Abstract
We introduced polypurine tract (PPT) mutations, which we had previously tested in an in vitro assay, into the viral clone NL4-3KFSΔnef. Each mutant was tested for single-round infectivity and virion production. All of the PPT mutations had an effect on replication; however, mutation of the 5′ end appeared to have less of an effect on infectivity than mutation of the 3′ end of the PPT sequence. Curiously, a mutation in which the entire PPT sequence was randomized (PPTSUB) retained 12% of the infectivity of the wild type (WT) in a multinuclear activation of galactosidase indicator assay. Supernatants from these infections contained viral particles, as evidenced by the presence of p24 antigen. Two-long terminal repeat (2-LTR) circle junction analysis following PPTSUB infection revealed that the mutant could form a high percentage of normal junctions. Quantification of the 2-LTR circles using real-time PCR revealed that number of 2-LTR circles from cells infected with the PPTSUB mutant was 3.5 logs greater than 2-LTR circles from cells infected with WT virus. To determine whether the progeny virions from a PPTSUB infection could undergo further rounds of replication, we introduced the PPTSUB mutation into a replication-competent virus. Our results show that the mutant virus is able to replicate and that the infectivity of the progeny virions increases with each passage, quickly reverting to a WT PPT sequence. Together, these experiments confirm that the 3′ end of the PPT is important for plus-strand priming and that a virus that completely lacks a PPT can replicate at a low level.
After entry of the human immunodeficiency virus type 1 (HIV-1) virion into the host cell, its single-stranded RNA genome is converted into a double-stranded DNA intermediate through the process of reverse transcription (reviewed in reference 11). This process, catalyzed by a virally encoded reverse transcriptase (RT), requires a primer for each strand of DNA to be synthesized. The primer for minus-strand DNA synthesis is provided by a cellular tRNA (tRNA3Lys in the case of HIV-1), which is selectively incorporated into the virion during budding (29, 34, 42; reviewed in references 11 and 43). This primer binds to the viral RNA at a region known as the primer-binding site (PBS) (37, 59, 72) to initiate minus-strand synthesis. The plus-strand DNA primer is provided by a 15-nucleotide (nt) purine-rich viral RNA sequence known as the polypurine tract (PPT; 5′-AAAAGAAAAGGGGGG-3′) (reviewed in reference 56), which is generated from viral RNA by the RNase H activity of RT (18, 41, 45, 47, 54, 55, 58, 64). The PPT is highly conserved in most retroviruses and has been shown to be selectively used as the site of plus-strand initiation (18, 21, 26, 41, 45, 47, 51, 54, 55, 67). Cleavage site specificity is required for proper generation and removal of the PPT (18, 26, 38, 41, 45, 47, 50, 52, 53, 54, 57, 58, 64, 75). Previous studies have demonstrated that certain residues within the PPT and its overall helical structure are important determinants for specific cleavage and extension (21, 26, 33, 41, 51, 55, 60, 63). The sites of plus- and minus-strand initiation are important because they ultimately define the ends of the full-length proviral DNA, which are recognized by the viral integrase (5, 62; reviewed in reference 11).
Much of the work that has previously been done to study the specific requirements for cleavage and extension of the PPT has been done using in vitro systems. Early systems used the Moloney murine leukemia virus (Mo-MLV) as the model substrate in reactions using purified RT. Cleavage specificity of the Mo-MLV PPT was most affected by changes in PPT residues −1, −2, −4, or −7, with nt −7 being the most significant for proper cleavage (55). In similar studies using HIV-1 substrates, the critical residues were found to be at nt −2 and −4 (53).
We have also determined the sequence requirements of the HIV-1 PPT using a simplified in vitro assay (summarized in Table 1). In this assay, 15- or 20-nt RNA oligomers containing specific PPT mutations were annealed to complementary 35-nt DNA templates to test for the oligomer's ability to be extended by HIV-1 RT. The results from this study demonstrated that the 5′ end of the PPT sequence could be mutated without loss of plus-strand priming, while the 3′ end of the PPT was sensitive to relatively small changes in the sequence (51). It should be noted that this assay primarily determines the sequence requirements for primer extension.
TABLE 1.
Summary of PPT mutations and results from our previous in vitro assaya
| Mutation | Sequence | Cleavage/extension |
|---|---|---|
| −15+1LTR | ||
| WT PPT | AAAAGAAAAGGGGGGACTGG | ++++/++++ |
| 5′ | GCGCGAAAAGGGGGGACTGG | ++++/++++ |
| 3′ | AAAAGAAAAGGCCCCACTGG | −/− |
| 6Gs | TCATACCATGGGGGGACTGG | ++++/++++ |
| GA | GAGAGAGAGAGAGAGACTGG | +++/++ |
| CG | AAAAGAAAAGGCGCGACTGG | ++/+ |
| GC | AAAAGAAAAGGGCGCACTGG | ++/+ |
| DWN | ACTGGAAGGGCTAATTCACT | −/− |
Summary of our previous study (51) using 15- and 20-nt RNA primers containing specific mutations in the PPT sequence annealed to a complementary 35-nt minus-strand DNA template. Changes from the WT PPT sequence (shown in bold) are underlined. The relative amounts of cleavage and extension are represented by + and −. The italic G in the GA oligonucleotide represents an imprecise removal of the RNA primer which results in a base of RNA remaining on the DNA.
The sequence requirements for plus-strand initiation have also been studied in the context of viral systems. Using the Mo-MLV virus, the requirements for plus-strand initiation were examined using pools of PPT mutants, “purine PPTs,” in which the PPT sequence was randomized with a guanine or an adenine in positions −1 to −11 (60). After several rounds of replication, the most efficient PPT sequences were selected. The results of this study showed that viruses that maintained the −1 to −6 sequence (6Gs) had a selective advantage during viral replication. Viruses with mutations in the 3′ end of the PPT were able to replicate, but the entire 3′ five or six bases were necessary for replication in the absence of the 5′ end of the PPT. It was also observed that the randomized PPT pools quickly evolved toward the sequences of the wild-type (WT) PPT (60).
In another in vivo study using HIV-1, the effect of mutations of the PPT on single-round infectivity in a multinuclear activation of galactosidase indicator (MAGI) assay was examined. Single and double mutations introduced into the PPT sequence resulted in reductions in infectivity from 20 to 45% of WT, whereas multiple mutations in the PPT decreased infectivity to 5%. The analysis of two-long terminal repeat (2-LTR) circle junctions was also used to determine the specificity of plus-strand initiation. In this study, it was discovered that the WT viruses produced correct junctions 55% of the time. Viruses with mutations within the PPT produced correct circle junctions 30 to 43% of the time. The nature of the insertions and/or deletions within the circle junctions varied for each mutant. However, the proportion of circle junctions containing insertions was higher for all of the PPT mutants than for the WT. A major conclusion from this work was that the 5′ end of the PPT sequence plays an important role in the proper generation and removal of the PPT during plus-strand initiation (44).
We were interested in determining how the HIV-1 PPT mutations that we previously tested in our model system would affect replication in a viral system. The results of the present study demonstrate that mutations in both the 5′ and 3′ halves of the PPT have an effect on replication, although mutations in the bases at the 3′ end of the PPT affect replication to a greater degree. We also show that a virus with a completely randomized PPT can replicate, albeit at a low level, and quickly reverts to WT during passage.
MATERIALS AND METHODS
Plasmid construction.
The plasmids used were pNL4-3KFSΔnef (30), pALTER-1 (Promega), and pNL4-3 (AIDS Reagent Program, catalog no. 114, 1). The KFS designation represents an insertion of multiple KpnI linkers into the env reading frame of pNL4-3, introducing a frameshift mutation (pNL4-3KFS, gift of Eric O. Freed, HIV Drug Resistance Program, National Cancer Institute, Frederick, MD [19]). The nef deletion was produced by insertion of tandem stop codons at the beginning of the nef reading frame to avoid the complication of inadvertently changing the activity of Nef, whose reading frame includes the PPT sequence. To construct pNL4-3KFSΔnef with a mutated 3′ PPT sequence, both plasmids were digested using BamHI and NcoI (Roche). The BamHI and NcoI fragment of pNL4-3KFSΔnef which contains the mutated PPT was subcloned into the pALTER-1 vector at the corresponding sites (pALTER-1-PPT). Mutations of the PPT (Table 2) were generated by using the Promega Altered Sites II system and confirmed by DNA sequencing. After digestion of the pALTER-1-PPT with BamHI and NcoI, the mutated fragment was religated into pNL4-3KFSΔnef. The replication-competent viruses were made by digesting pNL4-3KFSΔnef containing the PPT mutations and WT pNL4-3 with BamHI and NcoI. The BamHI and NcoI fragment of the PPT mutant was cloned into pNL4-3 at the corresponding sites. The resulting plasmid no longer contains the KFS frameshift that deletes the env gene, but it retains the nef deletion. The virus produced can undergo multiple rounds of replication.
TABLE 2.
PPT mutations used in this studya
| Mutation | Sequence |
|---|---|
| −15+1LTR | |
| WT PPT | AAAAGAAAAGGGGGGACTGGAAGG |
| 6Gs | TCATACCATGGGGGGACTGGAAGG |
| 4Cs | AAAAGAAAAGGCCCCACTGGAAGG |
| PPTSUB | TCATACCATACTTCCACTGGAAGG |
| ATT SITE | AAAAGAAAAGGGGGGTAGATCCCG |
| 6Cs | AAAAGAAAACCCCCCACTGGAAGG |
Some of the mutations tested in the in vitro assay were introduced into the NL4-3KFSΔnef plasmid. Changes from the WT PPT sequence (shown in bold) are underlined.
As an additional control, the PPTSUB mutant virus was compared to the following integration-defective viruses: integrase (IN)-minus SG3S-IN, having a translational stop codon (TAA) at position 1 to inhibit the entire translation of the IN coding region (designated S-IN), and the IN mutant SG3D116A, having an alanine substitution at position 116 of IN within the catalytic center, which abolishes IN activity (designated D116A). Both plasmids were a gift from John C. Kappes (University of Alabama at Birmingham) (39, 76).
Preparation of viral stocks.
HEK 293 cells (AIDS Reagent Program, catalog no. 103) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). To make viral particles that will undergo a single round of replication, pNL4-3KFSΔnef containing the PPT mutations was cotransfected in 293 cells with pIIIenv3-1 (gift of Eric O. Freed) (66), which produces HIV-1 envelope in trans. To make multiround replication-competent viral particles, the viral clones pNL4-3Δnef, pSG3S-IN, or pSG3D116A were transfected into 293 cells. The 293 cells were transfected using the QIAGEN Effectene transfection kit according to the manufacturer's protocol. Transfections were done in 75-cm2 flasks using 6 μg of viral plasmid and 0.5 μg of envelope plasmid per flask (envelope plasmid for single round only). Cells were incubated for 6 h at 37°C and then refed to remove the Effectene and any residual plasmid. Inoculated cells were then incubated for an additional 72 h before supernatants containing the pseudotyped or replication-competent viral particles were collected. The resulting viruses were filtered through 0.45-μm-pore-size filters and tested for p24 content by enzyme-linked immunosorbent assay (ELISA) (Immuno Diagnostics). Viral particles were treated with 20 μg of DNase I (Roche) per ml for 30 min at 37°C to remove any residual plasmid.
Single-round infectivity assay.
The relative infectivity of the PPT mutant viral particles was determined by MAGI assay (30, 31). One day prior to infection, P4-R5 MAGI cells (AIDS Reagent Program, catalog no. 3580) were seeded in a 24-well plate at 4 × 104 cells per well in 5% FBS-DMEM. Cells were infected with a total volume of 500 μl of infection cocktail (50 μl of DEAE-dextran [200 μg/ml in DMEM], 5 ng of the NL4-3KFSΔnef-pIIIenv3-1 PPT mutant in a volume of 500 μl of 5% FBS-DMEM; total volume/well, 500 μl). The cells were incubated for 2 h in 5% CO2 at 37°C. After adding 1 ml of 5% FBS-DMEM to each well, the plate was incubated for 48 h at 37°C. The cells were then fixed with 0.2% glutaraldehyde and 1% formaldehyde and stained in X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) solution (0.4 mg of X-Gal per ml dissolved in dimethylformamide, 4 mM potassium ferricyanide, 4 mM potassium ferrocyanide, 2 mM MgCl2 in phosphate-buffered saline). The infectivity was scored as the total number of blue cells per nanogram of p24 of virus added. The controls for this assay were uninfected cells.
To control for β-galactosidase expression induced by Tat expressed from unintegrated 2-LTR circles, the PPTSUB mutant virus was compared to the viruses S-IN and D116A. One day prior to infection, U373-MAGI-CXCR4CEM cells (AIDS Reagent Program, catalog no. 3596) were seeded in a six-well plate at 1.5 × 105 cells per well in 5% FBS-DMEM. Cells were infected with a total volume of 2 ml of infection cocktail (200 μl of DEAE-dextran [200 μg/ml in DMEM], 10 ng of PPTSUB NL4-3KFSΔnef-pIIIenv3-1, S-IN, and D116A integrase mutant viral particles in a volume of 2 ml of 5% FBS-DMEM; total volume/well was 2 ml). The cells were incubated for 2 h in 5% CO2 at 37°C followed by two washes with 1 ml of phosphate-buffered saline and then refed with 3 ml of 5% FBS-DMEM. After 48 h, the cells were fixed and stained as described above, and the infectivity was scored as the total number of blue cells per nanogram of p24 of virus added. The controls for this assay were heat-inactivated WT virus and uninfected cells.
Progeny virus detection.
To detect progeny viral particle production, the infectivity assay was carried out using 50 ng of virus as described above. After the 2-h incubation, the cells were washed twice with 1 ml phosphate-buffered saline and refed with 1 ml of medium. The supernatants were collected 48 h postinfection and tested for p24 antigen content using a p24 ELISA.
Propagation assay.
U373-MAGI-CXCR4CEM cells were seeded in T-25 flasks (7 × 105 cells per flask) in 10 ml of 5% FBS-DMEM 1 day prior to infection. After removal of the medium, each flask was inoculated with the infection cocktail (200 μl of DEAE-dextran [200 μg/ml in DMEM], 80 ng of WT PPT or PPTSUB NL4-3Δnef viral particles in a volume of 2 ml of 5% FBS-DMEM; total volume/well was 2 ml). The cells were incubated for 2 h in 5% CO2 at 37°C followed by two washes with 5 ml of phosphate-buffered saline and then refed with 10 ml of 5% FBS-DMEM. The cells were passaged every 72 h with 5% FBS-DMEM (0.2 mg of G418/ml, 100 U of penicillin-100 μg of streptomycin/ml [Atlanta Biological]) over 15 days. At each time point beginning on day 6, 2 × 105 infected cells and 2 × 105 uninfected cells were seeded to the plate. Infectivity was measured by testing the supernatants at each time point using a p24 ELISA.
Proviral DNA detection.
Forty-eight hours postinfection, cells infected with 50 ng of NL4-3KFSΔnef-pIIIenv3-1 PPT mutants (infection as described above for single-round assay) were harvested by trypsinization, and the proviral DNA was recovered from the P4 MAGI cells using the QIAGEN DNeasy tissue kit according to the manufacturer's protocol (50-μl eluate). Proviral DNA was detected by PCR using the following primers, designed to amplify proviral DNA synthesized (see Fig. 2A) during reverse transcription (nt correspond to the numbering in pNL4-3 [1]): minus-strand DNA after transfer forward primer MDP044 (nt 9028 to 9051), 5′-ACA AGG CAG CTG TAG ATC TTA GCC-3′; reverse primer MDP045 (nt 9307 to 9288), 5′-TCC ATT CCA TGC AGG CTC AC-3′; plus-strand strong-stop DNA forward primer MDP046 (nt 454 to 477), 5′-GGG TCT CTC TGG TTA GAC CAG ATC-3′; reverse primer MDP047 (nt 653 to 635), 5′-GTC CCT GTT CGG GCG CCA C-3′. The reaction mixtures contained 1× HotStart Taq Mastermix (QIAGEN), 30 pmol/μl each of forward and reverse primers, and 10 μl of DNA template. The reactions were amplified for 35 cycles of 30 s at 65°C followed by 45 s at 72°C. PCR products were run on a 2% agarose gel (FisherBiotech).
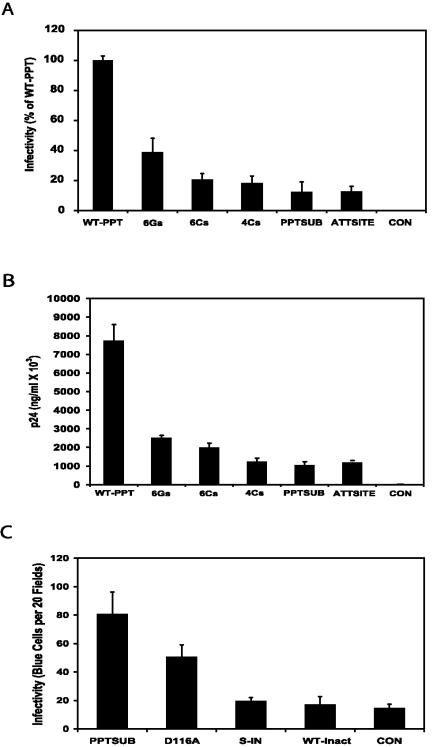
FIG. 1.
PPT mutant infectivity and progeny virus production. (A) A single-round infection with PPT mutants in NL4-3KFSΔnef was performed using 5 ng of p24 antigen. Infectivity was measured 48 h postinfection using a P4 MAGI cell assay. (B) The supernatants from a 5-ng single-round infection with the PPT mutants were collected and analyzed for p24 antigen 48 h postinfection. (C) Comparison of PPTSUB with IN mutants. Ten nanograms of p24 antigen was used in a single-round infection with the PPTSUB mutant and IN mutant. D116A, IN mutation that destroys enzymatic activity; S-IN, IN-minus mutant; WT-inact, inactivated WT virus; CON, uninfected cells. Infectivity was measured using a MAGI assay 48 h postinfection. Note that all of the assays described for panels A, B, and C were done in triplicate, with error bars representing 1 standard deviation.
2-LTR circle junction DNA analysis.
Extraction of 2-LTR circles from infected P4 MAGI cells (infection as described above for single-round assay) was performed using the QIAGEN QIAprep Spin Plasmid kit with the following modifications: for step 2, after addition and mixing of buffer P2, the tube was incubated for 5 min at room temperature; for step 3, upon addition and mixing of buffer N3, the tube was incubated on ice for 5 min; and for step 10, elution of proviral DNA with 50 μl of Tris-EDTA buffer by incubation for 5 min at 37°C and centrifugation for 1 min. This kit was used as a substitute for the traditional Hirt procedure (24). The primers used to analyze the 2-LTR circle junctions were as follows: 3′-U5 forward primer MDP037 (nt 522 to 547), 5′-GCC TCA ATA AAG CTT GCC TTG AGT GC-3′; and 5′-U3 reverse primer MDP038 (nt 132 to 110), 5′-CAG GAT CCA AAG GTC AGT GGA TAT CTG-3′ (25) (see Fig. 3A). The reaction mixtures contained 1× HotStart Taq Mastermix (QIAGEN), 30 pmol/μl each of forward and reverse primers, and 10 μl of DNA template. The reactions were amplified for 45 cycles for 2 min at 70°C followed by 7 min at 72°C. PCR products were run on a 3.5% agarose gel (FisherBiotech) to verify that normal 2-LTR circle junctions were formed (250-bp fragment); fragments larger than 250 bp represent circle junctions with additional bases within the junction. PCR products were cloned directly using the Invitrogen TOPO TA cloning kit for sequencing. Sequences were obtained using an ABI 3100 automated sequencer according to the manufacturer's recommendations. Results were analyzed using OMIGA 2.0 software (Accelrys Inc.).
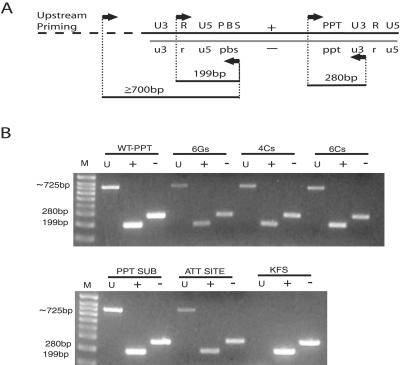
FIG. 2.
Detection of upstream priming in proviral DNA. (A) PCR strategy for detecting plus-strand, minus-strand, and upstream priming. Proviral DNA was detected by PCR using primers designed to amplify proviral DNA synthesized: minus-strand DNA after strand transfer (280-bp fragment), plus-strand strong-stop DNA (199-bp fragment), and upstream priming (700-bp fragment). (B) Gel electrophoresis of amplified linear proviral DNA. DNA extracted from cells infected with the WT PPT and each of the PPT mutants was analyzed using PCR primers specific for plus, minus, and upstream priming.
Quantitative results were obtained using the I-cycler real-time PCR instrument using the iQ SYBR green Supermix reagent (Bio-Rad) for detection. The primers used to amplify 2-LTR circle junctions are described above. The amount of DNA from each cell was normalized by detection of β-globin (forward primer, 5′-TCT ACC CTT GGA CCC AGA GG-3′; reverse primer, 5′-CTG AAG TTC TCA GGA TCC ACG-3′). Plasmid DNA positive for the 2-LTR circle junction was used as a standard for quantitation of 2-LTR circle junctions, and the human genomic DNA from the Perkin-Elmer kit was used for the standard for β-globin.
RESULTS
Infectivity of PPT mutants in a single-round infectivity assay.
To determine the effect of mutations of the PPT on infectivity, MAGI cells were infected with NL4-3KFSΔnef- pIIIenv3-1 containing each of the PPT mutations listed in Table 2, and the number of blue cells per nanogram of p24 antigen was determined. Each mutant was compared to infections with a virus containing the WT PPT, and the result was expressed as a percentage of WT PPT infectivity (Fig. 1A). The 6Gs mutant and the 5′ end mutation maintained 38% of WT PPT infectivity. In contrast, the two 3′ end mutations, 6Cs and 4Cs, resulted in 20% of WT infectivity. The PPTSUB mutant, containing a completely randomized PPT, and the ATT mutant, containing a completely randomized attachment site (att) for integrase, both maintained 12% of WT PPT infectivity. The control, a heat-inactivated WT PPT virus, showed no infectivity. All of the tissue culture supernatants from infected cells were positive for p24 antigen in amounts roughly proportional to the relative infectivity of the virus (Fig. 1B). This demonstrates that each mutant was able to produce progeny virions. Note that the mock infection with heat-inactivated virus had no detectable p24 antigen, which demonstrates that no p24 antigen was carried over from the initial inoculum.
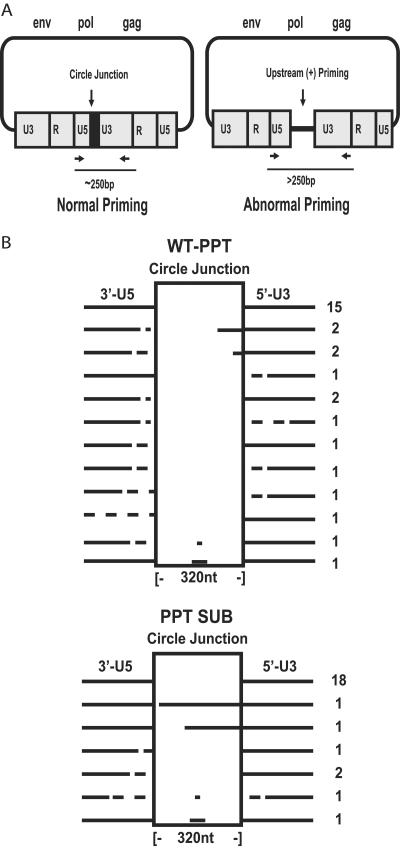
FIG. 3.
2-LTR circle junction analysis. (A) 2-LTR circle junction detection scheme. Primers designed to detect circle junctions were located in the 3′ R region (forward) and the 5′ U3 region (reverse). Normal circle junctions produce a 250-bp fragment. Circle junctions with fragments greater than 250 bp indicate upstream priming. (B) Analysis of 2-LTR circle junctions. DNA extracted from cells infected with the WT PPT and PPTSUB mutant was analyzed using PCR for 2-LTR circle junctions. The PCR products were cloned directly using the Invitrogen TOPO TA cloning kit for sequencing. Circle junctions are represented by solid lines against the vertical box; the vertical boxes are to accommodate the upstream priming or substitutions within the circle junction; deletions in the circle junction are indicated with dashed lines; and upstream priming and substitutions are indicated with solid lines within the vertical boxes.
Since the expression of Tat from 2-LTR circles could potentially transactivate the β-galactosidase reporter in the MAGI assay, we included additional controls to check for this possibility. The first is a virus containing the D116A point mutation in IN (designated D116A), which produces increased numbers of 2-LTR circles. The second is an IN-deleted virus (designated S-IN), which produces normal numbers of 2-LTR circles. The results are summarized in Fig. 1C. The relative number of blue cells produced by the D116A mutant was more than double the number of the other negative controls. However, this number of blue cells was less than that produced by the PPTSUB mutant. This suggests that the PPTSUB mutant virus retained some residual infectivity beyond what could be explained by the increase in 2-LTR circles alone. This is consistent with the observation that the PPTSUB infection resulted in the production of p24 antigen, which is produced only by integrated provirus.
Detection of upstream priming during plus-strand generation.
To determine if upstream priming occurred during any of the infections with mutant or WT virus, genomic DNA from cells infected with mutant and WT viruses was tested using PCR. Two sets of primers were used to detect normal plus- and minus-strand DNA formation during reverse transcription (70). A third set of primers was included to detect proviral DNA which had additional bases added to its 5′ end. This type of product would be formed if plus-strand priming had occurred at an alternative site(s) upstream of the 3′ PPT (Fig. 2A), especially in the absence of a functional PPT. All of the PPT mutants showed significant amounts of upstream priming, and surprisingly, upstream priming occurred in the WT sample as well (Fig. 2B). The control was the viral clone pNL4-3KFS, plasmid which contains only proviral DNA, with the correct 5′ end.
Use of 2-LTR circle junction analysis to determine sites of plus-strand initiation.
During a normal infection, infected cells contain both integrated viral DNA and unintegrated linear and circular viral DNA (11, 17, 25). The unintegrated forms are dead-end products of reverse transcription. One of the circular forms present is the 2-LTR circles, which are produced by the ligation of the unintegrated linear proviral DNA ends by the host cellular machinery (11, 35, 36, 65) and are relatively stable once formed (8, 49). The sequence of the circle junction has been used as a surrogate to determine the state of the linear proviral DNA prior to ligation (7, 8, 11, 17, 25, 35, 44, 49, 65). PCR amplification across the sequences of the 2-LTR junctions can be used to provide information on the site(s) of initiation (Fig. 3A). Priming of plus-strand DNA from sites upstream of the 3′ PPT will result in additional bases being added to the 5′ end of the proviral DNA. This, in turn, will result in 2-LTR junctions that contain additional DNA.
We used this type of analysis to determine if upstream priming was occurring at a higher frequency in cells infected with PPT mutant viruses than in those infected with WT viruses. PCR primers which would amplify a region across the circle junction were selected (Fig. 3A). The PCR products were then cloned and sequenced to determine the state of the circle junctions. Our results showed that 52% of the WT PPT junctions contained normal circle junctions (Fig. 3B). Upstream priming accounted for 14% of the WT PPT junctions, whereas deletions and additions within the junction accounted for 34%. Surprisingly, 72% of the PPTSUB mutant junctions were normal, while 8% showed upstream priming and 20% had deletions and/or additions within the junction. In those 2-LTR junctions that had additional sequences and therefore showed evidence of upstream priming, we determined the predicted sequences of the putative primers. The alternative primers for WT PPT or the PPTSUB mutants did not resemble the known functional PPT: WT primer 1 (W1), ACA ATG CTG CTT GTG CCT GC; WT primer 2 (W2), CAC CTC AGG TAC CTT TAA GA; PPTSUB primer 1 (P1), ACC TAG AAG ATT AAG ACA GG; and PPTSUB primer 2 (P2), TTT GCT ATA AGA TGG GTG GC (Fig. 4).
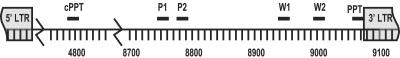
FIG. 4.
Alternative primers for plus-strand synthesis. The sites for alternative primers for plus-strand initiation were determined in cases where the 2-LTR junctions had additional sequences and showed evidence of upstream priming. Alternative primers were mapped to the 20-nt region upstream of the site of alternative plus-strand initiation. P1, PPTSUB primer 1; P2, PPTSUB primer 2; W1, WT primer 1; W2, WT primer 2.
Quantitation of the 2-LTR circles.
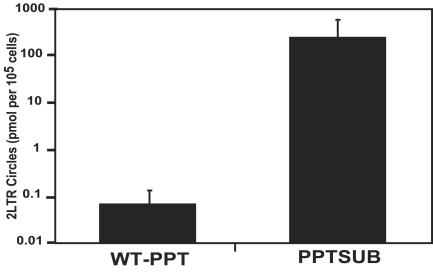
In the absence of a functional PPT, one might expect that the proviral DNA produced would not have the proper ends for efficient integration. In this case, less integration would occur and might increase the amount of 2-LTR circle DNA that is formed as a by-product of reverse transcription. To determine whether the absence of a functional PPT would affect the relative amount of 2-LTR circle DNA that was being formed, we quantitated 2-LTR circles using quantitative PCR. Our analysis revealed that infections with the PPTSUB mutant produced 2-LTR circles that were 3.5 logs greater in number than that of the wild-type infection (Fig. 5). These results are consistent with the finding that the accumulation of reverse transcription by-products are derived from unsuccessful integration events (12, 15, 61) and suggest that fewer proviruses are being integrated in cells infected with the PPTSUB mutant.
FIG. 5.
Quantification of 2-LTR circles. 2-LTR circle junctions were quantitated by real-time PCR using 2-LTR circle junction primers in the presence of SYBR green (40 cycles). β-Globin was used as a control for total cellular DNA added to each reaction mixture. Samples were run in triplicate, and the data shown are the means ± 1 standard deviation.
Propagation of the PPTSUB mutant.
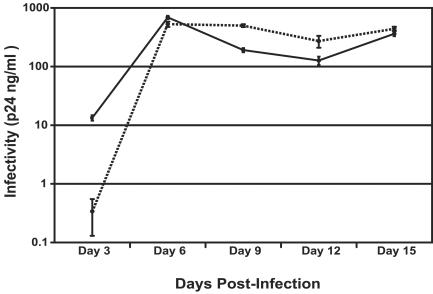
To determine if the progeny viruses produced from a PPTSUB infection could undergo further rounds of replication, we introduced the PPTSUB mutation into a replication-competent virus (NL4-3Δnef; see Materials and Methods for details) (1). To overcome the effects of decreased infectivity of the virus, this experiment was performed with a larger inoculum (80 ng of the p24 antigen). Our results showed that the mutant virus at day 3 was 1.5 logs less infectious than the WT virus, which appears to mirror the results of the single-round infectivity assay (Fig. 6). However, by day 6, the PPTSUB mutant was able to propagate as efficiently as the WT. Sequencing of proviral DNA at each time point revealed that the PPTSUB mutant sequence was retained by the virus at day 3 but quickly reverted to the WT PPT sequence by day 6 (data not shown).
FIG. 6.
Propagation of replication-competent PPT mutant virus. The PPTSUB mutant was introduced into the background of NL4-3Δnef (see the text) and propagated in a multiround infection using 80 ng of p24 antigen. At each time point beginning at day 6, 2 × 105 infected cells and 2 × 105 uninfected cells were seeded to the plate. WT PPT, solid line; PPTSUB, dotted line. Infectivity was measured using a p24 ELISA at each time point. The inactivated viral control showed no detectable p24 antigen. The measurements were done in triplicate, with error bars representing 1 standard deviation.
DISCUSSION
The goal of the present study was to determine the effect of PPT mutations, which we had previously tested in an in vitro assay, on plus-strand initiation in a viral system. We tested mutants in which both the 5′ and 3′ ends of the PPT were changed as well as a mutation that completely randomized the PPT sequence. All of the PPT mutants used in this study showed a significant reduction in infectivity. In general, mutation of the 3′ end had a more dramatic effect on infectivity than mutation of the 5′ end of the PPT. These results are in agreement with our previous study, which demonstrated that the identity of the bases at the 3′ end of the PPT is crucial for PPT priming activity (51). However, the data demonstrate that in vivo, mutation of the 5′ end of the PPT also significantly reduces infectivity. These results support the observation that the 5′ end of the PPT is important for correct processing of the PPT primer (44). Collectively, these data demonstrate that the entire PPT sequence is needed for proper cleavage and extension of plus-strand DNA to achieve maximal infectivity.
One of the more astonishing observations from this study is that the PPTSUB mutant retained a level of infectivity which was significantly above the level of background in MAGI assays (Fig. 1A). Two possibilities could account for this observation. First, a virus that lacks a PPT could produce sufficient Tat protein to activate the reporter gene in these cells during infection. Tat can be produced from the nonintegrated viral DNA, particularly in our study, from the increased number of 2-LTR circles and result in activation of the MAGI cells without integration actually occurring. There is precedent for transcription from proviral DNA which is not integrated (3, 6, 16, 74, 77). Second, some of the PPTSUB virus could become integrated and activate the MAGI cells as a consequence of a complete infectious cycle. To delineate these two possibilities, we compared the infectivity of the PPTSUB mutant to additional negative controls (Fig. 1C). The IN mutant D116A produced increased numbers of 2-LTR circles and showed an increase in numbers of blue cells in the MAGI assay over background levels. However, the PPTSUB mutant showed significantly higher numbers of blue cells, which suggests that this mutant has residual activity not explained by increases in 2-LTR DNA alone. In addition, our results show that p24 antigen is produced in the tissue culture medium of PPTSUB-infected cells (Fig. 1B). This suggests that a small number of virions are produced from these infections and supports the idea that at least some of the PPTSUB proviral DNA is being integrated.
Other studies have investigated the infectivity of viruses lacking a PPT. Aldrovandi et al. showed that HIV-1 viruses with deletions of nef, which also delete the PPT, failed to replicate (2). However, some propagation studies have shown that viruses with significant mutations in the PPT sequence are able to replicate to various degrees (46, 60). One of these studies used Mo-MLV to determine whether pyrimidines would be permitted at any of the positions in the PPT sequence. Pools of viruses that had PPTs randomized with any of the four bases (GATC PPT pool, subset of approximately 100 viruses) were tested. Replication of the mutant viruses was not detectable until 17 days postinfection (60). In another study, simian immunodeficiency virus was mutated to remove the T-rich region upstream of the PPT, the PPT, and the att sequences downstream of the PPT (TPI region; 13 point mutations were introduced to make the region dysfunctional) (46). The TPI mutant retained 12% of WT infectivity in a single-round simian MAGI assay (9). This finding is similar to our result for the PPTSUB mutant compared to our WT in a single-round MAGI assay. Together, these studies demonstrate that in the absence of a functional PPT some level of replication can occur.
To determine if DNA from infected cells contained provirus with additional sequences at the 5′ end, we analyzed genomic DNA using PCR primers specific for upstream priming events. We found that genomic DNA from cells infected with each of the mutants as well as with the WT PPT produced significant amounts of proviral DNA with additional sequences at the 5′ end. Other studies have also shown that there may be alternative sites of initiation for plus-strand synthesis (32). In addition, there are experiments that show the significant role of the central PPT (cPPT, identical to the 3′ PPT but located at the center of the HIV-1 genome) in the process of reverse transcription. It has been shown that the cPPT can also prime a portion of plus-strand synthesis and that mutating this sequence results in replication deficiency (10, 22, 27, 28, 68, 73; reviewed in reference 56). To determine where plus-strand synthesis was initiating in infections with our mutants, we analyzed 2-LTR circle junctions. Our results demonstrate that upstream priming did not initiate from the cPPT nor did the sites of initiation resemble an authentic HIV-1 PPT. Several studies have shown that nonspecific RNA oligomers can act as primers for DNA synthesis (14, 20, 48; reviewed in reference 56).
2-LTR circles are the products of failed integration, as evidenced by accumulation of the circles in significant numbers in infections with integration-defective viruses (7, 12, 13, 15, 61). Our study revealed a 3.5-log increase in the number of 2-LTR circles produced by the PPTSUB mutant compared with the WT PPT during virus infection; this suggests that there is a defect in the integration of these proviruses. Other reports have shown that retroviral vectors lacking a PPT and att site have the ability to complete reverse transcription and form provirus (4, 11). In this case, it was proposed that the provirus is integrated into the target cell by aberrant integration events or by some other unknown mechanism (4).
Since our PPTSUB mutant was able to produce p24 antigen from single-round infections, we wished to determine whether progeny virions were able to undergo further rounds of replication. When the PPTSUB mutation was inserted into a replication-competent virus (NL4-3Δnef), multiple rounds of replication could be induced (Fig. 6). Although the number of particles produced from each round of infection was low, the particles were infectious. This residual infectivity suggests that there may be an alternative mechanism to ensure that the proper ends of proviral DNA are formed for integration into the host cell genome or that provirus with the incorrect 5′ end may be able to integrate and produce virions. In the study cited above, it was shown that the simian immunodeficiency virus TPI mutant (46), like our PPTSUB virus, was able to replicate after extended incubation, and the titer dramatically increased after replication for 3 days postinfection (46).
Curiously, the PPTSUB mutant in our study appeared to revert completely back to the WT PPT sequence in the span of only 3 days. How could such a dramatic change in sequence have occurred? The specific sequence of the strictly conserved PPT could be the driving force for this rapid change. Thus, for Mo-MLV, after infection with the complete GATC PPT pool (PPT randomized with any of the four bases) (60), viral spread was detectable 3 days after that of the WT. Following a single passage of the virus, the pyrimidines were not detectable in most positions, suggesting quick reversion toward the sequences of the WT PPT. It is possible that the same strong selection is at work in this case in combination with rapid mutation rates or some form of recombination.
The results from this study confirm many of the features of plus-strand priming that have been reported using in vitro systems. A major surprise from this work is that a virus with a completely randomized PPT can still replicate to some degree. Our observation that the PPTSUB mutant appears to completely revert to the WT sequence after only a few rounds of replication was also observed in mutational studies of the PBS. Investigation of the sequence requirements for the interaction between tRNA3Lys and the PBS during reverse transcription provided strong evidence that reversion does occur (59). For example, deletions that eliminated the entire PBS (ΔPBS) or the first 9 (Δ1-9), the second 9 (Δ10-18), or 12 (Δ7-18) nt of the PBS were introduced into the PBS of the HIV-1 proviral genome. One other mutation in the PBS was the substitution of the deleted second 9 nt of the PBS with additional nt [Lys(1-9)]. Infectious virus was produced with the Δ10-18, Δ7-18, and Lys(1-9) mutants, although their detection was delayed compared to the WT virus. Sequence analysis of the PBS region of the proviral genomes revealed that the 18-nt PBS complementary to tRNA3Lys was present within 4 to 5 days. The authors also found nucleotide deletions and insertions 3′ of the PBS region. Reinfection with the revertant viruses maintained the deletions 3′ of the PBS, and the viruses replicated as efficiently as the WT (59). The quick reversion of these PBS mutants is likely due to the use of the tRNA3Lys as the template for regeneration of the WT PBS sequence during synthesis of plus-strand strong-stop DNA (11).
In the case of our PPTSUB mutant, no such mechanism is known to be present for regeneration of the PPT. The use of the cPPT as a primer for plus-strand synthesis is not evident in our study. The flanking sequences of the revertant PPTs did not resemble those that surround the cPPT (data not shown). Thus, recombination with the cPPT seems unlikely. One possibility is that the virus may be recombining with human endogenous retrovirus (HERV) sequences which constitute about 7 to 8% of the human genome (23, 69). It has been shown that partial and full-length proviral transcripts of HERVs or their translation products are present in human cells (40, 69, 71). Presently, there has been no detection of HERVs that are related to HIV-1 or HIV-2 in the human genome, but there are some DNA sequences related to the highly conserved domain of the HIV RT and gp41 (71). The PPT sequences of HERV-K and HERV-W are similar to that of HIV-1 and other retroviruses and retroelements. Alternatively, other unknown mechanisms might be in place to ensure the restoration of the PPT sequence. If this is the case, we were not able to determine the exact nature of such a mechanism using the techniques developed in this study.
In summary, we have shown that changes in both the 5′ and 3′ ends of the PPT can reduce the infectivity of HIV-1. However, changes of the 3′ end of the PPT reduced infectivity to a greater extent than mutation of the 5′ end. These results are in substantial agreement with previous in vitro data (44). Surprisingly, a virus containing a completely randomized PPT maintained a low level of infectivity and quickly reverted to a WT PPT sequence upon passage. Further understanding of these phenomena will require continued study of plus-strand formation using similar viral systems.
Acknowledgments
We thank Eric O. Freed for pNL4-3KFS and the HIV-1 envelope vector and for helpful advice in the early phase of this project. We also thank John Kappes for his gift of the two IN mutants. This work was supported by NIH grants G12-RR03034 and S06-GM08248 (M.D.P.) and in part by an award from the NIH Intramural AIDS Targeted Antiviral Program (J.G.L.). Lesa R. Miles is supported in part by NRSA fellowship 1F31GM65793.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldrovandi, G. M., L. Gao, G. Bristol, and J. A. Zack. 1998. Regions of human immunodeficiency virus type 1 nef required for function in vivo. J. Virol. 72:7032-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansari-Lari, M. A., L. A. Donehower, and R. A. Gibbs. 1995. Analysis of human immunodeficiency virus type 1 integrase mutants. Virology 211:332-335. [DOI] [PubMed] [Google Scholar]
- 4.Bowman, R. R., W. S. Hu, and V. K. Pathak. 1998. Relative rates of retroviral reverse transcriptase template switching during RNA- and DNA-dependent DNA synthesis. J. Virol. 72:5198-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, P. O., B. Bowerman, H. E. Varmus, and J. M. Bishop. 1989. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc. Natl. Acad. Sci. USA 86:2525-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brussel, A., and P. Sonigo. 2004. Evidence for gene expression by unintegrated human immunodeficiency virus type 1 DNA species. J. Virol. 78:11263-11271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckman, J. S., W. J. Bosche, and R. J. Gorelick. 2003. Human immunodeficiency virus type 1 nucleocapsid Zn2+ fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J. Virol. 77:1469-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butler, S. L., E. P. Johnson, and F. D. Bushman. 2002. Human immunodeficiency virus cDNA metabolism: notable stability of two-long terminal repeat circles. J. Virol. 76:3739-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chackerian, B., N. L. Haigwood, and J. Overbaugh. 1995. Characterization of a CD4-expressing macaque cell line that can detect virus after a single replication cycle and can be infected by diverse simian immunodeficiency virus isolates. Virology 213:386-394. [DOI] [PubMed] [Google Scholar]
- 10.Charneau, P., M. Alizon, and F. Clavel. 1992. A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J. Virol. 66:2814-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 12.Colicelli, J., and S. P. Goff. 1985. Mutants and pseudorevertants of Moloney murine leukemia virus with alterations at the integration site. Cell 42:573-580. [DOI] [PubMed] [Google Scholar]
- 13.Colicelli, J., and S. P. Goff. 1988. Sequence and spacing requirements of a retrovirus integration site. J. Mol. Biol. 199:47-59. [DOI] [PubMed] [Google Scholar]
- 14.DeStefano, J. J., R. A. Bambara, and P. J. Fay. 1993. Parameters that influence the binding of human immunodeficiency virus reverse transcriptase to nucleic acid structures. Biochemistry 32:6908-6915. [DOI] [PubMed] [Google Scholar]
- 15.Donehower, L. A., and H. E. Varmus. 1984. A mutant murine leukemia virus with a single missense codon in pol is defective in a function affecting integration. Proc. Natl. Acad. Sci. USA 81:6461-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engelman, A., G. Englund, J. M. Orenstein, M. A. Martin, and R. Craigie. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 69:2729-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farnet, C. M., and W. A. Haseltine. 1991. Circularization of human immunodeficiency virus type 1 DNA in vitro. J. Virol. 65:6942-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finston, W. I., and J. J. Champoux. 1984. RNA-primed initiation of Moloney murine leukemia virus plus strands by reverse transcriptase in vitro. J. Virol. 51:26-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freed, E. O., E. L. Delwart, G. L. Buchschacher, Jr., and A. T. Panganiban. 1992. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc. Natl. Acad. Sci. USA 89:70-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu, T. B., and J. Taylor. 1992. When retroviral reverse transcriptases reach the end of their RNA templates. J. Virol. 66:4271-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuentes, G. M., L. Rodriguez-Rodriguez, P. J. Fay, and R. A. Bambara. 1995. Use of an oligoribonucleotide containing the polypurine tract sequence as a primer by HIV reverse transcriptase. J. Biol. Chem. 270:28169-28176. [DOI] [PubMed] [Google Scholar]
- 22.Fuentes, G. M., L. Rodriguez-Rodriguez, C. Palaniappan, P. J. Fay, and R. A. Bambara. 1996. Strand displacement synthesis of the long terminal repeats by HIV reverse transcriptase. J. Biol. Chem. 271:1966-1971. [DOI] [PubMed] [Google Scholar]
- 23.Griffiths, D. J. 5. June 2001, posting date. Endogenous retroviruses in the human sequence. Genome Biol. [Online.] http://genomebiology.com/2001/2/6/reviews/1017. [DOI] [PMC free article] [PubMed]
- 24.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 25.Hong, T., K. Drlica, A. Pinter, and E. Murphy. 1991. Circular DNA of human immunodeficiency virus: analysis of circle junction nucleotide sequences. J. Virol. 65:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huber, H. E., and C. C. Richardson. 1990. Processing of the primer for plus strand DNA synthesis by human immunodeficiency virus 1 reverse transcriptase. J. Biol. Chem. 265:10565-10573. [PubMed] [Google Scholar]
- 27.Hungnes, O., E. Tjotta, and B. Grinde. 1991. The plus strand is discontinuous in a subpopulation of unintegrated HIV-1 DNA. Arch. Virol. 116:133-141. [DOI] [PubMed] [Google Scholar]
- 28.Hungnes, O., E. Tjotta, and B. Grinde. 1992. Mutations in the central polypurine tract of HIV-1 result in delayed replication. Virology 190:440-442. [DOI] [PubMed] [Google Scholar]
- 29.Jiang, M., J. Mak, A. Ladha, E. Cohen, M. Klein, B. Rovinski, and L. Kleiman. 1993. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J. Virol. 67:3246-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan, M., M. Garcia-Barrio, and M. D. Powell. 2001. Restoration of wild-type infectivity to human immunodeficiency virus type 1 strains lacking nef by intravirion reverse transcription. J. Virol. 75:12081-12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klarmann, G. J., H. Yu, X. Chen, J. P. Dougherty, and B. D. Preston. 1997. Discontinuous plus-strand DNA synthesis in human immunodeficiency virus type 1-infected cells and in a partially reconstituted cell-free system. J. Virol. 71:9259-9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kvaratskhelia, M., S. R. Budihas, and S. F. Le Grice. 2002. Pre-existing distortions in nucleic acid structure aid polypurine tract selection by HIV-1 reverse transcriptase. J. Biol. Chem. 277:16689-16696. [DOI] [PubMed] [Google Scholar]
- 34.Levin, J. G., and J. G. Seidman. 1981. Effect of polymerase mutations on packaging of primer tRNAPro during murine leukemia virus assembly. J. Virol. 38:403-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, L., J. M. Olvera, K. E. Yoder, R. S. Mitchell, S. L. Butler, M. Lieber, S. L. Martin, and F. D. Bushman. 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20:3272-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, L., K. Yoder, M. S. Hansen, J. Olvera, M. D. Miller, and F. D. Bushman. 2000. Retroviral cDNA integration: stimulation by HMG I family proteins. J. Virol. 74:10965-10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, X., J. Mak, E. J. Arts, Z. Gu, L. Kleiman, M. A. Wainberg, and M. A. Parniak. 1994. Effects of alterations of primer-binding site sequences on human immunodeficiency virus type 1 replication. J. Virol. 68:6198-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim, D., M. Orlova, and S. P. Goff. 2002. Mutations of the RNase H C helix of the Moloney murine leukemia virus reverse transcriptase reveal defects in polypurine tract recognition. J. Virol. 76:8360-8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, H., X. Wu, M. Newman, G. M. Shaw, B. H. Hahn, and J. C. Kappes. 1995. The Vif protein of human and simian immunodeficiency viruses is packaged into virions and associates with viral core structures. J. Virol. 69:7630-7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lower, R., J. Lower, and R. Kurth. 1996. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl. Acad. Sci. USA 93:5177-5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo, G. X., L. Sharmeen, and J. Taylor. 1990. Specificities involved in the initiation of retroviral plus-strand DNA. J. Virol. 64:592-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mak, J., M. Jiang, M. A. Wainberg, M. L. Hammarskjöld, D. Rekosh, and L. Kleiman. 1994. Role of Pr160gag-pol in mediating the selective incorporation of tRNALys into human immunodeficiency virus type 1 particles. J. Virol. 68:2065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marquet, R., C. Isel, C. Ehresmann, and B. Ehresmann. 1995. tRNAs as primer of reverse transcriptases. Biochimie 77:113-124. [DOI] [PubMed] [Google Scholar]
- 44.McWilliams, M. J., J. G. Julias, S. G. Sarafianos, W. G. Alvord, E. Arnold, and S. H. Hughes. 2003. Mutations in the 5′ end of the human immunodeficiency virus type 1 polypurine tract affect RNase H cleavage specificity and virus titer. J. Virol. 77:11150-11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitra, S. W., M. Chow, J. Champoux, and D. Baltimore. 1982. Synthesis of murine leukemia virus plus strong stop DNA initiates at a unique site. J. Biol. Chem. 257:5983-5986. [PubMed] [Google Scholar]
- 46.Münch, J., N. Adam, N. Finze, N. Stolte, C. Stahl-Hennig, D. Fuchs, P. Ten Haaft, J. L. Heeney, and F. Kirchhoff. 2001. Simian immunodeficiency virus in which nef and U3 sequences do not overlap replicates efficiently in vitro and in vivo in rhesus macaques. J. Virol. 75:8137-8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Omer, C. A., R. Resnick, and A. J. Faras. 1984. Evidence for involvement of an RNA primer in initiation of strong-stop plus DNA synthesis during reverse transcription in vitro. J. Virol. 50:465-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palaniappan, C., G. M. Fuentes, L. Rodriguez-Rodriguez, P. J. Fay, and R. A. Bambara. 1996. Helix structure and ends of RNA/DNA hybrids direct the cleavage specificity of HIV-1 reverse transcriptase RNase H. J. Biol. Chem. 271:2063-2070. [PubMed] [Google Scholar]
- 49.Pierson, T. C., T. L. Kieffer, C. T. Ruff, C. Buck, S. J. Gange, and R. F. Siliciano. 2002. Intrinsic stability of episomal circles formed during human immunodeficiency virus type 1 replication. J. Virol. 76:4138-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Powell, M. D., W. A. Beard, K. Bebenek, K. J. Howard, S. F. Le Grice, T. A. Darden, T. A. Kunkel, S. H. Wilson, and J. G. Levin. 1999. Residues in the alphaH and alphaI helices of the HIV-1 reverse transcriptase thumb subdomain required for the specificity of RNase H-catalyzed removal of the polypurine tract primer. J. Biol. Chem. 274:19885-19893. [DOI] [PubMed] [Google Scholar]
- 51.Powell, M. D., and J. G. Levin. 1996. Sequence and structural determinants required for priming of plus-strand DNA synthesis by the human immunodeficiency virus type 1 polypurine tract. J. Virol. 70:5288-5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pullen, K. A., and J. J. Champoux. 1990. Plus-strand origin for human immunodeficiency virus type 1: implications for integration. J. Virol. 64:6274-6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pullen, K. A., A. J. Rattray, and J. J. Champoux. 1993. The sequence features important for plus strand priming by human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 268:6221-6227. [PubMed] [Google Scholar]
- 54.Rattray, A. J., and J. J. Champoux. 1987. The role of Moloney murine leukemia virus RNase H activity in the formation of plus-strand primers. J. Virol. 61:2843-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rattray, A. J., and J. J. Champoux. 1989. Plus-strand priming by Moloney murine leukemia virus. The sequence features important for cleavage by RNase H. J. Mol. Biol. 208:445-456. [DOI] [PubMed] [Google Scholar]
- 56.Rausch, J. W., and S. F. Le Grice. 2004. ‘Binding, bending and bonding’: polypurine tract-primed initiation of plus-strand DNA synthesis in human immunodeficiency virus. Int. J. Biochem. Cell Biol. 36:1752-1766. [DOI] [PubMed] [Google Scholar]
- 57.Rausch, J. W., D. Lener, J. T. Miller, J. G. Julias, S. H. Hughes, and S. F. Le Grice. 2002. Altering the RNase H primer grip of human immunodeficiency virus reverse transcriptase modifies cleavage specificity. Biochemistry 41:4856-4865. [DOI] [PubMed] [Google Scholar]
- 58.Resnick, R., C. A. Omer, and A. J. Faras. 1984. Involvement of retrovirus reverse transcriptase-associated RNase H in the initiation of strong-stop (+) DNA synthesis and the generation of the long terminal repeat. J. Virol. 51:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rhim, H., J. Park, and C. D. Morrow. 1991. Deletions in the tRNALys primer-binding site of human immunodeficiency virus type 1 identify essential regions for reverse transcription. J. Virol. 65:4555-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robson, N. D., and A. Telesnitsky. 2000. Selection of optimal polypurine tract region sequences during Moloney murine leukemia virus replication. J. Virol. 74:10293-10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roth, M. J., P. Schwartzberg, N. Tanese, and S. P. Goff. 1990. Analysis of mutations in the integration function of Moloney murine leukemia virus: effects on DNA binding and cutting. J. Virol. 64:4709-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roth, M. J., P. L. Schwartzberg, and S. P. Goff. 1989. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell 58:47-54. [DOI] [PubMed] [Google Scholar]
- 63.Schultz, S. J., M. Zhang, C. D. Kelleher, and J. J. Champoux. 2000. Analysis of plus-strand primer selection, removal, and reutilization by retroviral reverse transcriptases. J. Biol. Chem. 275:32299-32309. [DOI] [PubMed] [Google Scholar]
- 64.Smith, J. K., A. Cywinski, and J. M. Taylor. 1984. Initiation of plus-strand DNA synthesis during reverse transcription of an avian retrovirus genome. J. Virol. 49:200-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith, J. S., S. Y. Kim, and M. J. Roth. 1990. Analysis of long terminal repeat circle junctions of human immunodeficiency virus type 1. J. Virol. 64:6286-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sodroski, J., W. C. Goh, C. Rosen, K. Campbell, and W. A. Haseltine. 1986. Role of the HTLV-III/LAV envelope in syncytium formation and cytopathicity. Nature 322:470-474. [DOI] [PubMed] [Google Scholar]
- 67.Sorge, J., and S. H. Hughes. 1982. Polypurine tract adjacent to the U3 region of the Rous sarcoma virus genome provides a cis-acting function. J. Virol. 43:482-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stetor, S. R., J. W. Rausch, M. J. Guo, J. P. Burnham, L. R. Boone, M. J. Waring, and S. F. Le Grice. 1999. Characterization of (+) strand initiation and termination sequences located at the center of the equine infectious anemia virus genome. Biochemistry 38:3656-3667. [DOI] [PubMed] [Google Scholar]
- 69.Stoye, J. P. 2001. Endogenous retroviruses: still active after all these years? Curr. Biol. 11:R914-R916. [DOI] [PubMed] [Google Scholar]
- 70.Tang, S., T. Murakami, B. E. Agresta, S. Campbell, E. O. Freed, and J. G. Levin. 2001. Human immunodeficiency virus type 1 N-terminal capsid mutants that exhibit aberrant core morphology and are blocked in initiation of reverse transcription in infected cells. J. Virol. 75:9357-9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Urnovitz, H. B., and W. H. Murphy. 1996. Human endogenous retroviruses: nature, occurrence, and clinical implications in human disease. Clin. Microbiol. Rev. 9:72-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wakefield, J. K., H. Rhim, and C. D. Morrow. 1994. Minimal sequence requirements of a functional human immunodeficiency virus type 1 primer binding site. J. Virol. 68:1605-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Whitwam, T., M. Peretz, and E. Poeschla. 2001. Identification of a central DNA flap in feline immunodeficiency virus. J. Virol. 75:9407-9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wiskerchen, M., and M. A. Muesing. 1995. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J. Virol. 69:376-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wohrl, B. M., and K. Moelling. 1990. Interaction of HIV-1 ribonuclease H with polypurine tract containing RNA-DNA hybrids. Biochemistry 29:10141-10147. [DOI] [PubMed] [Google Scholar]
- 76.Wu, X., H. Liu, H. Xiao, J. A. Conway, E. Hunter, and J. C. Kappes. 1997. Functional RT and IN incorporated into HIV-1 particles independently of the Gag/Pol precursor protein. EMBO J. 16:5113-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu, Y., and J. W. Marsh. 2003. Early transcription from nonintegrated DNA in human immunodeficiency virus infection. J. Virol. 77:10376-10382. [DOI] [PMC free article] [PubMed] [Google Scholar]