243-022-00827-y. [DOI] [Google Scholar]
- 10.Li R.M., Liu T., Zhang Y.M., Huang J., Xu C.B. Efficient Extraction of Vanadium from Vanadium-Titanium Magnetite Concentrate by Potassium Salt Roasting Additives. Minerals. 2018;8:25. doi: 10.3390/min8010025. [DOI] [Google Scholar]
- 11.Moskalyk R.R., Alfantazi A.M. Processing of vanadium: A review. Miner. Eng. 2003;16:793–805. doi: 10.1016/S0892-6875(03)00213-9. [DOI] [Google Scholar]
- 12.Zhang Y., Zhou J.P., Sun D.Q., Li H.M. Two pass laser welding of TC4 Titanium alloy to 301L stainless steel via pure V interlayer. J. Mater. Res. Technol. 2020;9:1400–1404. doi: 10.1016/j.jmrt.2019.11.066. [DOI] [Google Scholar]
- 13.Yu J., Hu N., Xiao H., Gao P., Sun Y. Reduction behaviors of vanadium-titanium magnetite with H2 via a fluidized bed. Powder Technol. 2021;385:83–91. doi: 10.1016/j.powtec.2021.02.038. [DOI] [Google Scholar]
- 14.Lee J.C., Kurniawan, Kim E.Y., Chung K.W., Kim R., Jeon H.S. A review on the metallurgical recycling of vanadium from slags: Towards a sustainable vanadium production. J. Mater. Res. Technol. 2021;12:343–364. doi: 10.1016/j.jmrt.2021.02.065. [DOI] [Google Scholar]
- 15.Rack H.J., Qazi J.I. Titanium alloys for biomedical applications. Mater. Sci. Eng. C. 2006;26:1269–1277. doi: 10.1016/j.msec.2005.08.032. [DOI] [Google Scholar]
- 16.Geetha M., Singh A.K., Asokamani R., Gogia A.K. Ti based biomaterials, the ultimate choice for orthopaedic implants—A review. Prog. Mater. Sci. 2009;54:397–425. doi: 10.1016/j.pmatsci.2008.06.004. [DOI] [Google Scholar]
- 17.Tang X., Huang Y., Li Y., Yang Y., Cheng X., Jiao G., Dai H. The response of bacterial communities to V and Cr and novel reducing bacteria near a vanadium-titanium magnetite refinery. Sci. Total Environ. 2022;806:151214. doi: 10.1016/j.scitotenv.2021.151214. [DOI] [PubMed] [Google Scholar]
- 18.Gan C.-D., Tang Q.-X., Wang H., Yang J.-Y., Nikitin A. Shewanella oneidensis MR-1 and oxalic acid mediated vanadium reduction and redistribution in vanadium-containing tailings. J. Hazard. Mater. 2023;451:131077. doi: 10.1016/j.jhazmat.2023.131077. [DOI] [PubMed] [Google Scholar]
- 19.Yang J., Chen T., Lu L., Tang Y. In Vitro Health Risk Assessment of Ingesting Metal-Enriched Soils and Dusts in a Chinese Mining City. Hum. Ecol. Risk Assess. Int. J. 2015;21:2005–2021. doi: 10.1080/10807039.2015.1004158. [DOI] [Google Scholar]
- 20.Turner T.R., James E.K., Poole P.S. The plant microbiome. Genome Biol. 2013;14:209. doi: 10.1186/gb-2013-14-6-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S.-S., Liu J.-M., Sun J., Sun Y.-F., Liu J.-N., Jia N., Fan B., Dai X.-F. Diversity of culture-independent bacteria and antimicrobial activity of culturable endophytic bacteria isolated from different Dendrobium stems. Sci. Rep. 2019;9:10389. doi: 10.1038/s41598-019-46863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z., Zhu Y., Li N., Liu H., Zheng H., Wang W., Liu Y. High-throughput sequencing-based analysis of the composition and diversity of endophytic bacterial community in seeds of saline-alkali tolerant rice. Microbiol. Res. 2021;250:126794. doi: 10.1016/j.micres.2021.126794. [DOI] [PubMed] [Google Scholar]
- 23.Wu Z., Su Q., Cui Y., He H., Wang J., Zhang Y., Zhao Y., Abul H., Yang Y., Long Y. Temporal and spatial pattern of endophytic fungi diversity of Camellia sinensis (cv. Shu Cha Zao) BMC Microbiol. 2020;20:270. doi: 10.1186/s12866-020-01941-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hassani M.A., Durán P., Hacquard S. Microbial interactions within the plant holobiont. Microbiome. 2018;6:58. doi: 10.1186/s40168-018-0445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aswani R., Jishma P., Radhakrishnan E.K. 2—Endophytic bacteria from the medicinal plants and their potential applications. In: Kumar A., Singh V.K., editors. Microbial Endophytes. Woodhead Publishing; Cambridge, UK: 2020. pp. 15–36. [Google Scholar]
- 26.Li H., Yan C., Tang Y., Ma X., Chen Y., Chen S., Lin M., Liu Z. Endophytic bacterial and fungal microbiota in different cultivars of cassava (Manihot esculenta Crantz) J. Microbiol. 2020;58:614–623. doi: 10.1007/s12275-020-9565-x. [DOI] [PubMed] [Google Scholar]
- 27.Afzal M., Khan Q.M., Sessitsch A. Endophytic bacteria: Prospects and applications for the phytoremediation of organic pollutants. Chemosphere. 2014;117:232–242. doi: 10.1016/j.chemosphere.2014.06.078. [DOI] [PubMed] [Google Scholar]
- 28.Tardif S., Yergeau É., Tremblay J., Legendre P., Whyte L.G., Greer C.W. The Willow Microbiome Is Influenced by Soil Petroleum-Hydrocarbon Concentration with Plant Compartment-Specific Effects. Front. Microbiol. 2016;7:186190. doi: 10.3389/fmicb.2016.01363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liotti R.G., da Silva Figueiredo M.I., da Silva G.F., de Mendonça E.A.F., Soares M.A. Diversity of cultivable bacterial endophytes in Paullinia cupana and their potential for plant growth promotion and phytopathogen control. Microbiol. Res. 2018;207:8–18. doi: 10.1016/j.micres.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 30.Pereira S.I.A., Monteiro C., Vega A.L., Castro P.M.L. Endophytic culturable bacteria colonizing Lavandula dentata L. plants: Isolation, characterization and evaluation of their plant growth-promoting activities. Ecol. Eng. 2016;87:91–97. doi: 10.1016/j.ecoleng.2015.11.033. [DOI] [Google Scholar]
- 31.Knoth J.L., Kim S.H., Ettl G.J., Doty S.L. Biological nitrogen fixation and biomass accumulation within poplar clones as a result of inoculations with diazotrophic endophyte consortia. New Phytol. 2013;201:599–609. doi: 10.1111/nph.12536. [DOI] [PubMed] [Google Scholar]
- 32.Shi Y.W., Lou K., Li C. Growth promotion effects of the endophyte Acinetobacter johnsonii strain 3-1 on sugar beet. Symbiosis. 2011;54:159–166. doi: 10.1007/s13199-011-0139-x. [DOI] [Google Scholar]
- 33.Rungin S., Indananda C., Suttiviriya P., Kruasuwan W., Jaemsaeng R., Thamchaipenet A. Plant growth enhancing effects by a siderophore-producing endophytic streptomycete isolated from a Thai jasmine rice plant (Oryza sativa L. cv. KDML105) Antonie Leeuwenhoek. 2012;102:463–472. doi: 10.1007/s10482-012-9778-z. [DOI] [PubMed] [Google Scholar]
- 34.Barnawal D., Bharti N., Tripathi A., Pandey S.S., Chanotiya C.S., Kalra A. ACC-Deaminase-Producing Endophyte Brachybacterium paraconglomeratum Strain SMR20 Ameliorates Chlorophytum Salinity Stress via Altering Phytohormone Generation. J. Plant Growth Regul. 2016;35:553–564. doi: 10.1007/s00344-015-9560-3. [DOI] [Google Scholar]
- 35.Zouari I., Jlaiel L., Tounsi S., Trigui M. Biocontrol activity of the endophytic Bacillus amyloliquefaciens strain CEIZ-11 against Pythium aphanidermatum and purification of its bioactive compounds. Biol. Control. 2016;100:54–62. doi: 10.1016/j.biocontrol.2016.05.012. [DOI] [Google Scholar]
- 36.Wicaksono W.A., Jones E.E., Casonato S., Monk J., Ridgway H.J. Biological control of Pseudomonas syringae pv. actinidiae (Psa), the causal agent of bacterial canker of kiwifruit, using endophytic bacteria recovered from a medicinal plant. Biol. Control. 2018;116:103–112. doi: 10.1016/j.biocontrol.2017.03.003. [DOI] [Google Scholar]
- 37.Xie J., Shu P., Strobel G., Chen J., Wei J., Xiang Z., Zhou Z., SWg P.A. promotes disease protection and seedling growth. Biol. Control. 2017;113:9–17. doi: 10.1016/j.biocontrol.2017.06.010. [DOI] [Google Scholar]
- 38.Xu W.F., Wang F., Zhang M., Ou T., Wang R.L., Strobel G., Xiang Z.H., Zhou Z.Y., Xie J. Diversity of cultivable endophytic bacteria in mulberry and their potential for antimicrobial and plant growth-promoting activities. Microbiol. Res. 2019;229:126328. doi: 10.1016/j.micres.2019.126328. [DOI] [PubMed] [Google Scholar]
- 39.Strobel G.A. Endophytes as sources of bioactive products. Microbes Infect. 2003;5:535–544. doi: 10.1016/S1286-4579(03)00073-X. [DOI] [PubMed] [Google Scholar]
- 40.Rangjaroen C., Lumyong S., Sloan W.T., Sungthong R. Herbicide-tolerant endophytic bacteria of rice plants as the biopriming agents for fertility recovery and disease suppression of unhealthy rice seeds. BMC Plant Biol. 2019;19:580. doi: 10.1186/s12870-019-2206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaiero J.R., McCall C.A., Thompson K.A., Day N.J., Best A., Dunfield K.E.J.A.J.O.B. Inside the root microbiome: Bacterial root endophytes and plant growth promotion. Am. J. Bot. 2013;100:1738–1750. doi: 10.3732/ajb.1200572. [DOI] [PubMed] [Google Scholar]
- 42.Xia F., Chen X., Guo M.-Y., Bai X.-H., Liu Y., Shen G.-R., Li Y.-L., Lin J., Zhou X.-W. High-throughput sequencing-based analysis of endogenetic fungal communities inhabiting the Chinese Cordyceps reveals unexpectedly high fungal diversity. Sci. Rep. 2016;6:33437. doi: 10.1038/srep33437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Magoč T., Salzberg S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edgar R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 47.Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Douglas G.M., Beiko R.G., Langille M.G.I. Predicting the Functional Potential of the Microbiome from Marker Genes Using PICRUSt. Methods Mol. Biol. 2018;1849:169–177. doi: 10.1007/978-1-4939-8728-3_11. [DOI] [PubMed] [Google Scholar]
- 49.Douglas G.M., Maffei V.J., Zaneveld J.R., Yurgel S.N., Brown J.R., Taylor C.M., Huttenhower C., Langille M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020;38:685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J., et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021;49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He Z., Yue H.-R., Xue X. Study of the High Temperature Metallurgical Properties of On-Site Samples with Vanadium–Titanium Magnetite. Trans. Indian Inst. Met. 2018;71:2001–2013. doi: 10.1007/s12666-018-1333-9. [DOI] [Google Scholar]
- 53.Gan C.D., Cui S.F., Wu Z.Z., Yang J.Y. Multiple heavy metal distribution and microbial community characteristics of vanadium-titanium magnetite tailing profiles under different management modes. J. Hazard. Mater. 2022;429:128032. doi: 10.1016/j.jhazmat.2021.128032. [DOI] [PubMed] [Google Scholar]
- 54.Afzal I., Shinwari Z.K., Sikandar S., Shahzad S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 2019;221:36–49. doi: 10.1016/j.micres.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 55.White J.F., Kingsley K.L., Zhang Q., Verma R., Obi N., Dvinskikh S., Elmore M.T., Verma S.K., Gond S.K., Kowalski K.P. Review: Endophytic microbes and their potential applications in crop management. Pest Manag. Sci. 2019;75:2558–2565. doi: 10.1002/ps.5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma P., Kumar S. Bioremediation of heavy metals from industrial effluents by endophytes and their metabolic activity: Recent advances. Bioresour. Technol. 2021;339:125589. doi: 10.1016/j.biortech.2021.125589. [DOI] [PubMed] [Google Scholar]
- 57.Li Q., Xiang P., Li L., Zhang T., Wu Q., Bao Z., Tu W., Zhao C. Phosphorus mining activities alter endophytic bacterial communities and metabolic functions of surrounding vegetables and crops. Plant Soil. 2023;497:155–174. doi: 10.1007/s11104-023-05961-4. [DOI] [Google Scholar]
- 58.Jose J., Giridhar R., Anas A., Bharathi P.A.L., Nair S. Heavy metal pollution exerts reduction/adaptation in the diversity and enzyme expression profile of heterotrophic bacteria in Cochin estuary, India. Environ. Pollut. 2011;159:2775–2780. doi: 10.1016/j.envpol.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 59.Zhang B., Wang S., Diao M., Fu J., Xie M., Shi J., Liu Z., Jiang Y., Cao X., Borthwick A.G.L. Microbial Community Responses to Vanadium Distributions in Mining Geological Environments and Bioremediation Assessment. J. Geophys. Res. Biogeosci. 2019;124:601–615. doi: 10.1029/2018JG004670. [DOI] [Google Scholar]
- 60.Tang Q.-X., Gan C.-D., Yang J.-Y., Huang Y. Dynamics of vanadium and response of inherent bacterial communities in vanadium-titanium magnetite tailings to beneficiation agents, temperature, and illumination. Environ. Pollut. 2023;330:121743. doi: 10.1016/j.envpol.2023.121743. [DOI] [PubMed] [Google Scholar]
- 61.Li Q., Xiong Z., Xiang P., Zhou L., Zhang T., Wu Q., Zhao C. Effects of uranium mining on soil bacterial communities and functions in the Qinghai-Tibet plateau. Chemosphere. 2024;347:140715. doi: 10.1016/j.chemosphere.2023.140715. [DOI] [PubMed] [Google Scholar]
- 62.Guan N., Li J., Shin H.-D., Du G., Chen J., Liu L. Microbial response to environmental stresses: From fundamental mechanisms to practical applications. Appl. Microbiol. Biotechnol. 2017;101:3991–4008. doi: 10.1007/s00253-017-8264-y. [DOI] [PubMed] [Google Scholar]
- 63.Xiang P., Liao W., Xiong Z., Xiao W., Luo Y., Peng L., Zou L., Zhao C., Li Q. Effects of polystyrene microplastics on the agronomic traits and rhizosphere soil microbial community of highland barley. Sci. Total Environ. 2023;907:167986. doi: 10.1016/j.scitotenv.2023.167986. [DOI] [PubMed] [Google Scholar]
- 64.Xiao S., Zhang Q., Chen X., Dong F., Chen H., Liu M., Ali I. Speciation Distribution of Heavy Metals in Uranium Mining Impacted Soils and Impact on Bacterial Community Revealed by High-Throughput Sequencing. Front. Microbiol. 2019;10:1867. doi: 10.3389/fmicb.2019.01867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang H., Wang S., Liu Z., Li Y., Wang Q., Zhang X., Li M., Zhang B. Community assembly and microbial interactions in an alkaline vanadium tailing pond. Environ. Res. 2024;246:118104. doi: 10.1016/j.envres.2024.118104. [DOI] [PubMed] [Google Scholar]
- 66.Bouskill N.J., Barker-Finkel J., Galloway T.S., Handy R.D., Ford T.E. Temporal bacterial diversity associated with metal-contaminated river sediments. Ecotoxicology. 2010;19:317–328. doi: 10.1007/s10646-009-0414-2. [DOI] [PubMed] [Google Scholar]
- 67.Jia T., Liang X., Guo T., Wu T., Chai B. Bacterial community succession and influencing factors for Imperata cylindrica litter decomposition in a copper tailings area of China. Sci. Total Environ. 2022;815:152908. doi: 10.1016/j.scitotenv.2021.152908. [DOI] [PubMed] [Google Scholar]
- 68.Balíková K., Vojtková H., Duborská E., Kim H., Matús P., Urík M. Role of Exopolysaccharides of Pseudomonas in Heavy Metal Removal and Other Remediation Strategies. Polymers. 2022;14:4253. doi: 10.3390/polym14204253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fakhar A., Gul B., Gurmani A.R., Khan S.M., Ali S., Sultan T., Chaudhary H.J., Rafique M., Rizwan M. Heavy metal remediation and resistance mechanism of Aeromonas, Bacillus, and Pseudomonas: A review. Crit. Rev. Environ. Sci. Technol. 2022;52:1868–1914. doi: 10.1080/10643389.2020.1863112. [DOI] [Google Scholar]
- 70.Saha J., Chaki M.G., Karmakar S., Chatterjee A., Pal A. Effect of different heavy metals on lipase production by a multiple heavy metal-resistant Pseudomonas aeruginosa strain isolated from arable land. Biologia. 2023;78:2975–2985. doi: 10.1007/s11756-023-01465-9. [DOI] [Google Scholar]
- 71.Tripathi M., Kumar S., Makarana G., Goel R. Metal-Tolerant Bioinoculant Pseudomonas putida KNP9 Mediated Enhancement of Soybean Growth under Heavy Metal Stress Suitable for Biofuel Production at the Metal-Contaminated Site. Energies. 2023;16:4508. doi: 10.3390/en16114508. [DOI] [Google Scholar]
- 72.Pan X., Zhang S., Zhong Q., Gong G., Wang G., Guo X., Xu X. Effects of soil chemical properties and fractions of Pb, Cd, and Zn on bacterial and fungal communities. Sci. Total Environ. 2020;715:136904. doi: 10.1016/j.scitotenv.2020.136904. [DOI] [PubMed] [Google Scholar]
- 73.Jiang Y., Zhang B., He C., Shi J., Borthwick A.G.L., Huang X. Synchronous microbial vanadium (V) reduction and denitrification in groundwater using hydrogen as the sole electron donor. Water Res. 2018;141:289–296. doi: 10.1016/j.watres.2018.05.033. [DOI] [PubMed] [Google Scholar]
- 74.Liu H., Wang C., Xie Y., Luo Y., Sheng M., Xu F., Xu H. Ecological responses of soil microbial abundance and diversity to cadmium and soil properties in farmland around an enterprise-intensive region. J. Hazard. Mater. 2020;392:122478. doi: 10.1016/j.jhazmat.2020.122478. [DOI] [PubMed] [Google Scholar]
- 75.Sibanda T., Selvarajan R., Msagati T., Venkatachalam S., Meddows-Taylor S. Defunct gold mine tailings are natural reservoir for unique bacterial communities revealed by high-throughput sequencing analysis. Sci. Total Environ. 2019;650:2199–2209. doi: 10.1016/j.scitotenv.2018.09.380. [DOI] [PubMed] [Google Scholar]
- 76.Wu B., Luo H., Wang X., Liu H., Peng H., Sheng M., Xu F., Xu H. Effects of environmental factors on soil bacterial community structure and diversity in different contaminated districts of Southwest China mine tailings. Sci. Total Environ. 2021;802:149899. doi: 10.1016/j.scitotenv.2021.149899. [DOI] [PubMed] [Google Scholar]
- 77.Lo K.-H., Lu C.-W., Liu F.-G., Kao C.-M., Chen S.C. Draft genome sequence of Pseudomonas sp. A46 isolated from mercury-contaminated wastewater. J. Basic Microbiol. 2022;62:1193–1201. doi: 10.1002/jobm.202200106. [DOI] [PubMed] [Google Scholar]
- 78.Yang C., Zeng Z., Wang Y., He G., Hu Y., Gao D., Dai Y., Li Q., Zhang H. Ecological risk assessment and identification of the distinct microbial groups in heavy metal-polluted river sediments. Environ. Geochem. Health. 2023;45:1311–1329. doi: 10.1007/s10653-022-01343-4. [DOI] [PubMed] [Google Scholar]
- 79.Park M.-K., Park Y.-J., Kim M., Kim M.-C., Ibal J.C., Ung K.G., Lee G.-D., Tagele S.B., Kwon H.-J., Kang M.-S., et al. Complete genome sequence of a plant growth-promoting bacterium Pseudarthrobacter sp. NIBRBAC000502772, isolated from shooting range soil in the Republic of Korea. Korean J. Microbiol. 2020;56:390–393. [Google Scholar]
- 80.Wang T.-J., Su N.-N., Lei P., Qiu M.-Y., Chen Z.-J., Yao L.-G., Han H. Community Structure of Heavy Metal Immobilized Bacteria in the Lettuce (Lactuca sativa L.) Rhizosphere in Soil Polluted by Heavy Metals and Its Effects on Reducing Heavy Metal Accumulation in Lettuce. Huan Jing Ke Xue = Huanjing Kexue. 2019;40:5133–5141. doi: 10.13227/j.hjkx.201906055. [DOI] [PubMed] [Google Scholar]
- 81.Mano H., Morisaki H. Endophytic bacteria in the rice plant. Microbes Environ. 2008;23:109–117. doi: 10.1264/jsme2.23.109. [DOI] [PubMed] [Google Scholar]
- 82.Jacquiod S., Nunes I., Brejnrod A., Hansen M.A., Holm P.E., Johansen A., Brandt K.K., Priemé A., Sorensen S.J. Long-term soil metal exposure impaired temporal variation in microbial metatranscriptomes and enriched active phages. Microbiome. 2018;6:1–14. doi: 10.1186/s40168-018-0606-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Longhi C., Maurizi L., Conte A.L., Marazzato M., Comanducci A., Nicoletti M., Zagaglia C. Extraintestinal Pathogenic Escherichia coli: Beta-Lactam Antibiotic and Heavy Metal Resistance. Antibiotics. 2022;11:328. doi: 10.3390/antibiotics11030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Briffa J., Sinagra E., Blundell R. Heavy metal pollution in the environment and their toxicological effects on humans. Heliyon. 2020;6:e04691. doi: 10.1016/j.heliyon.2020.e04691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Meng X.L., Tian X., Nie G.X., Wang J.L., Liu M., Jiang K.Y., Wang B.J., Guo Q.Q., Huang J.R., Wang L. The transcriptomic response to copper exposure in the digestive gland of Japanese scallops (Mizuhopecten yessoensis) Fish Shellfish. Immunol. 2015;46:161–167. doi: 10.1016/j.fsi.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 86.Yao Q., Yang A.J., Hu X., Zou H.T., Chen J.F., Li Q., Lv S.H., Yu X.G., Li C.X. Effects of antimony exposure on DNA damage and genome-wide variation in zebrafish (Danio rerio) liver. Aquat. Toxicol. 2023;259:106524. doi: 10.1016/j.aquatox.2023.106524. [DOI] [PubMed] [Google Scholar]
- 87.Kaur J., Reinhardt D.P., Chapter, Vishwakarma A., Sharpe P., Shi S., Ramalingam M., editors. Stem Cell Biology and Tissue Engineering in Dental Sciences. Academic Press; Boston, MA, USA: 2015. pp. 25–45. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are contained within the article.